Summary
Heterozygous missense variants and in-frame indels in SMC3 are a cause of Cornelia de Lange syndrome (CdLS), marked by intellectual disability, growth deficiency, and dysmorphism, via an apparent dominant-negative mechanism. However, the spectrum of manifestations associated with SMC3 loss-of-function variants has not been reported, leading to hypotheses of alternative phenotypes or even developmental lethality. We used matchmaking servers, patient registries, and other resources to identify individuals with heterozygous, predicted loss-of-function (pLoF) variants in SMC3, and analyzed population databases to characterize mutational intolerance in this gene. Here, we show that SMC3 behaves as an archetypal haploinsufficient gene: it is highly constrained against pLoF variants, strongly depleted for missense variants, and pLoF variants are associated with a range of developmental phenotypes. Among 14 individuals with SMC3 pLoF variants, phenotypes were variable but coalesced on low growth parameters, developmental delay/intellectual disability, and dysmorphism, reminiscent of atypical CdLS. Comparisons to individuals with SMC3 missense/in-frame indel variants demonstrated an overall milder presentation in pLoF carriers. Furthermore, several individuals harboring pLoF variants in SMC3 were nonpenetrant for growth, developmental, and/or dysmorphic features, and some had alternative symptomatologies with rational biological links to SMC3. Analyses of tumor and model system transcriptomic data and epigenetic data in a subset of cases suggest that SMC3 pLoF variants reduce SMC3 expression but do not strongly support clustering with functional genomic signatures of typical CdLS. Our finding of substantial population-scale LoF intolerance in concert with variable growth and developmental features in subjects with SMC3 pLoF variants expands the scope of cohesinopathies, informs on their allelic architecture, and suggests the existence of additional clearly LoF-constrained genes whose disease links will be confirmed only by multilayered genomic data paired with careful phenotyping.
Keywords: Cornelia de Lange syndrome, SMC3, loss-of-function, cohesin, CdLS3, LoF
To date, heterozygous missense variants and in-frame indels have been reported in association with “atypical” Cornelia de Lange syndrome. Here, we describe variable developmental phenotypes in individuals with heterozygous, predicted loss-of-function SMC3 variants, and investigate the functional effects of this variant type.
Introduction
The cohesin complex, a multimeric structure with the ability to entrap DNA, is integral to several dynamic genome processes.1,2 Specifically, cohesin facilitates sister chromatid cohesion during cell division,3 DNA repair,4 three-dimensional chromatin architecture,5 and transcriptional control of developmental genes.6,7 SMC3 (structural maintenance of chromosomes 3), the sole subunit shared by mitotic, interphase, and meiotic cohesin, is a ubiquitously expressed protein that binds with SMC1A/B to form the legs of the isosceles triangle commonly used to represent the closed heterotrimeric cohesin “ring,” with RAD21/REC8 forming the base.
In mouse models, homozygous Smc3 loss8,9 or conditional Smc3 depletion in oocytes10 results in embryonic lethality. In embryonic and adult mice, homozygous Smc3 loss in the blood compartment causes myeloid-based hematopoietic failure.11,12 However, Smc3 heterozygosity in mice is survivable and associated with behavioral phenotypes, neuronal/synaptic differences in the cerebral cortex, decreased body weight, craniofacial dysmorphism, and changes in gene expression,8,9 suggesting the potential for a similar phenomenon in humans.13
Missense variants and in-frame indels in SMC3 have been identified among patients with mild/atypical Cornelia de Lange syndrome (CdLS).13,14,15 Typical (classic) CdLS is caused by constitutional or mosaic heterozygous loss-of-function (LoF) pathogenic variants in NIPBL, whose protein product loads cohesin onto chromatin and facilitates its functioning. Classic CdLS is distinguished from atypical CdLS, which is a spectrum, by greater overall severity with a more characteristic facial appearance and the presence of major malformations. Variable intellectual disability, behavioral abnormalities, hirsutism, growth failure, gastroesophageal dysfunction, and short first metacarpals are seen in both typical and atypical cases. SMC3-related CdLS is marked principally by intellectual disability, facial dysmorphism, microcephaly, and postnatal growth delay.13,14,15,16
The reported missense variants and in-frame indels in SMC3 generally fall within important protein regions, including the antiparallel coiled-coil, hinge, and head domains.14 Tested pathogenic variants appear to act as dominant-negative alleles in human cell lines, where they increase the affinity of SMC hinge dimers for DNA, promoting genome instability and impairing genomic spatial organization.17 In silico modeling is also consistent with a dominant-negative effect.14 Furthermore, proteomic analyses reveal that missense-mutated SMC3 proteins are incorporated into the cohesin complex normally but prompt dysregulation of the c-MYC transcription factor, a feature of CdLS in the early prenatal period.18 Finally, SMC3 missense variants in CdLS patients confer a DNA methylation episignature grouping with that of other CdLS genes.19
Genotype-phenotype correlations are known for other cohesin genes, for example, NIPBL (missense in mild CdLS vs. LoF in severe CdLS20,21,22) and SMC1A (missense in atypical CdLS vs. LoF in SMC1A-related developmental and epileptic encephalopathy23). Thus, it has been proposed that the absence of SMC3 predicted LoF (pLoF) variants in previously described CdLS cases could be due to those variants either causing a different phenotype or being lethal for the developing embryo.24 Somatic SMC3 pLoF variants have been found in myeloid malignancies,25 including acute myeloid leukemia (AML)26 and Down syndrome acute megakaryoblastic leukemia.27,28 This is in line with LoF alleles in cohesin genes being a common event in several tumor types, not as initiating events but as subsequent drivers.29 Population genetic data suggest that germline SMC3 pLoF variants are considerably depleted in the general population (https://gnomad.broadinstitute.org/gene/ENSG00000108055); however, the consequence of heterozygous germline pLoF variants in humans is unknown because only a single case is documented in the literature.15
Here, we describe the clinical phenotypes of 14 individuals with germline pLoF variants in SMC3, including 4 frameshifts, 5 stop gains, 4 multigene deletions, and 1 predicted damaging splice variant. Most individuals have developmental delay, low growth parameters, and/or mild dysmorphism—reminiscent of atypical CdLS—although the penetrance of each of these features is incomplete and most individuals were not diagnosed clinically with CdLS. Comparisons of quantitative growth and developmental data between pLoF carriers and published cases of SMC3 pathogenic missense variants and in-frame indels reveal a genotype-phenotype correlation, with pLoF conferring overall milder, albeit overlapping, parameters. Our findings suggest that a broader class of undiscovered Mendelian conditions stemming from LoF variants in highly constrained genes likely remains uncataloged owing to generic, variable, and/or incompletely penetrant phenotypes and will require large dataset analyses, deep phenotyping, and functional experimentation to solve.
Subjects and methods
Case recruitment
Anonymized genetic and phenotypic information was collected, subjects were enrolled, and database searches performed under Boston Children’s Hospital institutional review board-approved protocol 00040134 or Children’s Hospital of Philadelphia protocol 16–013231. Multiple sources were queried to identify individuals with pLoF variants in SMC3: ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/)30; GeneMatcher/Matchmaker Exchange (https://genematcher.org/)31; the Baylor Genetics, GeneDx, Indiana University School of Medicine Genetic Testing, and Invitae clinical laboratories; a database of patients at Boston Children’s Hospital32; an in-house data portal built upon seqr33 and drawing from the Broad Institute Center for Mendelian Genomics and GREGoR Consortium projects; DECIPHER (https://www.deciphergenomics.org)34; and one previously published individual with an SMC3 nonsense variant referred for routine CdLS screening due to CdLS-like features and developmental delay.15
Growth and developmental milestone comparisons
Growth measurements were converted to Z scores using British Growth Survey data (https://www.rcpch.ac.uk/resources/growth-charts). Statistical comparisons and plot generation were undertaken using R.
Subject variant identification
Variants were identified via exome sequencing, microarray, or gene panel. These methods are listed for each case in Table S1, as is the confirmation status of variants by orthogonal methods.
Gene annotations
RefSeq transcript NM_005445 (Gencode: ENST00000361804.5) via (https://genome.ucsc.edu/) was used as the gene model for analyses. The Ensembl Variant Effect Predictor (https://useast.ensembl.org/Tools/VEP) was used to corroborate subjects’ variant nomenclature. Regional missense constraint for SMC3 was based on gnomAD data and assessed at a threshold of p = 0.001 as in https://www.biorxiv.org/content/10.1101/148353v1 and Wright et al.35 The region of potential escape from nonsense mediated decay was considered to be the final 55 nt of the penultimate exon of SMC3 (codon 1,176 and beyond). GRCh38/hg38 is the default genome build used unless specified otherwise.
UK Biobank (UKBB) pLoF sequence variant curation
The 10-110575403-TGTGA-T splice site variant, present in 5 individuals in the UKBB, was removed from the list of SMC3 pLoF variants in the UKBB; this is because it did not result in an alteration to the first 5 nt of the 5′ splice site.
Copy-number variant (CNV) detection among UK Biobank samples
UKBB SMC3 deletions were called using Genome Analysis Toolkit (GATK)-genomic CNV (gCNV) with genomic interval selection, sample processing, and defragmentation performed as in (https://www.biorxiv.org/content/10.1101/2022.08.25.504851v1). In brief, exome sequencing reads were first mapped to a predefined set of genomic intervals curated to capture the exonic regions of canonical protein-coding genes, and coverage was collected across well-captured intervals for CN inference. Samples with similar global read depth profiles were then clustered into batches to be jointly processed by GATK-gCNV, which outputs CNV calls across the set of well-captured intervals. These raw calls were subsequently defragmented, and to ensure that calls were not merged across high-quality CN = 2 regions, CNVs were refragmented to preserve any CN = 2 segments spanning at least 3 exome sequencing probes with quality score (QS) ≥100. Finally, the resulting callset was refined using the recommended QS, sample-level, and site frequency filters from (https://www.biorxiv.org/content/10.1101/2022.08.25.504851v1), along with a filter to remove CNVs covering fewer than 3 exons.
