Abstract
Background
The expression of Egl-9 family hypoxia-inducible factor 3 (EGLN3) is notably decreased in various malignancies, including gastric cancer (GC). While the predominant focus has been on the hydroxylase activity of EGLN3 for its antitumour effects, recent findings have suggested nonenzymatic roles for EGLN3.
Methods
This study assessed the clinical significance of EGLN3 expression in GC and explored the connection between EGLN3 DNA promoter methylation and transcriptional silencing. To investigate the effect of EGLN3 on GC cells, a gain-of-function strategy was adopted. RNA sequencing was conducted to identify the key effector molecules and signalling pathways associated with EGLN3.
Results
EGLN3 expression was significantly reduced in GC tissues, correlating with poorer patient prognosis. EGLN3 hypermethylation disrupts transcriptional equilibrium, contributing to deeper tumour invasion and lymph node metastasis, thus exacerbating GC progression. Conversely, restoration of EGLN3 expression in GC cells substantially inhibited cell proliferation and metastasis. EGLN3 was also found to impede the malignant progression of GC cells by downregulating Jumonji C domain-containing protein 8-mediated activation of the NF-κB pathway, independent of its hydroxylase activity.
Conclusions
EGLN3 has the potential to hinder the spread of GC cells through a nonenzymatic mechanism, thereby shedding light on the complex nature of GC progression.

Subject terms: Gastric cancer, DNA methylation, Cell signalling
Background
Gastric cancer (GC) is the fifth most prevalent malignancy and the fourth leading cause of cancer-related fatalities worldwide. It imposes a substantial disease burden owing to its frequent late-stage diagnosis and the absence of efficient therapeutic modalities [1–3]. Consequently, the prognosis of patients remains dismal [2]. Endeavours to comprehend the initiation and advancement of GC have aimed to identify valuable biomarkers with clinical applicability. However, the notably heterogeneous nature of GC complicates our understanding of this lethal malignancy, necessitating further investigation to unravel its underlying biology.
The Egl-9 family hypoxia-inducible factor 3 (EGLN3) belongs to the group of 2-oxoglutarate- and ferrous iron- dependent dioxygenases, known for their proficient hydroxylation of proline residues in substrates [4, 5]. Multiple substrates of EGLN3 have been identified, with hypoxia-inducible factors (HIFs) extensively documented among them [4–8]. Previous research has shown that under normoxic conditions, EGLN3 recognises and hydroxylates two proline residues (P402 and P564) located in the N-terminal and C-terminal domains of the oxygen-dependent degradation domains of HIFα, denoted NODD and CODD, respectively. Subsequently, HIFα undergoes proteasome-mediated degradation [6–11]. However, the enzymatic activity of EGLN3 diminishes in oxygen-deficient environments, resulting in rapid accumulation of HIFα proteins and activation of downstream signalling pathways [5, 12]. The pivotal role of EGLN3 as a regulator of HIFα is underscored by its abnormal downregulation in a diverse spectrum of cancers [13, 14]. Recent studies have revealed additional substrates in addition to the HIF family. EGLN3 disrupts fatty acid oxidation by directly catalysing the hydroxylation of acetyl-CoA carboxylase 2, consequently inhibiting cellular proliferation and disease progression in acute myeloid leukaemia [15]. Moreover, EGLN3 hydroxylates proline residue 359 within the deubiquitinating enzyme-binding domain of p53, enhancing its binding affinity to the deubiquitinating enzymes USP7 and USP10 and thereby preventing p53 degradation [16]. Notably, EGLN3 operates in a hydroxylase-independent manner under specific conditions. For instance, it competitively binds to the inhibitor of κB kinase β (IKKβ) and disrupts the IKKβ-Hsp90 interaction necessary for the activation of the IKKβ/NF-κB signalling pathway [17]. These findings underscore the antineoplastic properties of EGLN3; nevertheless, the precise mechanisms of EGLN3 in different cancer types require further validation.
Anomalous DNA promoter CpG island hypermethylation patterns play a pivotal role in carcinogenesis and serve as epigenetic markers for gene silencing [18, 19]. In this study, we determined that EGLN3 was silenced via CpG island hypermethylation in GC cells. Furthermore, our investigations revealed EGLN3 as an independent prognostic factor for patients with GC, and its restoration significantly curtails malignant progression in GC cells. Importantly, we demonstrated that EGLN3 exerts its tumoricidal effect independent of its enzymatic activity in GC cells.
Materials and methods
Patient specimens
Tissue samples were collected from 109 patients with GC who underwent curative gastrectomy at Tianjin Medical University Cancer Hospital (Tianjin, China), Xijing Hospital of Air Force Medical University (Xi’an, China), and Renji Hospital of Shanghai Jiao Tong University School of Medicine (Shanghai, China) from January 2004 to September 2007. All these patients enrolled did not receive neoadjuvant therapy before gastrectomy. Freshly collected GC (N = 8) and normal gastric mucosa specimens (N = 4) were collocated from patients receiving curative gastrectomy in February 2022 at Tianjin Medical University Cancer Hospital (Tianjin, China). All experiments used these samples were approved by the Institutional Research Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (no. bc2020091) (Tianjin, China).
Cell lines and cell culture
Human GC cell lines (NCI-N87, SNU-1, AGS, SNU-16 and KATO III) and HEK293T were obtained from American Type Culture Collection (VA, USA). MKN-45, HGC-27, SGC-7901, MGC-803, BGC-823 and human immortalised gastric epithelial cells (GES-1) were from the National Infrastructure of Cell Line Resource (Beijing, China). All cell lines were cultured in RPMI-1640 supplemented with 10% foetal bovine serum (FBS), except for HEK293T, AGS and KATO III cells (HEK293T cells were cultured in DMEM containing 10% FBS, AGS in F12K with 10% FBS and KATO III cells in Iscove’s Modified Dulbecco’s Medium (IMDM) with 20% FBS). Cells were maintained in a cell incubator with 5% CO2 at 37 °C. All cells in this study were STR-validated.
Antibodies and reagents
Antibodies against the following proteins were used for western blotting: EGLN3 (1:1000, Novus; NB100-139), JMJD8 (1:250, Santa Cruz; sc-515520), HIF1α (1:1000, Abcam; ab216842), Phospho-Histone H3 (Ser10) (1:1000, CST; D7N8E), Lamin-B1 (1:1000, Proteintech; 66095-1-AP), Vinculin (1:1000, CST; E1E9V), β-actin (1:1000, Proteintech; 20536-1-AP), NF-κB P65 (1:1000, CST; D14E12) and Phospho-P65 (Ser536) (1:1000, CST; 93H1). The following antibodies were used for immunohistochemistry (IHC): EGLN3 (1:100, Novus; NB100-303).
MassARRAY analysis and next-generation sequencing
Genomic DNA of GC tissues and cells were extracted using QIAamp DNA Mini Kit (Qiagen, Valencia, CA, USA). The methylation of EGLN3 promoter was detected using MassARRAY analyser (Agena Bioscience, San Diego, CA, USA) and next-generation sequencing (NGS) platform (Sango Biotech, Shanghai, China).
Lentivirus packaging and cell infection
Lentivirus was produced in HEK293T cells simultaneously transfected with the indicated lenti-vectors, psPAX2 (Plasmid #12260) and pMD2.G (Plasmid #12259). The intact EGLN3 or JMJD8 ORF was cloned into pLVX-IRES-puro vector or pLVX-IRES-neo vector, respectively. Short hairpin RNA (shRNA) sequences targeting EGLN3 or JMJD8 were engineered in pSIH1-puro vector. Empty pLVX-IRES-puro vector, pLVX-IRES-neo vector and pSIH1-puro vector were used as corresponding negative controls. The shRNA sequences were as follows: CTACGTCAAGGAGAGGTCTAA (shE3), gcCCAATGAGACTGACATCAA (shDNMT1), ccGGCTCTTCTTTGAGTTCTA (shDNMT3A), AGATGACGGATGCCTAGAG (shDNMT3B), ACAGGTTGCTGGCTTCGTTTG (shJ8#1) and ACTTGCCCTTCCAGGAGTATG (shJ8#2). Cells were incubated with the indicated lentivirus and polybrene (1 μl/ml) for 24 h. G418 (400 μg/ml) or puromycin (2 μg/ml) was used to establish stable cells. The vector pLVX-IRES-puro/neo and pSIH-H1-puro were generously provided by Prof. Zhihua Liu from the National Cancer Centre/Cancer Hospital (Beijing, China).
