Abstract
Chlamydia psittaci produces a collection of proteins, termed IncA, IncB, and IncC, that are localized to the chlamydial inclusion membrane. In this report we demonstrate that IncA is also produced by Chlamydia trachomatis. C. trachomatis IncA is structurally similar to C. psittaci IncA and is also localized to the inclusion membrane. Immunoblot analysis demonstrated that sera from C. trachomatis-infected patients and from experimentally infected monkeys both recognized C. trachomatis IncA.
Chlamydiae depend heavily on their host cells for energy and essential nutrients, including amino acids and nucleoside triphosphates. Unlike species of the bacterial parasites Shigella, Listeria, and Rickettsia, which have direct access to the nutrient-rich environment of the host cytoplasm (8, 20, 21), chlamydiae are sequestered in a membrane-bound vacuole, termed an inclusion. Living within a vacuole presents some unique challenges not faced by organisms in the cytoplasm. One of these challenges includes the acquisition of nutrients from the host cell. Heinzen and Hackstadt (6) showed that the inclusion membrane is not passively permeable to molecules as small as 520 Da by microinjection studies of fluorescent tracer molecules. Therefore, nutrient acquisition is likely mediated through transport mechanisms at the inclusion membrane.
Another key to chlamydial pathogenesis and survival is their ability to avoid fusion with lysosomal compartments in order to persist and replicate within the host cell. Several experiments have shown the mature chlamydial inclusion to be nonfusogenic with markers from the endosomal-lysosomal pathway. Electron microscopic analysis showed that ferritin-labeled lysosomes do not fuse with the inclusion (23). Neither fluid-phase markers nor markers of the early or late endosomes are associated with the chlamydial inclusion (7, 15, 19). However, chlamydiae do sequester and modify host cell lipids and apparently reside in an exocytic arm of the host vesicular trafficking network (4, 5, 22). Modification of the vesicle to intersect an exocytic pathway requires chlamydial protein synthesis, which suggests that the chlamydiae synthesize proteins that determine the vesicular interactions of the inclusion (16).
It is thought that both acquisition of nutrients and avoidance of lysosomal fusion may be mediated by chlamydial proteins secreted into the inclusion membrane. This led to the identification and characterization of IncA, a Chlamydia psittaci protein that is present uniquely in infected cells, is localized to the inclusion membrane (12), is exposed to the host cell cytoplasm, and is phosphorylated by the host cell (13). Two additional inclusion membrane proteins, termed IncB and IncC, were recently identified in C. psittaci (1).
Despite considerable effort, incA, incB, and incC were never detected in Chlamydia trachomatis by conventional laboratory methods. The failure of these approaches led to the concern that C. psittaci IncA, IncB, and IncC might not directly model inclusion development in the human pathogenic species of the chlamydiae. With the completion of the C. trachomatis genome project (17), incA has been identified in this species. This report describes our characterization of IncA from C. trachomatis.
Organisms.
C. trachomatis LGV-434, serovar L2, and C. trachomatis serovar D were cultivated in HeLa 229 cells as previously described (3). The trachoma biovar strains (serovars A, B, Ba, and C), the genital strains (serovars D, D-, E, F, G, H, I, Ia, J, and K), and the LGV biovar strains (serovars L1, L2, L2a, and L3) were also cultivated in HeLa cells. Specific strains studied included A/G-17/OT, B/TW-5/OT, Ba/Ap-2/OT, C/TW-3/OT, D/UW-3/Cx, Da/TW-448/Cx, D-/MT 157/Cx, E/UW-5/Cx, F/UW-6/Cx, G/UW-57/Cx, H/UW-4/Cx, I/UW-12/Ur, Ia/UW-202/NP, I-/MT 518/Cx, J/UW-36/Cx, K/UW-31/Cx, L1/440/Bu, L2/434/Bu, L2a/UW-396/Bu, L3/404/Bu, and C. psittaci GPIC.
Antiserum production.
A maltose-binding protein (MBP)-IncA fusion protein was produced by using the pMAL-c2 vector system from New England Biolabs as described previously (1). C. trachomatis serovar D incA was amplified with 5′-AGCCATAGGATCTGGTTTCAGCGA-3′ and 5′-GCGCGGATCCTAGGAGCTTTTTGTAGAGGGTGA-3′ and then cloned into pMAL-c2.