Infertility cohort missense burden analysis
Comparisons were made to gnomAD version 2.1.1 missense SMC3 variants with Combined Annotation Dependent Depletion (CADD) score >25 (Table S4) as assigned by the Variant Effect Predictor (https://useast.ensembl.org/info/docs/tools/vep/index.html). The cumulative allele frequency was transformed to a fraction with a denominator representing the mean number of alleles across these variants, for the purpose of chi-square analysis.
Gene expression analyses
SMC3 gene expression data were derived from Cancer Cell Line Encyclopedia (CCLE) samples, restricted to hematopoietic and lymphoid tumors, obtained via the Xena Functional Genomics Explorer (http://xenabrowser.net). Nonsense, frameshift, or splice site variants were considered pLoF. Samples containing >1 SMC3 variant were categorized by the most damaging variant class. Samples solely containing missense or silent SMC3 variants were removed, as were samples lacking SMC3 genotype or expression data.
Perturb-Seq data were derived from Replogle et al.36 and obtained via figshare (https://plus.figshare.com/articles/dataset/_Mapping_information-rich_genotype-phenotype_landscapes_with_genome-scale_Perturb-seq_Replogle_et_al_2022_-_commonly_requested_supplemental_files/21632564/1) and Genome-Wide Perturb-Seq (https://gwps.wi.mit.edu/). Briefly, this Perturb-Seq experiment involved single-cell sequencing following pooled, multiplexed CRISPR interference (CRISPRi) in 3 parallel approaches, each using a different CRISPRi guide library: K562 (chronic myelogenous leukemia, female) cells receiving a genome-wide guide library against all expressed genes and harvested on day 8 after transduction; K562 cells receiving a DepMap essential gene guide library and harvested on day 6 after transduction; and human telomerase reverse transcriptase RPE1 (retinal pigment epithelium, female) cells receiving a DepMap essential gene guide library and harvested on day 7 after transduction. Each library vector construct encoded 2 guides (single-guide RNAs) per target gene.
Smc3+/− mouse cortex differentially expressed genes (DEGs) from RNA sequencing were obtained from Fujita et al.8 Nipbl+/− whole-brain RNA sequencing data were obtained from Kean et al.37 In comparisons of these datasets, a denominator of 30,686 genes (see Figure 3D) was established from among the 55,228 total genes mapped in Kean et al.37 (GEO: GSE203014) based on those having ≥10 reads across all 22 samples, which was the cutoff used in the original analysis.
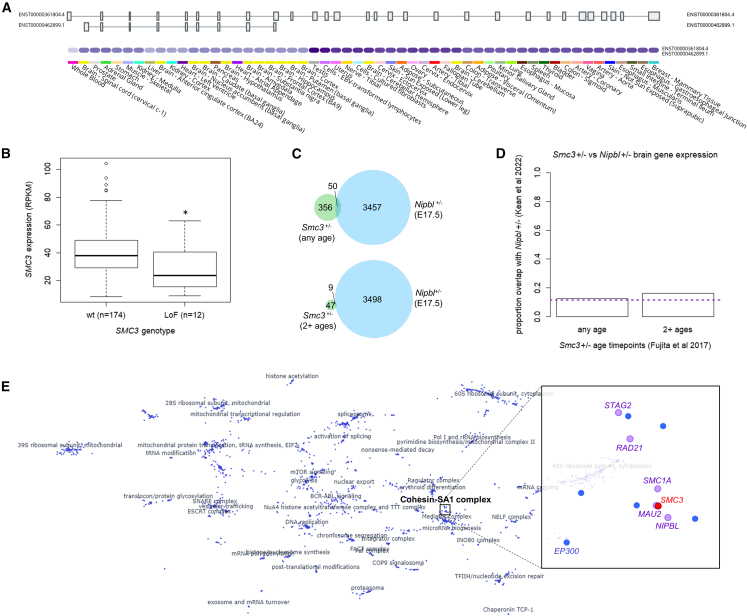
Figure 3.
Gene expression in WT and SMC3+/− tissues
(A) GTEx data demonstrate that only a single, full-length SMC3 isoform (ENST00000361804.4) is expressed at an appreciable level in any adult human tissue represented (https://gtexportal.org/home/gene/SMC3).
(B) pLoF SMC3 variants (LoF; nonsense, frameshift indel, splice site) in hematopoietic and lymphoid tumors correlate with decreased SMC3 gene expression, to a mean of 71.6% of WT (p = 0.018, Mann-Whitney U test). Data derived from the CCLE (http://xenabrowser.net). Plot is standard boxplot.
(C and D) Comparison of genes with altered expression in early postnatal Smc3+/− mouse cortex8 and those altered in a late prenatal Nipbl+/− mouse whole brain.37
(C) Fifty of 406 (12.3%) DEGs at any age (postnatal day [P]1, P3, P7, P14, P21) in Smc3+/− mice overlap with 3,507 DEGs in E17.5 Nipbl+/− mice (top); 9 of 58 (15.5%) DEGs in 2+ ages in Smc3+/− mice overlap with those same 3,507 genes (bottom).
(D) Neither of these is significantly different from chance (3,507/30,686 total genes; 11.4%) (any age p = 0.632, chi-square test of proportions; 2+ ages p = 0.441). See subjects and methods for derivation of denominator of 30,686 genes and discussion for a commentary on methodological differences between the 2 models.
(E) Minimal displacement embedding of pseudobulk-averaged, z normalized expression profiles from a genome-wide Perturb-Seq experiment in K562 CML cells36 (plot generated via https://gwps.wi.mit.edu/). Each dot is 1 of 1,973 genes with strong transcriptomic signatures. The SMC3 knockdown transcriptional disturbances group tightly with those of other cohesin genes (pink) and even an epigenetic regulator occasionally mutated in CdLS-like subjects (EP300).
Statistical analyses were conducted in R. The key gene lists are found in Table S5.
DNA methylation analysis
DNA from blood was subjected to array-based methylation analysis via the Manchester EpiPro project (https://mft.nhs.uk/nwglh/test-information/rare-disease/epipro-project/). The genome-wide DNA methylation profile was compared to the EpiSign Knowledge Database, a collection of DNA methylation signatures specific for rare disorders.19
Results
Case series and subject phenotypes
Fourteen individuals with SMC3 pLoF variants were identified. Most of the subjects underwent genetic testing as part of routine clinical assessment for growth retardation, developmental delay, and/or dysmorphic features. In only 4 cases was there clinical suspicion of CdLS (individuals 1, 7, 8, and 14). Some cases were identified via diagnostic laboratories, research repositories, hospital-based databases, and GeneMatcher. Subjects possessed frameshift indel, deletion, stop gain, or splice variants (Tables 1 and S1; Figure 1). Of these 14 variants, 7 were de novo, 1 was maternally inherited, 2 were paternally inherited, 1 was not maternally inherited, and 3 were of unknown inheritance. Of the subjects, 11 were male and 3 were female. Adjudication of the 10 SNVs using an advanced LoF curation framework39 flagged 2 as potentially not having an LoF effect (case 1, splice variant with potential rescue; case 10, escape from nonsense mediated decay) (Table S1). Missense variants were specifically excluded because missense variant effects (e.g., hypomorphic vs. nullimorphic vs. dominant negative) are unable to be reliably predicted.
Table 1.