Immunohistochemistry and in situ hybridisation assay
EGLN3 staining was performed following the IHC procedure described previously [20]. The staining was evaluated based on the average intensity of tumour cells: -, negative; +, weakly positive; ++, medium positive; +++, strong positive. The intensity scores of ++ and +++ were regarded as high expression of EGLN3.
JMJD8 mRNA was detected following the manufacturer’s instructions of RNAscope 2.5 HD Reagent Kit (Advanced Cell Diagnostics, Newark, CA, USA) in the freshly collected tissues.
RNA extraction and quantitative PCR
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, MA, USA). The cDNAs were generated using GoScript™ Reverse Transcription Kit (Promega, Madison, WI, USA). The mRNA levels of all genes were determined on the QuantStudio 5 real-time PCR system (Applied Biosystems, Foster City, CA, USA) using TB Green Premix Ex TaqTM II (TaKaRa, Japan). β-actin was used for data normalisation, and the 2−ΔΔCt method was used to evaluate the relative abundance of the detected genes. The primer sequences are listed in Supplementary Table S1.
Protein extraction and western blotting
Cells were washed with pre-chilled PBS buffer and lysed with radioimmuno-precipitation assay (RIPA) buffer (Boster Biological Technology, China) supplemented with protease inhibitors (MCE, NJ, USA) and phosphatase inhibitors (MCE, USA). BCA Protein Assay Kit (Thermo Scientific, MA, USA) was used to quantify protein concentration. Denatured proteins were electrophoresed by vertical SDS-PAGE system (Bio-Rad, CA, USA) and transferred to polyvinylidene fluoride membranes. Membranes were blocked with 5% nonfat milk buffer for 1.5 h and then incubated with primary antibodies overnight at 4 °C. Following washing with TBST, the membrane was incubated with secondary antibody for 2 h and was visualised using chemiluminescence reagent (Thermo Scientific, USA).
Co-immunoprecipitation assay
To assess the interaction between EGLN3 and JMJD8, HEK293T cells were transfected with EGLN3-Myc and JMJD8-Flag plasmids. The intact EGLN3-Myc and JMJD8-Flag were cloned into pcDNA3.1(+) vector respectively. Harvested cells were lysed in 1%NP-40 lysis buffer [50 mM Tris-HCl (pH 7.5), 150 mM NaCl and 1% NP-40] containing protease inhibitor cocktail (Selleck). Immunoprecipitation was performed by incubating 500 μg of total protein with anti-Flag magnetic beads (M8823, Sigma), anti-Myc-Tag mAb magnetic beads (M04711, MBL) or mouse IgG (M07511, MBL) pre-cleaned with 1% NP-40 lysis buffer at 4 °C overnight. Bounded proteins were eluted with 0.05%NP-40 lysis buffer for four times and the elution was analyzed by western blot analysis with anti-Flag or Myc antibodies. The primary antibodies used for immunoprecipitation were as follows: anti-Flag (1:1000) (F1804, Sigma), anti-Myc (1:1000) (PTM-5390, PTM).
Cell proliferation analysis
Cell proliferation was examined by CCK8 and EdU assay. The cell suspension was inoculated in a 96-well plate with 2000 cells/well. CCK8 (Zeta Life, China) was added to the cell suspension at a ratio of 1:10 and incubated at 37 °C for 2 h. The absorbance at 450 nm was measured using a microplate reader (BioTek, VT, USA).
The EdU assay kit was purchased from Keygen Biotech Company (KGA330-50, China). 5 × 105 cells were inoculated in 6-well culture plate and cultured for 24 h to achieve logarithmic growth. The cells were labelled by 50 µM EdU and incubated for 2 h. After fixation with 4% paraformaldehyde and permeabilization with 1% saponin, the collected cells were stained with Click-iT reaction cocktail for 30 min. The cell proliferation rate was detected by BD FACSCanto™ II Flow Cytometer (BD Biosciences, CA, USA).
Cell migration and invasion analysis
For the migration assay, the serum-free cell suspension was seeded in the top chambers (4 × 104 cells/well) of 24-well Transwell plates (8-μm pore size, Corning, NY, USA), and culture medium containing 20% FBS was added to the bottom chambers. Invasion assay was carried out using chambers coated with Matrigel (BD Biosciences, USA). After 24 h of incubation, the inserts were fixed with 4% paraformaldehyde for 30 min, and stained with 0.4% crystal violet for 30 min. The cell numbers were calculated by averaging the counts from three random fields.
Simulation of hypoxia environment
The concentrated storage solution (100 mM) of cobalt chloride (CoCl2) was prepared by dissolving in sterile distilled water, and the final required concentration was diluted with the corresponding cell culture medium. SNU-1 and NCI-N87 cells were cultured in a standard incubator for 24 h and treated with CoCl2 at a concentration of 100 μM for 12 h.
RNA expression profile
Total RNA was extracted from GC cells overexpressing EGLN3 and empty vector. Sample sequencing and analysis were performed by Beijing Novogene Technology (Beijing, China). The data is deposited under GSE200646 in GEO database.
Animal experiments
Female 4-week-old BALB/c nude mice and NOD/SCID mice were purchased from Gempharmatech Company (Jiangsu, China). NCI-N87 cells (2 × 106/100 μl) were injected subcutaneously into the back of these athymic nude mice. The growth of tumours was measured with caliper every 2 days, and the tumour was collected and weighed at the 4 weeks after the implantation. The calculation formula of average tumour volume (TV) is: TV (mm3) = 1/2 × length × Width2.
For lung metastasis assays, 8 × 105 luciferase-labelled GC cells were injected into the caudal vein of 5-week-old female NOD/SCID mice for monitoring and imaging using a bioluminescence imaging system (Calliper Life Science, IVIS spectrum, MA, USA). After 12 weeks of injection, lung tissues were collected and nodules were counted. All experimental animal procedures were carried out with the approval of the Institutional Animal Care and Research Advisory Committee of Tianjin Medical University (AE-2021073) (Tianjin, China).
Statistical analysis
Each experiment, except the IHC and animal studies, was independently performed three times. All data are presented as the mean values ± SD. Data analysis was performed using Prism 8.0 software (GraphPad Software), in which *, **and *** indicated P < 0.05, P < 0.01, P < 0.001, respectively.
Results
Low expression of EGLN3 indicated poor GC results
To elucidate the clinical significance of EGLN3 expression in GC, we analysed EGLN3 mRNA levels in TCGA GC specimens. This analysis revealed a significant downregulation of EGLN3 in GC tissues (n = 415) compared to normal tissues (n = 35) (P < 0.0001) (Fig. 1a). Parallel observation of diminished EGLN3 mRNA expression was made in a separate dataset (GSE29272) sourced from the Gene Expression Omnibus repository (P < 0.0001) (Fig. 1b). Furthermore, within a set of 30 pairs of tumour and adjacent nontumor tissues, EGLN3 mRNA levels were consistently lower in GC tissues than in their adjacent nontumor counterparts (P < 0.0001) (Fig. 1c).
