Abstract
Isoniazid is metabolized by the genetically polymorphic arylamine N-acetyltransferase type 2 (NAT2). A greater number of high-activity alleles are related to increased acetylation capacity and in some reports to low efficacy and toxicity of isoniazid. The objective of this study was to assess individual isoniazid exposure based on NAT2 genotype to predict a personalized therapeutic dose. Isoniazid was administered to 18 healthy Caucasians (age 30 ± 6 years, body weight 74 ± 10 kg, five women) in random order as a 200-mg infusion, a 100-mg oral, and a 300-mg oral single dose. For the assessment of NAT2 genotype, common single nucleotide polymorphisms identifying 99.9% of variant alleles were characterized. Noncompartmental pharmacokinetics and compartmental population pharmacokinetics were estimated from isoniazid plasma concentrations until 24 h postdose by high-pressure liquid chromatography. The influence of NAT2 genotype, drug formulation, body weight, and sex on dose-normalized isoniazid pharmacokinetics was assessed by analysis of variance from noncompartmental data and confirmed by population pharmacokinetics. Eight high-activity NAT2*4 alleles were identified. Sex had no effect; the other factors explained 93% of the variability in apparent isoniazid clearance (analysis of variance). NAT2 genotype alone accounted for 88% of variability. Individual isoniazid clearance could be predicted as clearance (liters/hour) = 10 + 9 × (number of NAT2*4 alleles). To achieve similar isoniazid exposure, current standard doses presumably appropriate for patients with one high-activity NAT2 allele may be decreased or increased by approximately 50% for patients with no or two such alleles, respectively. Prospective clinical trials are required to assess the merits of this approach.
Isoniazid is a pivotal agent in the treatment of tuberculosis in combination with other drugs or alone as a prophylactic agent. The genetically polymorphic arylamine N-acetyltransferase type 2 (NAT2), a hepatic phase II drug-metabolizing enzyme, is responsible for isoniazid metabolism (Fig. 1), and individuals can be classified as “rapid acetylators” or “slow acetylators” according to their isoniazid N-acetylation capacity (1, 11). Traditionally, “rapid” and “slow acetylators” have been differentiated based on phenotyping tests, performed with a number of substrates such as caffeine and isoniazid. More recently, genetic methods enabled the identification of 36 NAT2 alleles (last revision as of 5 June 2003, see http://www.louisville.edu/medschool/pharmacology/NAT2.html). Patients phenotyped as “rapid acetylators” carry one (“intermediate acetylator”) or two high-activity alleles (NAT2*4 and NAT2*12), whereas “slow acetylators” have two low-activity variants.
FIG. 1.
Metabolism of isoniazid. NAT2, arylamine N-acetyltransferase type 2.
Early studies conducted with healthy volunteers and tuberculosis patients have shown the polymorphic elimination of isoniazid with mean elimination half-lives of 80 min for rapid acetylators and 180 min for slow acetylators (26). Seifart et al. reported that plasma concentrations of isoniazid and metabolic ratios of N-acetylisoniazid over isoniazid preferably at 3 h postdose were useful to distinguish between fast, intermediate, and slow acetylator phenotypes after a 5-mg/kg oral standard dose (25). Evans et al. found that patients with the highest plasma isoniazid levels were generally slow acetylators (11).
Most existing studies indicate that not only isoniazid concentrations but also efficacy and toxicity of isoniazid are linked to the activity of the NAT2 enzyme. In an early study by Ellard et al., rapid acetylators responded to a lesser extent than slow acetylators (56% and 76% of patients, respectively) to once-weekly treatment and also in the case of initial daily treatment (85% and 100% of patients, respectively) (8). This has not been confirmed in studies using more modern multidrug treatment schemes, but these studies were differently designed and had insufficient numbers of patients. In general, rapid and slow acetylators have been reported to respond equally well to standard daily isoniazid treatment for tuberculosis (16, 27).
However, it is well established that slow acetylators are more likely to develop polyneuropathy during isoniazid therapy if no pyridoxal is coadministered. Hepatotoxicity, the most relevant adverse effect of isoniazid, occurs in approximately 10% of all patients receiving standard drug doses. Slow acetylators are at greater risk to develop hepatotoxicity compared to rapid acetylators (4, 7, 10, 17, 23, 27). The association between NAT2 genotypes and severe liver injury has been noted in Japanese tuberculosis patients treated with standard dose of isoniazid plus rifampin (23). Therefore, the authors suggested that NAT2 genotyping prior to therapy could be useful to predict adverse reactions in patients with tuberculosis (17, 23).
