Abstract
The protein inverse folding problem, designing amino acid sequences that fold into desired protein structures, is a critical challenge in biological sciences. Despite numerous data-driven and knowledge-driven methods, there remains a need for a user-friendly toolkit that effectively integrates these approaches for in-silico protein design. In this paper, we present DIProT, an interactive protein design toolkit. DIProT leverages a non-autoregressive deep generative model to solve the inverse folding problem, combined with a protein structure prediction model. This integration allows users to incorporate prior knowledge into the design process, evaluate designs in silico, and form a virtual design loop with human feedback. Our inverse folding model demonstrates competitive performance in terms of effectiveness and efficiency on TS50 and CATH4.2 datasets, with promising sequence recovery and inference time. Case studies further illustrate how DIProT can facilitate user-guided protein design.
Keywords: Computational Protein design, Interactive design toolkit, Protein inverse folding, Non-autoregressive decoding
1. Introduction
Proteins, composed of amino acid chains that fold into various structures, are fundamental to numerous biological functions. Predicting the structure of a given protein sequence is a well-explored problem, with a range of algorithms developed, including energy function based methods [1], co-evolutionary based methods [2], and end-to-end based methods [3,4]. The success of these methods underscores the potential of computational approaches in the protein-related domain.
An equally important but more challenging problem is the inverse folding problem, which involves designing amino acid sequences that conform as closely as possible to a given protein structure [5]. Such a task aids in the design of refined mutant proteins and serves as a core step in de novo protein design for bioengineering [6]. Early works [[7], [8], [9], [10]] [[7], [8], [9], [10]] [[7], [8], [9], [10]] focused on designing de novo proteins by analyzing energy function terms and mutating to achieve low energy states that fit the structure. The advent of end-to-end deep learning models for protein folding prediction has inspired the development of similar models for the inverse folding problem [[11], [12], [13]] [[11], [12], [13]] [[11], [12], [13]]. These models, typically conditional generative models, use the target structure as the condition to model the conditional distribution of the amino acid sequence. They aim to fit the unknown conditional distribution of the amino acid sequences to all the coordinates group backbone atoms of length , i.e., fit .
Since all the possible states of grow exponentially with the sequence length , the practical training object is usually to maximize the conditional likelihood of a residue given structure and parts of sequences . The whole sequence distribution for protein design can be indirectly given by the following equation.
The factorization form implicates the autoregressive sampling pattern to decode the final sequence, which means sampling the residue at the th position refers to the , and use the result as the condition to sample the next residue iteratively. However, these autoregressive models suffer from two key issues. First, the teacher forcing problem [14] arises when using autoregressive models. These models are trained to predict each token based on the correct previous tokens. But during inference, the model must predict based on its own previous predictions. This discrepancy between training and inference can lead to errors that propagate through the sequence. This is particularly problematic in the inverse folding problem, where the order of the residue sequence does not reflect the dependency among residues. Second, the efficiency of autoregressive models is another limitation. The time complexity of autoregressive generation, which corresponds to its left-to-right, one-by-one decoding behavior, can significantly increase the time required for in silico design processes, particularly for long residue chains.
Non-autoregressive generation, a collection of parallel generation algorithms, offers a solution to these problems. By simultaneously generating and refining the whole sequence, these algorithms can address the teacher forcing problem and improve generation efficiency through parallelization. These methods have shown promise in the domain of speech generation, as evidenced by works such as FastSpeech [15], MaskPredict [16], and Improved Non-Autoregressive [17].
Inspired by these works, we apply a non-autoregressive generative model to the protein design problem, addressing both the teacher forcing gap and low generation efficiency by generating and refining the entire sequence at once. In silico experiments suggest that our model achieves a 54.4 % sequence recovery rate on TS50 and 50.6 % in CATH4.2 (details in Appendix A). Building on this, we develop DIProT, a deep learning based interactive protein design toolkit that allows users to specify the target structure and fix parts of the sequence they want to preserve. This facilitates user-guided, in-silico protein design and evaluation. The web server of DIProT is available at http://bioinfo-xwwang-thu.cn/DIProT.
