Abstract
Previously, we reported the isolation of 10 vancomycin-resistant gram-positive anaerobic bacilli carrying the vanB ligase gene from nine hemodialysis patients (S. A. Ballard et al., Antimicrob. Agents Chemother. 49:77-81, 2005; T. P. Stinear et al., Lancet 357:855-856, 2001). In the present study, the molecular and evolutionary relationship of the vanB resistance element within these 10 anaerobes and two vancomycin-resistant Enterococcus faecium strains were examined. PCR analysis and nucleotide sequencing demonstrated that all 12 isolates carried the vanB operon associated with an element identical to Tn1549 and Tn5382 of Enterococcus. Restriction fragment length polymorphism analysis of the vanB operon in these isolates revealed two distinct patterns, and sequencing showed that minor base differences existed. PCR amplification of the joint region of a circular intermediate was demonstrated in nine of these organisms, a finding indicative of an ability to excise and circularize, an intermediate step in transposition and conjugative transfer. Southern hybridization with a vanB-vanXB probe suggests that there is one insert of the transposon in all isolates. Sequence analysis of the integration site revealed distinct sequences: the Tn1549/5382 element within E. faecium was inserted within the host chromosome, whereas nucleotide sequences surrounding the Tn1549/5382 element in the 10 anaerobes showed no significant homology to sequences in the GenBank database. We demonstrate considerable similarity between the Tn1549/5382 element identified in 10 anaerobe isolates with that found in enterococci. The homology and potential to transpose suggest a recent horizontal transfer event may have occurred. However, the original direction of transposition and the mechanism involved remains unknown.
The emergence of resistance to glycopeptide antibiotics (vancomycin and teicoplanin) in enterococci is one of the most serious infection control issues currently facing large hospitals in Australia, as elsewhere (13, 20, 24). There are currently six different vancomycin resistance (van) gene operons (vanA, -B, -C, -D, -E, and -G) mediating glycopeptide resistance. The two genotypes of greatest clinical importance are vanA and vanB, with three alleles of the vanB ligase gene (vanB1, vanB2, and vanB3), based on sequence similarity (12, 16, 21). Both the vanA and the vanB operons are associated with transposons, and the transfer of resistance among enterococci appears to be mediated via acquisition of these elements (32). Although the vanA operon is located on the transposon, Tn1546, three transposons have been described that contain the vanB operon: Tn1547, Tn5382, and Tn1549 (4, 6, 11, 23). Sequencing data suggest that Tn1549 is probably quasi-identical to Tn5382 and specifically linked to the vanB2 allele (8, 11, 32).
There has been increasing evidence of vanB-mediated resistance among organisms other than enterococci. The vanB genotype has been described in Streptococcus bovis isolated from a human stool swab and in S. gallolyticus and S. lutetiensis isolated from fecal samples from veal calves (9, 18, 22). Transfer of vanB from S. bovis, S. gallolyticus, and S. lutetiensis to enterococci has also been reported (9, 18, 22). Recently, we isolated 10 anaerobic organisms (Clostridium sp., n = 8; Eggerthella lenta, n = 1; Ruminococcus sp., n = 1) from the bowel flora of nine hemodialysis patients who were undergoing screening for fecal vancomycin-resistant enterococci (VRE) colonization (5, 28). These organisms all contained a vanB ligase gene identical to that observed in VRE. The aim of the present study was to characterize the vanB resistance element and its transposon in these 10 anaerobic organisms in order to gain insight into the potential transmissibility of these elements between bowel flora and also to help explain the role they may play in the emergence of new strains of VRE.
(This study was presented in part previously [S. A. Ballard, K. K. Pertile, P. D. R. Johnson, and M. L. Grayson, 44th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C1-950, 2004].)
MATERIALS AND METHODS
Organisms and susceptibility testing.
The vanB locus of 10 anaerobic bacilli and two clinical carriage isolates of vancomycin-resistant E. faecium was examined (Table 1). Organisms were cultured as described previously, (5). Susceptibility to vancomycin, teicoplanin, and benzylpenicillin were determined by E-test (AB Biodisk, Dalvägen, Sweden) according to the manufacturer's instructions. Isolates resistant to benzyl-penicillin were tested for the production of β-lactamase by using Cefinase disks (BD Diagnostics, Franklin Lakes, N.J.) as described by the manufacturer. The control strains Enterococcus faecalis containing vanB (ATCC 51299), E. faecalis (ATCC 29212), Bacteroides fragilis (ATCC 25285), Peptostreptococcus anaerobius (ATCC 27337), Clostridium perfringens (ATCC 13124), Staphylococcus aureus (ATCC 29213), and Haemophilus influenzae (ATCC 10211) were included where appropriate.
TABLE 1.