In spite of this supporting evidence that NAT2 genotype and the rate of acetylation may affect the metabolism of isoniazid, therapeutic response, and incidence of adverse reactions, NAT2 genotyping and/or phenotyping is not used in clinical practice. The analysis of individual variability in isoniazid exposure and isoniazid dose adjustment based on NAT2 genotype has not been elucidated in prospective clinical trials. The objective of the present study was to assess individual isoniazid exposure based on NAT2 genotype to predict a personalized therapeutic dose.
MATERIALS AND METHODS
Volunteers.
Eighteen healthy Caucasian volunteers (13 male and 5 female, age 30 ± 6 years, body weight 74 ± 10 kg) participated in the open randomized change-over study for which a positive vote of the Ethics Committee of the University of Cologne, Germany, was available. The participants had given their written informed consent. Isoniazid was administered in random order as a 200-mg intravenous (Tebesium-s-250, HEFA PHARMA Vertriebs GmbH & Co. KG, Germany) short infusion (20 min), a 100-mg oral (ISOZID, FATOL Arzneimittel GmbH, Germany), and a 300-mg (with 60 mg vitamin B6) oral (ISOZID COMP. N, FATOL Arzneimittel GmbH, Germany) single dose under standardized conditions.
NAT2 genotype with respect to common single nucleotide polymorphisms comprising 99.9% of central European genotypes was characterized by restriction fragment length polymorphism-PCR as previously described (2). Specifically, the subjects were screened for the presence of the following mutations: G191A, C282T, T341C, C481T, G590A, A803G, and G857A.
Quantification of isoniazid.
Plasma isoniazid concentrations up to 24 h postdose were determined by high-pressure liquid chromatography with electrochemical detection (LLOQ: 0.050 μg/ml). Samples (0.5 ml) were mixed with internal standard (N-methyldopamin) solution and HCl. After addition of 350 μl 1 M perchloric acid, the mixture was shaken and centrifuged; 20 μl of the supernatant was chromatographed on a reversed phase column (Spherimage-80 ODS2 5-μm particle size, precolumn 5 by 4 mm, analytical column 120 by 4 mm; Knauer, Berlin, Germany), eluted with an isocratic solvent system consisting of 7.05 g NaH2PO4, 0.750 g 1-octansulfonic acid sodium salt hydrate, 0.245 g ethylenediamine tetraacetate, and methanol [840:170, vol/vol, pH 2.8 using phosphoric acid (85%)] and monitored by electrochemical detection. Isoniazid and internal standard were eluted after approximately 2.9 and 7.5 min, respectively. Calibration was performed by weighted (1/concentration2) linear regression. The interday precision of the calibration standards ranged from 7.2 to 7.4%, and the analytical recovery of the spiked quality control standards of isoniazid in plasma was 100.8% (8.00 μg/ml), 97.1% (2.00 μg/ml), and 100.5% (0.100 μg/ml), respectively.
Noncompartmental analysis.
Noncompartmental pharmacokinetic parameters were calculated using WinNonlin Professional 4.0.1, Pharsight Corp., Mountain View, CA. The parameters were derived individually for each subject from the concentrations of isoniazid in plasma. Absolute bioavailability (F) was calculated as the AUC ratio for the oral preparations over that for the intravenous preparation, corrected for the individual dose: F = (AUC0-∞ [oral]/dose)/(AUC0-∞ [intravenous]/dose). Apparent clearance was calculated as dose/AUC0-∞. All other statistical calculations were done with SPSS 11.0 for Windows (SPSS Inc., Chicago, IL) and Excel 97 (Microsoft Corp., Redmond, WA). The Kruskal-Wallis test was used to examine the impact of NAT2 genotype on dose-normalized isoniazid pharmacokinetics. Subsequently, the quantitative influence of NAT2 genotype, drug formulation, body weight and sex on apparent clearance of isoniazid was assessed by analysis of variance and analysis of covariance.
Compartmental population pharmacokinetic analysis.