Fig. 1. EGLN3 expression decreases in GC tissues and GC cell lines, which indicates poor patient outcome.
a EGLN3 mRNA level is significantly downregulated in tumour tissues (n = 415) compared with normal tissues (n = 35) from TCGA GC database (P < 0.0001). b The mRNA level of EGLN3 in tumours (n = 134) is dramatically reduced compared with their paired adjacent normal tissues (n = 134) in one dataset (GSE29272) deposited in the Gene Expression Omnibus repository database (P < 0.0001). c EGLN3 mRNA expression is significantly downregulated in in-house GC tissues by quantitative PCR (P < 0.001). d EGLN3 staining in GC tissues is weaker than that in non-tumour adjacent tissues. Representative IHC images are shown here. Scale bars, 200 μm. A total of 109 tumour and 109 non-tumour adjacent tissues were analyzed. ***, P < 0.001. e Higher EGLN3 expression is indicative of better overall survival rate (P = 0.0045). f The mRNA levels of EGLN3 are downregulated in most GC cell lines relative to GES1 cells detected by quantitative PCR. g EGLN3 protein levels are decreased in most GC cell lines relative to GES1 cells detected by western blot.
Subsequently, immunohistochemical (IHC) examination of tissue microarrays from 109 GC patients corroborated these findings, with EGLN3 staining exhibiting significantly reduced intensity in GC tissues compared to that in adjacent nontumor tissues (n = 109, P < 0.001) (Fig. 1d). Further investigation into the association between EGLN3 expression and the clinical characteristics of these 109 GC patients revealed a correlation between lower EGLN3 protein expression and depth of invasion (pT stage) (P < 0.05) and lymph node metastasis (pN stage) (P < 0.05) (Table 1).
Table 1.
Analysis of EGLN3 expression in GC tissues and associated clinicopathological factors.
| Characteristics | EGLN3 | P value | |
|---|---|---|---|
| Low | High | ||
| Age, years | 0.259 | ||
| <65 | 46 | 21 | |
| ≥65 | 33 | 9 | |
| Gender | 0.227 | ||
| Male | 57 | 25 | |
| Female | 22 | 5 | |
| Size, cm | 0.484 | ||
| ≤4 cm | 22 | 11 | |
| >4 cm | 57 | 19 | |
| Tumour location | 0.282 | ||
| Upper 1/3 | 15 | 3 | |
| Middle 1/3 | 10 | 3 | |
| Lower 1/3 | 36 | 20 | |
| More than 2/3 stomach | 18 | 4 | |
| pT stagea | 0.035 | ||
| pT1-2 | 3 | 5 | |
| pT3-4 | 76 | 25 | |
| pN stagea | 0.014 | ||
| pN0 | 12 | 11 | |
| pN+ | 67 | 19 | |
| Lauren type | 0.106 | ||
| Intestinal | 17 | 11 | |
| Diffuse | 62 | 19 | |
apT and pN stage is defined using the American Joint Committee on Cancer (AJCC) Staging System, 8th Edition.
Kaplan‒Meier analysis demonstrated a significant link between low EGLN3 expression and poor prognosis in patients with GC (Fig. 1e). Moreover, through Cox regression analysis, EGLN3 emerged as an independent prognostic factor in patients with GC (Table 2). Collectively, these results underscore the potential of EGLN3 expression as a determinant for improving the prognosis of GC.
Table 2.
Univariate and multivariate Cox regression analyses for overall survival of gastric cancer patients.
| Predictor | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR(95% CI) | P | HR(95% CI) | P | |
| Age, years | ||||
| ≥65 vs <65 | 1.214(0.786–1.875) | 0.381 | ||
| Gender | ||||
| Female vs Male | 1.430(0.897–2.280) | 0.133 | ||
| Size, cm | ||||
| >4 cm vs ≤4 cm | 1.707(1.054–2.765) | 0.030 | 1.549(0.949–2.527) | 0.080 |
| Tumour location | ||||
| Middle 1/3 vs Upper 1/3 | 0.867(0.397–1.893) | 0.721 | ||
| Lower 1/3 vs Upper 1/3 | 0.701(0.379–1.297) | 0.258 | ||
| More than 2/3 stomach vs Upper 1/3 | 1.058(0.524–2.139) | 0.874 | ||
| pT stagea | ||||
| pT3-4 vs pT1-2 | 0.935(0.430–2.030) | 0.864 | ||
| pN stagea | ||||
| pN+ vs pN0 | 2.095(1.177–3.730) | 0.012 | 1.729(0.956–3.125) | 0.070 |
| Lauren type | ||||
| Diffuse vs Intestinal | 1.095(0.662–1.809) | 0.724 | ||
| Expression of EGLN3 | ||||
| Low vs High | 2.077(1.234–3.496) | 0.006 | 1.905(1.122–3.235) | 0.017 |
apT and pN stage is defined using the American Joint Committee on Cancer (AJCC) Staging System, 8th Edition.
Promoter hypermethylation gave rise to low EGLN3 expression in GC
In vitro analysis revealed a significant reduction in EGLN3 expression at both the mRNA and protein levels in most GC cell lines in comparison to the immortalised human gastric epithelial cell line GES-1 (Fig. 1f, g). Promoter DNA hypermethylation serves as an epigenetic marker for gene silencing. To establish the connection between transcriptional silencing of EGLN3 and DNA promoter hypermethylation, we examined the methylation pattern of the promoter CpG sites of EGLN3 in GC cell lines. The region selected for MassARRAY methylation analysis within the EGLN3 promoter spanned from +481 to +866 bp relative to the transcription start site, including 37 CpG dinucleotides (Fig. 2a). DNA methylation levels of EGLN3 were notably elevated in GC cell lines compared to those of GES-1 cells, with particular emphasis on CpGs 3, 29, and 31 (+495, +733, and +750 bp relative to the TSS), which exhibited the most pronounced differences (Fig. 2a). Importantly, EGLN3 hypermethylation was inversely correlated with low gene expression in GC cell lines (P < 0.05) (Fig. 2b).
Fig. 2. Promoter hypermethylation inhibits EGLN3 expression in GC cells.
a DNA methylation sequencing spans the promoter region of EGLN3 from +481 to +866, including 37 CpG islands. b EGLN3 methylation inversely correlates with its expression in GC cell lines (R2 = 0.5026, P < 0.05). c Comparison of DNA methylation level of EGLN3 between GC tissues and non-tumour adjacent tissues (N = 6) (P < 0.05). d EGLN3 promoter methylation was correlated with poor overall survival in GC patients (P = 0.0067, log-rank test). e EGLN3 mRNA expression markedly increased in GC cell lines treated with 5-Aza compared with control cells by quantitative real-time PCR. f The EGLN3 mRNA levels were examined after knocking down DNMT1, DNMT3A and DNMT3B, respectively. g The DNA methylation level of EGLN3 (CpG3, 29, 31) was reduced, followed by the knockdown of DNMT1.
Treatment with 5-aza-2’-deoxycytidine (5-Aza), a demethylating agent, or vehicle (DMSO) in two GC cell lines (NCI-N87 and SNU-1) revealed a significant increase in EGLN3 mRNA expression in cells treated with 5-Aza (Fig. 2e). This effect was accompanied by a notable decrease in DNA methylation levels at specific CpG sites in 5-Aza-treated cells (Fig. S1A). To elucidate the effect of DNA methyltransferases (DNMTs), including DNMT1 (a maintenance methyltransferase), DNMT3A, and DNMT3B (de novo methyltransferases targeting previously unmethylated CpG sites) [21], on EGLN3 methylation and expression, we conducted further investigations. Notably, DNMT1 knockdown resulted in a significant reduction in EGLN3 methylation levels at specific sites (CpG 3, 29, and 31; +495, +733, and +750 bp relative to the TSS, respectively) (Fig. 2g), concomitant with an increase in EGLN3 mRNA expression (Fig. 2f). DNMT1 is the primary enzyme responsible for maintaining the methylation patterns of EGLN3.