Characteristics of isolates
| Isolate | Genus and speciesa | MIC (μg/ml)
|
Genotypeb | Circular intermediatec | Reference | ||
|---|---|---|---|---|---|---|---|
| Vancomycin | Teicoplanin | Benzylpenicillin | |||||
| MLG055 | Clostridium sp. | >256 | 0.094 | >256 | vanB2 | − | 28 |
| MLG480 | Clostridium sp. | 8 | 0.023 | >256 | vanB1 | + | 5 |
| MLG043 | Eggerthella lenta | >256 | 0.064 | 4 | vanB2 | + | 28 |
| MLG101 | Clostridium symbiosum | 96 | 0.125 | 0.094 | vanB2 | + | 28 |
| MLG245 | Clostridium sp. | >256 | 0.75 | 0.125 | vanB2 | + | 28 |
| MLG080-1 | Clostridium bolteae* | >256 | 4 | >256 | vanB2 | + | 5 |
| MLG080-3 | Ruminococcus sp.* | >256 | 0.47 | 0.75 | vanB2 | + | 5 |
| MLG392 | Clostridium hathewayi | 24 | 3 | 0.75 | vanB2 | + | 5 |
| MLG661 | Clostridium hathewayi | 16 | 0.25 | 0.75 | vanB2 | + | 5 |
| MLG856-1 | Clostridium hathewayi† | 96 | 1.5 | 0.75 | vanB2 | + | 5 |
| MLG856-2 | Enterococcus faecium† | 32 | 0.38 | 64 | vanB2 | − | 5 |
| MLG229 | Enterococcus faecium | 24 | 0.75 | 48 | vanB2 | − | 5 |
* and †, both strains were isolated from the same patient.
Based on the sequence identity of the vanB ligase gene.
Presence (+) or absence (−) of a circular intermediate of Tn1549/5382 within isolates.
PCR amplification and restriction fragment length polymorphism (RFLP) analysis.
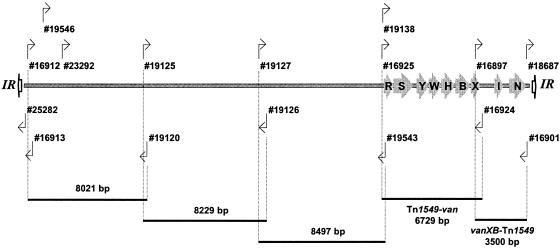
DNA was prepared from pure isolates of anaerobes and enterococci by using the DNeasy Tissue Kit (Qiagen Pty., Ltd., Victoria, Australia) and the modified protocol for isolation of genomic DNA from gram-positive bacteria as described in the manufacturer's instructions. Sequences of oligonucleotide primers used are listed in Table 2; the relative positions of primers and amplicons generated are depicted in Fig. 1. PCR was performed in a GeneAmp PCR System 2700 (Applied Biosystems, Foster City, Calif.) with known positive, negative, and multiple sterile water (no DNA) controls included in each run. The Expand Long Template PCR system (Roche Applied Science, Mannheim, Germany) was utilized, and PCR elongation times and temperatures were adjusted according to the expected size of the amplicon and nucleotide sequence of the primers, respectively, as recommended by the manufacturer. Inverse PCR was performed by using restricted and religated genomic DNA as a template with the following enzymes and internal divergent primer pairs: EcoRV, 16924 and 18687; EcoRV, 16913 and 19138; SfuI, 16913 and 23292; PstI, 16913 and 19127; DdeI, 16913 and 19546; and DdeI, 25282 and 19546. Circular intermediates of Tn1549 were demonstrated by PCR of the joint region by using the primers 18687 and 16913. RFLP analysis was achieved by restriction of the Tn1549-van amplicon (generated by using the primers 16925 and 16924) with RcaI (Roche Diagnostics) and DraI (New England Biolabs, Beverly, Mass.) for 3 h at 37°C. RFLP fragments and PCR amplicons were resolved by electrophoresis on agarose-Tris-acetate-EDTA gels containing 0.5 μg of ethidium bromide/ml. Gels were visualized and photographed by using a Fluor-S MultiImager (Bio-Rad Laboratories, Hercules, Calif.) and Quantity One version 4.2 (Bio-Rad Laboratories).
TABLE 2.