The software NONMEM V version 1.1 was used (NONMEM Project Group, University of California at San Francisco, 1998). An exponential error model was chosen for the interindividual variability of the kinetic parameters whereas a combined additive and proportional error model was used for the residual variability. Fitting was performed with the “first-order conditional estimates” algorithm, taking interactions between the parameters into account. The most simple model, i.e., a one-compartment model with first-order absorption, a bioavailability of F = 1 for oral preparations and no intraindividual variation in any parameter, was stepwise expanded to more complex models if these were superior as assessed by the plausibility of the parameter estimates and their 95% confidence intervals (which were not allowed to include zero and/or unity), by goodness-of-fit plots, and by a significant (P < 0.05) reduction of the objective function provided by NONMEM. Covariates tested included sex, body weight and NAT2 genotype, with the effect of NAT2 genotype on clearance being modeled as a fractional contribution of each high and low activity NAT2 allele to overall clearance.
RESULTS
Four different NAT2 alleles were detected in the study population. Eight wild-type high-activity NAT2*4 alleles were identified. The following NAT2 alleles related to a decreased activity of the enzyme (2, 13, 14) were found: NAT2*5A (n = 2), NAT2*5B (n = 16), and NAT2*6A (n = 10). Three of the 18 subjects had two high-activity alleles, two had one high- activity allele, and 13 subjects had none (Table 1).
TABLE 1.
NAT2 genotypes observed in the study population
| No. of subjects | Genotype | Predicted acetylator phenotype |
|---|---|---|
| 1 | NAT2*5A/NAT2*5B | Slow |
| 1 | NAT2*5A/NAT2*6A | Slow |
| 4 | NAT2*5B/NAT2*5B | Slow |
| 6 | NAT2*5B/NAT2*6A | Slow |
| 1 | NAT2*6A/NAT2*6A | Slow |
| 1 | NAT2*4/NAT2*5B | Intermediate |
| 1 | NAT2*4/NAT2*6A | Intermediate |
| 3 | NAT2*4/NAT2*4 | Rapid |
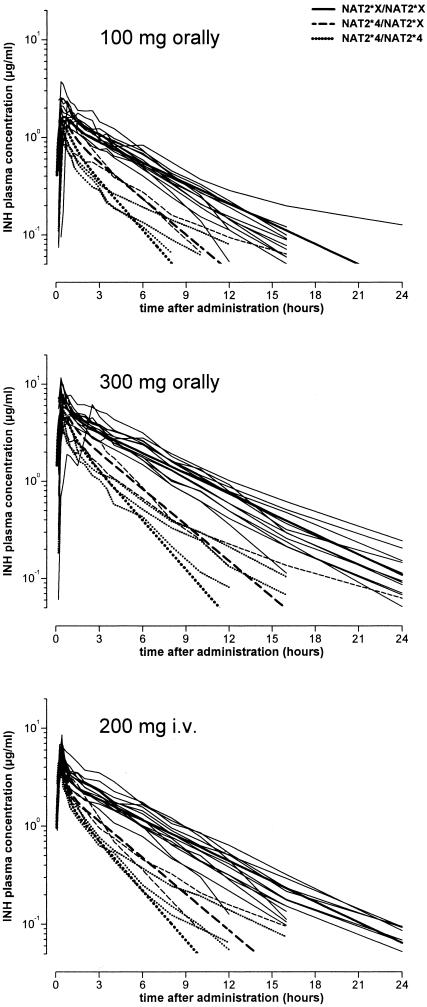
For all three treatments, concentrations observed in rapid acetylators were considerably lower than those found in slow acetylators. Concentrations versus time profiles depending on NAT2 genotype are presented in Fig. 2.
FIG. 2.
Concentration versus time profiles for each isoniazid preparation according to the number of low-activity (NAT2*X) and wild-type high-activity (NAT2*4) alleles for NAT2. Thin lines represent individual data, and thick lines are the population estimates for the NAT2 genotype. The 300-mg isoniazid dose was administered in combination with 60 mg vitamin B6.