To determine whether the low expression of EGLN3 in GC tissues could be attributed to DNA hypermethylation, we conducted MassARRAY methylation analysis of the EGLN3 promoter in GC tissue samples. This analysis revealed higher methylation levels in tumour tissues than in adjacent nontumor tissues at specific sites (CpG 3, 29, and 31; +495, +733, and +750 bp relative to the TSS) (N = 6) (P < 0.05) (Fig. 2a and 2c). Subsequently, we investigated EGLN3 methylation in the tumour tissues of 70 patients with GC. Aberrant hypermethylation of CpG sites in the EGLN3 promoter was observed in 52 tumour tissues (52 70; 74.29%). The associations between EGLN3 hypermethylation and the clinicopathological features of GC are shown in Table 3. EGLN3 hypermethylation was correlated with the depth of invasion (pT stage) (P < 0.05) and lymph node metastasis (pN stage) (P < 0.05). Kaplan–Meier survival analysis demonstrated that GC patients with elevated DNA methylation levels at specific sites (CpG 3, 29, and 31; +495, +733, and +750 bp relative to the TSS) exhibited a poorer prognosis than those with lower levels of methylation (P = 0.0067, log-rank test) (Fig. 2d). The heightened DNA methylation status of EGLN3 is a critical prognostic marker for patients with GC.
Table 3.
Analysis of EGLN3 Methylation in GC tissues and associated clinicopathological factors.
| Characteristics | EGLN3 Methylation | P value | |
|---|---|---|---|
| Low | High | ||
| Age, years | 0.216 | ||
| <65 | 16 | 39 | |
| ≥65 | 2 | 13 | |
| Gender | 0.329 | ||
| Male | 14 | 34 | |
| Female | 4 | 18 | |
| Size, cm | 0.685 | ||
| ≤4 cm | 3 | 6 | |
| >4 cm | 15 | 46 | |
| Tumour location | 0.694 | ||
| Upper 1/3 | 6 | 10 | |
| Middle 1/3 | 5 | 20 | |
| Lower 1/3 | 6 | 19 | |
| More than 2/3 stomach | 1 | 3 | |
| pT stagea | 0.023 | ||
| pT1-2 | 5 | 3 | |
| pT3-4 | 13 | 49 | |
| pN stagea | 0.015 | ||
| pN0 | 6 | 4 | |
| pN+ | 12 | 48 | |
| Lauren type | 0.718 | ||
| Intestinal | 2 | 10 | |
| Diffuse | 16 | 42 | |
apT and pN stage is defined using the American Joint Committee on Cancer (AJCC) Staging System, 8th Edition.
EGLN3 attenuated GC cell proliferation and metastasis
EGLN3 exhibits aberrant silencing in GC tissues, and its reduced expression is associated with poor survival in GC patients. This suggests a potential effect on the malignant behaviour of GC cells. To further investigate this, lentiviral vectors were used to ectopically express EGLN3, and its effects on GC cell proliferation were examined (Fig. 3a). The Cell Counting Kit-8 (CCK8) assay revealed that the re-expression of EGLN3 significantly suppressed the proliferation of NCI-N87 and SNU-1 cells compared to control cells (Fig. 3b). Subsequently, shRNAs were used to construct GC cells with EGLN3 knockdown (Fig. 3a). In the absence of EGLN3, cell proliferation was promoted (Fig. S3A). EdU incorporation assays demonstrated that EGLN3 reduced the number of replicating cells (Fig. 3c and S3B). Furthermore, the level of phosphorylated histone H3 (S10), which is indicative of cells in the S phase, was notably reduced in GC cells overexpressing EGLN3 (Fig. 3d). Additionally, in vivo experiments involved subcutaneous injection of EGLN3-overexpressing and control NCI-N87 cells into BALB/c nude mice, with EGLN3 significantly suppressing the growth of injected GC cells in vivo (Fig. 3e).
Fig. 3. The tumour suppression effect of EGLN3 in GC is independent on its catalytic activity.
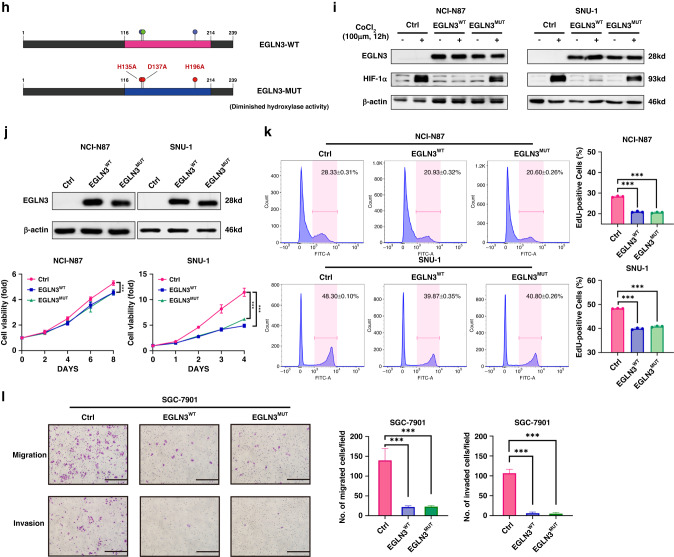
a EGLN3 restoration and knockdown in GC cells are verified by immunoblot assay. b Restored EGLN3 markedly inhibits the propagation in NCI-N87 and SNU-1 cells as shown by CCK8 assay. c The numbers of replicating cells dramatically decrease in NCI-N87 and SNU-1 cells with EGLN3 re-expression as demonstrated by EdU incorporation assay. d The phosphorylated histone H3 (S10) level is significantly reduced in NCI-N87 and SNU-1 cells with EGLN3 overexpression. e EGLN3 overexpression in NCI-N87 cells markedly suppresses tumour growth in vivo. f The migration and invasion ability of SGC-7901 and AGS cells are detected after EGLN3 overexpression or knockdown, respectively. Histograms display the average cell number/field. Scale bar, 100 µm. g Tumour metastasis is assessed by bioluminescent imaging. Bioluminescent images (left) and quantification (right) of metastatic lesions in NOD/SCID mice models injected with luciferase-NCI-N87 cells. Representative images (left) display the macroscopic appearance of lung metastases in mice models. Statistical quantification (right) of lung metastasis nodules (n = 5) produced by injecting NCI-N87 cells into NOD/SCID mice via tail vein for 12 weeks. All in vitro experiments were independently repeated three times. h Schematics of mutations paralyzing EGLN3 enzyme activity. Three-point mutations (H153A, D137A and H196A), which have been reported to markedly eliminate hydroxylase activity, is simultaneously introduced into EGLN3WT. i In contrast to EGLN3WT, EGLN3mut could not decrease HIF-1α protein level in the CoCl2-treated GC cells. The HIF-1α protein was induced by treatment with 100 μM CoCl2 for 12 h. j EGLN3mut impedes the proliferation in NCI-N87 and SNU-1 cells in vitro as shown by CCK8 assay. k The number of EdU-tagged cells is dramatically reduced in NCI-N87 and SNU-1 cells with re-expressed EGLN3WT or EGLN3mut, as compared with control cells. l Transwell assays are carried out to assess re-expressed EGLN3WT and EGLN3mut cells migration and invasion ability. Histograms display the average cell number each view. Scale bar, 100 µm.
Given that metastasis is the most aggressive function of GC, migration and invasion assays were conducted to assess the metastatic potential of GC cells. Stable cell lines with ectopic expression or knockdown of EGLN3 were established in SGC-7901 (low endogenous EGLN3 expression cells) and AGS (high endogenous EGLN3 expression cells) cells, respectively (Fig. 3a). Ectopic expression of EGLN3 in SGC-7901 cells markedly attenuated cell migration and invasion, whereas EGLN3 knockdown in AGS cells significantly enhanced metastasis (Fig. 3f). To validate the effects of EGLN3 on GC cell metastasis in vivo, luciferase-labelled EGLN3-overexpressing and control GC cells were injected into the tail vein of NOD/SCID mice, and cell metastasis was monitored using bioluminescence imaging (Fig. 3g). The number of metastatic lung nodules was quantified after three months. Compared to the control group, EGLN3 overexpression substantially inhibited the lung metastasis of GC cells (Fig. 3g). Collectively, these findings underscore the inhibitory role of EGLN3 in the malignant progression of GC cells.