Oligonucleotide primers used in this study
| Primera | Nucleotide sequence | Locationb (nt) |
|---|---|---|
| 16897 | 5′-ATAGAGCGAGCCGAGTTGATTACA-3′ | 30163-30186 |
| 16901 | 5′-TCAAAAGTAGTAAATGGAGTAGTAAAACCG-3′ | 33663-33634 |
| 16912* | 5′-ACGCCATGCTATTTACTTCCGGC-3′ | 247-269 |
| 16913* | 5′-GTTCTTATTCCGCAGGTGGTGATT-3′ | 557-534 |
| 16924 | 5′-CAAAATACAAAATGGGCGTGGGAA-3′ | 30665-30642 |
| 16925 | 5′-AAAGAAGCGAGTATCCCAATGACG-3′ | 23936-23959 |
| 18687 | 5′-GTACATTATGGGACATTCCAATATCACC-3′ | 33526-33553 |
| 19120 | 5′-TCAAAAGCAAATGGAACTCG-3′ | 8268-8249 |
| 19125 | 5′-CCACAAAGCGTTGTTAGATC-3′ | 7994-8013 |
| 19126 | 5′-TTTGTCATAGCCTGCCTCTG-3′ | 16223-16204 |
| 19127 | 5′-AGACCCTAAAGACCGCCATC-3′ | 15676-15696 |
| 19138 | 5′-TTGAAACTACAGGGAAACTA-3′ | 23988-24007 |
| 19543 | 5′-AAAACTTGGTGTATGCCTCA-3′ | 24173-24154 |
| 19546 | 5′-CGGGAGAACACAACGAAAGT-3′ | 1269-1288 |
| 23292 | 5′-ACATCGACCCCAAGTTTGAAAA-3′ | 2555-2576 |
| 25282 | 5′-TCCGAAAGTAGTAAATTGGTA-3′ | 90-70 |
FIG. 1.
Schematic diagram of Tn1549 flanked by imperfect IRs. Coding regions for vanRB (R), vanSB (S), vanYB (Y), vanW (W), vanHB (H), vanB (B), vanXB (X), and the excisase (I) and integrase (N) genes are shown with arrows. The regions amplified and the positions of the primers used are indicated.
DNA sequencing and analysis.
The complete nucleotide sequence of the vanB operon and right end of the transposon in each of strains MLG043 (Eggerthella lenta), MLG245 (Clostridium sp.), and MLG229 (E. faecium) were obtained by directly sequencing the Tn1549-van amplicon and the vanXB-Tn1549 amplicon (see Fig. 1). Contiguous nucleotide sequences obtained were compared to the complete sequence of Tn1549 (GenBank accession no. AF192329) and, where sequence data were available, with Tn5382 (GenBank accession numbers AF310956 and AF063010). Sequences flanking the transposon and including the left and right inverted repeat (IR) of the transposon in all isolates was obtained by using template generated by inverse PCR as described above. PCR products to be sequenced were purified by using a QIAquick PCR purification kit (Qiagen). Sequences of PCR products were determined by cycle sequencing with the same primers used in PCR and by primer walking by using the ABI Prism BigDye Terminator v3.0 Ready Reaction Cycle sequencing kit (Applied Biosystems) with an ABI Prism 3100 genetic analyzer. Chromatograms were read, and contiguous sequences were constructed by using Vector NTI Advance version 8.0 (Informax, Inc., Bethesda, Md.). Sequences were compared for homology with the GenBank, EMBL, DDBJ, and PDB databases by using the BLASTN local alignment search tool (2) and the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov). Multiple sequence alignment was performed by using Vector NTI Advance version 8.0 and the CLUSTALW algorithm (29).
Southern hybridization, pulsed-field gel electrophoresis (PFGE), and ribotyping.
To determine whether the transposon had integrated discreetly and assess the number of elements within the genome of host organisms, Southern hybridization of the right integration site was used. A 502-bp amplicon spanning the 3′ end of vanXB and the adjacent Tn1549 sequence was amplified from genomic DNA extracted from MLG245 and labeled with digoxigenin (DIG) by PCR by using the primers 16897 and 16924 and a PCR DIG probe synthesis kit (Roche Applied Science) according to the manufacturer's recommendations. Genomic DNA samples to be probed were digested with EcoRI for 1 to 3 h at 37°C. Restricted DNA was electrophoresed on a 1% Tris-acetate EDTA (TAE) agarose gel overnight and then transferred to a positively charged nylon membrane and probed under high stringency using the technique of Southern (25). Probe-target hybrids were detected by chemiluminescence by using CDP-Star (Roche Diagnostics) according to the manufacturer's recommendations.
Automated ribotyping was performed by using the RiboPrinter microbial characterization system in accordance with the manufacturers instructions (DuPont Qualicon, Inc., Wilmington, Del.). Clostridium hathewayi isolates (MLG392, MLG661, and MLG856-1) were riboprinted by using EcoRI and enterococcal isolates (MLG229 and MLG856-2) were riboprinted by using a 1:1 mix of BamHI and AseI. Enterococcal isolates were also typed by PFGE of SmaI-digested genomic DNA. Agarose plugs were prepared as described previously (19), with the following modification. Plugs were pretreated with 6 mM Tris, 1 M NaCl, 100 mM EDTA (pH 7.6), 0.2% deoxycholate, 0.5% N-lauroylsarcosine, 1 mg of lysozyme/ml, and 50 μg of lysostaphin/ml for 4 h at 37°C, followed by 0.5 M EDTA, 1% Sarkosyl, and 1 mg of proteinase K/ml overnight at 56°C. DNA was restricted overnight at room temperature with 30 U of SmaI and then subjected to electrophoresis according to the Harmony study PFGE protocol (19).
