Abstract
The human pathogen Candida albicans is responsible for a large proportion of infections in immunocompromised individuals, and the emergence of drug-resistant strains is of medical concern. Resistance to antifungal azole compounds is often due to an increase in drug efflux or an alteration of the pathway for synthesis of ergosterol, an important plasma membrane component in fungi. However, little is known about the transcription factors that mediate drug resistance. In Saccharomyces cerevisiae, two highly related transcriptional activators, Upc2p and Ecm22p, positively regulate the expression of genes involved in ergosterol synthesis (ERG genes). We have identified a homologue in C. albicans of the S. cerevisiae UPC2/ECM22 genes and named it UPC2. Deletion of this gene impaired growth under anaerobic conditions and rendered cells highly susceptible to the antifungal drugs ketoconazole and fluconazole. Conversely, overexpression of Upc2p increased resistance to ketoconazole, fluconazole, and fluphenazine. Azole-induced expression of the ERG genes was abolished in a Δupc2 strain, while basal levels of these mRNAs remained unchanged. Importantly, the purified DNA binding domain of Upc2p bound in vitro to putative sterol response elements in the ERG2 promoter, suggesting that Upc2p increases the expression of the ERG genes by directly binding to their promoters. These results provide an important link between changes in the ergosterol biosynthetic pathway and azole resistance in this opportunistic fungal species.
The recent increase in numbers of severe fungal infections caused by opportunistic organisms has become an imminent medical concern. In particular, the human pathogen Candida albicans is responsible for a large proportion of infections in immunocompromised individuals; including patients undergoing cancer treatment, transplant patients, and those infected with the human immunodeficiency virus (reviewed in references 35 and 40). The development of multidrug resistance in clinical isolates has challenged effective treatment of these infections. Specifically, the extensive and repetitive use of antifungal azole derivatives such as fluconazole has allowed C. albicans to utilize many mechanisms of resistance in order to ensure its survival. For instance, changes in the ergosterol biosynthetic pathway including the overexpression or mutation of the azole drug target ERG11, as well as overexpression of multidrug transporters, have proven to be responsible for acquired multidrug resistance in this and other fungal species (28, 31-33, 42).
Ergosterol is the main component of the fungal plasma membrane and plays many important roles within the cell (28, 30). It helps maintain membrane integrity and fluidity, as well as ensuring proper function of several membrane-bound enzymes (26). Accordingly, many antifungal drug classes target enzymes in the ergosterol biosynthetic pathway. In Candida, mutations or changes in levels of expression of ERG genes can lead to alterations in drug sensitivity (20, 21, 34, 44).
Alternatively, the overexpression of drug efflux pumps is a widespread and well-documented mechanism that confers multidrug or pleiotropic drug resistance (PDR) in C. albicans. In general, membrane-associated transporter proteins are responsible for ridding cells of a myriad of different compounds, including hormones, ions, lipids, chemotherapeutic drugs, peptides, antibiotics, and antifungals (7, 24). There are at least two groups of multidrug transporters involved in PDR in C. albicans: the major facilitator superfamily (e.g., Mdr1p) (28, 40), and the ABC (ATP-binding cassette) transporter family, which includes the Cdr1p and Cdr2p proteins.
In budding yeast, the transcriptional regulation of these transporter genes is based on an intricate system involving many different regulators (3, 4, 12, 25, 27, 29, 46). One prominent family of transcription factors that mediate PDR consists of zinc cluster proteins. They are characterized by a highly conserved Zn(II)2Cys6 zinc finger motif within the N-terminal DNA binding domain (DBD). Well known Pdr1p and Pdr3p zinc cluster proteins positively regulate the expression of several multidrug ABC transporter genes, including PDR5, SNQ2, and YOR1. Yrr1p regulates SNQ2 and YOR1 expression (12, 25), while Stb5p positively regulates both SNQ2 and, to a lesser extent, PDR5 (4). In addition, we have identified another protein, Rdr1p, which acts as a transcriptional repressor of PDR5 (16). Two additional zinc cluster proteins in S. cerevisiae play important regulatory roles within the ergosterol biosynthesis pathway. Upc2p and Ecm22p are two highly homologous proteins that target the ERG2 and ERG3 genes by acting through sterol response elements (SREs) in their promoters (36, 39). Upc2p is also involved in the anaerobic expression of the DAN/TIR genes that encode mannoproteins (1, 9). Moreover, the upc2-1 mutant is responsible for the overexpression of the ABC transporter genes AUS1 and PDR11, which are required for sterol uptake (41). We have shown that a Δupc2 strain is hypersensitive to ketoconazole, whereas a Δecm22 strain is susceptible to cycloheximide (4), suggesting roles for these two factors in PDR.