The clear difference in plasma concentrations of isoniazid according to the number of high activity (i.e., NAT2*4) NAT2 alleles was reflected in noncompartmental AUC values and, for the low isoniazid dose, also in Cmax values (Table 2). A linear relationship between isoniazid clearance and the number of highly active NAT2*4 alleles was apparent (Fig. 3). The analysis of covariance indicated the following results for this parameter: the number of high-activity NAT2 alleles accounted for 88% of overall variability; isoniazid preparation and body weight explained only 2% and 3% of overall variation, respectively, whereas sex had no significant effect. Combined, these parameters explained 93% of the variation in apparent noncompartmental isoniazid clearance. Considering only NAT2 genotype, individual isoniazid clearance could be predicted as apparent clearance (liters/hour; noncompartmental approach) = 10.00 + (number of NAT2*4 alleles) × 8.9.
TABLE 2.
Pharmacokinetic parameters for each isoniazid preparation according to the number of “wild-type” high-activity (NAT2*4) and low-activity (NAT2*X) allelesa
| Dose (mg) | No. of NAT2*4 alleles (n)b | AUC/dose (h/liter) | Cmax/dose (1/liter) | tmax (h) | t1/2 (h) | Bioavailability (%) | Vdss (liters) |
|---|---|---|---|---|---|---|---|
| 100, oral | 0 (13) | 0.091 (0.067-0.132)** | 0.021 (0.015-0.037)* | 0.50 (0.33-1.00) | 3.58 (1.70-10.40) | 90.5 (79.2-113.3) | |
| 1 (2) | 0.048** | 0.017* | 1.25 | 2.20 | 87.4 | ||
| 0.048** | 0.009* | 0.50 | 6.02 | 92.8 | |||
| 2 (3) | 0.031 (0.027-0.037)** | 0.012 (0.0117-0.015) | 0.33 | 3.82 (2.55-4.75) | 78.4 (74.7-89.3) | ||
| 300, oral | 0 (13) | 0.107 (0.077-0.154)** | 0.026 (0.015-0.039) | 0.50 (0.33-2.50) | 4.40 (2.44-5.86) | 107.2 (91.8-115.9) | |
| 1 (2) | 0.066** | 0.026 | 0.50 | 2.52 | 118.8 | ||
| 0.048** | 0.015 | 0.33 | 6.57 | 92.0 | |||
| 2 (3) | 0.041 (0.036-0.042)** | 0.018 (0.017-0.027) | 0.33 (0.33-0.50) | 3.39 (2.46-3.93) | 100.1 (98.2-105.9) | ||
| 200, i.v. | 0 (13) | 0.102 (0.066-0.138)** | 0.034 (0.019-0.041) | 4.18 (2.08-5.02) | 51.8 (30.0-71.1) | ||
| 1 (2) | 0.055** | 0.031 | 1.96 | 43.6 | |||
| 0.051** | 0.029 | 5.32 | 98.2 | ||||
| 2 (3) | 0.040 (0.036-0.042)** | 0.026 (0.019-0.029) | 4.27 (2.76-4.38) | 77.5 (77.1-109) |
The parameters were derived directly from the data using the standard noncompartmental approach. AUC, area under the concentration-time curve; Cmax, maximal plasma concentration; tmax, time of occurrence of Cmax; t1/2, apparent elimination half-life; Vdss, volume of distribution at steady state; i.v., intravenous. The 300-mg isoniazid dose was administered in combination with 60 mg vitamin B6. *, P < 0.05; **, P < 0.01 in the Kruskal-Wallis test comparing genotypes. For all other parameters, there was no significant difference. Values are medians (range).
For n = 2, both values are shown.
FIG. 3.
Individual clearance of isoniazid in relationship to the number of high-activity NAT2*4 alleles. Clearance was calculated as dose/AUC from the noncompartmental analysis and corresponds to apparent clearance for the oral administrations and to true clearance for the intravenous administration. Values are slightly shifted along the x axis to improve clarity.
Compartmental population pharmacokinetic modeling gave very similar results (Table 3). The model describing the data best was a two-compartment model with different bioavailability for each oral preparation and with intraindividual variation in each parameter except in bioavailability and intercompartmental clearance (Table 3). Sex had no significant effect, whereas body weight had a significant effect on clearance and on both central and peripheral volume of distribution. NAT2 genotype had a major effect on isoniazid clearance (change in NONMEM objective function by −47, P < 0.001), with a pronounced difference mediated by the individual NAT2 alleles (Table 3). Transformation of the fractional clearance attributable to each allele gave the equation clearance (liters/hour; compartmental population pharmacokinetic approach) = 10.00 + (number of NAT2*4 alleles) × 9.2.
TABLE 3.