Clinical features and variant details of heterozygous SMC3 predicted LoF variants
| Case no. |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |
| Variant type | Splice | Frameshift | Nonsense | Nonsense | Nonsense | Frameshift | Frameshift | Nonsense | Nonsense | Frameshift | Whole-gene del | Whole-gene del | Whole-gene del | Whole-gene del |
| Variant | c.430-1G>T p.(?) | c.461dup p.(Arg155GlufsTer12) | c.661C>T p.(Arg221Ter) | c.778C>T p.(Gln260Ter) | c.1078C>T p.(Arg360Ter) | c.1474_1478del p.(Lys492AlafsTer5) | c.1539del p.(Asn513LysfsTer4) | c.1561C>T p.(Arg521Ter) | c.2899C>T p.(Arg967Ter) | c.3646_3647dup p.(Gly1217MetfsTer39)a | arr[GRCh37] 10q25.1q25.2(111490480_113668388)x1 | arr[GRCh37] 10q25.1q25.3(106709945_118460100)x1 | arr[GRCh37]10q25.1q25.2(108508297_113338552)x1 | arr[GRCh37] 10q25.2(112323003_112579895)x1 |
| Inheritance | De novo | U | Mat | De novo | Pat | De novo | U | U | De novo | De novo | De novo | De novo | Not mat | Pat |
| Sex | M | M | M | M | M | M | M | F | M | M | M | M | F | F |
| Age | 18 y | 15 y | 9 y | 15 mo | 2 yr 8 mo | 9 y | 4 y | 8 y | 3 y | 17 y | 3 y | <1 y | 31 y | 8 mo |
| Developmental | Late sitting, walking, talking; ID, speech deficit | Global DD | Speech delay (m) | Late sitting, walking, talking; LD/ID (m), EHCP | Late sitting, walking, talking; no ID or LD | Speech delay, ID, DD | Late walking, LD, DD | Global DD | Late sitting (m), walking, talking; normal development later | Late walking, talking; speech deficit; ID | ||||
| Behavioral/psychiatric | Autism | Self-injurious behavior when frustrated | Anger, frustration, autistic traits, inattentiveness, hyperactivity | ADD, autism | ADHD | |||||||||
| Growth parameters | Low ht, wt; microcephaly | Low ht, wt; failure to thrive | Low wt, microcephaly | High ht, wt | Low ht, wt | Low ht, wt; microcephaly; low birth weight | Microcephaly | Low ht | Neonatal microcephaly | ?Relative macrocephaly | Low ht | Low ht, wt; microcephaly | ||
| Integumentary | Hirsutism, hypoplastic finger creases | Bruising susceptibility | Persistent fetal fingertip pads, decreased 4th finger creases | Hirsutism | Fragile stretchy skin, areas of hair loss | Hairy lumbar region | ||||||||
| Head shape | Brachycephaly | Brachycephaly (m) | Brachycephaly, plagiocephaly, triangular face, prominent forehead as newborn | Dolichocephaly | ||||||||||
| Facial dysmorphism | Characteristic facies of CdLS, high nasal bridge, face asymmetry (m) | Abnormal facial shape | Flat facial profile, anteverted nares | Smooth philtrum, beaked nasal tip, high nasal bridge | Depressed nasal bridge, long/featureless philtrum | Depressed nasal bridge, anteverted nostrils, broad/bulbous nasal tip | Depressed nasal bridge, broad/bulbous nasal tip | Broad/bulbous nasal tip | ||||||
| Mouth/palate/jaw | Cleft palate, retained primary dentition, small teeth, microretrognathia, small mouth with limited opening | Translucent thin enamel | Thin lips, downturned corners of mouth, small widely spaced teeth, sharp Cupid’s bow | Micrognathia/retrognathia | Thin upper lip, micrognathia | Thin upper lip, micrognathia/retrognathia (m) | High arched palate, ?poor dentition, retrognathia | |||||||
| Ear | Hearing loss, cupped right ear, thick left ear | Hearing loss | Preauricular skin tag | Small ears | Skin tag of right preauriculum and right cheek | Immature pinnae, anteriorly rotated, overfolded helix | ||||||||
| Eye | Synophrys, long eyelashes, ptosis (m) | Coats disease, bilateral | Visual impairment, strabismus, nystagmus | Arched eyebrows | Arched thick eyebrows, long eyelashes, nystagmus | Upslanting palpebral fissures | Prominent eyes, wide appearing palpebral fissures, arched eyebrows | Synophrys, long eyelashes | Laterally flared eyebrows, long eyelashes (m), upslanting (m) palpebral fissures | Arched eyebrows, synophrys, long eyelashes | Myopia, downslanting palpebral fissures | Myopia, deep set eyes, downslanting palpebral fissures | Upslanting palpebral fissures | |
| Cardiovascular | Patent ductus arteriosus | Tachycardia, bradycardia | PDA (resolved) | Neonatal stroke, possible LVH | Aortic coarctation | Atrial septal defect | Thoracic aortic aneurysm, bicuspid aortic valve | |||||||
| Respiratory/choanae | Choanal atresia, bilateral | Shortness of breath, chest pain | ||||||||||||
| Gastrointestinal | Resolved reflux, feeding problems into adulthood requiring G-tube, constipation | Gastroparesis, Hirschsprung disease, constipation, failure to thrive, G-tube | Poor weight gain | Hirschsprung disease, constipation, abdominal distension, esophageal atresia, tube feeding | Gastroenteric anomaly, volvulus | Feeding difficulty | Reflux | |||||||
| Renal | Hydronephrosis | Chronic kidney disease (from stroke) | Left double collecting system | |||||||||||
| Endocrine | Congenital hypothyroidism with DUOX2 variants, neonatal hypoglycemia | Low IGF-1 | Advanced bone age | |||||||||||
| Reproductive/Genitourinary | Cryptorchidism | Phimosis, resolved left cryptorchidism | Micropenis | |||||||||||
| Musculoskeletal | Small hands and feet, limited PIP extension fingers 2–5, proximal 2, 4 cutaneous finger syndactyly, ulnar deviation (m), elbow restriction, restricted pronation/supination, kyphoscoliosis | Hip dysplasia | Torticollis | Hypermobile joints, large sandal gap, 2, 3 toe syndactyly (m) | Arthropathy, abnormal skeletal survey, low muscle bulk, thin habitus | Small hands, short 5th fingers with clinodactyly, bilateral elbow restriction, persistent fetal fingertip pads | Single transverse palmar crease | Long narrow fingers that tend to overlap, deep plantar creases | Pectus carinatum, kyphosis, bilateral pes planus | Broad great toes (m) | ||||
| Neurologic | Gait disturbance | Hypotonia (m) | Seizures | Brain MR: “extraaxial fluid spaces and mild prominence of ventricles,” 1 febrile seizure | ||||||||||
| Hematologic/immune | Myelodysplastic syndrome, then AML | Leukopenia, resolved | Bone marrow failure, resolved; subclinical immunodeficiency | |||||||||||
| Other genetic findings | VUS dup 1q22q23.1 | DUOX2 P/LP variants, SCN4A VUSs | Several other variants, AOH | Short telomeres, RTEL1 VUS | VUS dup at 12q | The 10q25 del also includes ATRNL1 | Inherited chr16 del | Inherited inv(10) | ||||||
Variant nomenclature is based on GenBank: NM_005445.4 (MANE Select). ADD, attention-deficit disorder; AOH, absence of heterozygosity; DD, developmental delay; EHCP, educational health and care plan; G-tube, gastrostomy tube; ht, height; ID, intellectual disability; IGF-1, insulin-like growth factor 1; inv(10), inversion invoving chromosome 10; LD, learning disability/difficulty; LP, likely pathogenic; LVH, left ventricular hypertrophy; m, mild; Mat, maternal; MR, magnetic resonance; P, pathogenic; Pat, paternal; PDA, patent ductus arteriosus; PIP, proximal interphalangeal; U, unknown/unreported inheritance; wt, weight.
Figure 1.
SMC3 pLoF variants
(A) pLoF SNVs mapped to the SMC3 protein sequence. Subject cases are in black (top). The c.430-1G>T splice variant (individual 1) immediately precedes exon 8, which, if skipped, would result in a shift of reading frame; however, splicing may be rescued (see text). The p.(Gly1217MetfsTer39) variant (individual 10) is in the final exon and is predicted to escape nonsense-mediated decay. gnomAD pLoF variants passing quality filters are in gray (bottom). UKBB pLoF variants are in brown (bottom). Domains via UniProt.38 Arrowhead denotes the C-terminal end (equivalent to codon 211) of the minor transcript described in Figure 3. The diamond denotes the point after which nonsense-mediated decay may be escaped (codon 1,176). The 3′ region homologous to the SMC3P1 pseudogene (codons 974–1,217) is in orange.
(B) Somatic SMC3 pLoF SNVs among 13,714 tumors from the NCI GDC collection (https://portal.gdc.cancer.gov/genes/ENSG00000108055).
(C) Regional missense constraint over SMC3, via an analysis of gnomAD. The transition point is in codon 390 (exon 13 of 29).
(D) Case deletions (based on genome build GRCh37/hg19). Of the OMIM phenotype-associated genes in this region (green), none are validated dominant haploinsufficient disease genes for disorders that include developmental delay or dysmorphic features.
A summary of the subjects’ clinical features is provided in Table 1 and further noted in Table S1 and Note S1. No feature was shared by all of the subjects; however, the features present in more than half of the subjects included developmental delay and/or intellectual disability, low growth, and facial dysmorphism. The majority of the typical CdLS facial features and associated malformations were rare or absent in our cohort. No low anterior hairline or major limb malformations were reported. Only 2 individuals had a long/featureless philtrum, 1 had ptosis, 3 had confirmed dental anomalies, 1 had downturned corners of the mouth. Three individuals had synophrys, 4 had arched eyebrows, and 5 had long eyelashes. Three individuals had brachycephaly, 3 had a thin upper lip, and 3 had a depressed nasal bridge. Four individuals had cardiac malformations, and 2 experienced gastroesophageal reflux. Hirsutism was described in 3 cases. Interestingly, 5 of 14 individuals had a degree of micrognathia/retrognathia. Intellectual disability was present in 4 individuals and learning disability in an additional one. A delay in reaching developmental milestones was detected in 10 individuals. Autistic features were present in 3 individuals. Photographs were not available for any subject with dysmorphic features.
We compared standardized postnatal growth parameters and age at reaching developmental milestones between carriers of SMC3 pLoF variants and 15 cases with pathogenic missense variants or in-frame indels in SMC3, as previously published.14 A difference in standardized birth weight, birth head circumference, postnatal (i.e., at time of enrollment) weight, and postnatal head circumference was suggested between the 2 groups, pLoF variants being associated with less pronounced growth delay than missense/in-frame indel variants; however, these differences did not reach a significance threshold of p ≤ 0.05 (Figure 2A). Postnatal height appears equivalently low. Growth parameters (weight and head circumference) seem to worsen between birth and later ages, as has been seen in individuals with SMC3 missense and in-frame indel variants.14 When comparing the age at achieving developmental milestones, the patients with pLoF variants were able to sit unaided, walk unaided, and speak at a younger age than patients with missense/in-frame indels, although only the difference in age at sitting was significant (p = 0.012, Mann-Whitney U test [Wilcoxon]) (Figure 2B). pLoF subjects’ mean growth (including height, weight, and head circumference) and developmental milestones were delayed compared with the population means (Figure 2). Thus, in general, SMC3 pLoF variants are associated with a milder growth and developmental phenotype than are missense variants; however, the spectra are variable and appear to overlap.