Nucleotide accession numbers.
The nucleotide sequences of Tn1549/5382 in MLG229, MLG043, and MLG245 were submitted to GenBank under accession numbers AY655721, AY655718, and AY655720, respectively. Nucleotide sequences flanking the insertion of Tn1549/5382 in all isolates were assigned GenBank accession numbers AY786558 to AY786569 and AY841983 to AY841993. Nucleotide sequences of vanRB to vanSB in MLG392, MLG661, and MLG856-1 are listed under GenBank accession numbers AY841994 to AY841996.
RESULTS
Susceptibility testing of isolates.
The 10 vancomycin-resistant anaerobic species and their susceptibilities to vancomycin, teicoplanin, and benzylpenicillin are shown in Table 1. The MICs of vancomycin were 8 to >256 μg/ml, and all strains were susceptible to teicoplanin (MICs of <4 μg/ml), features consistent with the VanB phenotype. Three anaerobes—MLG080-1, MLG055, and MLG480—were highly resistant to benzylpenicillin (MIC of >256 μg/ml), whereas enterococcal isolates MLG229 and MLG856-2 were moderately resistant (MICs of 48 and 64 μg/ml, respectively). Only MLG080-1 demonstrated detectable β-lactamase. All VRE isolates and ATCC control strains had MICs within quality control ranges.
Vancomycin-resistant anaerobic gram-positive bacilli carry a Tn1549/5382 element.
PCR amplification of the Tn1549-van and vanXB-Tn1549 amplicons confirmed the presence of the vanB operon and a genetic linkage between this operon and Tn1549/5382 in all 10 anaerobes and the two E. faecium isolates, MLG229 and MLG856-2 (data not shown). No variation in the expected size of amplicons was observed, and sequencing of the ends of both amplicons confirmed the identity of the products obtained. In comparison, both E. faecalis ATCC 51299 and ATCC 29212 were negative for these amplicons. Presumably, the vanB gene of E. faecalis ATCC 51299 is not located on a Tn1549/5382-like element, whereas ATCC 29211 is sensitive to vancomycin and does not contain vanB.
Characterization and sequencing of the vancomycin resistance element in two anaerobes.
Using the sequence of Tn1549 (GenBank accession number AF192329), a series of overlapping PCRs were designed to characterize the vancomycin resistance element present within two anaerobes, MLG043 (Eggerthella lenta) and MLG245 (Clostridium sp.), and a local E. faecium carriage isolate (MLG229) isolate (Fig. 1). All three strains demonstrated amplicons indicative of the presence of the entire Tn1549/5382 element, and sequencing of the ends of these amplicons confirmed the location and identity of the amplicons (data not shown). Based on the size of amplicons obtained the Tn1549/5382 homologue present in each of these three isolates has a minimum size of 33 kb. Moreover, the nucleotide sequence from the start of the vanB operon to the right-end IR (IRR) in the three isolates has >99% identity to the same region in the published sequences of both Tn1549 and Tn5382. Sequence analysis of all three isolates also demonstrated the presence of the excisase and integrase genes and the IRR (CATATAATTTT) at the same position as observed for Tn1549 and Tn5382. The vanB operon within these three isolates differed by 65 nucleotides (nt) from the vanB operon of Tn1549 and by only 5 nt compared to the vanB operon within Tn5382. The nucleotide sequences of the vanB operons within MLG245 (Clostridium sp.) and MLG229 (E. faecium) were 100% identical to each other, and the deduced amino acid sequences of the open reading frames (ORFs) demonstrated the presence of VanRB, VanSB, VanYB, VanW, VanHB, VanB, and VanXB. MLG043 (Eggerthella lenta) demonstrated three nucleotide differences with MLG245 and MLG229; one of these included a transversion at nt 3641 (G→A) resulting in an in-frame TGA termination codon within vanW. This was immediately followed by a GTG start codon, leading to two ORFs spanning the region expected to encode vanW.
RFLP analysis of the vanB operon within isolates.