Despite the wealth of information on transcription factors that mediate PDR in budding yeast, little is known of regulators that modulate azole drug resistance in Candida. In this study, we have identified a homologue of UPC2/ECM22 in C. albicans. Deletion of this gene renders cells susceptible to ketoconazole and fluconazole. In addition, drug-induced expression of some ERG genes is abolished in cells lacking Upc2p. These results provide an important link between changes in the ergosterol biosynthetic pathway and azole resistance in this opportunistic fungal species.
MATERIALS AND METHODS
Plasmid and strain construction.
All experiments were performed with the C. albicans strain SGY-243 or derivatives (Table 1). The SGY-243 Δupc2 strain was constructed using four different knockout (KO) cassette plasmids. The 3′ end of the first KO cassette was generated by amplifying 1,000 bp located at −30 and +1000 relative to the stop codon of the open reading frame (ORF) of UPC2 (orf19.391) with the oligonucleotides AGTACCTGCAGGCAAGGTGATAATGGGTTTA and CGGGATCCCTGTCATCATCATAAATGGC, respectively. The PCR product was cut with HindIII and PstI and then subcloned into pMB-7 (14) cut with the same enzymes. The 5′ end of the cassette comprised 1,012 bp (−999 to +13 relative to the ATG) amplified with AGTACGAGCTCCCTCCTGCAAAATAAACACG and ATCTAGGAGCTCCTGTCATCATCATAAATGGC. The PCR product was cut with SacI and then cloned into the modified pMB-7 plasmid cut with SacI, already containing the 3′ fragment. The second, third, and fourth KO cassette plasmids all retained the 5′ end of the original KO cassette, while varying at the 3′ end to create three different internal KO cassettes (see Fig. 2A). A region located from +1584 and +2099 bp (relative to the ATG) was amplified with AGATTACTCGAGGACCGTGAAAATTCGGCTTA and ATCTAGAAGCTTTTACTGGTAAGGACGCTTGG to make up the 3′ fragment of the second KO cassette, and a region located between +1005 and +1531 was amplified with AGATTACTCGAGGTCCATTCCTGCAAATCCAC and ATCTAGAAGCTTCTCCCAAGTCGACAGATATA to comprise the 3′ end of the third KO cassette, while the region from +535 to +1005 was amplified with AGATTACTCGAGATCTTTCTGGGTTAGGTGAC and ATCTAGAAGCTTAGGTGATATGTTAGGTAACG for the 3′ end of the fourth KO cassette. The PCR products were digested with HindIII and XhoI and then cloned into the original KO cassette plasmid cut with HindIII and SalI. All of the KO plasmids were linearized with PvuII before transforming them into the appropriate strains.
TABLE 1.
Strains used in this study
| Strain | Genotypea | Reference |
|---|---|---|
| SGY-243 (wild type) | ade2/ade2 Δura3::ADE2/Δ::ADE2 | 15, 23 |
| upc2Δ1 | UPC2/UPC2/UPC2/upc2Δ5-701::hisG | This study |
| upc2Δ2 | UPC2/UPC2/upc2Δ5-528::hisG/upc2Δ5-701::hisG | This study |
| upc2Δ3 | UPC2/upc2Δ4-335::hisG/upc2Δ5-334::hisG/upc2Δ5-701::hisG | This study |
| Δupc2 | upc2Δ4-178::hisG/upc2Δ5-177::hisG/upc2Δ4-528::hisG/upc2Δ5-701::hisG | This study |
| CSM2-2 | SGY-243 RP10::(pCaEXP) URA3 MET3 prom | This study |
| CSM2-3 | SGY-243 RP10::(pCaEXP) URA3 MET3 prom | This study |
| CSM1-2 | SGY-243 RP10::(pCaEXP) URA3 MET3 prom-UPC2 | This study |
| CSM1-3 | SGY-243 RP10::(pCaEXP) URA3 MET3 prom-UPC2 | This study |
| CSM13-1 | Δupc2 RP10::(pCaEXP) URA3 MET3 prom | This study |
| CSM13-2 | Δupc2 RP10::(pCaEXP) URA3 MET3 prom | This study |
| CSM15-1 | Δupc2 RP10::(pCaEXP) URA3 MET3 prom-UPC2 | This study |
| CSM15-2 | Δupc2 RP10::(pCaEXP) URA3 MET3 prom-UPC2 | This study |
prom, promoter.