EGLN3 exhibited catalytic activity-independent mechanisms of tumour suppression in GC
Previous studies have consistently demonstrated that EGLN3 typically exerts its antineoplastic functions by catalysing the incorporation of a single oxygen atom into substrates such as hypoxia-inducible factor 1α (HIF-1α) in various cancers [13, 22–24]. Consequently, we sought to determine whether EGLN3 functions similarly in GC cells. To investigate this, we introduced three point-mutations (H153A, D137A and H196A), which have been reported to substantially abolish hydroxylase activity [25], into wild-type EGLN3 (EGLN3WT) (Fig. 3h).
HIF-1α serves as a pivotal substrate subject to EGLN3 hydroxylation, and hydroxylated HIF-1α is promptly targeted for degradation via ubiquitination [6–11]. Thus, the enzymatic activity of EGLN3 could be assessed by evaluating its effect on HIF-1α levels. In comparison to EGLN3WT, mutated EGLN3 (EGLN3mut) was incapable of reducing HIF-1α protein levels in GC cells treated with CoCl2, providing conclusive evidence of EGLN3mut’s loss of enzymatic activity (Fig. 3i).
Remarkably, EGLN3mut inhibited the proliferation of GC cells in vitro, with no significant difference observed between EGLN3mut-restored and EGLN3WT-restored GC cells when assessing the effect of EGLN3mut on GC cell viability (Fig. 3j, k). Moreover, the number of invading and migrating cells was significantly decreased in cells re-expressing either EGLN3WT or EGLN3mut compared to control cells (Fig. 3l). Collectively, these findings established that EGLN3 inhibits GC cell proliferation and metastasis independently of its hydroxylase activity.
EGLN3 reduced JMJD8 expression
As the IHC assay indicated the nuclear localisation of EGLN3 and the presence of exogenously expressed EGLN3 in GC cells (Fig. 4a and Fig. 3d), this strongly suggests that EGLN3 might exert an influence on the gene expression profile. To investigate the molecular mechanisms underlying the antiproliferative function of EGLN3, we performed RNA sequencing to explore its effects (Fig. 4b). This analysis revealed 63 upregulated and 46 downregulated genes when comparing EGLN3-restored NCI-N87 cells with control cells (|log2(FC)| > 0.5 and P < 0.05) (Fig. 4c).
Fig. 4. EGLN3 reduces the expression of JMJD8.
a EGLN3 is enriched in nucleus as shown by IHC assay. b Clustered heatmap of transcriptomes of EGLN3 restoration GC cells and control group. c The volcano plots based on the RNA sequencing results of EGLN3-overexpressing and control NCI-N87 cells. There are 63 upregulated and 46 downregulated genes between EGLN3-restored and control NCI-N87 cells (|log2(FC)| > 0.5 and p < 0.05). d JMJD8 protein is reduced as the result of EGLN3WT or EGLN3mut expression. e JMJD8 mRNA level in GC is significantly higher than normal gastric tissue from TCGA GC cohort. f RNAscope in situ hybridisation method detects that JMJD8 is highly expressed in GC specimens (n = 8) compared to normal gastric mucosa tissues (n = 4). g The disease specific survival is prolonged in patients with lower JMJD8 expression relative to those with higher JMJD8 level from TCGA GC database (P < 0.05).
Among these differentially expressed genes, we observed a reduction in JMJD8 protein levels resulting from the expression of EGLN3WT or EGLN3mut (Fig. 4d). Previous studies have identified JMJD8 as a significant biomarker that promotes the progression of various cancers, including breast and lung cancers [26, 27]. The regulation of JMJD8 by EGLN3 is independent of the hydroxylase mechanism, suggesting that JMJD8 may play a pivotal role in the nonenzymatic regulation mediated by EGLN3. An examination of JMJD8 expression in the TCGA GC cohort revealed significantly higher JMJD8 mRNA levels in GC than in normal gastric tissues (Fig. 4e).
Given that the available primary antibody against JMJD8 is not suitable for IHC assays, we employed RNAscope in situ hybridisation to detect JMJD8 mRNA expression in GC specimens and normal gastric mucosa. The results demonstrated elevated JMJD8 expression in GC tissues compared to that in normal tissues (Fig. 4f). Furthermore, patients with lower JMJD8 expression exhibited extended disease-specific survival compared with those with higher JMJD8 levels (Fig. 4g). Collectively, these findings suggest that JMJD8 may play a critical role in GC progression.
EGLN3 inhibited the malignant evolution of GC dependent on JMJD8 depletion
Although previous reports have established the role of JMJD8 in promoting malignant tumour progression, its function in GC has not yet been elucidated. To investigate the role of JMJD8 in GC cells, we used lentiviral vectors encoding shRNAs to deplete endogenous JMJD8 in NCI-N87 and SNU-1 cells. The efficiency of JMJD8 knockdown was confirmed by immunoblotting (Fig. 5a). Notably, JMJD8-depleted GC cells exhibited significantly slower growth than control cells (Fig. 5b). Similarly, JMJD8 deficiency led to a reduction in the number of EdU-positive cells (Fig. 5c). Consistently, GC cells with high JMJD8 expression exhibited more malignant proliferative characteristics (Fig. S4A). Furthermore, JMJD8 depletion markedly suppressed the growth of NCI-N87 cells in vivo (Fig. 5d).
Fig. 5. JMJD8 knockdown inhibits the propagation in GC cells and the tumour suppression effect of EGLN3 is dependent on JMJD8 depletion.
a The knockdown efficiency of JMJD8 is verified by western blotting assay, and the levels of phosphorylated histone H3 (S10) are significantly decreased in JMJD8 silenced NCI-N87 and SNU-1 cells. b JMJD8-depleted NCI-N87 and SNU-1 cells grew much slowly compared with control cells as shown by CCK8 assay. c The number of the EdU-positive cells dramatically decreases in JMJD8 silenced NCI-N87 and SNU-1 cells with JMJD8. d Deficiency of JMJD8 markedly suppresses the growth of NCI-N87 cells in vivo. e, f The migration and invasion ability of SGC-7901 and AGS cells is detected after JMJD8 knockdown or overexpression, respectively. Histograms display the average cell number/field. Scale bar, 100 µm. All in vitro experiments were independently repeated three times. g EGLN3 and JMJD8 overexpression in NCI-N87 and SNU-1 cells is verified by immunoblot assay. h EGLN3 overexpression alone inhibits the GC cells propagation in vitro, whereas additional overexpression JMJD8 mitigates the inhibitory effect of EGLN3 on the proliferation. i The number of EdU-stained GC cells with EGLN3/JMJD8 overexpression increases relative to those with sole EGLN3 overexpression. j JMJD8 overexpression markedly rescues the growth of tumour xenografts in vivo. k The migratory and invasive capabilities of GC cells are evaluated by using Transwell assays. Scale bar, 100 µm. All in vitro experiments were independently repeated three times.
Stable cell lines with ectopic expression or knockdown of JMJD8 were established using SGC-7901 and AGS cells, respectively (Fig. 5e). Knockdown of JMJD8 in SGC-7901 cells significantly attenuated cell migration and invasion, whereas ectopic expression of JMJD8 in AGS cells notably increased cell metastasis (Fig. 5f). Collectively, these findings demonstrate that JMJD8 accelerates GC cell progression.