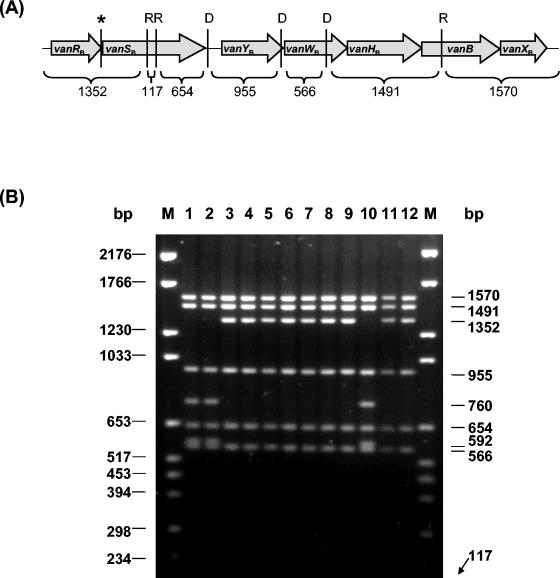
Restriction analysis of long PCR amplicons, which cover 5,959 bp of the 6,436 bp of the gene cluster (vanRB to vanXB), has been used previously to investigate the molecular heterogeneity of vanB alleles in enterococci and streptococci (7, 9). Using the restriction enzymes BspHI and DraI, two restriction profiles have been identified that differentiate vanB1 isolates (RFLP-1) from vanB2 or vanB3 isolates (RFLP-2) (7, 8). RFLP analysis of the Tn1549-van amplicon, using RcaI (an isoschizomer of BspHI) and DraI, was therefore used in the present study to identify any such variations within the vanB gene cluster found in our anaerobes. The Tn1549-van amplicon used in the present study comprised the entire vanB operon and contained flanking DNA derived from Tn1549/5382 to give an amplicon size of 6,729 bp. These longer amplicons were specifically chosen to enable differentiation of all fragments, which is not possible when shorter amplicons are used. According to the prototype vanB operon in Tn1549, the enzymes RcaI and DraI were expected to cut the Tn1549-van amplicon six times, yielding seven distinct fragments (Fig. 2A). RFLP analysis of Tn1549-van amplicons of all of the vancomycin-resistant isolates are shown in Fig. 2B. MLG043, MLG055, MLG080-1, MLG080-3, MLG101, MLG245, MLG480, MLG856-2, and MLG229 gave the profile expected. The RFLP profile for MLG392, MLG661, and MLG856-1, identified as C. hathewayi, gave a unique vanB RFLP pattern in which the 1,352-bp fragment (spanning vanRB to within vanSB) has been replaced with a 760- and a 592-bp product. Sequencing of the Tn1549-van amplicons of these three isolates in the region spanning vanRB to vanSB revealed three base substitutions. The first base change was a T→G at the end of the vanRB gene (nt 24705 of Tn1549). This generated a new RcaI restriction site (TCATGA) and a termination codon instead of leucine as the last amino acid of VanRB. Two other missense base substitutions were also identified in the vanSB gene, resulting in transition changes. A base change of T→C at nt 24833 of Tn1549 changed the amino acid from a leucine into a serine and a base change of C→T at nt 25250 of Tn1549 changed the amino acid proline into a leucine.
FIG. 2.
(A) Schematic diagram of the Tn1549-van PCR product. RcaI (R) and DraI (D) sites were deduced from the sequence of Tn1549 (GenBank accession no. AF192329). The sizes of the expected fragments are indicated in base pairs. The position of an additional RcaI site in C. hathewayi strains is indicated by an asterisk. Arrows denote the relative positions and sizes of coding regions within the operon. (B) RFLP analysis of Tn1549-van PCR amplicons. Amplicons were digested with RcaI (R) and DraI (D). Lanes: 1, MLG392; 2, MLG661; 3, MLG043; 4, MLG055; 5, MLG080-1; 6, MLsG080-3; 7, MLG101; 8, MLG245; 9, MLG480; 10, MLG856-1; 11, MLG856-2; 12, MLG229; M, Molecular DNA Marker VI (Roche Diagnostics). The sizes of the molecular markers are shown to the left of the gel. The sizes of the fragments obtained are shown to the right of the gel.
Characterization of the integration site of Tn1549/5382 within the genome of isolates.
Southern hybridization of the right integration site of Tn1549/5382 homologues demonstrated a single fragment in all isolates, suggesting a single insertion of Tn1549/5382 within the genome of the host organism (data not shown). EcoRI fragment sizes ranged from 8 to 20 kb, encompassing a minimum of 2 kb downstream of the right-end imperfect IR (IRR), and were variable with the exception of C. hathewayi and E. faecium isolates.
The vanXB-Tn1549 probe hybridized to a 9.4-kb EcoRI fragment in all three C. hathewayi isolates (MLG392, MLG661, and MLG856-1 [data not shown]), and flanking sequences generated by inverse PCR at both the left and right ends of Tn1549/5382 were identical. A BLASTn search of flanking sequences showed no homology with known sequences present within the GenBank database, although a single ORF, identified downstream of the transposon, showed 39% identity (41 of 103 amino acids) with helix-turn-helix DNA transcriptional regulators. Riboprints of these isolates were identical. All three organisms were isolated at different times and from different patients, but two of the patients were at the same hospital.
The vanXB-Tn1549 probe hybridized to a 10-kb EcoRI fragment in both E. faecium strains (MLG229 and MLG856-2), which contained identical nucleotide sequences downstream of the IRR. A BLASTn search of the sequence indicated insertion of the element within the chromosome between an ISEf1 element and the gene Efae1387 (nt 6729 of GenBank accession no. NZAAAK01000210). The flanking sequence was also homologous to the complementary strand of section six of eleven of the complete genome of E. faecalis V583 (GenBank accession no. AE16952). Both PFGE and ribotyping suggested that these two isolates were clonal.