MBP-IncA was used as antigen for the production of monospecific antibody in New Zealand White rabbits (12). Antiserum against C. trachomatis serovar L2 was produced in cynomolgus monkeys (Macaca fascicularis). Monkeys were anesthetized and infected urethrally with C. trachomatis elementary bodies (EBs) three times over the course of 6 months. Symptoms of infection were monitored over time. Antisera from infected monkeys were tested for reactivity to chlamydiae by enzyme-linked immunosorbent assay (reference 18 and unpublished data) and immunoblotting. Human sera that demonstrated high titers of antibody to C. trachomatis or Chlamydia pneumoniae by microimmunofluorescence assay were selected from stored serum specimens at the University of Washington. Negative control antisera were taken from patients who had no detectable reactivity by microimmunofluorescence against any of the C. trachomatis serovars listed above or C. pneumoniae TWAR. Antilipopolysaccharide monoclonal antibody was produced as described previously (2).
Immunoblotting and immunofluorescence microscopy.
Polyacrylamide gel electrophoresis and immunoblotting were performed as previously described (11, 12). Chlamydiae grown in HeLa cells on sterile glass coverslips were methanol fixed 30 h postinfection and stained as previously described (12). Immunostained coverslips were visualized with the 63× objective of a Zeiss microscope equipped with an epifluorescence condenser and an MC 63 C photomicrographic camera.
Sequence analysis of C. trachomatis incA.
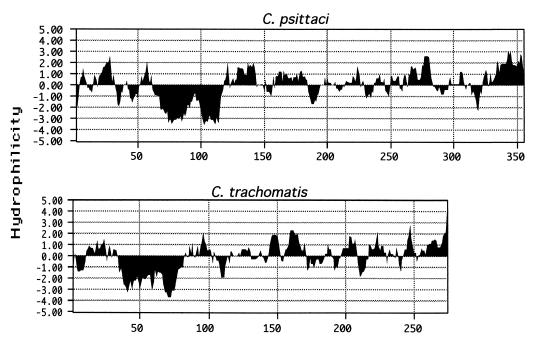
All sequence analysis was conducted by using methods described by Bannantine et al. (1). C. trachomatis incA was identified by limited homology in the C. trachomatis genome sequence database (17). A BLAST search of the amino acid sequence showed C. psittaci IncA to be the strongest match in the database, but that match was weak, with an E value of only 2 × 10−5. The 30-kDa size of IncA from C. trachomatis is smaller than that of C. psittaci IncA, and their identity and similarity were only 21 and 41%, respectively. Weak homology at the nucleotide sequence level explained why C. trachomatis incA was not detected by Southern hybridization or PCR amplification with probes and primers from the C. psittaci genomic sequence. Although IncA sequence identity between C. trachomatis and C. psittaci is low, comparison of their hydropathy plots shows similar large hydrophobic regions near the N-terminal ends (Fig. 1). Such a long hydrophobic region, with its unique bilobed shape, may be useful in predicting other chlamydial proteins in the inclusion membrane since it is also present in IncB and IncC (1). The location of the hydrophobic domain is near the C-terminal end in IncB and IncC. To show that this hydrophobic domain is not fortuitous, several open reading frames (ORFs) identified in the C. trachomatis genome project have been screened by hydropathy plot analysis, and only tested ORFs that encode proteins with similar secondary structure are localized to the inclusion membrane (13a). Primers were designed from the serovar D incA sequence, and they amplified incA from serovar L2 as well as D. The sequence from these two serovars is highly conserved: only 5 of 273 amino acids are different. The same primers did not amplify a product with C. pneumoniae genomic DNA as a template.
FIG. 1.
Comparison of IncA proteins from C. psittaci and C. trachomatis by hydropathy plot analysis. A hydropathy profile of each protein shows a unique bilobed hydrophobic domain in the N-terminal half. Profiles were determined by the algorithm developed by Kyte and Doolittle (9), with a window size of seven amino acids. The vertical axis displays relative hydrophilicity, with negative scores indicating relative hydrophobicity.
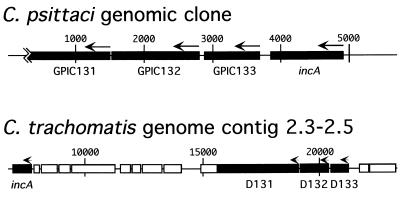
The region surrounding incA is not conserved between C. trachomatis and C. psittaci. In previous work, we and others have isolated four independent C. psittaci genomic clones that collectively define a group of four physically linked genes as shown in Fig. 2 (12, 13a). The completion of the C. trachomatis genome sequence has allowed a comparison of the arrangement of these genes in C. psittaci and C. trachomatis. Each of the four ORFs is present in the C. trachomatis genome, but the physical linkage has been disrupted. In C. psittaci, incA is immediately upstream of an ORF designated GPIC133 (Fig. 2), with an intergenic region of 157 bp (see orf2 in reference 12). ORF 133 is present in both C. psittaci and C. trachomatis and is relatively conserved, with 58% identity between the deduced amino acid sequences. The incA coding sequence in C. trachomatis is downstream and separated from ORF 133 (D133) by 12,678 bp, with incA located at contig 2.3 in the genome and D133 located at contig 2.5. Note the scale difference between the two genomic segments in Fig. 2.