To further investigate whether the inhibitory effect of EGLN3 on GC progression is mediated by silencing JMJD8 expression, EGLN3 and JMJD8 were simultaneously overexpressed in GC cells (Fig. 5g). Functionally, CCK8 assays revealed that EGLN3 overexpression alone attenuated cellular growth in vitro, and additional overexpression of JMJD8 mitigated the inhibitory effect of EGLN3 on proliferation (Fig. 5h). Moreover, EdU-stained GC cells overexpressing both EGLN3 and JMJD8 increased in number relative to those with EGLN3 overexpression alone, with no significant difference observed between the control and EGLN3/JMJD8-overexpressing cells (Fig. 5i). In accordance with the in vitro findings, JMJD8 overexpression markedly rescued the growth of tumour xenografts in vivo, which was impaired by EGLN3 (Fig. 5j). Furthermore, migration and invasion assays demonstrated that JMJD8 overexpression reversed the inhibitory effects of EGLN3 on GC cell migration and invasion (Fig. 5k). These results suggest that JMJD8 weakens the suppressive effect of EGLN3 on the malignant evolution of GC.
EGLN3 inhibited JMJD8-mediated NF-κB signalling activation
Our study underscores the inhibitory role of EGLN3 in the proliferation and motility of GC cells, primarily achieved through the reduction of JMJD8 expression. Importantly, this effect was reversed by restoration of JMJD8 expression and was not contingent on the hydroxylase function of EGLN3. To elucidate the nonenzymatic mechanism by which EGLN3 suppresses GC, we investigated its signalling pathways.
KEGG enrichment analysis of transcriptome sequencing data revealed significant enrichment of NF-κB signalling pathways (Fig. 6a), suggesting that EGLN3 may play a role in the regulation of NF-κB signalling pathways. Notably, both wild-type and mutant EGLN3 expression attenuated activation of the NF-κB signalling pathway (Fig. 6b), indicating that the influence of EGLN3 on the NF-κB signalling pathway is independent of its hydroxylase function.
Fig. 6. EGLN3 attenuate GC cell malignant characteristics via inhibiting JMJD8/NF-κB signalling activation.
a KEGG enrichment analysis of DEGs in transcriptome. b Phospho-NF-κB P65 is reduced as the result of EGLN3WT or EGLN3mut expression. c GESA analysis based on the TCGA GC cohort showed that high JMJD8 expression positively correlated with NF-κB signalling pathways. d JMJD8 knockdown decreases NF-κB P65 phosphorylation levels. e The Co-IP detection results showed an interaction between EGLN3 and JMJD8 in cells. f JMJD8 overexpression markedly rescues the phosphorylation levels of NF-κB P65. g The diagram for the mechanisms of EGLN3 in the suppressive effects of GC cell progression.
As per existing reports, JMJD8 is indispensable for IKK kinase activation and positively regulates the NF-κB signalling pathway in a TNF-dependent manner [28]. JMJD8 functioned as an activator of NF-κB signalling in GC (Fig. 6d and Fig. S2B). Gene set enrichment analysis of the TCGA database further substantiated the significant correlation between JMJD8 expression and NF-κB signalling pathways (Fig. 6c). Subsequent experiments revealed that increased JMJD8 expression led to elevated phosphorylation of NF-κB P65 (RelA) (Fig. 6f and Fig. S2C).
Moreover, immunoprecipitation analysis revealed a protein‒protein interaction between EGLN3 and JMJD8 (Fig. 6e). This suggests that EGLN3 may modulate JMJD8 through direct protein-protein interactions. In summary, our findings indicate that EGLN3 hinders the malignant progression of GC by inhibiting JMJD8-mediated activation of the NF-κB signalling pathway, and this effect is independent of its hydroxylase activity (Fig. 6g).
Discussion
Previous studies have consistently revealed that EGLN3 expression is aberrantly reduced in multiple cancer types, and its restoration effectively mitigates malignant phenotypes [22–24]. DNA methylation, a pivotal epigenetic mechanism governing gene transcription, has long been linked to the silencing of tumour suppressor genes. In previous studies, associations between EGLN3 expression and DNA methylation have been reported in prostate and colorectal cancers [29, 30]. In our study, we investigated the DNA methylation status of EGLN3 in GC and established a correlation between EGLN3 DNA methylation and clinicopathological characteristics. Our findings indicate that DNA hypermethylation of EGLN3 in GC is notably associated with T-stage and lymph node metastasis, culminating in an unfavourable prognosis for patients with GC. Downregulation of EGLN3 in GC tissues and cells, coupled with DNA promoter hypermethylation, disrupts transcriptional homoeostasis, thereby fostering the progression of GC.
EGLN3 exerts its antitumour effects through a dual mechanism, including both hydroxylase-dependent and hydroxylase-independent pathways. HIFα is a well-explored substrate of EGLN3 that serves as a pivotal regulator of HIFα protein stability. In various cancers, EGLN3 deficiency has been shown to lead to the accumulation of HIF1α/HIF2α, consequently upregulating factors such as TGFα and promoting epithelial-mesenchymal transition in lung cancer [13]. In pancreatic cancer, elevated HIF1α levels resulting from EGLN3 depletion have been linked to increased invasion via the induction of angiogenesis factors [14]. Additionally, in hepatocellular carcinoma (HCC), EGLN3 attenuates VEGF secretion and inhibits HCC cell proliferation by enhancing HIF1α degradation [24]. Under hypoxic conditions, GC cells secrete exosomal miR-301a-3p to silence EGLN3 expression and boost HIF1α-mediated metastasis [31].
A hypoxic environment is a common characteristic of tumour development; however, it is not an inevitable state experienced by all solid tumours [32, 33]. In the later stages, many solid tumours exhibit a complex environment with both hypoxic and normoxic regions. For instance, glioblastomas typically exhibit distinct layers, including a necrotic core layer, an intermediate/hypoxic layer, and an outer layer characterised by an adequate oxygen supply, vascularisation, and high proliferation, including the invasive tumour front [34]. In the later stages, many solid tumours develop a complex environment with both hypoxic and normoxic regions. Although HIF1α protein expression is classically regulated by cellular oxygen levels, there are alternative mechanisms through which HIF1α can remain stable in normoxic tumour environments [35, 36]. These unconventional mechanisms can enhance HIF1α stability independently of the oxygen levels. These findings provide valuable insight into the pathogenesis and progression of GC.
In our study, we demonstrated that the restoration of EGLN3 significantly impeded the progression of GC cells, consistent with previous research [31]. Intriguingly, mutations that inhibit hydroxylase activity do not compromise the antitumour effect of EGLN3 under oxygen-rich conditions, suggesting a novel nonenzymatic mechanism through which EGLN3 operates in GC cells. Currently, the evidence supporting the nonenzymatic function of EGLN3 is limited. As previously reported, EGLN3 disrupts IKKβ/NF-κB activation by binding to IKKβ [17]. In our investigation, we observed a reduction in the expression of JMJD8 due to EGLN3, irrespective of hydroxylase activity. Coimmunoprecipitation results further highlighted the interaction between EGLN3 and JMJD8 within cells, providing further evidence for the nonenzymatic molecular underpinnings of EGLN3 and JMJD8 expression.
JMJD8, a member of the Jumonji C domain-containing oxygenase family [37], possesses a highly conserved JmjC domain that contains a specific sequence motif (HXD/EXnH) known for its ability to sequester ferrous ions [38, 39]. Notably, JMJD8’s ion-sequestering motif sequence HXHXnH differs from that of other family members (HXD/EXnH), potentially endowing it with distinct functional properties compared to other JMJDs [40]. In endothelial cells, JMJD8 expression has been observed to increase during cellular differentiation [41]. Furthermore, it interacts with pyruvate kinase M2 (PKM2) to restore endothelial cell metabolism, thereby enhancing angiogenic sprouting [41]. JMJD8 has also been shown to stimulate the TNF-induced NF-κB signalling pathway, and its role in cancer through the modulation of NF-κB signalling has been elucidated accordingly [28, 42]. Recent studies have revealed an association between JMJD8 and Akt signalling, demonstrating that JMJD8 can stimulate the PI3K/Akt pathway by maintaining EGFR stability and binding with Akt1 to enhance Akt activation [26, 43].