2. Results
2.1. Design process and Utilization of DIProT for interactive Protein design
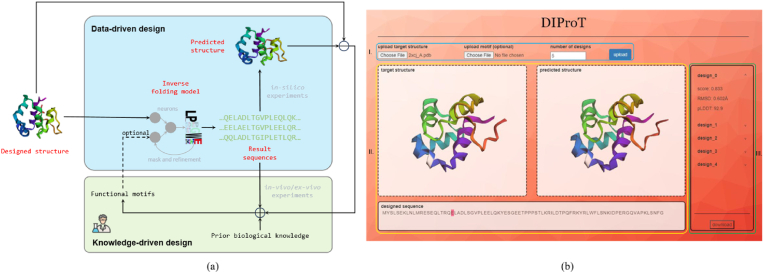
Protein design is a complex task that often involves multiple iterations of trial and error [18,19], as depicted in Fig. 1(a). The process normally starts with the selection of a reference structure with a high potential for the desired function. This structure can originate from natural proteins with similar functions or be based on domain knowledge in biochemistry and structural biology [20,21]. Amino acid sequences are then generated through mutation or protein design algorithms, and experimentally validated to assess their structural proximity to the reference and their functional efficacy. Insights from these experimental results guide the identification of conserved regions and the refinement of other regions, leading to subsequent iterations that potentially increase the success rate.
Fig. 1.
Protein design process and toolkit overview. (a) A typical protein design process that involves a computational design (blue) and a wet-lab validation (green) cycle. The computational design step relies on deep learning models or big data analysis, which are data-driven methods. The wet-lab validation step depends on the experimenters' prior knowledge and experience, which are knowledge-driven methods. Functional motifs from the knowledge-driven step can be used as inputs for the data-driven step, which can bridge the two steps and enable iterative refinement of the design results. (b) The graphical user interface (GUI) of DIProT. (i) Buttons for uploading the designed structure (in.pdb format), the functional motif (in.json format), and selecting the number of designs. (ii) Canvas for displaying the uploaded structure (on the left) and the predicted structure (on the right) for the designed sequence (at the bottom). (iii) Designs and their evaluation metrics. When a design is clicked, the corresponding sequence and predicted structure will be shown in the respective areas in (ii).
However, this process is time-consuming and labor-intensive. One of the most challenging steps is the generation of candidate sequences, a problem known as inverse folding. Existing algorithms to tackle this problem [11,13,22] often require low-level command-line programming environments and complicated parameter tuning, posing significant challenges. To address these inconveniences and fit the entire design loop, we developed DIProT, a toolkit with the following features.
-
1.
An efficient inverse-folding model based on the non-autoregressive decoding paradigm, enabling cost-effective and rapid in silico experiments.
-
2.
A user-friendly graphical user interface (GUI) that integrates multiple algorithms to facilitate a fast and intuitive feedback design loop (Fig. 1(b)).
-
3.
The ability to generate and rank candidate sequences given the reference structure and an optional sequence motif to be preserved, powered by deep generative models.
-
4.
Detailed atom-level structure comparisons between the predicted structure of generated sequences and the reference structure, assisting users in filtering sequences with low structural similarity or other undesired features and identifying conserved motifs for preservation in subsequent generation rounds.
Fig. 1(b) provides a visual representation of the DIProT process, using the redesign of a phage protein (PDB ID: 2xcj_A) as an example. In this example, the user uploads the full structure of 2xcj_A as the reference structure (in.pdb format), with no motif file, and specifies “5” for the number of designs to be generated. Motif file is an optional component in the design process. Specifically, the file should be in.json format, where keys represent fixed positions and values represent fixed amino acid types. It forms a vital bridge between data-driven design and knowledge-driven design, as shown in Fig. 1(a). It should be noted that DIProT only considers the coordinates of the main chain atoms of the reference structure as input. This means users can upload a full protein structure to generate a similar or improved version or a file with only main chain information for De Novo protein design.
Once the reference structure is uploaded and the number of designs specified, DIProT generates several sequences matching the reference structure using the inverse folding model. The in silico evaluations are then automatically applied to them, primarily contributed by an efficient structure prediction model, ESMFold [4]. As shown in Fig. 1(b)(iii), the designs are then ranked using the in silico metrics: (1) score, the likelihood of this sequence provided by the inverse folding model (higher means better). (2) RMSD, the root mean square deviation between the designed (predicted by ESMFold) and reference structure (lower means better). (3) pLDDT, ESMFold's local distance difference test score when predicting the structure (higher means better). These metrics provide a comprehensive evaluation of the protein designs, considering both the sequence and structural aspects.
The reference structure, the specific design sequence, and its prediction structure are visualized for users to interact with them (Fig. 1(b)(ii)). Visualization aids in the understanding and interpretation of the results, allowing users to intuitively check the potential success of the design and identify fragments that might cause failure.