FIG. 2.
ORF map of the chromosomal region surrounding incA in C. psittaci and C. trachomatis. ORFs 131, 132, 133, and incA are labeled. Note the scale difference between the maps. ORF 133 is immediately downstream of incA in C. psittaci, whereas it is upstream and separated by at least 10 kb in C. trachomatis. Base pairs are indicated above each map, and arrows indicate the direction of transcription. The ORF designation is preserved from the C. trachomatis serovar D genome database designations. Pustell protein matrix analysis was used to confirm that GPIC131 and GPIC132 correspond to D131 and D132, respectively.
Immunoblot analysis of infected cells and purified C. trachomatis EBs was performed with rabbit anti-MBP-IncA as a probe. A 27-kDa band was present only in the infected cells and not in lysates of EBs or uninfected cells (data not shown).
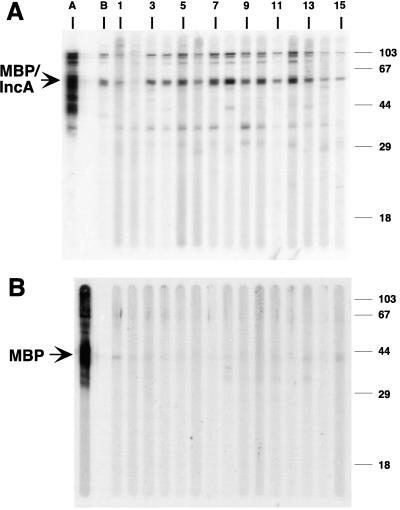
In order to determine if IncA was recognized by sera from convalescent animals and humans, purified MBP-IncA fusion protein was loaded onto a preparative sodium dodecyl sulfate-polyacrylamide gel and used to examine reactivity with sera from patients and monkeys infected with C. trachomatis. The majority of the sera from chlamydia-infected patients (10 of 11) and all monkey convalescent-phase sera recognized the IncA protein (Fig. 3A) but not the MBP portion of the fusion (Fig. 3B). IncA was faintly recognized by sera from one of the C. pneumoniae-infected patients (Fig. 3A, lane 1).
FIG. 3.
Preparative immunoblot analysis of a purified MBP-C. trachomatis IncA fusion protein (A) and purified MBP (B), each probed with antisera from chlamydia-infected patients and monkeys. Lane A, anti-MBP; lane B, monkey convalescent-phase sera; lanes 1 and 2, sera from C. pneumoniae-infected patients; lanes 3 to 13, sera from C. trachomatis-infected patients; lanes 14 and 15, negative control sera.
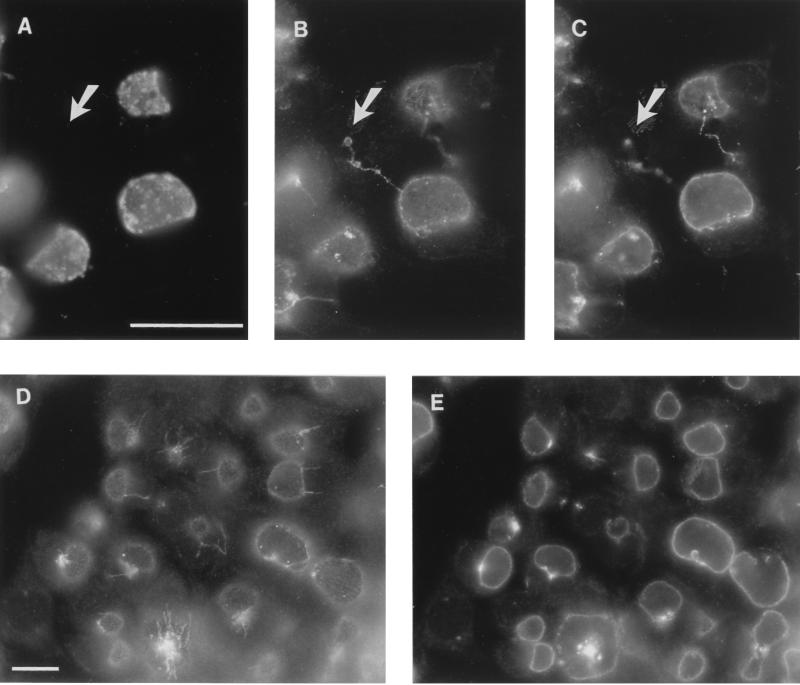
Antisera against IncA and a monoclonal antibody against chlamydial lipopolysaccharide were used to immunostain methanol-fixed layers of C. trachomatis-infected HeLa cells. Anti-IncA reacted with the membrane of the inclusion but not the chlamydial developmental forms (Fig. 4A to C). Antigenic fibers extending away from the inclusion, which are similar in structure to those found in C. psittaci-infected cells (12), were also present in C. trachomatis-infected cells (Fig. 4B to D). Their function and origins remain unknown. Also evident in Fig. 4B and C are antigenic fibers that traverse between otherwise apparently separate cells. It is likely that these are daughter cells in which inclusions can either divide with the dividing cell (10) or stay in one daughter cell and leave the other uninfected. C. psittaci IncA can also be found in fibers that extend between pairs of infected cells (data not shown). One major difference between these two processes is that in C. psittaci (strain GPIC), each daughter cell usually remains infected. In C. trachomatis, however, uninfected progeny cells are common. Because IncA is also found in fibers that extend to the uninfected daughter cells (Fig. 4B and C), the result is a cell lacking chlamydial developmental forms but containing chlamydial antigen.
FIG. 4.