Figure 2.
Growth and developmental milestones associated with SMC3 pLoF compared with pathogenic missense/in-frame indels
(A) Growth measurements adjusted for age and sex (Z score) are shown for SMC3 pLoF and missense/in-frame indel cases. Postnatal measurements are at the time of study enrollment/sampling.
(B) Age at which developmental milestones were reached for the 2 categories of SMC3 variants (LoF vs. pathogenic missense/in-frame indel). The 25th/50th/75th centiles of population data (Denver II, via DECIPHER) are shown at the right of each figure. Missense/in-frame indel data were obtained from Reference 14. Plots are standard boxplots. Statistical comparisons are the Mann-Whitney U test (Wilcoxon).
Interestingly, several subjects had other or additional phenotypes with potential biological links to SMC3 (see discussion), including bone marrow failure, leukopenia, AML, and Coats retinal telangiectasis; however, these were limited to a single affected individual each. Additional phenotypic and laboratory details, where available, are in Note S1.
SMC3 mutational constraint
We were intrigued by a few subjects who were nonpenetrant for or had mild presentations of the above growth and developmental phenotypes, given the initial metrics that suggested substantial mutational intolerance in this gene. Thus, we sought to further characterize LoF constraint in SMC3. Consistent with in vitro40,41 and model organism studies9,10,42,43 indicating that SMC3 is an essential developmental gene, biallelic pLoF variants have never been described in a human being, including our subjects.
Regarding heterozygous variants, the gnomAD version 2.1.1 (https://gnomad.broadinstitute.org/) probability of LoF intolerance (pLI) score (evidence for depletion of functional variation from expectation within each gene) and the LoF observed/expected (o/e) upper bound fraction (LOEUF; a more continuous metric that directly measures the ratios of o/e variation) both place SMC3 among the group of genes predicted to be most highly intolerant to LoF variants in a population cohort of adults without severe pediatric disease (pLI = 1; observed/expected ratio of SNV pLoF variants passing filters = 0/79.5; LOEUF 0.04).44 In agreement with this, an estimation of the probability of haploinsufficiency of SMC3 based on a machine learning model trained on a broad case-control CNV burden analysis in >950,000 individuals and gene-level features was 0.998 (maximum score = 1.00)45; of interest, the probability of triplosensitivity is also very high (0.999). A review of multiple additional databases of control and affected subjects revealed rarity and considerable apparent selection against such alleles (Table S2): No pLoF structural variants are present in gnomAD-SV, and our own CNV analysis of UKBB exome data identified only 2 deletions involving SMC3 among 196,869 subjects (Figure S2). pLoF sequence and structural variants are similarly rare or absent in the Database of Genomic Variants and DECIPHER (Table S2).
To gain insights into the few heterozygous SMC3 pLoF SNV carriers in control populations, who presumably would be nonpenetrant or mildly affected, we cataloged these alleles in gnomAD and the UKBB (Figure 1A; Table S3). The overall frequency of pLoF variants in gnomAD is 6.37E−5 (1/15,699) and in UKBB it is 3.32E−5 (1/30,102), and all are singletons. Adjudication of the 9 gnomAD pLoF variants using the LoF curation framework described above39 flagged 2 as potentially not having an LoF effect and an additional 2 as uncertain or potential technical artifacts (e.g., homopolymer repeat region, lack of read data) (Table S3). Interestingly, after removing the 2 predicted non-LoF variants, gnomAD SMC3 pLoF carriers show, on average, lower than expected allele balance (p = 0.020, Mann-Whitney U test [Wilcoxon]), potentially consistent with mosaicism ± clonal hematopoiesis (Figure S1).
Mapping case pLoF SNVs to the SMC3 coding sequence suggested possible clustering toward the 5′ half of the transcript (Figure 1A). To identify gene-level features that would provide precedent for or inform the biological rationale for such a cluster, we first referenced GTEx (Genotype-Tissue Expression; https://gtexportal.org/home/gene/SMC3) and found that the full-length, canonical transcript is the only isoform expressed at an appreciable level in any adult human tissue (Figure 3A). Further supporting this being the sole isoform, normalized by-exon expression as measured by pext46 (obtained via gnomAD) was homogeneous (score = 1) across all of the exons in every GTEx tissue. Next, we investigated whether the existence of SMC3P1, a homologous pseudogene at 2q11.2 that is likely the result of retrotransposition of approximately the last 5 exons of SMC3 (Figure 1A), may be impairing variant mapping in that region, resulting in an artifactual depletion of 3′ variants. However, per-base mean depth of coverage among gnomAD exomes and genomes is similar across the entirety of SMC3 (https://gnomad.broadinstitute.org/gene/ENSG00000108055?dataset=gnomad_r2_1), and SMC3 pLoF variants in gnomAD/UKBB (Figure 1A) and somatic SMC3 pLoF variants in tumors from the National Cancer Institute Genomic Data Commons NCI GDC collection (Figure 1B) show an even spread across the gene. Finally, we calculated regional missense constraint scores across SMC3 (Figure 1C); although constraint scores are slightly higher at the 5′ end, the entire gene is substantially missense constrained (o/e = 0.28 [0.25–0.32]; Z = 6.4). These data fail to demonstrate precedent for or inform a biological rationale for any 5′ clustering of pLoF SNVs in subjects.
Potential explanations for mildly affected individuals
Having made a strong case for intolerance to heterozygous LoF of SMC3, we continued to seek to explain the few subjects with mild or even apparently nonpenetrant developmental and/or growth phenotypes. One potential explanation is mosaicism. Thus, we assessed inheritance and allele balance among our enrolled subjects. Variants were inherited in 3 instances (individuals 3, 5, and 14), and thus by definition germline mutations in those probands. No case variants were signed out by clinical labs as having allele fractions suggestive of mosaicism. Furthermore, in 1 subject with normal growth and development (individual 9), we identified the mutant allele in 3 separate tissues (bone marrow, skin biopsy, and blood) with a near-50% allele balance (Note S1).
Another potential explanation for the existence of mildly affected individuals with LoF variants in this considerably loss-intolerant gene would be if SMC3 were a dominant infertility/subfertility gene. Testis and ovary are the adult human tissues with highest relative expression of SMC3 (https://gtexportal.org/home/gene/SMC3); however, SMC3 has never been identified as a human infertility gene, including the most recent large study of primary ovarian insufficiency,47 and we identified 1 maternally transmitted SMC3 pLoF variant (individual 3). We tested an alternative hypothesis that heterozygous LoF of SMC3 may cause male infertility. We evaluated whole-exome sequencing data from >1,000 individuals with nonobstructive azoospermia in the GEMINI Phase I cohort48 and >1,200 additional exomes in a Phase II cohort (some of which were normozoospermic male controls and female infertility cases). No SMC3 pLoF variants and only 1 missense variant with a CADD score ≥25 were identified (Table S4). In addition, we assessed rare (gnomAD minor allele frequency <1%) coding variants in the Male Reproductive Genomics (MERGE) cohort,49 consisting of 2,100 exomes of infertile men with severe oligo-, crypto-, or azoospermia. One pLoF and 3 missense variants with CADD ≥25 were identified (Table S4). MERGE missense variants were not enriched when compared with gnomAD variants with CADD ≥25 (p = 0.612, 2-sample chi-square test with continuity correction). Finally, among our subjects are 2 paternally inherited variants.
Functional effect
We next sought to determine whether pLoF SMC3 variants act as true LoF alleles and have broader functional genomic effects. Because subject cell lines and RNA were not available, pLoF SMC3 variants were identified among hematopoietic and lymphoid tumors from the CCLE with available RNA sequencing data; these led to a significant decrease in SMC3 gene expression compared to tumors wild-type (WT) for SMC3 (p = 0.018, Mann-Whitney U test) (Figure 3B).
To determine whether decreasing SMC3 expression is likely to effect a molecular (transcriptomic) signature and to what extent this matches that of other CdLS/cohesinopathy genes, we analyzed Perturb-Seq data from Replogle et al.36 This resource used multiplexed CRISPRi followed by single-cell RNA sequencing. In a genome-wide experiment in K562 chronic myelogenous leukemia cells, cells receiving SMC3-targeting guide RNAs reduced SMC3 expression to 68.5% residual and exhibited a transcriptomic signature of several hundred up- and downregulated genes (Table S6). The expression profile of SMC3 knockdown was highly correlated with the knockdown profiles of other cohesin ring components (SMC1A, RAD21, STAG2) and the cohesin loading machinery (NIPBL, MAU2) (Figure 3E). This correlation held true in separate Perturb-Seq experiments targeting only essential genes in K562 or RPE1 cells (Figure S3). The data were also able to show a threshold above which SMC3 knockdown, despite cohesin being involved in chromosome segregation, does not result in chromosome instability, similar to other cohesin genes (Table S6) and previous studies involving SMC3.50
Smc3+/− mice were previously shown to possess differences in P1-21 cortical gene expression,8 in addition to behavioral differences. Despite substantial methodologic differences—for example, age at tissue harvest, which is associated with significant changes in cohesin level8—we ventured to compare these DEGs to those observed in a study of E17.5 Nipbl+/− mouse whole brain, a model of typical CdLS.37 No significant overlap was observed (Figure 3C).