The inverse folding model plays a key role in DIProT's design process. To have a better understanding of how it works and demonstrate its effectiveness and efficiency, the following sections will describe its implementation and performance.
2.2. Technical Realization
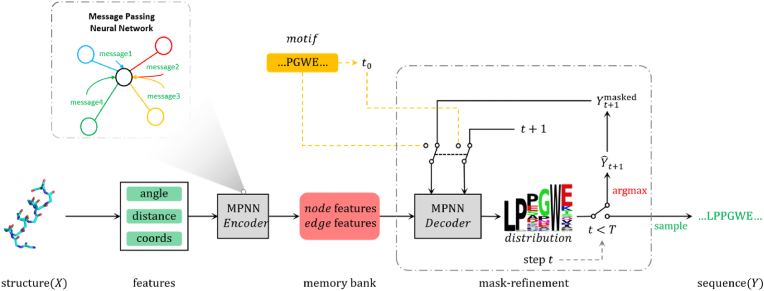
The overall inference pipeline for DIProT is depicted in Fig. 2. Given a protein structure, we represent it as a graph and extract features from the coordinates of its main chain atoms. These features are then embedded into a high-dimensional latent space and updated using an MPNN-based encoder [23], forming a memory bank. An MPNN-based decoder then generates the protein sequence from the memory bank, the previously decoded sequence, and the current decoding step. The decoder outputs a latent representation for all nodes, which is then processed by a classification head to obtain a distribution over 20 amino acids for each residue. If the decoding step reaches a pre-defined maximum, we sample a design sequence from the predicted distribution. Otherwise, we mask positions with low prediction confidence and proceed to the next decoding step. Masking residues with lower prediction confidence can provide purer sequence information for the next step of decoding, thereby facilitating the correction of previously incorrectly generated sequences (see Appendix A, Figure A2 for a case study). The mask-and-refinement algorithm allows for the direct use of functional motifs as pre-generated sequences, if available. Detailed implementation and principles are provided in Appendix A.
Fig. 2.
DIProT overview. Inference process for DIProT. We use a non-autoregressive method to generate protein sequences from protein structures. We first encode the input structure features into node embeddings and edge embeddings using an MPNN-based [23] encoder. Then we iteratively update the node embeddings and edge embeddings using an MPNN-based decoder and decode the node embeddings into amino acid probabilities using a softmax layer. We repeat this process for a fixed number of iterations (T = 20 in our experiments) and sample the amino acid at each position from the final probabilities.
2.3. In-silico performance of DIProT
2.3.1. Sequence level evaluation
In assessing DIProT's design performance, we first evaluated the native sequence recovery rate and perplexity on the CATH4.2 and TS50 datasets (see Appendix A for dataset details). Sequence recovery measures how well a computational method can reproduce a protein's natural amino acid sequence given its structure, while perplexity evaluates the model's prediction of the native sequence. The results, presented in Table 1 and Table 2, demonstrate that DIProT performs comparably or better than existing methods.
Table 1.
Sequence recovery and perplexity on CATH4.2 test set. DIProT is compared with ProteinMPNN, GVP-GNN, and Structured Transformer. As a non-autoregressive method, DIProT shows comparable performance to the state-of-the-art.
Table 2.
Sequence recovery on TS50 dataset. The same training set as used by GVP-GNN was used for evaluation on TS50, as there is no standard training set for this dataset. We retrained ProteinMPNN using the same training set and its open-source code [31].
Although sequence recovery is a widely used metric to evaluate a design model, it is not enough to judge its practical effectiveness. Therefore, we performed additional evaluations for both singleton and dataset cases, as depicted in Fig. 3.
Fig. 3.
Sequence level experiment results. (a) Sequence logo for a designed protein sequence (PDB id: 1h4a_X). To facilitate visualization, we only show a segment from position 31 to 45. The height of each letter indicates the predicted probability of the corresponding amino acid at that position. The letters are colored by the same groups as in BLOSUM62 (see subfigure (b)). The native sequence is displayed on the bottom, and at each position, we color its conservation analysis result predicted by ConSurf [32]. (b) Confusion matrix of DIProT's prediction (left) and BLOSUM62 substitution matrix (right). The 20 amino acids are grouped into 6 categories based on their similarity and interchangeability in BLOSUM62 (colored by different colors). Our model's prediction shows a similar pattern of mutual substitution as BLOSUM62. (c) Sequence recovery and diversity metrics for DIProT with different sampling temperatures. Higher sampling temperature leads to slightly lower sequence recovery but greatly higher sequence diversity. (d) Inference time of DIProT (red) and ProteinMPNN (blue) for different chain lengths. DIProT is more than 10 times faster than ProteinMPNN in inference speed, and the efficiency advantage of DIProT is more obvious for longer proteins.
