Abstract
We analyzed the didanosine (ddI) arm of the randomized, placebo-controlled Jaguar trial in order to define a genotypic score for ddI associated with virologic response. In this arm, 111 patients experiencing virologic failure received ddI in addition to their current combination therapy for 4 weeks. The impact of mutations in the reverse transcriptase gene on the virologic response to ddI was studied in univariate analysis. Genotypic score was constructed using step-by-step analyses first including only mutations associated to poorer virologic response (scored as +1), while secondarily, mutations associated to a better response (scored as −1) were also eligible. Eight mutations were associated with a reduced response to ddI, M41L, D67N, T69D, L74V, V118I, L210W, T215Y/F, and K219Q/E, and two mutations were associated with a better response, K70R and M184V/I. The best prediction of the virologic response to ddI was obtained with a composite score comprising mutations added and subtracted (set II, M41L + T69D + L74V+ T215Y/F + K219Q/E − K70R − M184V/I; P = 4.5 × 10−9) and by comparing that to only mutations added (set I, M41L + T69D + L74V + L210W + T215Y/F + K219Q/E; P = 1.2 × 10−7). Patients had a human immunodeficiency virus RNA reduction of 1.24, 0.84, 0.61, 0.40, and 0.07 log10 copies/ml when they were ranked as having a genotypic score II of −2, −1, or 0 or 1 and 2 mutations or more, respectively. In conclusion, we developed and validated a genotypic score, taking into account mutations negatively and positively impacting the virologic response to ddI.
Resistance to antiretroviral drugs is associated with clinical progression and is an increasing problem for the management of human immunodeficiency virus (HIV)-infected patients. Selection of drug resistance mutations in a patient's virus strains is associated with resistance to specific antiretroviral agents and may confer cross-resistance, limiting treatment options and reducing the efficacy of subsequent regimens. It is now widely recognized that there is significant cross-resistance among nucleoside reverse transcriptase inhibitors (NRTIs).
Didanosine (ddI) is a 2′,3′-dideoxyinosine analogue reverse transcriptase inhibitor used in antiretroviral combination therapy for the treatment of HIV-infected patients. The in vitro and in vivo patterns associated with resistance to ddI have been described in several studies. Resistance to ddI is frequently associated with a mutation at codon 74 (L74V), which confers an approximate 4- to 10-fold reduction in ddI susceptibility (22). Among patients treated with ddI monotherapy for 12 months following prolonged zidovudine monotherapy, the L74V mutation was observed in approximately 65% of cases (21, 22). In addition to the L74V mutation, ddI resistance can also occur from a mutation at codon 65 (K65R) (25). This mutation has been isolated from several patients receiving long-term treatment with ddI and resulted in a three- to fivefold decrease in ddI susceptibility in vitro. Other in vitro studies indicate that a mutation at codon 75 (V75T) confers an approximate fivefold reduction in sensitivity to ddI as well as cross-resistance to zalcitabine and stavudine (13). However, the V75T mutation has not been reported in clinical isolates from patients failing ddI therapy. A mutation at codon 184 (M184V), typically associated with lamivudine resistance, has been introduced by site-directed mutagenesis, and the recombinant viruses thus generated displayed a fivefold decrease in susceptibility in ddI in vitro (6). However, several recent studies showed that the M184V mutation may not affect the clinical response to ddI (24; J. J. Eron, R. J. Bosch, L. Petch, S. Fiscus, and I. Franck, Abstr. XI Int. HIV Drug Resist. Workshop, abstr. 123, 2002; A. Pozniak, B. G. Gazzard, M. Peeters, R. Hoetelmans, and N. M. Graham, Abstr. XI Int. HIV Drug Resist. Workshop, abstr. 152, 2002). Like other NRTIs, the Q151M mutation also confers resistance to ddI (5- to 40-fold).
Reverse transcriptase mutations frequently identified in viruses from patients who have received zidovudine (M41L, D67N, K70R, L210W, T215Y or F, and K219Q or E) were thought initially to be associated only with zidovudine resistance (8, 11, 14). However, the response to other NRTIs after zidovudine failure is often muted, and zidovudine mutations can emerge during therapy with stavudine or ddI in the absence of zidovudine (3). Moreover, it has been shown that an increasing number of thymidine analogue mutations displayed a progressive reduction in drug susceptibility for all NRTIs (23). Cross-resistance to ddI resulting from prior NRTI therapy was studied in the control arm of the Narval ANRS (Agence Nationale de Recherches sur le SIDA) 088 trial and showed that the presence of at least three thymidine analogue mutations (TAMs) including the T215Y/F mutation significantly impaired the virologic response to ddI (D. Costagliola, D. Descamps, V. Calvez, B. Masquelier, A. Ruffault, F. Telles, J. L. Meynard, and F. Brun-Vézinet, Abstr. 8th Conf. Retrovir. Opportun. Infect., abstr. 450, 2001). This result suggests that TAMs could impact the response to ddI.