Immunofluorescence microscopy with anti-IncA demonstrating that IncA is localized to the inclusion membrane in C. trachomatis-infected cells. Serovar L2-infected HeLa cells were fixed in methanol 25 h postinfection and stained with anti-major outer membrane protein (A) and/or anti-MBP-IncA (B to E). Panels A to C represent a single image, with panel C photographed in a different focal plane. Note the fibers extending between the two inclusions in different cells as well as from one infected cell to an apparently uninfected cell (uninfected cell at tip of arrow). Note also the antigenic fibers extending from several inclusions in one focal plane (D) and IncA in inclusions at different stages of maturation in another focal plane (E). Bars in panels A and D represent 10 μm for panels A to C and panels D and E, respectively.
In addition to the LGV biovar strain (serovar L2) shown in Fig. 4, several other C. trachomatis serovars of clinical interest were analyzed by immunofluorescence microscopy for staining with anti-MBP-IncA (Table 1). Anti-MBP-IncA labeled the inclusion membranes of all serovars tested.
TABLE 1.
Chlamydia strains used for reactivity with anti-C. trachomatis IncA
| Biovar, strain, or cell type | Serovars | Immuno-fluorescence staining |
|---|---|---|
| Trachoma | A, B, Ba, C | + |
| Oculogenital | D, Da, D-, E, F, G, H, I, Ia, I-, J, K | + |
| LGV | L1, L2, L2a, L3 | + |
| C. psittaci GPIC | − | |
| Uninfected HeLa cells | − |
The inclusion membrane mediates all contact between the host cell and chlamydiae; therefore, the acquisition of nutrients and the nonfusogenic nature of the chlamydial inclusion may be elucidated by studying chlamydial proteins that reside in the inclusion membrane. Because the routing of transport vesicles throughout the cell is mediated by proteins present on the transport vesicle membrane (14), IncA as well as IncB and IncC are excellent candidate proteins for mediating inclusion trafficking within infected cells. We undertook these studies to define the presence and intracellular location of IncA in all of the major C. trachomatis serovars and to assess whether an antibody response to IncA was present in infected patients and primates. We speculate that C. pneumoniae also produces Inc-like proteins and are initiating an investigation into this system. Finally, we continue to pursue questions surrounding the role of the Inc proteins in the chlamydial infection process as well as their role as possible protective antigens in the host response to chlamydial infection.
Nucleotide sequence accession number.
The nucleotide sequence of C. trachomatis LGV-434, serotype L2, incA has been deposited in the GenBank database under accession no. AF067958.
Acknowledgments
We gratefully acknowledge Richard Stephens and Claudia Fenner at the University of California, Berkeley, for their efforts in the completion of the C. trachomatis genome project. We thank Linda Cles of the University of Washington for providing the human serum samples. We also thank Harlan Caldwell and Michael Parnell for assistance with the production of primate convalescent-phase sera.
A portion of this work was supported by Public Health Service (PHS) grants AI-31448 and N01-AI-75329 to W.E.S. and PHS grant AI42869-01 to D.D.R.
Footnotes
Technical paper 11411 of the Oregon State University Extension and Experiment Station.