Our study underscores the pivotal role of JMJD8 in sustaining the viability of GC tumour cells. Examination of the TCGA GC cohort revealed that heightened JMJD8 expression tended to result in a poor prognosis for patients. Silencing JMJD8 has been shown to suppress the malignant progression of GC cells both in vitro and in vivo. A rescue assay further confirmed that JMJD8 restoration partially counteracted the inhibitory effect of EGLN3 on the malignant progression of GC cells.
To investigate the underlying molecular mechanisms, we conducted an analysis of EGLN3-mediated regulation of the NF-κB pathway. The upregulation of EGLN3 resulted in a significant reduction in the phosphorylation of NF-κB P65/RelA independent of its enzymatic function. The pivotal role of NF-κB in GC progression is well established, as previous studies have demonstrated a substantial increase in GC progression associated with NF-κB activation [44], particularly in the context of Helicobacter pylori infection [45]. GC cell-derived exosomes have also been shown to affect the immunomodulatory function of mesenchymal stem cells through NF-κB signalling, thereby maintaining an inflammatory environment conducive to tumour growth [46].
NF-κB is a pleiotropic transcription factor found in nearly all cell types and consists of various subunits, including NF-κB1 P50/P105, NF-κB2 P52/P100, c-Rel, RelA/P65, and RelB. These proteins regulate gene expression and play critical roles in numerous biological processes, such as immunity, inflammation, differentiation, cell growth, and tumorigenesis [47, 48]. The most prevalent form appears to be the heterodimeric P65-P50 complex [48]. NF-κB complexes remain inactive in the cytoplasm and bind to members of the IκB (NF-κB inhibitor) family. In the conventional activation pathway, IκB phosphorylation leads to ubiquitination and subsequent proteasomal degradation, releasing the NF-κB/Rel complexes. These active complexes then undergo further posttranslational modifications (e.g. phosphorylation, acetylation, and glycosylation) and translocate to the nucleus [47, 48]. The inhibitory effect of IκB on cytoplasmic NF-κB primarily involves its interaction with NF-κB P65.
Prior research has established an association between JMJD8 and the NF-κB signalling pathway [28]. In the current study, we elucidated protein‒protein interactions between EGLN3 and JMJD8. JMJD8 has been identified as an activator of the NF-κB signalling pathway in GC. Notably, EGLN3 attenuates the malignant progression of GC by inhibiting JMJD8-mediated NF-κB signalling activation independent of its hydroxylase activity. These findings further shed light on the inhibitory role of EGLN3 in GC progression.
This study had certain limitations that should be acknowledged. First, the mechanisms underlying the translocation of EGLN3 into the nucleus and its regulatory role in gene expression, including JMJD8, were not elucidated in this study. Second, although preliminary evidence supports the role of JMJD8 in GC biology, a comprehensive understanding of its clinical relevance is hindered by the lack of suitable antibodies for IHC application. Further investigation is warranted to uncover the precise mechanisms by which JMJD8 contributes to GC progression.
In summary, our study revealed that aberrantly low expression of EGLN3 is indicative of poor prognosis in GC patients. Furthermore, restoration of EGLN3 significantly impeded the progression of GC cells. Notably, it has been demonstrated that the antitumour effect of EGLN3 is not contingent on its hydroxylase activity but likely hinges on its regulatory role in gene expression. Specifically, EGLN3 inhibits GC cell proliferation and metastasis by suppressing JMJD8/NF-κB signalling activation. This discovery not only underscores the multifaceted functions of EGLN3 in GC biology but also provides valuable insights into potential therapeutic avenues for this malignancy.
Supplementary information
Acknowledgements
Thanks for all help.
Author contributions
FC and JD were major contributors to the design of this study and drafted the manuscript. GM, YN, CD and JD supported the research techniques. NZ, XY, PW, and MZ were responsible for tissue collection. RZ, HL and JD were responsible for clinical databases. JD designed this study. All authors read and approved the final manuscript.
Funding
This work was jointly funded by the Tianjin Key Medical Discipline (Specialty) Construction Project (TJYXZDXK-009A), the Distinguished professor of Tianjin (JTZB [2019] No.120), the Programs of National Natural Science Foundation of China (No. 81572372), the National Natural Science Foundation of China (81974373).
Data availability
GSE200646 (GEO database).
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
The use of specimens from patients were approved by the Institutional Research Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (no. bc2020091, date: 27 July 2020) (Tianjin, China). Informed consent was obtained for the collection of all samples in accordance with the Declaration of Helsinki. All experimental animal procedures were carried out with the approval of the Institutional Animal Care and Research Advisory Committee of Tianjin Medical University (Tianjin, China) under grant no. AE-2021073 (date: 11 October 2021).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Fenglin Cai, Xiuding Yang, Gang Ma, Jingyu Deng.
Contributor Information
Cheng Dong, Email: dongcheng@tmu.edu.cn.
Jingyu Deng, Email: dengery@126.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02546-x.
References
- 1.Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–48. doi: 10.1016/S0140-6736(20)31288-5. [DOI] [PubMed] [Google Scholar]
- 2.Thrift AP, El-Serag HB. Burden of gastric cancer. Clin Gastroenterol Hepatol. 2020;18:534–42. doi: 10.1016/j.cgh.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Pescador N, Cuevas Y, Naranjo S, Alcaide M, Villar D, Landázuri MO, et al. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem J. 2005;390:189–97. doi: 10.1042/BJ20042121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaakkola PM, Rantanen K. The regulation, localization, and functions of oxygen-sensing prolyl hydroxylase PHD3. Biol Chem. 2013;394:449–57. doi: 10.1515/hsz-2012-0330. [DOI] [PubMed] [Google Scholar]
- 6.Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–72. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 7.Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–8. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 8.Place TL, Domann FE. Prolyl-hydroxylase 3: evolving roles for an ancient signalling protein. Hypoxia (Auckl) 2013;2013:13–17. doi: 10.2147/HP.S50091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/S0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 10.Flashman E, Bagg EA, Chowdhury R, Mecinović J, Loenarz C, McDonough MA, et al. Kinetic rationale for selectivity toward N- and C-terminal oxygen-dependent degradation domain substrates mediated by a loop region of hypoxia-inducible factor prolyl hydroxylases. J Biol Chem. 2008;283:3808–15. doi: 10.1074/jbc.M707411200. [DOI] [PubMed] [Google Scholar]
- 11.Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci USA. 2001;98:9630–5. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, et al. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–65. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- 13.Dopeso H, Jiao HK, Cuesta AM, Henze AT, Jurida L, Kracht M, et al. PHD3 controls lung cancer metastasis and resistance to EGFR inhibitors through TGFα. Cancer Res. 2018;78:1805–19. doi: 10.1158/0008-5472.CAN-17-1346. [DOI] [PubMed] [Google Scholar]
- 14.Chiba N, Sunamura M, Nakagawa M, Koganezawa I, Yokozuka K, Kobayashi T, et al. Overexpression of hydroxyproline via EGLN/HIF1A is associated with distant metastasis in pancreatic cancer. Am J Cancer Res. 2020;10:2570–81. [PMC free article] [PubMed] [Google Scholar]