However, most of algorithms used to define genotypic resistance to ddI in NRTI-experienced patients are based on limited data. The Jaguar study was a randomized multicenter, double-blind, placebo-controlled trial evaluating the efficacy of adding ddI to an ongoing failing highly active antiretroviral therapy (HAART) regimen. The use of an add-on study provides the most rigorous evaluation of the intrinsic activity of a drug and the genotypic determinants of the virologic response to this given drug. Using the Jaguar trial data set, we defined a clinically relevant genotype interpretation for resistance to ddI.
(This work was presented during the XIII International HIV Drug Resistance Workshop, Tenerife, Spain, 8 to 12 June 2004 [abstract 132].)
MATERIALS AND METHODS
Study population.
The Jaguar study (AI454-176) was a randomized multicenter, double-blind, placebo-controlled trial evaluating the efficacy of adding ddI to an ongoing failing HAART regimen (18). Subjects were recruited from 29 clinical centers located throughout France and must have been treated for at least 3 months with a stable antiretroviral drug regimen that did not include ddI, although ddI could have been part of treatment previously.
Laboratory methods. (i) Plasma HIV-1 RNA assay.
Plasma HIV type 1 (HIV-1) RNA was measured in a central laboratory at day 0, week 2, and week 4 by the Amplicor HIV-1 Monitor kit (Cobas 1.5 test; Roche Diagnostic Systems) with a detection limit of 50 copies/ml.
(ii) Genotyping.
Plasma HIV-1 RNA was genotyped using the consensus method of the French ANRS (2). Genotyping was performed at baseline and at week 4 in all patients by using an ABI 3100 Genetic Analyzer (PE Applied Biosystems). The sequences were analyzed using the Sequence Navigator software (PE Applied Biosystems) and reported as amino acid changes with respect to the sequence of the wild-type virus HXB2. Mutations associated with NRTI resistance were identified according to the consensus statement from the 2003 International AIDS Society (IAS) Resistance-USA Panel (http://www.iasusa.org).
Statistical analyses.
The endpoint for the analysis was the magnitude of reduction in HIV-1 RNA levels in log10 copies/ml from day 0 to week 4. The crude method is used to compute viral load reductions; i.e., HIV-1 RNA levels <50 copies/ml at week 4 were fixed at 50 copies/ml. Although censored methods are preferable to compute HIV-1 RNA reductions, the small percentage (11%) of patients censored by the limit of quantification ensures that the naïve method provides unbiased estimates (4, 5, 17). We analyzed the impact of each IAS mutation on the virologic response to ddI by comparing reduction in plasma HIV-1 RNA in patients with or without the specific mutation using a Wilcoxon-Mann-Whitney rank test. Among the 2003 IAS NRTI mutations, those that were present in at least 5% of patients and for which P was < 0.20 in the above-mentioned univariate analysis were retained for further analysis. Let k be the number of mutations retained from the univariate analysis. Such a procedure was applied only to patients randomized in the ddI group.
We used the backward elimination technique with a nonparametric test to select two final sets of mutations associated with the week-4 virologic response. The backward elimination technique begins with all k mutations retained from the univariate analysis. The objective is to compare HIV-1 RNA reductions in patients having no mutations to those with k mutations with the hypothesis that an increased number of mutations will result in a poorer virologic response. Under such conditions, the Wilcoxon-Mann-Whitney rank test, with an unspecified alternative hypothesis, is not appropriate. We used the nonparametric Jonckheere-Terpstra (JT) test for ordered alternatives that allow testing of a specific a priori sequence (10). As an ordered alternative is specified, the preferable P value for a JT test is one sided. When the number of groups is greater than 3 or when the samples are of unequal size, the large-sample size approximation to the sampling distribution is used. From the initial set of k mutations, the first step is to remove one mutation. One by one, all combinations of k−1 mutations are investigated, and the combination providing the lower P value with the JT test is retained. In the second step, mutations are again removed one by one to compare the combinations of k−2 mutations, the combination providing the lower P value is again retained, and so on. The procedure ends when removing a mutation does not provide a lower P value than the previous P value.