Finally, we sought to determine whether SMC3 pLoF variants result in changes to the epigenome matching those of previously described subjects with CdLS, including individuals with missense and in-frame indel variants in SMC3, although largely driven by other CdLS genes.19 Methylation analysis of blood DNA from 2 available subjects (individual 5 with p.Arg360Ter and another individual with p.Arg879Ter, from whom only methylation data, but no clinical information, were available) did not match that episignature (Figure 4).
Figure 4.
Global DNA methylation pattern of SMC3 pLoF compared with that of CdLS
Multidimensional scaling plot of global methylation signature (episignature) of blood-derived DNA. SMC3 pLoF subject 5 (p.Arg360Ter, purple) and an additional individual (c.2635C>T, p.(Arg879Ter), orange) do not plot with individuals clinically diagnosed with CdLS (red), instead plotting among the control population (green).
Discussion
Although heterozygous SMC3 missense and in-frame indel variants are a cause of atypical CdLS, and somatic SMC3 pLoF variants are found in cancers, the consequence of germline SMC3 LoF variants in humans has remained solely in the realm of speculation. The present study was aimed at resolving this question.
Clinical phenotype
The SMC3 pLoF cases presented here demonstrate that this variant type also contributes to a disease phenotype. Published SMC3 missense and in-frame indel cases with atypical CdLS demonstrate growth retardation, including small head size, developmental delay, and characteristic facial features.14 We found that pLoF patients share these features, although by quantitative comparisons appear on average to have a milder, although overlapping, phenotype than those with missense/in-frame indel variants and would for the most part not be considered to have CdLS. This may explain the near-absence of SMC3 pLoF variants among previously sequenced CdLS cohorts (Figure 5). Furthermore, the phenotype is variable, with some subjects showing more severe growth or developmental delays and others being less severe or even nonpenetrant for ≥1 features. Of note, Smc3+/− mice have demonstrated neuronal, behavioral, growth, and craniofacial phenotypes.8,9
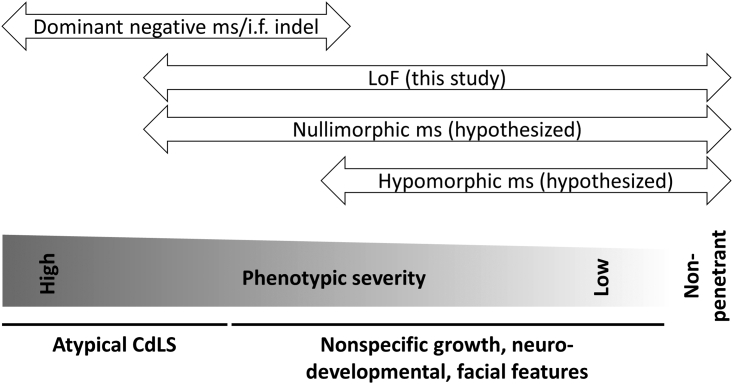
Figure 5.
Hypothesized phenotypic spectra of SMC3 variant types
Missense (ms)/in-frame (i.f.) indel variants, via an apparent dominant-negative mechanism as supported by prior literature, carry the most severe SMC3-associated phenotype, with many subjects appearing to have atypical CdLS. LoF variants generally carry a less severe phenotype, with only some individuals being recognized to have CdLS and some individuals lacking key phenotypic features (growth retardation, developmental delay, characteristic facial dysmorphism). We hypothesize that nullimorphic missense and hypomorphic missense variants also exist.
The pathogenesis of CdLS in previously reported SMC3 missense/in-frame cases has been suggested to be dominant negative, involving aberrant pairing of mutant SMC3 with other components of the cohesin ring.17,18 Our data suggest that, by contrast, a relative shortage of SMC3 has a less severe impact on cohesin function, which suggests that allele-specific therapy to knock down dominant-negative alleles, even completely, should be explored further. Nullimorphic and hypomorphic SMC3 missense variants likely also exist and will be identified as such with further study (Figure 5).
Deletions involving SMC3
In the 4 subjects with deletions at 10q25.2, SMC3 is not the only gene involved in the rearrangement (Figure 1D), and it is therefore possible that haploinsufficiency for other genes in the region is causative for some aspects of their phenotype. A small number of genes in these intervals have been formally associated with dominant human disease, among which only heterozygous SHOC2 and SMC3 pathogenic variants cause dysmorphic features and developmental delay; however, the only known pathogenic SHOC2 variant that causes Noonan-like syndrome with loose anagen hair (MIM: 607721) is missense. One additional constrained gene (pLI = 1), deleted only in individual 12, is ATRNL1. An intragenic deletion of this gene was previously found in a boy with developmental delay, autism spectrum disorder, and facial dysmorphism.51 Previously reported 10q25.2 deletions detected by cytogenetic methods do not seem to be associated with CdLS-like facial features.52,53
Missense variants in RBM20, which is deleted in all 4 deletion cases, are associated with a form of dilated cardiomyopathy.54 No subjects in our study with deletions encompassing RBM20 had any sign of cardiomyopathy. The small deletion that only contains SMC3 and RBM20, detected in individual 14, was of particular interest because it was also detected in the girl’s father, who was reported to be healthy with no mention of heart disease.
Potential explanations for mildly affected individuals despite LoF constraint
Multiple population genomic analyses suggested that SMC3 is substantially LoF, missense, and even duplication constrained. Given substantial LoF constraint, it was potentially surprising that we identified some individuals as having milder features as well as pLoF individuals in gnomAD and UKBB (albeit at a very low allele frequency). Of note, constraint metrics are indicative of overall selective pressure rather than observable phenotypic severity,55 and pLoF cases are seen in gnomAD for other, similarly LoF-constrained disease genes (e.g., ASXL3, ARID1B, and AUTS2).56 Nevertheless, we sought to investigate potential alternative explanations.
Obligatory postzygotic mosaicism is seen in disorders for which germline pathogenic variation is lethal.57 Mosaicism restricted to the blood lineage may be related to clonal hematopoiesis in aging,58 which can be caused by somatic variants in a number of cohesin and related genes (e.g., RAD21, SMC1A, STAG2, CTCF).59 At least 1 report has also found SMC3 pLoF variants in isolation with clonal hematopoiesis.60 We found no evidence for mosaicism in our subjects; however, we did discover a trend toward lower allele frequency in gnomAD subjects carrying SMC3 pLoF variants, potentially consistent with mosaicism ± clonal hematopoiesis.
In mice, sufficient SMC3 protein is required in oocytes for early embryonic development,10 and heterozygous depletion of Smc3 in female mice has deleterious effects on both the integrity and transmission of zygotic chromosomes.9,10,12 Furthermore, conditional knockout of Hdac8, an SMC3 recycling factor, causes subfertility.61 Finally, several cohesin or cohesin-related genes have been implicated in infertility in humans (REC8, SMC1B, STAG3, SGO2).62 However, there is no published evidence of SMC3 being involved in female infertility, and our own analyses identified only rare SMC3 pLoF variants among male infertility (azoospermia) cases and no statistical enrichment of predicted damaging missense variants.
Finally, the skewed sex ratio (11 male, 3 female) of our subjects is curious, suggesting that the possibility of sex-biased expressivity deserves future investigation.
Case-control phenotype associations
SMC3 deletions or pLoF (frameshift, stop gain, splice site) variants have not previously been found to be associated with disease in large case-control studies. For example, there were no control deletions or case deletions <1 Mb in a large study generating a CNV “morbidity map” of individuals with developmental delay,63 and only 1 protein truncating variant (included in the present paper) was found among Deciphering Developmental Disorders (DDD) study data.64 The UKBB demonstrates only 1 moderately significant phenotypic association for pLoF variants (mean corpuscular volume) (https://app.genebass.org/gene/ENSG00000108055).65 Intriguingly, in an analysis of >31,000 individuals with neurodevelopmental phenotypes and their family members, de novo variants in SMC3 were found to be associated with developmental delay (false discovery rate = 3.46E−7), although driven mostly by missense variants.66 The lack of phenotypic associations with SMC3 pLoF variants in large cohorts could be because of a lack of true association and/or lack of power owing to the rarity of these variants.
The possibility of pleiotropic phenotypes
Several subjects had intriguing “other” phenotypes with rational potential links to SMC3. (1) Individual 9 had cytopenias and somewhat low telomere length, while being otherwise remarkably healthy, with no intellectual disability, facial dysmorphisms, or other phenotypes consistent with CdLS (Table 1; Note S1). Individual 7 also had leukopenia. This is of interest because of poor hematopoietic replicative potential across serial transplantation experiments in Smc3+/− mice and experiments showing that SMC3 mutant human cells are out-competed by normal cells12; however, other studies found increased self-renewal.11,67 Poor hematopoietic renewal could also result from telomere dysfunction, and some basic science studies potentially implicate cohesin in telomere biology,68 although CdLS patients (mostly with NIPBL variants) do not have short telomeres.69 Of note, individual 9 also had a variant of uncertain clinical significance (VUS) in RTEL1, a telomere biology disorder gene (MIM: 608833), which is absent from gnomAD and affects a conserved amino acid, although it was transmitted from his father with normal telomere length. (2) Subject 1 with AML is also interesting because of the SMC3 variants seen as secondary hits in this and other types of malignancy.29 (3) Individual 2 had bilateral Coats disease, an ultrarare, idiopathic retinal telangiectasia leading to intraretinal and subretinal exudates.70,71 Syndromic associations with Coats disease include facioscapulohumeral muscular dystrophy (FSHD); ∼1% of FSHD1 patients have Coats disease, ∼1,000 times higher than the general population.72 FSHD1 is caused by an autosomal dominantly inherited loss of D4Z4 repeats on chromosome 4q35, contracting the array from 11 to 100 to 1–10 repeats and promoting DUX4 retrogene misexpression.72,73 The remaining 5% of FSHD cases (FSHD2) may be attributed to genetic pathogenic variants that lead to repeat-independent hypomethylation of D4Z4 in genes such as SMCHD1 (involved in genome organization) and DNMT3B (a chromatin modifier), also prompting misexpression of DUX4.74,75 Based on the involvement of SMCHD1 and DNMT3B in FSHD, perhaps other chromatin modifiers or cohesin genes such as SMC3 may affect 4q35 chromatin decompaction, causing DUX4 misexpression and leading to FSHD-related phenotypes. Unfortunately, additional functional testing to explore this hypothesis (D4Z4 repeat length, D4Z4 methylation, 4qA vs. B haplotype, telomere length, and genome-wide methylation analyses) were not able to be performed for this subject.