Nucleotide sequencing of the region flanking the left and right IRs of Tn1549/5382 in the remaining seven anaerobes (MLG043,MLG055, MLG101, MLG245, MLG480, MLG080-1, and MLG080-3) revealed unique sequences with no significant nucleotide homologies with known sequences in GenBank. Identified ORFs showed homology with various housekeeping genes (data not shown) that are common among many species, suggesting an insertion within the chromosome. However, this location requires further study, especially given previous reports of conjugal transfer of Tn1549/5382 in association with plasmids (30). The IRL (AAAATTTTAGG) and IRR (CATATAATTTT) was present in the same position as observed for Tn1549 and Tn5382 in all 12 isolates.
Circular intermediate of Tn1549/5382 located within isolates.
The existence of a nonreplicative circular intermediate for both Tn1549 and Tn5382 has previously been demonstrated in E. faecalis and E. faecium, respectively, by using amplification strategies with primers designed to polymerize outward from the ends of the transposon (6, 11). Using this strategy, an 818-bp PCR amplicon, encompassing the region where the two ends of the transposon join, was detected in nine of the vancomycin-resistant isolates (Table 1). In contrast, MLG856-2, MLG229, and MLG055 did not demonstrate the presence of a circular intermediate. After sequencing of the amplicons in seven of the nine isolates (insufficient amounts of amplicon for MLG392 and MLG043 were obtained for direct sequencing), we were able to identify both imperfect IRs, IRR and IRL, at each end of the transposon separated by a 5- to 6-bp coupling sequence.
DISCUSSION
In this study we have demonstrated the presence of the vanB operon on a Tn1549/5382 element, a situation essentially identical to that found in enterococci, in a collection of 10 anaerobic bacillus strains (eight Clostridium spp., one Eggerthella lenta, and one Ruminococcus sp.). This is the first characterization of vancomycin resistance via this transposon type in these species. These diverse organisms form an important and predominant part of the microflora of the human gastrointestinal tract, yet little is known about their influence on the normal physiology and pathology of the host (1, 31). The discovery of a reservoir of Tn1549/5382 within bowel flora and the increasingly diverse nature of organisms with which this transposon type has been associated (Enterococcus spp., Streptococcus spp., Clostridium spp., Ruminococcus spp., and Eggerthella spp.) may have important implications for the emergence of vancomycin-resistant pathogens, such as VRE (28, 5, 18, 9).
All our anaerobes displayed resistance to vancomycin, but not teicoplanin, a finding consistent with the VanB phenotype. Interestingly, MLG043 demonstrated an in-frame termination codon (TGA) with vanW, which appears to have little impact on the expression of resistance with the organism, for which the vancomycin MIC was >256 μg/ml. It remains to be seen how vanW is translated in MLG043, but there are rare exceptions to the universal code involving the codon UGA; in some unique contexts it encodes selenocysteine, and it encodes tryptophan in Mycoplasma (26). Previously, integration of Tn5382 within the genome of some isolates of E. faecium has been demonstrated immediately adjacent to pbp5, a gene encoding resistance to penicillin (6, 14). Cotransfer of penicillin-ampicillin resistance and vancomycin resistance between E. faecium isolates via a large chromosomally located element containing both pbp5 and Tn5382 has also been demonstrated (6). For this reason the 10 anaerobes and 2 E. faecium isolates were examined by E-test for phenotypic resistance to benzylpenicillin. Although both E. faecium isolates (MLG229 and MLG856-2) and three anaerobes (MLG055, MLG480, and MLG080-1) displayed resistance to benzylpenicillin, sequences flanking the Tn1549/5382 element of these isolates did not encode the pbp5 gene, suggesting that in these isolates vancomycin and penicillin resistance is unlikely to be linked.
RFLP analysis of the Tn1549-van amplicon was used in the present study to identify variations within the vanB gene cluster found in our anaerobes. Seven of the ten anaerobes and both E. faecium isolates revealed a RFLP profile similar to the vanB2 (RFLP-2) profile of Dahl et al. (7), whereas a unique RFLP profile was detected for the C. hathewayi isolates (MLG392, MLG661, and MLG856-1). Subsequent sequencing of the left end of the operon in these latter isolates revealed three base substitutions in the vanRB-to-vanSB region, one of which was responsible for an additional RcaI restriction site. Changes within these genes may affect the transcriptional activation of proceeding genes (3, 16). However, there was no apparent phenotypic effect on vancomycin and teicoplanin resistance in these three isolates compared to E. faecium. It is interesting that MLG856-1 (C. hathewayi) and MLG856-2 (E. faecium) were isolated from the same patient (Table 1), and yet both showed different RFLP profiles for Tn1549-van amplicons. This finding suggests that, despite the similar origins of these strains, it is unlikely that the Tn1549/5382 homologue present in these isolates resulted from genetic exchange between the two organisms. The vanB2 subtype has been universally linked with the Tn1549/5382 family of elements and has a 3.6% base-pair difference from the vanB1 ligase gene of E. faecalis V583 (8, 9, 11, 12, 17). However, we have observed an unusual anomaly with the vanB subtype of the Tn1549/5382 element of MLG480. The RFLP profile of this organism corresponds to the profile expected for vanB2, and yet the nucleotide sequence of the vanB ligase gene from this isolate is only 1% different from vanB1 but 3% different from vanB2, suggesting that the gene may belong to the vanB1 subtype (5). Dahl et al. (7) postulated that the vanB1 cluster might be older in evolutionary terms than vanB2, leading to the possibility that the vanB ligase gene of MLG480 might provide a tenuous link between the evolution of vanB subtypes.