We generate 20 sequences referencing the structure of a human gamma-crystallin D protein (PDB id: 1h4a_X) and draw a sequence logo (Fig. 3(a)). This shows that DIProT recommends sequences that maintain the original amino acid in positions like 35 and 39, conservative substitution in positions like 34 and 42, and suggests high diversity substitution in positions like 31 and 38. The prediction of conservation sites aligns with ConSurf's results, a widely-used tool for estimating evolutionary conservation based on phylogenetic relations between homologous sequences [32].
After running the inference process for each reference protein structure in our hold-out test set, we obtained a confusion matrix of the amino acid from the original protein. The matrix can be partitioned into 6 groups corresponding to the conservative substitution group indicated by BLOSUM62 (Fig. 3(b)). This shows that DIProT can learn substitution patterns among different types of amino acids without explicit instruction during model training, demonstrating its powerful sequence-generation capability.
We also assessed DIProT's ability to recommend diverse sequences at different sampling temperatures. Fig. 3(c) shows how sampling temperature affects sequence diversity and recovery, proving that DIProT can generate more diverse sequences with only a slight trade-off in sequence recovery.
2.3.2. Structure level evaluation
As protein function is determined by its structure, and structure by sequence, it's essential to evaluate designs on a structural level. To determine whether the designed sequences can fold to these reference structures as input, we use a structure-prediction model, ESMFold [4], to fold the designs. ESMFold has similar prediction precision compared with AlphaFold2 [3] but is more efficient. We mainly consider two metrics: RMSD and pLDDT. As described above, RMSD reflects the structural similarity between the predicted and native structures, while pLDDT reflects ESMFold's prediction confidence.
Similar to sequence similarity evaluation, we evaluate DIProT on a dataset level, comparing its results with random mutation. Fig. 4 shows that DIProT’ designs have lower RMSD (median = 1.31 Å vs. 16.38 Å) and higher pLDDT (median = 84.36 vs. 31.67) than randomly mutated sequences with the same sequence recovery. This suggests that DIProT can capture the deeper relationship between protein sequence and structure, beyond just optimizing sequence similarity. To rule out the effect of ESMFold prediction errors, we compare the prediction results of ESMFold for the native sequences and designed sequences (Fig. 4(b)). The results show a high correlation, further supporting that DIProT can generate sequences structurally similar to native ones.
Fig. 4.
Structural level experiment results. (a) Comparison of sequence recovery, ESMFold pLDDT, and RMSD metrics for our inverse folding model's designs (red) and randomly mutated sequences (blue). We use proteins in TS50 dataset for evaluation (see Appendix A). For each protein, we generate 20 designed sequences and 20 mutant sequences. The designed sequences are created by sampling from DIProT's prediction distribution at a temperature of 0.1. The mutant sequences are created by randomly introducing mutations into the native sequences, ensuring that the sequence recovery between the mutant and the native ranges from 20 % to 80 %. (b) Correlation between DIProT designed sequences and native sequences using pLDDT and RMSD metrics on the TS50 dataset. The designed sequences show a positive correlation with native sequences in terms of pLDDT ( = 0.48) and RMSD ( = 0.63). The results illustrate that DIProT can generate structurally similar sequences for natural proteins that ESMFold can predict accurately and that ESMFold's predictions are poor for unsuccessful designs. (c) A redesigned case for DIProT on a sugar-binding protein structure (PDB ID: 2xr6). The native structure, ESMFold's prediction of the native sequence, and ESMFold's prediction of the designed sequence are shown in cyan, red, and blue, respectively.
As shown in Fig. 4(c), we used DIProT to redesign a sugar-binding protein (PDB ID: 2xr6) and employed ESMFold to predict its structure. The result shows that despite a low sequence recovery (50.8 %), the structure corresponding to the designed sequence closely resembles the native structure (RMSD = 0.503 Å). Although there are still deviations in the structure corresponding to some positions (such as a small region at the N-terminus), it is a challenge not only for the inverse folding models but also for the folding prediction models to recover such parts. Indeed, ESMFold's analysis of the native sequence also shows certain deviations in the prediction results for such parts.