Functional effect of heterozygous SMC3 pLoF variants
We analyzed hematologic and lymphoid cancer transcriptome data, which suggested that pLoF SMC3 alleles act as LoF alleles at the RNA level. This is in line with in vitro data showing that the p.(Arg245Ter) variant causes loss of cohesin assembly.67 Analysis of Perturb-Seq data demonstrated impeccable grouping of the transcriptomic signature from SMC3 knockdown with those of other cohesin ring and cohesin loading genes. An analysis of mouse brain transcriptome data failed to find significant overlap between DEGs in Smc3+/− and Nipbl+/− mice (a model of classic CdLS); however, substantial methodological differences existed between the construction, tissue sampling, RNA preparation, and analyses of these 2 models. DNA methylation in human blood of 2 SMC3 pLoF cases did not cluster with CdLS subjects on a robust epigenetic analysis platform, yet future work is needed to identify whether SMC3+/− LoF yields its own methylation signature, either distinct from or an attenuated version of the CdLS signature.
Comparison with SMC1A LoF phenotype
Because of the shared functions of SMC1A and SMC3 (both are binding partners required to form the core cohesin ring), it is of interest to compare the phenotypes resulting from their loss of function. The SMC1A LoF phenotype, SMC1A-related developmental and epileptic encephalopathy, results in early-onset intractable epilepsy and severe intellectual disability in females and is presumed to be developmentally lethal in most males.76 This contrasts with the overall milder SMC3 pLoF phenotype described above. This difference could be due to moonlighting functions of SMC1A, or perhaps more obviously because of SMC1A being X-linked and subject to incomplete X-inactivation (reviewed in references77,78,79). Whether SMC1A is a limiting reagent (and thus potentially more dosage sensitive) for cohesin assembly in the developing brain is unknown.80 The understanding of dosage sensitivity and phenotypic liability thresholds is in its infancy, and cohesins will make an intriguing set of conditions to use in further work on this topic.
Conclusions
Here, we demonstrate that heterozygous SMC3 pLoF variants are depleted at the population level, yet they are survivable, and provide evidence that they are associated with developmental phenotypes. Specifically, they are associated with variable developmental delay, growth deficiency, and/or facial dysmorphism, although not all individuals displayed these features. On average, these variants bear a phenotype milder than but overlapping with that of SMC3 missense/in-frame indel variants present in CdLS cohorts. Variants in SMC3 have been seen in cancers; however, our data do not suggest that pLoF patients are at risk for cancer; there are not enough data to suggest an association.
There are limitations to our attempt to aggregate a consensus phenotype for this variant type that are not unique among initial descriptions of novel syndromes, including a moderate number of subjects, variable depth of clinical data, the potential for ascertainment bias, co-occurrence of additional genetic variants, non-LoF variant effects, and a lack of clinical histories across the full lifespan. Further clarity will be realized with additional cases over time, buoyed by even larger-scale genomic data from emerging cohorts numbering in the millions of participants (see, for example, reference81).
Finally, our work suggests the existence of additional considerably haploinsufficient genes, LoF of which yield yet-undiscovered mild-to-moderate or nonspecific phenotypes that will be ascertained only by careful hybrid studies marrying genomic and patient-level data.
Data and code availability
The published article and its supplement list all of the datasets analyzed during this study.
Acknowledgments
The authors thank: the patients and families who participated in this study; Dr. Jason Flannick, Emma Sherrill, Minh Nguyen, Aubrie Soucy, Heidi Schulz, and the Cornelia de Lange Syndrome Foundation for general support of the project; and Drs. Jennifer Gerton, Zuzana Tothova, and Kaitlin Samocha for helpful discussion. P.M.B. is supported by award K08NS117891 from the National Institute of Neurological Disorders and Stroke (NINDS) and an award from the Boston Children’s Hospital Office of Faculty Development. K.N.W.F. and the Boston Children's Hospital Clinic for Cornelia de Lange Syndrome and Related Disorders are supported by gifts from Peter and Kathy Wagner and Julie and Frank Mairano. M.E.T., P.M.B., L.W., and D.B.-I. were supported by grants from the National Institutes of Health (NIH) (U01HG011755, R01MH115957, HD081256, and P50HD104224). D.B.-I. is supported by T32HG002295 from the US National Human Genome Research Institute. S.S. is supported by award K23NS119666 from the NINDS. T.B.H. is supported by the Deutsche Forschungsgemeinschaft (German Research Foundation) (awards 418081722 and 433158657). F.T. was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) in the framework of the Clinical Research Unit “Male Germ Cells” (CRU326, project no. 329621271). Some sequencing and analysis was provided by the Broad Institute Center for Mendelian Genomics and was funded by NIH grants UM1HG008900 and R01HG009141. K.M.B. is supported by the National Eye Institute (R01EY026904, R01EY012910, and P30EY014104) and the Foundation Fighting Blindness (EGI-GE-1218-0753-UCSD). D.F.C. was supported in part by NIH grants R01HD078641 and P50HD096723. S.B. is supported by the NIHR Manchester Biomedical Research Centre (NIHR203308). A.S. is supported by RC2DK122533. M.S.-B. was supported by the National Heart, Lung, and Blood Institute (R01HL143295). Analysis by E.G., S.T., and A.O.-L. was supported by NIH grants U01HG011755 and R01HG009141. This work was supported by the Boston Children’s Hospital Rare Disease Cohorts Initiative (CRDC). This study makes use of data generated by the DECIPHER community. A full list of centers who contributed to the generation of the data is available from https://deciphergenomics.org/about/stats and via e-mail from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by Wellcome (grant no. WT223718/Z/21/Z).
Author contributions
Author contributions, using CRediT taxonomy, are listed in Table S7.
Declaration of interests
M.E.T. is supported by research funding and/or reagents from Illumina, Microsoft, Ionis Therapeutics, and Levo Therapeutics. M.P.N., J.H., and J.J. are employees of GeneDx.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xhgg.2024.100273.