Independent transposition of Tn1549 and Tn5382 has not yet been demonstrated. Instead, conjugative transfer of chromosomally located elements is associated with the transfer of large (>130-kb) elements that have yet to be delineated, whereas conjugative transfer of plasmid-located elements is attributed to the host plasmid (6, 11, 10). However, both Tn1549 and Tn5382 have previously been shown to excise discretely and form a circular intermediate, and unique insertions of the element provide indirect evidence for possible conjugative transposition of this element (10, 6, 11). Similarly, in our anaerobes the presence of a circular intermediate and unique sequences flanking the integrative site in nonclonal isolates demonstrates discrete excision and insertion of the Tn1549/5382 element with no evidence of a common larger element at either end.
The present study has demonstrated the presence of the vancomycin resistance Tn1549/5382 element in a collection of anaerobic bowel flora belonging primarily to the genus Clostridium. The extensive similarity and the potential to transpose as demonstrated by excision of the element, suggests a recent horizontal transfer event has occurred between enterococci and anaerobic bowel flora. However, the original direction of transposition and mechanism remains unknown. The ability of these diverse anaerobic organisms to exchange resistance determinants with enterococci and other potential pathogens in the human gut requires further study and has potentially important implications in terms of the evolution of VRE and possibly the infection control measures needed to control its spread. The infection control practice of monitoring fecal VRE carriage, which is currently being followed at many institutions, may be inadequate and require extension to focus on fecal vanB gene carriage (15, 20, 27). The discovery of a reservoir of the Tn1549/5382 element in bacteria that are part of the normal bowel flora supports this.
Acknowledgments
We thank E. A. Grabsch and S. Xie, Department of Microbiology, Austin Health, Melbourne, Victoria, Australia, for their generous assistance with Automated Ribotyping and PFGE analysis. We also thank J. Rood, Department of Microbiology, Monash University, Melbourne, Victoria, Australia, for scientific discussion of clostridia and transposable elements.
This project was supported in part by a quality improvement grant from the Department of Human Services, Victoria, Australia.
REFERENCES
- 1.Allen, S. D., C. L. Emery, and D. M. Lyerly. 2003. Clostridium, p. 835-856. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur, M., and R. Quintiliani, Jr. 2001. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob. Agents Chemother. 45:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur, M., C. Molinas, F. Depardieu, and P. Courvalin. 1993. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in enterococcus faecium BM4147. J. Bacteriol. 175:117-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballard, S. A., E. A. Grabsch, P. D. R. Johnson, and M. L. Grayson. 2005. Comparison of three PCR primer sets for identification of vanB gene carriage in feces and correlation with carriage of vancomycin-resistant enterococci: interference by vanB-containing anaerobic bacilli. Antimicrob. Agents Chemother. 49:77-81. [DOI] [PMC free article] [PubMed]
- 6.Carias, L. L., S. D. Rudin, C. J. Donskey, and L. B. Rice. 1998. Genetic linkage and cotransfer of a novel, vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J. Bacteriol. 180:4426-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl, K. H., G. S. Simonsen, (swsl)O. Olsvik, and A. Sundsfjord. 1999. Heterogeneity in the vanB gene cluster of genomically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob. Agents Chemother. 43:1105-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahl, K. H., E. W. Lundblad, T. P. Røkenes, (swsl)O. Olsvik, and A. Sundsfjord. 2000. Genetic linkage of the vanB2 gene cluster to Tn5382 in vancomycin-resistant enterococci and characterization of two novel insertion sequences. Microbiol. 146:1469-1479. [DOI] [PubMed] [Google Scholar]
- 9.Dahl, K. H., and A. Sundsfjord. 2003. Transferable vanB2 Tn5382-containing elements in fecal streptococcal strains from veal calves. Antimicrob. Agents Chemother. 47:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahl, K. H., T. P. Røkenes, E. W. Lundblad, and A. Sundsfjord. 2003. Nonconjugative transposition of the vanB-containing Tn5382-like element in Enterococcus faecium. Antimicrob. Agents Chemother. 47:786-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 12.Gold, H. H., S. Unal, E. Cercenado, C. Thauvin-Eliopoulos, G. M. Eliopoulos, C. B. Wennersten, and R. C. Moellering, Jr. 1993. A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob. Agents Chemother. 37:1604-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grayson, M. L., E. A. Grabsch, P. D. R. Johnson, D. Olden, M. Aberline, H. Y. Li, G. Hogg, M. Abbott, and P. Kerr. 1999. Outcome of a screening program for vancomycin-resistant enterococci (VRE) in a University Teaching Hospital. Med. J. Aust. 171:133-136. [DOI] [PubMed] [Google Scholar]
- 14.Hanrahan, J., C. Hoyen, and L. B. Rice. 2000. Geographic distribution of a large mobile element that transfers ampicillin and vancomycin resistance between Enterococcus faecium strains. Antimicrob. Agents Chemother. 44:1349-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hospital Infection Control Practices Advisory Committee. 1995. Recommendations for preventing the spread of vancomycin resistance. Infect. Control Hosp. Epidemiol. 16:105-113. [DOI] [PubMed] [Google Scholar]
- 16.Kak, V., and J. W. Chow. 2002. Acquired antibiotic resistances in enterococci, p. 355-383. In D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci. American Society for Microbiology, Washington, D.C.