Compartmental population pharmacokinetic parameters derived from the complete data set using the NONMEM softwarea
| Population estimate of pharmacokinetic parameter (unit) | Lower limit of 95% CI | Point estimate | Upper limit of 95% CI | Intraindividual coefficient of variation (%) |
|---|---|---|---|---|
| Absorption constant for oral administration (h−1) | 1.87 | 2.28 | 2.69 | 27.1 |
| Clearance per low-activity NAT2 allele (liters h−1)* | 4.65 | 5.00 | 5.35 | 11.0 |
| Clearance per high-activity NAT2*4 allele (liters h−1)* | 12.4 | 14.2 | 16.0 | 11.0 |
| Central volume of distribution (liters) | 18.0 | 22.1 | 26.2 | 37.3 |
| Intercompartmental clearance (liters h−1) | 35.5 | 44.3 | 53.1 | n.s. |
| Peripheral volume of distribution (liters) | 29.6 | 35.2 | 40.8 | 33.3 |
| Bioavailability for the 100-mg oral dose (%) | 87.5 | 91.4 | 95.3 | n.s. |
| Bioavailability for the 300-mg oral dose (%) | 104 | 108 | 112 | n.s. |
| Effect of body wt on clearance (% increase of clearance per kg of body wt above mean body wt) | 0.511 | 1.14 | 1.77 | n.a. |
| Effect of body wt on both volumes of distribution (% increase per kg of body wt above mean body wt) | 0.303 | 1.45 | 2.60 | n.a. |
The best model was a two-compartment model with a separate contribution of each NAT2 allele to isoniazid clearance, an effect of body weight on both clearance, and the two volumes of distribution and assuming intraindividual variation of all parameters except the intercompartmental clearance. The 300-mg isoniazid dose was administered in combination with 60 mg vitamin B6. n.s., inclusion of this element of intraindividual variation did not improve the model significantly; n.a., not applicable. *, overall clearance is calculated as the sum of clearance fractions for each allele; i.e., the point estimate is 10.0, 19.2, and 28.4 liters h−1 for individuals with no, one, and two high-activity NAT2*4 alleles, respectively.
DISCUSSION
The variability in pharmacokinetics of isoniazid in healthy volunteers after standard dosing and its relationship with genetic variants of N-acetyltransferase type 2 and demographic characteristics was described in the present study. The study clearly shows that overall isoniazid exposure increases with a lower individual number of highly active NAT2 alleles (Tables 2 and 3). Corresponding results were demonstrated in recent clinical trials. Carriers of the NAT2*4/*4 genotype (two high-activity NAT2 alleles) have been reported to show lower isoniazid concentrations and high acetylisoniazid/isoniazid ratios at 3 h postdose than those with other NAT2 genotypes (15). In a study by Parkin et al., where plasma concentrations had been measured at short time intervals in tuberculosis patients, differences in the disposition kinetics of isoniazid have been observed between subjects with different NAT2 genotypes. Slow acetylators had four- to sixfold higher serum isoniazid concentrations over the interval 2 to 6 h after oral administration than rapid acetylators, prompting the authors to propose an individualized isoniazid dosing regimen (24). Likewise, a study based on urinary excretion in healthy volunteers and in patients showed that isoniazid is acetylated more extensively in subjects with a higher number of active NAT2 alleles (19).
Different mutations in the NAT2 gene associated with slow acetylation phenotype that had been identified in the trial group had been shown to account for the majority of the slow acetylator genotype in human population (NAT2*5A, NAT2*5B, and NAT2*6A) (3). Previous investigations of the functional characteristics of NAT2 single nucleotide polymorphisms have revealed different molecular mechanisms for slow acetylator phenotype (i.e., the reduction of mRNA and NAT2 protein expression in the presence of NAT2*5B and NAT2*5D alleles and protein instability in the cases of NAT2*6D and NAT2*14G) and various activity of NAT2 enzyme between different alleles (12, 20). The question arises whether the multiple slow acetylator phenotypes may lead to pharmacokinetic variability of isoniazid within the group of slow acetylators and to overlapping of slow and rapid acetylator groups. However, the current studies including the present investigation show that most of the overall variability can indeed be explained by grouping rapid and slow acetylation alleles.