Supplemental information
References
- 1.Hoencamp C., Rowland B.D. Genome control by SMC complexes. Nat. Rev. Mol. Cell Biol. 2023;24:633–650. doi: 10.1038/s41580-023-00609-8. [DOI] [PubMed] [Google Scholar]
- 2.Peters J.M., Tedeschi A., Schmitz J. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 3.Nasmyth K., Haering C.H. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 4.Nagao K., Adachi Y., Yanagida M. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature. 2004;430:1044–1048. doi: 10.1038/nature02803. [DOI] [PubMed] [Google Scholar]
- 5.Rao S.S.P., Huang S.C., Glenn St Hilaire B., Engreitz J.M., Perez E.M., Kieffer-Kwon K.R., Sanborn A.L., Johnstone S.E., Bascom G.D., Bochkov I.D., et al. Cohesin Loss Eliminates All Loop Domains. Cell. 2017;171:305–320.e24. doi: 10.1016/j.cell.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 7.Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 8.Fujita Y., Masuda K., Bando M., Nakato R., Katou Y., Tanaka T., Nakayama M., Takao K., Miyakawa T., Tanaka T., et al. Decreased cohesin in the brain leads to defective synapse development and anxiety-related behavior. J. Exp. Med. 2017;214:1431–1452. doi: 10.1084/jem.20161517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White J.K., Gerdin A.K., Karp N.A., Ryder E., Buljan M., Bussell J.N., Salisbury J., Clare S., Ingham N.J., Podrini C., et al. Genome-wide generation and systematic phenotyping of knockout mice reveals new roles for many genes. Cell. 2013;154:452–464. doi: 10.1016/j.cell.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yueh W.T., Singh V.P., Gerton J.L. Maternal Smc3 protects the integrity of the zygotic genome through DNA replication and mitosis. Development. 2021;148 doi: 10.1242/dev.199800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viny A.D., Ott C.J., Spitzer B., Rivas M., Meydan C., Papalexi E., Yelin D., Shank K., Reyes J., Chiu A., et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. J. Exp. Med. 2015;212:1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang T., Glover B., Hadwiger G., Miller C.A., di Martino O., Welch J.S. Smc3 is required for mouse embryonic and adult hematopoiesis. Exp. Hematol. 2019;70:70–84.e6. doi: 10.1016/j.exphem.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deardorff M.A., Kaur M., Yaeger D., Rampuria A., Korolev S., Pie J., Gil-Rodríguez C., Arnedo M., Loeys B., Kline A.D., et al. Mutations in cohesin complex members SMC3 and SMC1A cause a mild variant of cornelia de Lange syndrome with predominant mental retardation. Am. J. Hum. Genet. 2007;80:485–494. doi: 10.1086/511888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil-Rodríguez M.C., Deardorff M.A., Ansari M., Tan C.A., Parenti I., Baquero-Montoya C., Ousager L.B., Puisac B., Hernández-Marcos M., Teresa-Rodrigo M.E., et al. De novo heterozygous mutations in SMC3 cause a range of Cornelia de Lange syndrome-overlapping phenotypes. Hum. Mutat. 2015;36:454–462. doi: 10.1002/humu.22761. [DOI] [PubMed] [Google Scholar]
- 15.Ansari M., Poke G., Ferry Q., Williamson K., Aldridge R., Meynert A.M., Bengani H., Chan C.Y., Kayserili H., Avci S., et al. Genetic heterogeneity in Cornelia de Lange syndrome (CdLS) and CdLS-like phenotypes with observed and predicted levels of mosaicism. J. Med. Genet. 2014;51:659–668. doi: 10.1136/jmedgenet-2014-102573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan B., Pehlivan D., Karaca E., Patel N., Charng W.L., Gambin T., Gonzaga-Jauregui C., Sutton V.R., Yesil G., Bozdogan S.T., et al. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J. Clin. Invest. 2015;125:636–651. doi: 10.1172/JCI77435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Revenkova E., Focarelli M.L., Susani L., Paulis M., Bassi M.T., Mannini L., Frattini A., Delia D., Krantz I., Vezzoni P., et al. Cornelia de Lange syndrome mutations in SMC1A or SMC3 affect binding to DNA. Hum. Mol. Genet. 2009;18:418–427. doi: 10.1093/hmg/ddn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gimigliano A., Mannini L., Bianchi L., Puglia M., Deardorff M.A., Menga S., Krantz I.D., Musio A., Bini L. Proteomic profile identifies dysregulated pathways in Cornelia de Lange syndrome cells with distinct mutations in SMC1A and SMC3 genes. J. Proteome Res. 2012;11:6111–6123. doi: 10.1021/pr300760p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aref-Eshghi E., Kerkhof J., Pedro V.P., Groupe DI France. Barat-Houari M., Ruiz-Pallares N., Andrau J.C., Lacombe D., Van-Gils J., Fergelot P., et al. Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders. Am. J. Hum. Genet. 2020;106:356–370. doi: 10.1016/j.ajhg.2020.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillis L.A., McCallum J., Kaur M., DeScipio C., Yaeger D., Mariani A., Kline A.D., Li H.h., Devoto M., Jackson L.G., Krantz I.D. NIPBL mutational analysis in 120 individuals with Cornelia de Lange syndrome and evaluation of genotype-phenotype correlations. Am. J. Hum. Genet. 2004;75:610–623. doi: 10.1086/424698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tonkin E.T., Wang T.J., Lisgo S., Bamshad M.J., Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat. Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 22.Yan J., Saifi G.M., Wierzba T.H., Withers M., Bien-Willner G.A., Limon J., Stankiewicz P., Lupski J.R., Wierzba J. Mutational and genotype-phenotype correlation analyses in 28 Polish patients with Cornelia de Lange syndrome. Am. J. Med. Genet. 2006;140:1531–1541. doi: 10.1002/ajmg.a.31305. [DOI] [PubMed] [Google Scholar]
- 23.Oguni H., Nishikawa A., Sato Y., Otani Y., Ito S., Nagata S., Kato M., Hamanaka K., Miyatake S., Matsumoto N. A missense variant of SMC1A causes periodic pharmaco-resistant cluster seizures similar to PCDH19-related epilepsy. Epilepsy Res. 2019;155 doi: 10.1016/j.eplepsyres.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 24.Mannini L., Liu J., Krantz I.D., Musio A. Spectrum and consequences of SMC1A mutations: the unexpected involvement of a core component of cohesin in human disease. Hum. Mutat. 2010;31:5–10. doi: 10.1002/humu.21129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Nardo M., Pallotta M.M., Musio A. The multifaceted roles of cohesin in cancer. J. Exp. Clin. Cancer Res. 2022;41:96. doi: 10.1186/s13046-022-02321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han C., Gao X., Li Y., Zhang J., Yang E., Zhang L., Yu L. Characteristics of Cohesin Mutation in Acute Myeloid Leukemia and Its Clinical Significance. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.579881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labuhn M., Perkins K., Matzk S., Varghese L., Garnett C., Papaemmanuil E., Metzner M., Kennedy A., Amstislavskiy V., Risch T., et al. Mechanisms of Progression of Myeloid Preleukemia to Transformed Myeloid Leukemia in Children with Down Syndrome. Cancer Cell. 2019;36:123–138.e10. doi: 10.1016/j.ccell.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida K., Toki T., Okuno Y., Kanezaki R., Shiraishi Y., Sato-Otsubo A., Sanada M., Park M.j., Terui K., Suzuki H., et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat. Genet. 2013;45:1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- 29.Jann J.C., Tothova Z. Cohesin mutations in myeloid malignancies. Blood. 2021;138:649–661. doi: 10.1182/blood.2019004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landrum M.J., Kattman B.L. ClinVar at five years: Delivering on the promise. Hum. Mutat. 2018;39:1623–1630. doi: 10.1002/humu.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rockowitz S., LeCompte N., Carmack M., Quitadamo A., Wang L., Park M., Knight D., Sexton E., Smith L., Sheidley B., et al. Children's rare disease cohorts: an integrative research and clinical genomics initiative. NPJ Genom. Med. 2020;5:29. doi: 10.1038/s41525-020-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pais L.S., Snow H., Weisburd B., Zhang S., Baxter S.M., DiTroia S., O'Heir E., England E., Chao K.R., Lemire G., et al. A web-based analysis and collaboration tool for rare disease genomics. Hum. Mutat. 2022;43:698–707. doi: 10.1002/humu.24366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright C.F., Quaife N.M., Ramos-Hernández L., Danecek P., Ferla M.P., Samocha K.E., Kaplanis J., Gardner E.J., Eberhardt R.Y., Chao K.R., et al. Non-coding region variants upstream of MEF2C cause severe developmental disorder through three distinct loss-of-function mechanisms. Am. J. Hum. Genet. 2021;108:1083–1094. doi: 10.1016/j.ajhg.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Replogle J.M., Saunders R.A., Pogson A.N., Hussmann J.A., Lenail A., Guna A., Mascibroda L., Wagner E.J., Adelman K., Lithwick-Yanai G., et al. Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq. Cell. 2022;185:2559–2575.e28. doi: 10.1016/j.cell.2022.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kean C.M., Tracy C.J., Mitra A., Rahat B., Van Winkle M.T., Gebert C.M., Noeker J.A., Calof A.L., Lander A.D., Kassis J.A., Pfeifer K. Decreasing Wapl dosage partially corrects embryonic growth and brain transcriptome phenotypes in Nipbl(+/-) embryos. Sci. Adv. 2022;8:eadd4136. doi: 10.1126/sciadv.add4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singer-Berk M., Gudmundsson S., Baxter S., Seaby E.G., England E., Wood J.C., Son R.G., Watts N.A., Karczewski K.J., Harrison S.M., et al. Advanced variant classification framework reduces the false positive rate of predicted loss-of-function variants in population sequencing data. Am. J. Hum. Genet. 2023;110:1496–1508. doi: 10.1016/j.ajhg.2023.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robison B., Guacci V., Koshland D. A role for the Smc3 hinge domain in the maintenance of sister chromatid cohesion. Mol. Biol. Cell. 2018;29:339–355. doi: 10.1091/mbc.E17-08-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang J., Shi X., Li Y., Kim B.J., Jia J., Huang Z., Yang T., Fu X., Jung S.Y., Wang Y., et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Banerji R., Skibbens R.V., Iovine M.K. Cohesin mediates Esco2-dependent transcriptional regulation in a zebrafish regenerating fin model of Roberts Syndrome. Biol. Open. 2017;6:1802–1813. doi: 10.1242/bio.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghiselli G. SMC3 knockdown triggers genomic instability and p53-dependent apoptosis in human and zebrafish cells. Mol. Cancer. 