Two genotypic scores were computed according to two analyses. In the first analysis, only variants providing poorer virologic response than that of wild-type codons were retained for the procedure described above. In the second analysis, variants involving a better virologic response were also retained for that procedure. In the first analysis, mutations are simply added, while in the second analysis, mutations providing better virologic response, whether retained by the procedure, are subtracted from the summation of mutations providing poorer virologic response.
Finally, we used the bootstrap resampling method to assess the robustness of the score obtained (1, 20). Multivariate analyses of the score were performed on 500 samples drawn from the 101 patients by sampling with replacement. We report the number of times the score had a P value of <0.05, <0.001, and <0.0001 and the median reductions in viral load in the resistance groups.
RESULTS
Patient characteristics.
In this study, 168 patients were randomized to receive either ddI (n = 111) or placebo (n = 57) during 4 weeks. Patients randomized in the two arms had no significant difference in age, baseline CD4 cell count, and baseline HIV-1 RNA (Table 1). At baseline, patients had a median (range) of 4 (0 to 9) NRTI mutations and a median (range) of 3 (0 to 6) TAMs with no significant difference between both treatment groups. The distribution of NRTI mutations at baseline is presented in Fig. 1. Week-4 genotypes were performed in order to detect the acquisition of additional mutations. No new L74V nor K65R mutations were detected in the ddI group, and the distribution of mutations remained well balanced between groups, except the V118I mutation that emerged more frequently in the ddI group (5 patients, 6%) than in the placebo group (0 patients). However, there was no relationship between the acquisition of additional mutations at week 4 and a weaker virologic response to ddI at week 4.
TABLE 1.
Baseline patient characteristics
| Characteristica | ddI (n = 111) | Placebo (n = 57) | Total (n = 168) |
|---|---|---|---|
| No. (%) male | 95 (86) | 44 (77) | 139 (83) |
| Median age in yr (range) | 44 (25-76) | 43 (29-67) | 44 (25-76) |
| No. (%) at CDC stage C | 19 (17) | 11 (20) | 30 (18) |
| Median plasma HIV-1 RNA log10 copies/ml (range) | 13.8 (1.70-5.42) | 13.8 (2.62-5.51) | 13.8 (1.70-5.51) |
| Median CD4 cell count/mm3 (range) | 380 (122-1,590) | 363 (98-873) | 378 (98-1590) |
| No. (%) with previous ddI exposure | 72 (65) | 43 (75) | 115 (68) |
CDC stage C, AIDS status according to the Centers for Disease Control and Prevention classification.
FIG. 1.
Distribution and prevalence of nucleoside reverse transcriptase inhibitor mutations.
Virologic response to didanosine.
The median reduction in plasma HIV-1 RNA at week 4 in the patients exposed to ddI was 0.57 log10 copies/ml (interquartile range, 0.14 to 1.02). Table 2 shows the univariate analysis of the virologic response according to the presence of variant or wild-type codons for positions providing a P value of ≤0.20. Eight mutations, present in at least 5% of the samples, were associated with a reduced virologic response to ddI: M41L, D67N, T69D, L74V, V118I, L210W, T215Y/F, and K219Q/E (Table 2). Two mutations were significantly associated with a better virologic response: K70R and M184V/I.
TABLE 2.
Median decrease in HIV-1 RNA at week 4 and corresponding to a P value of ≤0.20 according to the presence or absence of the variant codon in the reverse transcriptase gene
| Site | Codon(s) | No. of isolates | Median decrease in HIV RNA | P value |
|---|---|---|---|---|
| M41 | M | 53 | −0.85 | <0.0001 |
| L | 49 | −0.28 | ||
| D67 | D | 67 | −0.66 | |
| N | 35 | −0.26 | 0.005 | |
| T69 | T | 93 | −0.62 | |
| D | 9 | −0.08 | 0.05 | |
| L74 | L | 93 | −0.62 | |
| V | 9 | −0.06 | 0.005 | |
| V118 | V | 83 | −0.62 | |
| I | 19 | −0.28 | 0.17 | |
| L210 | L | 74 | −0.66 | |
| W | 28 | −0.34 | 0.003 | |
| T215 | T | 47 | −0.88 | |
| Y, F | 55 | −0.38 | 0.0001 | |
| K219 | K | 77 | −0.62 | |
| Q, E | 25 | −0.44 | 0.20 | |
| K70 | K | 75 | −0.44 | |
| R | 27 | −0.94 | 0.014 | |
| M184 | M | 8 | −0.15 | |
| V, I | 94 | −0.61 | 0.042 |
Didanosine genotypic score.