REFERENCES
- 1.Bannantine J P, Rockey D D, Hackstadt T. Tandem genes of Chlamydia psittaci that encode proteins localized to the inclusion membrane. Mol Microbiol. 1998;28:1017–1026. doi: 10.1046/j.1365-2958.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- 2.Caldwell H D, Hitchcock P J. Monoclonal antibody against a genus-specific antigen of Chlamydia species: location of the epitope on chlamydial lipopolysaccharide. Infect Immun. 1984;44:306–314. doi: 10.1128/iai.44.2.306-314.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caldwell H D, Kromhout J, Schachter J. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun. 1981;31:1161–1176. doi: 10.1128/iai.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hackstadt T, Scidmore M A, Rockey D D. Lipid metabolism in Chlamydia trachomatis-infected cells: directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc Natl Acad Sci USA. 1995;92:4877–4881. doi: 10.1073/pnas.92.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hackstadt T, Rockey D D, Heinzen R A, Scidmore M A. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 1996;15:964–977. [PMC free article] [PubMed] [Google Scholar]
- 6.Heinzen R A, Hackstadt T. The Chlamydia trachomatis parasitophorous vacuolar membrane is not passively permeable to low-molecular-weight compounds. Infect Immun. 1997;65:1088–1094. doi: 10.1128/iai.65.3.1088-1094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinzen R A, Scidmore M A, Rockey D D, Hackstadt T. Differential interaction [sic] with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.High N, Mounier J, Prevost M C, Sansonetti P J. IpaB of Shigella flexneri causes entry into epithelial cells and escape from the phagocytic vacuole. EMBO J. 1992;11:1991–1999. doi: 10.1002/j.1460-2075.1992.tb05253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:1005–1032. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 10.Richmond S J. Division and transmission of inclusions of Chlamydia trachomatis in replicating McCoy cell monolayers. FEMS Microbiol Lett. 1985;29:49–52. [Google Scholar]
- 11.Rockey D D, Rosquist J L. Protein antigens of Chlamydia psittaci present in infected cells but not detected in the infectious elementary body. Infect Immun. 1994;62:106–112. doi: 10.1128/iai.62.1.106-112.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockey D D, Heinzen R A, Hackstadt T. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol Microbiol. 1995;15:617–626. doi: 10.1111/j.1365-2958.1995.tb02371.x. [DOI] [PubMed] [Google Scholar]
- 13.Rockey D D, Grosenbach D, Hruby D E, Peacock M G, Heinzen R A, Hackstadt T. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol Microbiol. 1997;24:217–228. doi: 10.1046/j.1365-2958.1997.3371700.x. [DOI] [PubMed] [Google Scholar]
- 13a.Rockey, D. D., and J. P. Bannantine. Unpublished data.
- 14.Rothman J E, Wieland F T. Protein sorting by transport vesicles. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 15.Scidmore M A, Fischer E R, Hackstadt T. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J Cell Biol. 1996;134:363–374. doi: 10.1083/jcb.134.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scidmore M A, Rockey D D, Fischer E R, Heinzen R A, Hackstadt T. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun. 1996;64:5366–5372. doi: 10.1128/iai.64.12.5366-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens R S, Kalman S, Fenner C, Davis R. Chlamydia genome project. 1997. http://chlamydia-www.berkeley.edu:4231 http://chlamydia-www.berkeley.edu:4231. . [Google Scholar]
- 18.Su H, Morrison R P, Watkins N G, Caldwell H D. Identification and characterization of T helper cell epitopes of the major outer membrane protein of Chlamydia trachomatis. J Exp Med. 1990;172:203–212. doi: 10.1084/jem.172.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taraska T, Ward D M, Ajioka R S, Wyrick P B, Davis-Kaplan S R, Davis C H, Kaplan J. The late chlamydial inclusion membrane is not derived from the endocytic pathway and is relatively deficient in host proteins. Infect Immun. 1996;64:3713–3727. doi: 10.1128/iai.64.9.3713-3727.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theriot J A. The cell biology of infection by intracellular bacterial pathogens. Annu Rev Cell Dev Biol. 1995;11:213–239. doi: 10.1146/annurev.cb.11.110195.001241. [DOI] [PubMed] [Google Scholar]
- 21.Tilney L G, Portnoy D A. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wylie J L, Hatch G M, McClarty G. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J Bacteriol. 1997;179:7233–7242. doi: 10.1128/jb.179.23.7233-7242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wyrick P B, Brownridge E A. Growth of Chlamydia psittaci in macrophages. Infect Immun. 1978;19:1054–1060. doi: 10.1128/iai.19.3.1054-1060.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]