The most important question is how differences of isoniazid exposure in genetically different individuals may alter the effectiveness and toxicity of isoniazid, especially in the case of dose-dependent toxicity such as neurological effects and liver disorders. Limited information exists regarding validated therapeutic ranges for isoniazid or the concentrations that cause toxic reactions. Mitchinson suggested that isoniazid maximal concentration to the MIC ratio could predict the clinical outcome of infections with Mycobacterium tuberculosis (22). The MIC of isoniazid (MIC90) for Mycobacterium tuberculosis is about 0.1 μg/ml, so a dose of 18.7 mg (16 times less than the usual therapeutic daily dose of 300 mg), which produced a mean 3-h concentration of 0.12 μg/ml, was reported to have a detectable early bactericidal activity (5). Nevertheless, the cumulative antibacterial effect of isoniazid seems to be related to the area under the concentration versus time curve and to maximal plasma concentration of the drug, as demonstrated in in vitro and laboratory animal studies (18). There is also evidence that the mean early bactericidal activity of isoniazid is lower in rapid acetylators than in slow acetylators and is influenced by plasma concentration of isoniazid (6). Accordingly, low plasma isoniazid levels in rapid acetylators are expected to be one of the reasons for occasional therapeutic failure or relapse (28).
On the other hand, isoniazid concentrations after a standard therapeutic dosing may be more toxic for slow acetylators because of relative overdosing, and dose adjusting may be useful to minimize relative overdosing. A significant association has been observed between NAT2 genotypes and severe hepatotoxicity in 77 Japanese tuberculosis patients undergoing treatment with isoniazid plus rifampin. Compared with rapid type, the relative risk of drug-induced hepatotoxicity was 4.0 (95% confidence interval, 1.9 to 6.1) for intermediate type and 28.0 (95% confidence interval, 26.0 to 30.0) for slow acetylator type (23). The authors suggested that NAT2 genotyping prior to therapy could be useful. According to the results of another study, patients with no high-activity NAT2 alleles had a higher risk of hepatotoxicity than those with at least one high-activity NAT2 allele, i.e., 26% versus 11% (17), so that NAT2 genotype was considered an important risk factor for antituberculosis drug-induced liver injury.
According to the results of a retrospective study in 102 Japanese tuberculosis patients, the incidence of adverse drug reactions upon standard treatment was high in slow acetylator-genotype patients (83.3%) and was low in the rapid acetylator genotype (0%) and intermediate acetylator genotype (2.4%) groups (15). In this study, there was a good association between a low value of N-acetylisoniazid over isoniazid ratio and the occurrence of adverse events, but there was no significant relationship between adverse reactions and plasma concentration of isoniazid at 3 h postdose. Possible reasons for this conflicting finding may be that (i) the 3-h plasma concentration does not necessarily reflect isoniazid exposure for the entire concentration versus time profile and (ii) the final chemical structure(s) causing hepatotoxicity is not isoniazid (alone). These data suggest that NAT2 genotyping may be superior to isoniazid therapeutic drug monitoring for an eventual dose individualization.
If one marketed preparation is to be replaced by another, extensive studies are required to show that there is no substantial pharmacokinetic difference, i.e., to prove bioequivalence defined as a ratio of mean bioavailability in the 0.80 to 1.25 range. Amazingly, for drugs such as isoniazid undergoing metabolism by a genetically polymorphic enzyme, this factor, which often is much more important than the preparation of the drug, usually is not taken into account. The data of the present study and published reports clearly suggest that determination of NAT2 genotype prior to isoniazid administration is clinically useful for the prediction of pharmacokinetic variability, and possible adjustment of isoniazid dosing regimen.
Conclusion.
The recommended standard daily dose of isoniazid in adults is 5.0 mg/kg (9, 21). This dose is probably balanced for the best risk/benefit ratio for all NAT2 genotypes and may be most appropriate for patients with a single rapid allele. Based on the concentration data presented, the standard dose could be roughly adjusted to 2.5 mg, 5.0 mg, and 7.5 mg/kg for subjects with none, one, or two rapid NAT2 alleles, respectively, to achieve similar isoniazid exposure. Probably a 50% increment of the recommended doses for all genotypes would be possible in order to achieve improved efficacy and safety, because relative overdosing of poor acetylators would be avoided. Prospective clinical trials are required to assess the merits of NAT2 genotyping and dose adjustment in isoniazid treatment.