The first analysis is a backward elimination procedure where only the eight mutations associated with poorer virologic response are included in the first step of the procedure (Table 2). The strongest association between the decrease in viral load and the ordered number of mutations was observed with the following set: I, M41L + T69D + L74V + L210W + T215Y/F + K219Q/E. The presence of the D67N or V118I mutations did not improve the association (larger P values). The final P value of the JT test indicates a strong association between the week-4 virologic response and the number of mutations from set I (Fig. 2a) (P = 1.2 × 10−7). Only one patient had five and six mutations among set I, having a poor impact on the corresponding JT test. Patients with no mutations had a median decrease in viral load of 0.96 log10 copies/ml, while those with 1, 2, or more than 2 mutations had an HIV-1 RNA reduction of 0.67 and 0.16 log10 copies/ml, respectively (Fig. 2b). We then defined a first genotypic score (score I) according to patients that had a virus harboring 0, 1 or 2, or more than 2 mutations (Fig. 2b) (P = 2.3 × 10−7; JT test).
FIG. 2.
(a) Median decrease in HIV-1 plasma RNA at week 4 according to the number of mutations among M41L plus T69D plus L74V plus L210W plus T215Y/F plus K219Q/E. (b) Median decrease in HIV-1 plasma RNA at week 4 according to the genotypic score I number of mutations among M41L plus T69D plus L74V plus L210W plus T215Y/F plus K219Q/E.
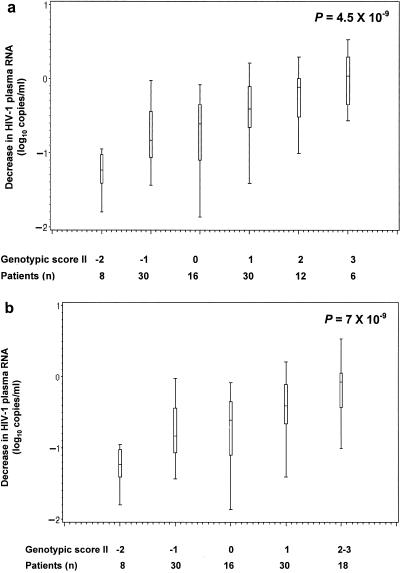
A second analysis was designed where all mutations described in Table 2, the eight mutations associated to poorer virologic response and the two mutations (K70R and M184V/I) providing better virologic response, were included in the beginning of the backward elimination technique. The second set of mutations retained (set II) defined a second genotypic score defined as follows: M41L + T69D − K70R + L74V − M184V/I + T215Y/F + K219Q/E. The final P value was lower than the one from the previous analysis, indicating a stronger association between the week-4 virologic response and set II (P = 4.5 × 10−9) compared with set I (P = 1.2 × 10−7). Patients with only both K70R and M184V/I mutations (score II, coded −2 in Fig. 3) had a median reduction in HIV-1 RNA of 1.24 log10 copies/ml. All 30 patients with score II, coded −1 in Fig. 3, had the M184V/I mutations and absence of the T69D, L74V, and T215Y/F mutations. Seven out of these 30 patients had the K70R mutation that was balanced in the computation of the score by the presence of the K219Q/E and M41L mutations in 6 and 1 patients, respectively. Patients with score II, coded −1, 0, 1, 2, or 3, had an HIV-1 RNA reduction of 0.84, 0.61, 0.40, and 0.07 log10 copies/ml, respectively (Fig. 3b). A strong association was founded between score II and the week-4 virologic response (P = 7 × 10−9; JT test).
FIG. 3.
(a) Median decrease in HIV-1 plasma RNA at week 4 according to the genotypic score II, computed as follows: M41L + T69D − K70R + L74V − M184V/I + T215Y/F + K219Q/E. (b) Median decrease in HIV-1 plasma RNA at week 4 according to the genotypic score II, computed as follows: M41L + T69D − K70R + L74V − M184V/I + T215Y/F + K219Q/E.
Bootstrap analyses.
In the 500 bootstrap samples, the P value of the JT test for the genotypic score I was <0.05 in 491/500 (98%) cases, <0.001 in 443/500 (89%) cases, and <0.0001 in 418/500 (84%) cases. The estimated median decrease in log10 copies/ml of HIV-1 RNA was 0.97, 0.66, and 0.18 in patients with 0, 1 or 2, and more than 2 mutations, respectively.
Bootstrap analysis with genotypic score II showed that 493/500 (99%) samples, 461/500 (92%) samples, and 432/500 (86%) samples had a P value of <0.05, <0.001, and <0.0001, respectively (JT test). The estimated median decrease in log10 copies/ml of HIV-1 RNA was 1.24, 0.81, 0.61, 0.41, and 0.06 in patients having a genotypic score II, coded as −2, −1, 0, 1, and 2 or 3, respectively.