- 17.McGregor, K. F., C. Nolan, H.-K. Young, M.-F. I. Palepou, L. Tysall, and N. Woodford. 2001. Prevalence of the vanB2 gene cluster in VanB glycopeptide-resistant enterococci in the United Kingdom and the Republic of Ireland and its association with a Tn5382-like element. Antimicrob. Agents Chemother. 45:367-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mevius, D., L. Devriese, P. Butaye, P. Vandamme, M. Verschure, and K. Veldman. 1998. Isolation of glycopeptide resistant Streptococcus gallolyticus strains with vanA, vanB, and both vanA and vanB genotypes from faecal samples of veal calves in The Netherlands. J. Antimicrob. Chemother. 42:275-276. [DOI] [PubMed] [Google Scholar]
- 19.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. E. Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Balkeum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombs, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padiglione, A., R. Wolfe, E. A. Grabsch, D. Olden, S. Pearson, C. Franklin, D. Spelman, B. Mayall, P. D. R. Johnson, and M. L. Grayson. 2003. Risk factors for the new detection of vancomycin-resistant enterococci (VRE) in acute-care hospitals that employ strict infection control procedures. Antimicrob. Agents Chemother. 47:2492-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel, R. J. R. Uhl, P. Kohner, M. K. Hopkins, J. M. Steckelberg, B. Kline, and F. R. Cockerill III. 1998. DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical enterococcus isolates. Antimicrob. Agents Chemother. 42:202-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poyart, C., C. Pierre, G. Quesne, B. Pron, P. Berche, and P. Trieu-Cuot. 1997. Emergence of vancomycin resistance in the genus Streptococcus: characterization of a vanB transferable determinant in Streptococcus bovis. Antimicrob. Agents Chemother. 41:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintiliani, R., Jr., and P. Courvalin. 1996. Characterization of Tn1547, a composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene 172:1-8. [DOI] [PubMed] [Google Scholar]
- 24.Rice, L. B. 2001. Emergence of vancomycin-resistant enterococci. Emerg. Infect. Dis. 7:183-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Snyder, L., and W. Champness. 2003. Molecular genetics of bacteria, 2nd ed. ASM Press, Washington, D.C.
- 27.Standing Committee on Infection Control. 1999. Guidelines for the management of patients with vancomycin-resistant enterococci (VRE) colonization/infection. Department of Health and Human Services, Washington, D.C.
- 28.Stinear, T. P., D. C. Olden, P. D. R. Johnson, J. K. Davies, and M. L. Grayson. 2001. Enterococcal vanB resistance locus in anaerobic bacteria in human faeces. Lancet 357:855-856. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multi-sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids. Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umeda, A., F. Garnier, P. Courvalin, and M. Galimand. 2002. Association between the vanB2 glycopeptide resistance operon and Tn1549 in enterococci from France. J. Antimicrobial. Chemother. 50:253-256. [DOI] [PubMed] [Google Scholar]
- 31.Wang, X., S. P. Heazlewood, D. O. Krause, and T. H. J. Florin. 2003. Molecular characterization of the microbial species that colonize human ileal and colonic mucosa by using 16S rDNA sequence analysis. J. Appl. Microbiol. 95:508-520. [DOI] [PubMed] [Google Scholar]
- 32.Weaver, K. E., L. B. Rice, and G. Churchward. 2002. Plasmids and transposons, p. 219-263. In D. B. Clewell, P. Courvalin, G. M. Dunny, B. E. Murray, and L. B. Rice (ed.), The enterococci. American Society for Microbiology, Washington, D.C.