Acknowledgments
We declare that we have no conflict of interest regarding this article.
The study was supported by a grant by Fatol Arzneimittel GmbH, Schiffweiler, Germany.
REFERENCES
- 1.Blum, M., A. Derrierre, D. M. Grant, M. Heim, and U. A. Meyer. 1991. Molecular mechanism of slow acetylation of drugs and carcinogens inhuman. Proc. Natl. Acad. Sci. USA 88:5237-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cascorbi, I., N. Drakoulis, J. Brockmoller, A. Maurer, K. Sperling, and I. Roots. 1995. Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am. J. Hum Genet. 57:581-592. [PMC free article] [PubMed] [Google Scholar]
- 3.Cascorbi, I., and I. Roots. 1999. Pitfalls in N-acetyltransferase 2 genotyping. Pharmacogenetics. 9:123-127. [DOI] [PubMed] [Google Scholar]
- 4.Clark, D. W. 1985. Genetically determined variability in acetylation and oxidation. Therapeutic implications. Drugs 29:342-375. [DOI] [PubMed] [Google Scholar]
- 5.Donald, P. R., F. A. Sirgel, F. J. Botha, H. I. Seifart, D. P. Parkin, M. L. Vandenplas, B. W. Van de Wal, J. S. Maritz, and D. A. Mitchison. 1997. The early bactericidal activity of isoniazid related to its dose size in pulmonary tuberculosis. Am. J. Respir. Crit. Care Med. 156:895-900. [DOI] [PubMed] [Google Scholar]
- 6.Donald, P. R., F. A. Sirgel, A. Venter, D. P. Parkin, H. I. Seifart, B. W. Van de Wal, C. Werely, P. D. van Helden, and J. S. Maritz. 2004. The influence of human N-acetyltransferase genotype on the early bactericidal activity of isoniazid. Clin. Infect. Dis. 39:1425-1430. [DOI] [PubMed] [Google Scholar]
- 7.Eichelbaum, M., H. K. Kroemer, and G. Mikus. 1992. Genetically determined differences in drug metabolism as a risk factor in drug toxicity. Toxicol. Lett Spec. No. 64-65:115-122. [DOI] [PubMed] [Google Scholar]
- 8.Ellard, G. A. 1976. Variations between individuals and populations in the acetylation of isoniazid and its significance for the treatment of pulmonary tuberculosis. Clin. Pharmacol. Ther. 19:610. [DOI] [PubMed] [Google Scholar]
- 9.Enarson, D. A., H. L. Rieder, T. Arnadottir, and A. Trebucq. 2000. Management of tuberculosis, a guide for low income countries, 5th ed. International Union Against Tuberculosis and Lung Disease, Paris, France.
- 10.Evans, D. A. 1989. N-acetyltransferase. Pharmacol. Ther. 42:157-234. [DOI] [PubMed] [Google Scholar]
- 11.Evans, D. A., K. A. Manley, and V. A. McKusick. 1960. Genetic control of isoniazid metabolism in man. Br. Med. J. 2:485-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fretland, A. J., M. A. Leff, M. A. Doll, and D. W. Hein. 2001. Functional characterization of human N-acetyltransferase 2 (NAT2) single nucleotide polymorphisms. Pharmacogenetics. 11:207-215. [DOI] [PubMed] [Google Scholar]
- 13.Grant, D. M., N. C. Hughes, S. A. Janezic, G. H. Goodfellow, H. J. Chen, A. Gaedigk, V. L. Yu, and R. Grewal. 1997. Human acetyltransferase polymorphisms. Mutat. Res. 376:61-70. [DOI] [PubMed] [Google Scholar]
- 14.Hein, D. W., M. A. Doll, A. J. Fretland, M. A. Leff, S. J. Webb, G. H. Xiao, U. S. Devanaboyina, N. A. Nangju, and Y. Feng. 2000. Molecular genetics and epidemiology of the NAT1 and NAT2 acetylation polymorphisms. Cancer Epidemiol. Biomarkers Prev. 9:29-42. [PubMed] [Google Scholar]