DISCUSSION
Resistance to ddI was initially related only to the L74V mutation, regarding the high rate of L74V mutation development among patients treated with ddI monotherapy (19, 24). More recently, progressive reduction in drug susceptibility for all NRTIs, including ddI, has been associated with an increasing number of TAMs (23). These results suggest that TAMs could impact the response to ddI and that resistance to this drug could be more complex than expected. One of the objectives of the Jaguar study was to assess the relationship between the virologic response at week 4 and baseline HIV reverse transcriptase genotype. To build up a resistance genotypic score from clinical studies for one individual drug, two ideal types of patient populations might be used. One is a population of patients receiving the same antiretroviral drug treatment besides the molecule to be studied, and the other is an add-on placebo-controlled study in which the only difference between the two groups is the drug to be studied.
Our results confirm that ddI continues to provide activity against viruses with the M184V/I mutation, as was suggested in previous studies (24; Eron et al., Abstr. XI Int. HIV Drug Resist. Workshop, abstr. 123, 2002; Pozniak et al., Abstr. XI Int. HIV Drug Resist. Workshop, abstr. 152, 2002). Moreover, in our study, the presence of the M184V/I mutation seems to have a positive impact on the virologic response to ddI since the median decrease in plasma HIV-1 RNA was −0.15 and −0.60 log10 in the absence and presence of M184V/I, respectively. A similar positive effect of the M184V/I mutation on the responsiveness to ddI has been recently observed (S. Capdepont, V. Aurillac-Lavignolle, M. Faure, M. Dupon, P. Morlat, J. M. Ragnaud, G. Chêne, H. Fleury, and B. Masquelier, Abstr. XIII Int. HIV Drug Resist. Workshop, abstr. 120, 2004). The fact that this effect has not been evidenced before might be related to the design of the previous studies, which were mainly switch studies, and to the particular design of the Jaguar trial, as an add-on study, which may have allowed evidence of such a positive effect because of the lack of interaction with other parameters.
The K70R mutation seems to have a positive impact as well, with a median decrease in plasma HIV-1 RNA of −0.44 and −0.94 log10 in the absence and presence of K70R, respectively. The K70R mutation is usually associated with D67N and K219Q/E in a specific pathway of accumulating TAMs (7, 16). Recent data have suggested that this pathway was associated with a lower reduction in susceptibility to zidovudine and stavudine (P. Flandre, N. T. Parkin, C. J. Petropoulos, and C. Chappey, Abstr. 11th Conf. Retrovir. Opportun. Infect., abstr. 645, 2004). Similarly, we have observed that the patients (n = 25) harboring the D67N/K70R/K219Q/T215F pattern had a better virologic response to ddI than the patients (n = 24) harboring the M41L/L210W/T215Y pattern (−0.95 versus −0.47 log10; P = 0.011). We could hypothesize that the difference in virologic response observed between the two pathways could be related to the fact that the K70R mutation, with its possible positive effect on response, is mainly observed within the context of the D67N/K70R/K219Q/T215F pathway. This positive effect has not been previously described and needs to be confirmed in further studies. But this could be compared to the concept of hypersusceptibility that has been described for mutants resistant to zidovudine that became phenotypically sensitive in vitro by the mutation M184V in the viral reverse transcriptase and for nonnucleoside reverse transcriptase inhibitors (9, 15).
The selection of additional NRTI mutations between baseline and week 4 was associated to neither baseline mutation profiles nor a reduced efficacy of ddI during the trial. However, the short treatment period of the present study could explain the low frequency of additional mutations at week 4, and a longer period of treatment should have been necessary to definitively avoid such a relationship.