- 15.German NJ, Yoon H, Yusuf RZ, Murphy JP, Finley LW, Laurent G, et al. PHD3 loss in cancer enables metabolic reliance on fatty acid oxidation via deactivation of ACC2. Mol cell. 2016;63:1006–20. doi: 10.1016/j.molcel.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez J, Herrero A, Li S, Rauch N, Quintanilla A, Wynne K, et al. PHD3 regulates p53 protein stability by hydroxylating proline 359. Cell Rep. 2018;24:1316–29. doi: 10.1016/j.celrep.2018.06.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue J, Li X, Jiao S, Wei Y, Wu G, Fang J, et al. Prolyl hydroxylase-3 is down-regulated in colorectal cancer cells and inhibits IKKbeta independent of hydroxylase activity. Gastroenterology. 2010;138:606–15. doi: 10.1053/j.gastro.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 18.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76:3446–50. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 19.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–92. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 20.Ma G, Jing C, Li L, Huang F, Ding F, Wang B, et al. MicroRNA-92b represses invasion-metastasis cascade of esophageal squamous cell carcinoma. Oncotarget. 2016;7:20209–22. doi: 10.18632/oncotarget.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grady WM, Yu M, Markowitz SD. Epigenetic alterations in the gastrointestinal tract: current and emerging use for biomarkers of cancer. Gastroenterology. 2021;160:690–709. doi: 10.1053/j.gastro.2020.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T, Li TS, Urata Y, Goto S, Ono Y, Kawakatsu M, et al. Increased expression of PHD3 represses the HIF-1 signaling pathway and contributes to poor neovascularization in pancreatic ductal adenocarcinoma. J Gastroenterol. 2015;50:975–83. doi: 10.1007/s00535-014-1030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang LR, Wu JX, Cai SL, Huang YX, Zhang XQ, Fu WK, et al. Prolyl hydroxylase domain 3 influences the radiotherapy efficacy of pancreatic cancer cells by targeting hypoxia-inducible factor-1α. OncoTargets Ther. 2018;11:8507–15. doi: 10.2147/OTT.S187615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi M, Dai WQ, Jia RR, Zhang QH, Wei J, Wang YG, et al. APC(CDC20)-mediated degradation of PHD3 stabilizes HIF-1a and promotes tumorigenesis in hepatocellular carcinoma. Cancer Lett. 2021;496:144–55. doi: 10.1016/j.canlet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 25.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–40. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 26.Zhang B, Zhang Y, Jiang X, Su H, Wang Q, Wudu M, et al. JMJD8 promotes malignant progression of lung cancer by maintaining EGFR stability and EGFR/PI3K/AKT pathway activation. J Cancer. 2021;12:976–87. doi: 10.7150/jca.50234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi J, Wang L, Du J, Wang M, Shen H, Liu Z, et al. ER-localized JmjC domain-containing protein JMJD8 targets STING to promote immune evasion and tumour growth in breast cancer. Dev Cell. 2023;58:760–78.e6. doi: 10.1016/j.devcel.2023.03.015. [DOI] [PubMed] [Google Scholar]
- 28.Yeo KS, Tan MC, Wong WY, Loh SW, Lam YL, Tan CL, et al. JMJD8 is a positive regulator of TNF-induced NF-κB signalling. Sci Rep. 2016;6:34125. doi: 10.1038/srep34125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rawluszko AA, Bujnicka KE, Horbacka K, Krokowicz P, Jagodziński PP. Expression and DNA methylation levels of prolyl hydroxylases PHD1, PHD2, PHD3 and asparaginyl hydroxylase FIH in colorectal cancer. BMC cancer. 2013;13:526. doi: 10.1186/1471-2407-13-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Place TL, Fitzgerald MP, Venkataraman S, Vorrink SU, Case AJ, Teoh ML, et al. Aberrant promoter CpG methylation is a mechanism for impaired PHD3 expression in a diverse set of malignant cells. PloS one. 2011;6:e14617. doi: 10.1371/journal.pone.0014617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia X, Wang S, Ni B, Xing S, Cao H, Zhang Z, et al. Hypoxic gastric cancer-derived exosomes promote progression and metastasis via MiR-301a-3p/PHD3/HIF-1α positive feedback loop. Oncogene. 2020;39:6231–44. doi: 10.1038/s41388-020-01425-6. [DOI] [PubMed] [Google Scholar]
- 32.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–39. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 33.Helmlinger G, Yuan F, Dellian M, Jain RK. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat Med. 1997;3:177–82. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 34.Peng G, Wang Y, Ge P, Bailey C, Zhang P, Zhang D, et al. The HIF1α-PDGFD-PDGFRα axis controls glioblastoma growth at normoxia/mild-hypoxia and confers sensitivity to targeted therapy by echinomycin. J Exp Clin Cancer Res. 2021;40:278. doi: 10.1186/s13046-021-02082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park JH, Kim TY, Jong HS, Kim TY, Chun YS, Park JW, et al. Gastric epithelial reactive oxygen species prevent normoxic degradation of hypoxia-inducible factor-1alpha in gastric cancer cells. Clin Cancer Res. 2003;9:433–40. [PubMed] [Google Scholar]
- 36.Lee YH, Bae HC, Noh KH, Song KH, Ye SK, Mao CP, et al. Gain of HIF-1α under normoxia in cancer mediates immune adaptation through the AKT/ERK and VEGFA axes. Clin Cancer Res. 2015;21:1438–46. doi: 10.1158/1078-0432.CCR-14-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yeo KS, Tan MC, Lim YY, Ea CK. JMJD8 is a novel endoplasmic reticulum protein with a JmjC domain. Sci Rep. 2017;7:15407. doi: 10.1038/s41598-017-15676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Accari SL, Fisher PR. Emerging roles of JmjC domain-containing proteins. Int Rev Cell Mol Biol. 2015;319:165–220. doi: 10.1016/bs.ircmb.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 39.Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 40.Oh S, Shin S, Janknecht R. The small members of the JMJD protein family: enzymatic jewels or jinxes? Biochim Biophys Acta Rev Cancer. 2019;1871:406–18. doi: 10.1016/j.bbcan.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boeckel JN, Derlet A, Glaser SF, Luczak A, Lucas T, Heumüller AW, et al. JMJD8 regulates angiogenic sprouting and cellular metabolism by interacting with pyruvate kinase M2 in endothelial cells. Arterioscler Thromb Vasc Biol. 2016;36:1425–33. doi: 10.1161/ATVBAHA.116.307695. [DOI] [PubMed] [Google Scholar]
- 42.Wang L, Jiang F, Ma F, Zhang B. MiR-873-5p suppresses cell proliferation and epithelial-mesenchymal transition via directly targeting Jumonji domain-containing protein 8 through the NF-κB pathway in colorectal cancer. J cell Commun Signal. 2019;13:549–60. doi: 10.1007/s12079-019-00522-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su Y, Wang X, Guo Z, Wang J. Aberrant JmjC domain-containing protein 8 (JMJD8) expression promotes activation of AKT and tumour epithelial-mesenchymal transition. Oncogene. 2020;39:6451–67. doi: 10.1038/s41388-020-01446-1. [DOI] [PubMed] [Google Scholar]
- 44.Lee BL, Lee HS, Jung J, Cho SJ, Chung HY, Kim WH, et al. Nuclear factor-kappaB activation correlates with better prognosis and Akt activation in human gastric cancer. Clin Cancer Res. 2005;11:2518–25. doi: 10.1158/1078-0432.CCR-04-1282. [DOI] [PubMed] [Google Scholar]
- 45.Soutto M, Bhat N, Khalafi S, Zhu S, Poveda J, Garcia-Buitrago M, et al. NF-kB-dependent activation of STAT3 by H. pylori is suppressed by TFF1. Cancer Cell Int. 2021;21:444. doi: 10.1186/s12935-021-02140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen Y, Xue C, Li X, Ba L, Gu J, Sun Z, et al. Effects of gastric cancer cell-derived exosomes on the immune regulation of mesenchymal stem cells by the NF-kB signalling pathway. Stem Cells Dev. 2019;28:464–76. doi: 10.1089/scd.2018.0125. [DOI] [PubMed] [Google Scholar]
- 47.Xia L, Tan S, Zhou Y, Lin J, Wang H, Oyang L, et al. Role of the NFκB-signaling pathway in cancer. OncoTargets Ther. 2018;11:2063–73. doi: 10.2147/OTT.S161109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zinatizadeh MR, Schock B, Chalbatani GM, Zarandi PK, Jalali SA, Miri SR, et al. The Nuclear Factor Kappa B (NF-kB) signaling in cancer development and immune. Dis Genes. 2021;8:287–97. doi: 10.1016/j.gendis.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GSE200646 (GEO database).