2.3.3. Time efficiency
Real-time performance is crucial for interactive design. To achieve this with the two deep-learning-based models in DIProT, we adopt two strategies. First, we use ESMFold as the folding prediction model. This end-to-end neural network does not require precomputed multiple sequence alignments [4], offering higher inference efficiency than AlphaFold. Second, we use a non-autoregressive decoding paradigm for the inverse folding model, which has a fixed number of iterations for generating sequences. In contrast, existing autoregressive models require as many iterations as the protein length. The non-autoregressive inverse folding model's inference time does not significantly change with protein length, while autoregressive methods' time increases linearly. This makes DIProT much faster than existing autoregressive models like ProteinMPNN (Fig. 3(d)).
3. Conclusion
Protein design is a critical and fascinating problem that has long captivated biologists. In this paper, we introduced a non-autoregressive inverse folding model for in-silico protein design. We combined this model with another protein folding prediction model, ESMFold, to develop the interactive protein design toolkit, DIProT. By integrating expert knowledge into a data-driven protein design model and providing a comprehensive analysis pipeline at both the sequence and structural levels, DIProT offers a human-in-the-loop design system that streamlines the user design workflow. It also provides visualization and user interaction of the design outcomes.
In this paper, we have not yet conducted wet-lab validations to evaluate DIPorT on the function level, in which motif sequence may play a crucial role in function. Looking forward, we plan to apply DIPorT to a specific protein design case, cooperating with its functional motif, to evaluate DIProT's design ability to incorporate the motif. We also plan to refine our inverse folding model and toolkit to tackle increasingly complex and diverse protein design challenges.
CRediT authorship contribution statement
Jieling He: Conceptualization, Methodology, Investigation, Validation, Writing – original draft. Wenxu Wu: Conceptualization, Methodology, Supervision, Writing – original draft. Xiaowo Wang: Conceptualization, Supervision, Funding acquisition, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 62250007, 62225307, 61721003), and a grant from the Guoqiang Institute, Tsinghua University (2021GQG1023).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2024.01.011.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Senior A.W., Evans R., Jumper J., Kirkpatrick J., Sifre L., Green T., et al. Improved protein structure prediction using potentials from deep learning. Nature. 2020;577:706–710. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- 2.Morcos F., Schafer N.P., Cheng R.R., Onuchic J.N., Wolynes P.G. Coevolutionary information, protein folding landscapes, and the thermodynamics of natural selection. Proc Natl Acad Sci. 2014;111:12408–12413. doi: 10.1073/pnas.1413575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Z., Akin H., Rao R., Hie B., Zhu Z., Lu W., et al. 2022. Language models of protein sequences at the scale of evolution enable accurate structure prediction 2022. 07.20.500902. [DOI] [Google Scholar]
- 5.Yue K., Dill K.A. Inverse protein folding problem: designing polymer sequences. Proc Natl Acad Sci USA. 1992;89:4163–4167. doi: 10.1073/pnas.89.9.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang P.-S., Boyken S.E., Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 7.Alford R.F., Leaver-Fay A., Jeliazkov J.R., O'Meara M.J., DiMaio F.P., Park H., et al. The Rosetta all-atom energy function for macromolecular modeling and design. J Chem Theory Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boas F.E., Harbury P.B. Potential energy functions for protein design. Curr Opin Struct Biol. 2007;17:199–204. doi: 10.1016/j.sbi.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 9.Lazaridis T., Karplus M. Effective energy function for proteins in solution. Proteins: Struct, Funct, Bioinf. 1999;35:133–152. doi: 10.1002/(SICI)1097-0134(19990501)35:2%3C133::AID-PROT1%3E3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 10.Pokala N., Handel T.M. Energy functions for protein design: Adjustment with protein–protein complex Affinities, models for the Unfolded state, and Negative design of Solubility and Specificity. J Mol Biol. 2005;347:203–227. doi: 10.1016/j.jmb.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 11.Dauparas J., Anishchenko I., Bennett N., Bai H., Ragotte R.J., Milles L.F., et al. Robust deep learning–based protein sequence design using ProteinMPNN. Science. 2022;378:49–56. doi: 10.1126/science.add2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Z., Tan C., Chacón P., Li S.Z. 2023. PiFold: toward effective and efficient protein inverse folding. [Google Scholar]