It is now widely recognized that correlation studies analyzing the virologic response in treatment-experienced patients according to the genotypic profile at baseline provide relevant information for establishing resistance algorithms (1, 12). A stepwise methodology for the development and validation of clinically relevant genotypic resistance scores for antiretroviral drugs has been proposed, and it has been shown for abacavir that this methodology provided the best prediction of the virologic response to abacavir compared to other available algorithms (1; C. Cabrera, A. Cozzi-Lepri, A. N. Phillips, C. Loveday, T. Puig, O. Kirk, M. Ait-Khaled, B. Ledergerber; J. Lundgren, B. Clotet, and L. Ruiz, Abstr. 9th Eur. AIDS Conf., abstr. F6/5, 2003). This approach used the Wilcoxon-Mann-Whitney test to select the best combination of mutations associated with the virologic response. This test, however, may not be the most appropriate to this setting because the groups, or samples, determined by the number of mutations should be treated as an ordered variable. Indeed, we expect better virologic response in patients with no mutation than in patients with one mutation, who are expected to have a better virologic response than patients with two mutations, and so on. The Jonckheere-Terpstra test used in this work is appropriate, as it is specifically designed to test the null hypothesis against an ordered alternative. In the previous approach, only mutations associated to poorer virologic response were eligible to be selected in the final set of mutations. The use of this procedure led to the selection of the following combination of mutations (set I) that was strongly associated with the week-4 virologic response (P = 1.2 × 10−7): M41L + L74V + T215Y/F + T69D + K219Q/E + L210W.
Another set of mutations, however, was more strongly associated with the virologic response. Allowing mutations associated with a better virologic response as well as mutations providing poorer virologic response to be eligible in the second and final set (set II) of mutations led to selection a genotypic score defined as follows: M41L − M184V/I + L74V + D67N − K70R + K219Q/E + T215Y/F + T69D. Set II was indeed more strongly associated with the week-4 virologic response (P = 8.5 × 10−9) compared with set I. Patients can be classified as having a genotypic score of −2, −1, 0 or 1, and 2 or more with a median HIV-1 RNA reduction of 1.24, 0.85, 0.50, and 0.11 log10 copies/ml, respectively. The two sets of mutations described above provide the lowest P values from the two step-by-step analyses carried out. Other combinations of mutations, however, led to very low P values, indicating strong association between the week-4 virologic response and other combinations of lower numbers of mutations. P values of 1.2 × 10−7 or 8.5 × 10−9 correspond to the tail of the normal distribution.
Unfortunately, some mutations, like the K65R mutation, were underrepresented in the population studied and could not be accounted for in the genotypic score, although they could have an impact on the response to ddI. However, recent studies showed that its prevalence is increasing and is probably associated with the use of tenofovir and that this mutation is able to reduce susceptibility to all NRTIs except those containing a 3′-azido moiety like zidovudine (U. Parikh, D. Koontz, J. Hammond, L. Bacheler, R. Schinazi, P. Meyer, W. Scott, and J. Mellors, Abstr. XII Int. HIV Drug Resist. Workshop, abstr. 136, 2003). In addition to the possibility in rare cases of selection of the K65R mutation under ddI therapy, this suggests that isolates harboring this mutation should be considered resistant to ddI. Similarly, the Q151M complex and the 69SS insertion were present in too few patients to assess the response.
We developed a new approach to investigate the association between genotype at baseline and virologic response to an antiretroviral drug, considering mutations that both negatively and positively impact the viral response and a nonparametric statistic test appropriate to test for ordered alternatives (Jonckheere-Terpstra test). This approach should improve the predictivity of the genotypic rules and should be validated in other data sets and for other antiretroviral drugs.
Acknowledgments
Participating members of the AI454-176 Jaguar Study Team were as follows: trial chair, J. M. Molina; trial coordinator and monitors, G. Leleu and M. Troccaz; trial statistician, N. Schmidely; trial virologists, V. Calvez and A. G. Marcelin; scientific committee, J. M. Molina, J. Pavie, G. Leleu, V. Calvez, N. Schmidely, M. Troccaz, F. Clavel, I. Dujardin, and H. Benech; data safety and monitoring board, P. Yéni, F. Ferchal, T. Grooters; participating centers and investigators (all in France), Hopital Lagny-Marne-La-Vallee, Lagny, F. David; Hopital Avicenne, Bobigny, M. Bentata; Hopital Saint-Jacques, Besançon, B. Hoen; Hopital Saint-Louis, Paris, J. M. Molina, B. Loze, J. Pavie, C. de Verdiere, S. Fournier, and G. Madelaine; Hopital Cochin, Paris, D. Salmon; Hopital Raymond Poincare, Garches, C. Perronne and P. De Truchis; Hopital Necker, Paris, B. Dupont, O. Lortholary, and L. Roudière; Hopital Pitie-Salpetriere, Paris, C. Katlama, A. Simon-Coutellier, V. Calvez, A. G. Marcellin, and M. Kirstetter; Hopital Bicetre, Kremlin Bicetre, J. F. Delfraissy; Paris Hopital Saint-Antoine, Paris, P. M. Girard; Hopital Saint-Andre, Bordeaux, P. Morlat; Hopital Pessac, Bordeaux, J. L. Pellegrin; Hopital Edouard Herriot, Lyon, J. M. Livrozet; Hopital Hotel-Dieu, Lyon, C. Trepo and L. Cotte; Hopital Sainte-Marguerite, Marseille, J. Gastaut and I. Poizot-Martin; Hopital Conception, Marseille, H. Gallais; Hopital Gui de Chauliac, Montpellier, J. Reynes; Hopital de l'Hotel-Dieu, Nantes, F. Raffi; Hopital de l'Archet, Nice, P. Dellamonica; Hopital Purpan, Toulouse, P. Massip and B. Marchou; Centre Hospitalo-Universitaire de Caen, Caen, C. Bazin; Hopital Saint-Etienne, St.-Etienne, B. Lucht; Hopital de Toulon, Toulon, A. Lafeuillade; Hopital d'Orleans, Orleans, D. Hocqueloux and T. Prazuc.