- 15.Hiratsuka, M., Y. Kushikawa, Y. Takekuma, M. Matsuura, K. Narahara, T. Inoue, et al. 2002. Genotyping of the N-acetyltransferase polymorphism in the prediction of adverse reactions to isoniazid in Japanese patients. Drug Metab. Pharmacokinet. 17:357-362. [DOI] [PubMed] [Google Scholar]
- 16.Horai, Y., and T. Ishizaki. 1987. Pharmacogenetics and its clinical implications: N-acetylation polymorphism. Ration. Drug Ther. 21:1-7. [PubMed] [Google Scholar]
- 17.Huang, Y. S., H. D. Chern, W. J. Su, J. C. Wu, S. L. Lai, S. Y. Yang, F. Y. Chang, and S. D. Lee. 2002. Polymorphism of the N-acetyltransferase 2 gene as a susceptibility risk factor for antituberculosis drug-induced hepatitis. Hepatology 35:883-889. [DOI] [PubMed] [Google Scholar]
- 18.Jayaram, R., R. K. Shandil, S. Gaonkar, P. Kaur, B. L. Suresh, B. N. Mahesh, R. Jayashree, V. Nandi, S. Bharath, E. Kantharaj, and V. Balasubramanian. 2004. Isoniazid pharmacokinetics-pharmacodynamics in an aerosol infection model of tuberculosis. Antimicrob Agents Chemother. 48:2951-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kita, T., Y. Tanigawara, S. Chikazawa, H. Hatanaka, T. Sakaeda, F. Komada, S. Iwakawa, and K. Okumura. 2001. N-Acetyltransferase2 genotype correlated with isoniazid acetylation in Japanese tuberculous patients. Biol. Pharm. Bull. 24:544-549. [DOI] [PubMed] [Google Scholar]
- 20.Leff, M. A., A. J. Fretland, M. A. Doll, and D. W. Hein. 1999. Novel human N-acetyltransferase 2 alleles that differ in mechanism for slow acetylator phenotype. J. Biol. Chem. 274:34519-34522. [DOI] [PubMed] [Google Scholar]
- 21.Management of Tuberculosis Training for Health Facility Staff. 2003, posting date. W.H.O. Report: Global tuberculosis control-surveillance, planning, financing-Global Tuberculosis Control (W.H.O./CDS/TB 2003.316). http://www.who.int/gbt/publications/globrep/index.htlm. [Online.]
- 22.Mitchison, D. A. 1984. Drug resistance in mycobacteria. Br. Med. Bull. 40:84-90. [DOI] [PubMed] [Google Scholar]
- 23.Ohno, M., I. Yamaguchi, I. Yamamoto, T. Fukuda, S. Yokota, R. Maekura, M. Ito, Y. Yamamoto, T. Ogura, K. Maeda, K. Komuta, T. Igarashi, and J. Azuma. 2000. Slow N-acetyltransferase 2 genotype affects the incidence of isoniazid and rifampicin-induced hepatotoxicity. Int. J. Tuberc. Lung Dis. 4:256-261. [PubMed] [Google Scholar]
- 24.Parkin, D. P., S. Vandenplas, F. J. Botha, M. L. Vandenplas, H. I. Seifart, P. D. van Helden, B. J. van der Walt, P. R. Donald, and P. P. van Jaarsveld. 1997. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am. J. Respir. Crit. Care Med. 155:1717-1722. [DOI] [PubMed] [Google Scholar]
- 25.Seifart, H. I., D. P. Parkin, F. J. Botha, P. R. Donald, and B. J. Van Der Walt. 2001. Population screening for isoniazid acetylator phenotype. Pharmacoepidemiol. Drug Safety 10:127-134. [DOI] [PubMed] [Google Scholar]
- 26.Tiitinen, H., M. J. Mattila, and A. W. Eriksson. 1968. Comparison of the isoniazid inactivation in Finns and Lapps. Ann. Med. Intern. Fenn. 57:161-166. [PubMed] [Google Scholar]
- 27.Weber, W. W., and D. W. Hein. 1985. N-acetylation pharmacogenetics. Pharmacol. Rev. 37:25-79. [PubMed] [Google Scholar]
- 28.Weiner, M., W. Burman, A. Vernon, D. Benator, C. A. Peloquin, A. Khan, S. Weis, B. King, N. Shah, and T. Hodge. 2003. Low isoniazid concentrations and outcome of tuberculosis treatment with once-weekly isoniazid and rifapentine. Am. J. Respir. Crit. Care Med. 167:1341-1347. [DOI] [PubMed] [Google Scholar]