2006;5:52. doi: 10.1186/1476-4598-5-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Collins R.L., Glessner J.T., Porcu E., Lepamets M., Brandon R., Lauricella C., Han L., Morley T., Niestroj L.M., Ulirsch J., et al. A cross-disorder dosage sensitivity map of the human genome. Cell. 2022;185:3041–3055.e25. doi: 10.1016/j.cell.2022.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cummings B.B., Karczewski K.J., Kosmicki J.A., Seaby E.G., Watts N.A., Singer-Berk M., Mudge J.M., Karjalainen J., Satterstrom F.K., O'Donnell-Luria A.H., et al. Transcript expression-aware annotation improves rare variant interpretation. Nature. 2020;581:452–458. doi: 10.1038/s41586-020-2329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ke H., Tang S., Guo T., Hou D., Jiao X., Li S., Luo W., Xu B., Zhao S., Li G., et al. Landscape of pathogenic mutations in premature ovarian insufficiency. Nat. Med. 2023;29:483–492. doi: 10.1038/s41591-022-02194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagirnaja L., Lopes A.M., Charng W.L., Miller B., Stakaitis R., Golubickaite I., Stendahl A., Luan T., Friedrich C., Mahyari E., et al. Diverse monogenic subforms of human spermatogenic failure. Nat. Commun. 2022;13:7953. doi: 10.1038/s41467-022-35661-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wyrwoll M.J., Köckerling N., Vockel M., Dicke A.K., Rotte N., Pohl E., Emich J., Wöste M., Ruckert C., Wabschke R., et al. Genetic Architecture of Azoospermia-Time to Advance the Standard of Care. Eur. Urol. 2023;83:452–462. doi: 10.1016/j.eururo.2022.05.011. [DOI] [PubMed] [Google Scholar]
- 50.Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D., et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark Z., Bruno D.L., Mountford H., Lockhart P.J., Amor D.J. De novo 325 kb microdeletion in chromosome band 10q25.3 including ATRNL1 in a boy with cognitive impairment, autism and dysmorphic features. Eur. J. Med. Genet. 2010;53:337–339. doi: 10.1016/j.ejmg.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 52.Irving M., Hanson H., Turnpenny P., Brewer C., Ogilvie C.M., Davies A., Berg J. Deletion of the distal long arm of chromosome 10; is there a characteristic phenotype? A report of 15 de novo and familial cases. Am. J. Med. Genet. 2003;123A:153–163. doi: 10.1002/ajmg.a.20220. [DOI] [PubMed] [Google Scholar]
- 53.Kehrer-Sawatzki H., Daumiller E., Müller-Navia J., Kendziorra H., Rossier E., du Bois G., Barbi G. Interstitial deletion del(10)(q25.2q25.3 approximately 26.11)--case report and review of the literature. Prenat. Diagn. 2005;25:954–959. doi: 10.1002/pd.1252. [DOI] [PubMed] [Google Scholar]
- 54.Lennermann D., Backs J., van den Hoogenhof M.M.G. New Insights in RBM20 Cardiomyopathy. Curr. Heart Fail. Rep. 2020;17:234–246. doi: 10.1007/s11897-020-00475-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fuller Z.L., Berg J.J., Mostafavi H., Sella G., Przeworski M. Measuring intolerance to mutation in human genetics. Nat. Genet. 2019;51:772–776. doi: 10.1038/s41588-019-0383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ropers H.H., Wienker T. Penetrance of pathogenic mutations in haploinsufficient genes for intellectual disability and related disorders. Eur. J. Med. Genet. 2015;58:715–718. doi: 10.1016/j.ejmg.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 57.Thorpe J., Osei-Owusu I.A., Avigdor B.E., Tupler R., Pevsner J. Mosaicism in Human Health and Disease. Annu. Rev. Genet. 2020;54:487–510. doi: 10.1146/annurev-genet-041720-093403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carlston C.M., O'Donnell-Luria A.H., Underhill H.R., Cummings B.B., Weisburd B., Minikel E.V., Birnbaum D.P., Exome Aggregation Consortium. Tvrdik T., MacArthur D.G., Mao R. Pathogenic ASXL1 somatic variants in reference databases complicate germline variant interpretation for Bohring-Opitz Syndrome. Hum. Mutat. 2017;38:517–523. doi: 10.1002/humu.23203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gudmundsson S., Carlston C.M., O’Donnell-Luria A. Interpreting variants in genes affected by clonal hematopoiesis in population data. Hum. Genet. 2023:1–5. doi: 10.1007/s00439-023-02526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moon I., Kong M.G., Ji Y.S., Kim S.H., Park S.K., Suh J., Jang M.A. Clinical, Mutational, and Transcriptomic Characteristics in Elderly Korean Individuals With Clonal Hematopoiesis Driver Mutations. Ann. Lab. Med. 2023;43:145–152. doi: 10.3343/alm.2023.43.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh V.P., Yueh W.T., Gerton J.L., Duncan F.E. Oocyte-specific deletion of Hdac8 in mice reveals stage-specific effects on fertility. Reproduction. 2019;157:305–316. doi: 10.1530/REP-18-0560. [DOI] [PubMed] [Google Scholar]
- 62.Yatsenko S.A., Rajkovic A. Genetics of human female infertilitydagger. Biol. Reprod. 2019;101:549–566. doi: 10.1093/biolre/ioz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coe B.P., Witherspoon K., Rosenfeld J.A., van Bon B.W.M., Vulto-van Silfhout A.T., Bosco P., Friend K.L., Baker C., Buono S., Vissers L.E.L.M., et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaplanis J., Samocha K.E., Wiel L., Zhang Z., Arvai K.J., Eberhardt R.Y., Gallone G., Lelieveld S.H., Martin H.C., McRae J.F., et al. Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–762. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karczewski K.J., Solomonson M., Chao K.R., Goodrich J.K., Tiao G., Lu W., Riley-Gillis B.M., Tsai E.A., Kim H.I., Zheng X., et al. Systematic single-variant and gene-based association testing of thousands of phenotypes in 394,841 UK Biobank exomes. Cell Genom. 2022;2 doi: 10.1016/j.xgen.2022.100168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu J.M., Satterstrom F.K., Peng M., Brand H., Collins R.L., Dong S., Wamsley B., Klei L., Wang L., Hao S.P., et al. Rare coding variation provides insight into the genetic architecture and phenotypic context of autism. Nat. Genet. 2022;54:1320–1331. doi: 10.1038/s41588-022-01104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mullenders J., Aranda-Orgilles B., Lhoumaud P., Keller M., Pae J., Wang K., Kayembe C., Rocha P.P., Raviram R., Gong Y., et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J. Exp. Med. 2015;212:1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Remeseiro S., Cuadrado A., Carretero M., Martínez P., Drosopoulos W.C., Cañamero M., Schildkraut C.L., Blasco M.A., Losada A. Cohesin-SA1 deficiency drives aneuploidy and tumourigenesis in mice due to impaired replication of telomeres. EMBO J. 2012;31:2076–2089. doi: 10.1038/emboj.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kline A.D., Calof A.L., Schaaf C.A., Krantz I.D., Jyonouchi S., Yokomori K., Gauze M., Carrico C.S., Woodman J., Gerton J.L., et al. Cornelia de Lange syndrome: further delineation of phenotype, cohesin biology and educational focus, 5th Biennial Scientific and Educational Symposium abstracts. Am. J. Med. Genet. 2014;164A:1384–1393. doi: 10.1002/ajmg.a.36417. [DOI] [PubMed] [Google Scholar]
- 70.Chang M.M., McLean I.W., Merritt J.C. Coats' disease: a study of 62 histologically confirmed cases. J. Pediatr. Ophthalmol. Strabismus. 1984;21:163–168. doi: 10.3928/0191-3913-19840901-03. [DOI] [PubMed] [Google Scholar]
- 71.Shields J.A., Shields C.L., Honavar S.G., Demirci H. Clinical variations and complications of Coats disease in 150 cases: the 2000 Sanford Gifford Memorial Lecture. Am. J. Ophthalmol. 2001;131:561–571. doi: 10.1016/s0002-9394(00)00883-7. [DOI] [PubMed] [Google Scholar]
- 72.Statland J.M., Sacconi S., Farmakidis C., Donlin-Smith C.M., Chung M., Tawil R. Coats syndrome in facioscapulohumeral dystrophy type 1: frequency and D4Z4 contraction size. Neurology. 2013;80:1247–1250. doi: 10.1212/WNL.0b013e3182897116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Campbell A.E., Belleville A.E., Resnick R., Shadle S.C., Tapscott S.J. Facioscapulohumeral dystrophy: activating an early embryonic transcriptional program in human skeletal muscle. Hum. Mol. Genet. 2018;27:R153–R162. doi: 10.1093/hmg/ddy162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lemmers R.J.L.F., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W.E., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R., et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van den Boogaard M.L., Lemmers R.J.L.F., Balog J., Wohlgemuth M., Auranen M., Mitsuhashi S., van der Vliet P.J., Straasheijm K.R., van den Akker R.F.P., Kriek M., et al. Mutations in DNMT3B Modify Epigenetic Repression of the D4Z4 Repeat and the Penetrance of Facioscapulohumeral Dystrophy. Am. J. Hum. Genet. 2016;98:1020–1029. doi: 10.1016/j.ajhg.2016.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bozarth X.L., Lopez J., Fang H., Lee-Eng J., Duan Z., Deng X. Phenotypes and Genotypes in Patients with SMC1A-Related Developmental and Epileptic Encephalopathy. Genes. 2023;14 doi: 10.3390/genes14040852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huisman S., Mulder P.A., Redeker E., Bader I., Bisgaard A.M., Brooks A., Cereda A., Cinca C., Clark D., Cormier-Daire V., et al. Phenotypes and genotypes in individuals with SMC1A variants. Am. J. Med. Genet. 2017;173:2108–2125. doi: 10.1002/ajmg.a.38279. [DOI] [PubMed] [Google Scholar]
- 78.Borck G., Zarhrate M., Bonnefont J.P., Munnich A., Cormier-Daire V., Colleaux L. Incidence and clinical features of X-linked Cornelia de Lange syndrome due to SMC1L1 mutations. Hum. Mutat. 2007;28:205–206. doi: 10.1002/humu.9478. [DOI] [PubMed] [Google Scholar]
- 79.Musio A. The multiple facets of the SMC1A gene. Gene. 2020;743 doi: 10.1016/j.gene.2020.144612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holzmann J., Politi A.Z., Nagasaka K., Hantsche-Grininger M., Walther N., Koch B., Fuchs J., Dürnberger G., Tang W., Ladurner R., et al. Absolute quantification of cohesin, CTCF and their regulators in human cells. Elife. 2019;8 doi: 10.7554/eLife.46269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.All of Us Research Program Investigators. Denny J.C., Rutter J.L., Goldstein D.B., Philippakis A., Smoller J.W., Jenkins G., Dishman E. The "All of Us" Research Program. N. Engl. J. Med. 2019;381:668–676. doi: 10.1056/NEJMsr1809937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article and its supplement list all of the datasets analyzed during this study.