REFERENCES
- 1.Brun-Vezinet, F., D. Descamps, A. Ruffault, B. Masquelier, V. Calvez, G. Peytavin, F. Telles, L. Morand-Joubert, J. L. Meynard, M. Vray, and D. Costagliola. 2003. Clinically relevant interpretation of genotype for resistance to abacavir. AIDS 17:1795-1802. [DOI] [PubMed] [Google Scholar]
- 2.Chaix, M. L., D. Descamps, M. Harzic, V. Schneider, C. Deveau, C. Tamalet, I. Pellegrin, J. Izopet, A. Ruffault, B. Masquelier, L. Meyer, C. Rouzioux, F. Brun-Vezinet, and D. Costagliola. 2003. Stable prevalence of genotypic drug resistance mutations but increase in non-B virus among patients with primary HIV-1 infection in France. AIDS 17:2635-2643. [DOI] [PubMed] [Google Scholar]
- 3.Coakley, E. P., J. M. Gillis, and S. M. Hammer. 2000. Phenotypic and genotypic resistance patterns of HIV-1 isolates derived from individuals treated with didanosine and stavudine. AIDS 14:F9-F15. [DOI] [PubMed] [Google Scholar]
- 4.Flandre, P., A. Alcais, D. Descamps, L. Morand-Joubert, and V. Joly. 2004. Estimating and comparing reduction in HIV-1 RNA in clinical trials using methods for interval censored data. J. Acquir. Immune Defic. Syndr. 35:286-292. [DOI] [PubMed] [Google Scholar]
- 5.Flandre, P., C. Durier, D. Descamps, O. Launay, and V. Joly. 2002. On the use of magnitude of reduction in HIV-1 RNA in clinical trials: statistical analysis and potential biases. J. Acquir. Immune Defic. Syndr. 30:59-64. [DOI] [PubMed] [Google Scholar]
- 6.Gu, Z., Q. Gao, X. Li, M. A. Parniak, and M. A. Wainberg. 1992. Novel mutation in the human immunodeficiency virus type 1 reverse transcriptase gene that encodes cross-resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine. J. Virol. 66:7128-7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanna, G. J., V. A. Johnson, D. R. Kuritzkes, D. D. Richman, A. J. Brown, A. V. Savara, J. D. Hazelwood, and R. T. D'Aquila. 2000. Patterns of resistance mutations selected by treatment of human immunodeficiency virus type 1 infection with zidovudine, didanosine, and nevirapine. J. Infect. Dis. 181:904-911. [DOI] [PubMed] [Google Scholar]
- 8.Harrigan, P. R., I. Kinghorn, S. Bloor, S. D. Kemp, I. Najera, A. Kohli, and B. A. Larder. 1996. Significance of amino acid variation at human immunodeficiency virus type 1 reverse transcriptase residue 210 for zidovudine susceptibility. J. Virol. 70:5930-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haubrich, R. H., C. A. Kemper, N. S. Hellmann, P. H. Keiser, M. D. Witt, D. N. Forthal, J. Leedom, M. Leibowitz, J. M. Whitcomb, D. Richman, and J. A. McCutchan. 2002. The clinical relevance of non-nucleoside reverse transcriptase inhibitor hypersusceptibility: a prospective cohort analysis. AIDS 16:F33-F40. [DOI] [PubMed] [Google Scholar]
- 10.Jonckheere, A. R. 1954. A distribution-free k-sample test against ordered alternatives. Biometrika 41:133-145. [Google Scholar]
- 11.Kellam, P., C. A. Boucher, and B. A. Larder. 1992. Fifth mutation in human immunodeficiency virus type 1 reverse transcriptase contributes to the development of high-level resistance to zidovudine. Proc. Natl. Acad. Sci. USA 89:1934-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kempf, D. J., J. D. Isaacson, M. S. King, S. C. Brun, J. Sylte, B. Richards, B. Bernstein, R. Rode, and E. Sun. 2002. Analysis of the virological response with respect to baseline viral phenotype and genotype in protease inhibitor-experienced HIV-1-infected patients receiving lopinavir/ritonavir therapy. Antivir. Ther. 7:165-174. [PubMed] [Google Scholar]