- 13.Hsu C., Verkuil R., Liu J., Lin Z., Hie B., Sercu T., et al. Learning inverse folding from millions of predicted structures. Int. Conf. Mach. Learn., PMLR. 2022:8946–8970. [Google Scholar]
- 14.Zhang W., Feng Y., Meng F., You D., Liu Q. Proc. 57th Annu. Meet. Assoc. Comput. Linguist. Association for Computational Linguistics; Florence, Italy: 2019. Bridging the gap between training and inference for neural machine Translation; pp. 4334–4343. [DOI] [Google Scholar]
- 15.Ren Y., Ruan Y., Tan X., Qin T., Zhao S., Zhao Z., et al. Fastspeech: fast, robust and controllable text to speech. Adv Neural Inf Process Syst. 2019;32 [Google Scholar]
- 16.Higuchi Y., Watanabe S., Chen N., Ogawa T., Kobayashi T. Mask CTC: non-autoregressive end-to-end ASR with CTC and mask predict. Proc Interspeech. 2020;2020:3655–3659. doi: 10.21437/Interspeech.2020-2404. [DOI] [Google Scholar]
- 17.Higuchi Y., Inaguma H., Watanabe S., Ogawa T., Kobayashi T. Icassp 2021-2021 IEEE Int. Conf. Acoust. Speech signal process. ICASSP, IEEE; 2021. Improved Mask-CTC for non-autoregressive end-to-end ASR; pp. 8363–8367. [Google Scholar]
- 18.Callaway E. Scientists are using AI to dream up revolutionary new proteins. Nature. 2022;609:661–662. doi: 10.1038/d41586-022-02947-7. [DOI] [PubMed] [Google Scholar]
- 19.Yu T., Boob A.G., Singh N., Su Y., Zhao H. In vitro continuous protein evolution empowered by machine learning and automation. Cell Syst. 2023;14:633–644. doi: 10.1016/j.cels.2023.04.006. [DOI] [PubMed] [Google Scholar]
- 20.Harbury P.B., Plecs J.J., Tidor B., Alber T., Kim P.S. High-resolution protein design with backbone freedom. Science. 1998;282 doi: 10.1126/science.282.5393.1462. [DOI] [PubMed] [Google Scholar]
- 21.Huang P.-.S. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing B., Eismann S., Suriana P., Townshend R.J.L., Dror R. 2020. Learning from protein structure with Geometric Vector Perceptrons. arXiv. [DOI] [Google Scholar]
- 23.Gilmer J., Schoenholz S.S., Riley P.F., Vinyals O., Dahl G.E. Int. Conf. Mach. Learn. PMLR; 2017. Neural Message Passing for Quantum Chemistry; pp. 1263–1272. [Google Scholar]
- 24.Ingraham J., Garg V., Barzilay R., Jaakkola T. Generative models for graph-based protein design. Adv Neural Inf Process Syst. 2019;32 [Google Scholar]
- 25.Qi Y., Zhang J.Z.H. DenseCPD: Improving the accuracy of neural-network-based computational protein sequence design with DenseNet. J Chem Inf Model. 2020;60:1245–1252. doi: 10.1021/acs.jcim.0c00043. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Chen Y., Wang C., Lo C.-C., Liu X., Wu W., et al. ProDCoNN: protein design using a convolutional neural network. Proteins: Struct, Funct, Bioinf. 2020;88:819–829. doi: 10.1002/prot.25868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S., Sun Z., Lin L., Liu Z., Liu X., Chong Y., et al. To improve protein sequence Profile prediction through image Captioning on Pairwise residue distance Map. J Chem Inf Model. 2020;60:391–399. doi: 10.1021/acs.jcim.9b00438. [DOI] [PubMed] [Google Scholar]
- 28.O'Connell J., Li Z., Hanson J., Heffernan R., Lyons J., Paliwal K., et al. SPIN2: predicting sequence profiles from protein structures using deep neural networks. Proteins: Struct, Funct, Bioinf. 2018;86:629–633. doi: 10.1002/prot.25489. [DOI] [PubMed] [Google Scholar]
- 29.Li Z., Yang Y., Faraggi E., Zhan J., Zhou Y. Direct prediction of profiles of sequences compatible with a protein structure by neural networks with fragment-based local and energy-based nonlocal profiles. Proteins: Struct, Funct, Bioinf. 2014;82:2565–2573. doi: 10.1002/prot.24620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leaver-Fay A., O'Meara M.J., Tyka M., Jacak R., Song Y., Kellogg E.H., et al. Scientific Benchmarks for guiding macromolecular energy function Improvement. Methods Enzymol. 2013;523:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. Cambridge, MA, USA: Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.ProteinMPNN. 2023 [Google Scholar]
- 32.ConSurf \vert Evolutionary conservation profiles of proteins. 2023. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.