- 13.Lacey, S. F., and B. A. Larder. 1994. Novel mutation (V75T) in human immunodeficiency virus type 1 reverse transcriptase confers resistance to 2′,3′-didehydro-2′,3′-dideoxythymidine in cell culture. Antimicrob. Agents Chemother. 38:1428-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larder, B. A., G. Darby, and D. D. Richman. 1989. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science 243:1731-1734. [DOI] [PubMed] [Google Scholar]
- 15.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 16.Marcelin, A. G., C. Delaugerre, M. Wirden, P. Viegas, A. Simon, C. Katlama, and V. Calvez. 2004. Thymidine analogue reverse transcriptase inhibitors resistance mutations profiles and association to other nucleoside reverse transcriptase inhibitors resistance mutations observed in the context of virological failure. J. Med. Virol. 72:162-165. [DOI] [PubMed] [Google Scholar]
- 17.Marschner, I. C., R. A. Betensky, V. DeGruttola, S. M. Hammer, and D. R. Kuritzkes. 1999. Clinical trials using HIV-1 RNA-based primary endpoints: statistical analysis and potential biases. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:220-227. [DOI] [PubMed] [Google Scholar]
- 18.Molina, J. M., A. G. Marcelin, J. Pavie, L. Heripret, C. M. de Boever, M. Troccaz, G. Leleu, V. Calvez, et al. 2005. Didanosine in HIV-1-infected patients experiencing failure of antiretroviral therapy: a randomized placebo-controlled trial. J. Infect. Dis. 191:840-847. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen, C., L. Bruun, L. R. Mathiesen, C. Pedersen, and J. Gerstoft. 1996. Development of resistance of zidovudine (ZDV) and didanosine (ddI) in HIV from patients in ZDV, ddI and alternating ZDV/ddI therapy. AIDS 10:625-633. [DOI] [PubMed] [Google Scholar]
- 20.Sauerbrei, W., and M. Schumacher. 1992. A bootstrap resampling procedure for model building: application to the Cox regression model. Stat. Med. 11:2093-2109. [DOI] [PubMed] [Google Scholar]
- 21.Shafer, R. W., M. J. Kozal, M. A. Winters, A. K. Iversen, D. A. Katzenstein, M. V. Ragni, W. A. Meyer III, P. Gupta, S. Rasheed, R. Coombs, et al. 1994. Combination therapy with zidovudine and didanosine selects for drug-resistant human immunodeficiency virus type 1 strains with unique patterns of pol gene mutations. J. Infect. Dis. 169:722-729. [DOI] [PubMed] [Google Scholar]
- 22.St. Clair, M. H., J. L. Martin, G. Tudor-Williams, M. C. Bach, C. L. Vavro, D. M. King, P. Kellam, S. D. Kemp, and B. A. Larder. 1991. Resistance to ddI and sensitivity to AZT induced by a mutation in HIV-1 reverse transcriptase. Science 253:1557-1559. [DOI] [PubMed] [Google Scholar]
- 23.Whitcomb, J. M., N. T. Parkin, C. Chappey, N. S. Hellmann, and C. J. Petropoulos. 2003. Broad nucleoside reverse-transcriptase inhibitor cross-resistance in human immunodeficiency virus type 1 clinical isolates. J. Infect. Dis. 188:992-1000. [DOI] [PubMed] [Google Scholar]
- 24.Winters, M. A., R. J. Bosch, M. A. Albrecht, and D. A. Katzenstein. 2003. Clinical impact of the M184V mutation on switching to didanosine or maintaining lamivudine treatment in nucleoside reverse-transcriptase inhibitor-experienced patients. J. Infect. Dis. 188:537-540. [DOI] [PubMed] [Google Scholar]
- 25.Zhang, D., A. M. Caliendo, J. J. Eron, K. M. DeVore, J. C. Kaplan, M. S. Hirsch, and R. T. D'Aquila. 1994. Resistance to 2′,3′-dideoxycytidine conferred by a mutation in codon 65 of the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 38:282-287. [DOI] [PMC free article] [PubMed] [Google Scholar]