FIG. 2.
Southern blot analysis of strains carrying deletion of the UPC2 gene. (A) Schematic view of the UPC2 gene and deleted alleles. The black rectangle corresponds to the probe used for Southern blot analysis, and the dotted rectangle corresponds to the UPC2 ORF. Deletions were performed as described in Materials and Methods. Expected lengths of ClaI fragments for wild-type and deleted alleles (upon looping out the URA3 marker by internal recombination) are also given. (B) Southern blot analysis of wild-type strain (WT) and upc2Δ1, upc2Δ2, upc2Δ3 andΔupc2 strains carrying deletion of one to four alleles of the UPC2 ORF, respectively. Lengths of fragments are given on the right part of the figure.
To construct the expression plasmid pCaEXP-UPC2, the plasmid pCaEXP described previously (8) was altered into pCaEXP-MCS, containing a multiple cloning site (BamHI, NotI, XhoI, KpnI, XbaI, PstI, and SphI). It was created by the insertion of annealed oligonucleotides GATCCATTAGCGGCCGCATCTCGAGGGGGTACCCTAGTCTAGACTAGCTG and GCTAGTCTAGACTAGGGTACCCCCTCGAGATGCGGCCGCTAATGCA into pCaEXP digested with BamHI and PstI. The UPC2 ORF was amplified with CGGGATCCATGATGATGACAGTGAAACA and AGATTACTCGAGCTATTTCATATTCATAAACCCAT and then digested with BamHI and XhoI and subcloned into pCaEXP-MCS cut with the same enzymes. The overexpression vector was linearized with StuI and integrated at the RP10 locus as described by Care et al. (8).
CAI4 (14) genomic DNA was used as a template in all PCRs. Transformations were carried out using a standard lithium acetate procedure (2), with the exception of a 3-h incubation period at 30°C, followed by a 45-min heat shock at 42°C. Cells were pelleted and resuspended in H2O before plating on selective media lacking uridine. For SGY-243 Δupc2, positive clones were screened by PCR and then confirmed by Southern blot analysis. The URA3 marker was removed using negative selection with 5-fluoroorotic acid obtained from Sigma (14).
Media and drug susceptibility assays.
Media were prepared according to Adams et al. (2), while yeast-peptone-dextrose (YPD) was supplemented with 0.0025% uridine (Sigma). Fluphenazine (Sigma), fluconazole (Pfizer), and ketoconazole (Medisca, Montreal, Canada) stock solutions were diluted in H2O (10 mg/ml), 50% ethanol (5 mg/ml), and 100% ethanol (0.5 mg/ml), respectively. Strains were grown overnight in 5 ml YPD supplemented with 0.0025% uridine or minimal media lacking uridine. Cells were serially diluted into four concentrations: optical density at 600 nm of 0.1, 0.02, 0.004, and 0.0008. Ten microliters of each dilution was spotted on the appropriate medium. Anaerobic conditions were obtained with an anaerobic jar (Becton Dickenson) and gas pack (BBL GasPak Ô; Becton Dickenson). Strains were grown at 30°C.
Southern and Northern blot analyses.
Genomic DNA was isolated as described previously (38), and approximately 1 μg was digested with ClaI. Digests were extracted twice with phenol-chloroform and then precipitated with 0.2 M NaCl and ethanol. Samples were electrophoresed on a 1% agarose-1× Tris-borate-EDTA gel and transferred to a nylon membrane (Hybond-N; Amersham). The probe, corresponding to nucleotides +3475 to +4389 relative to the ATG, was amplified with the oligonucleotides ATAGGTCGGCGATACTAGAA and ATCGATACTTCTTGCTCTAG (Fig. 2).
For RNA extraction, cells were grown in 100 ml YPD to an optical density at 600 nm of 1.0 and split into two 250-ml flasks. Fluconazole was added to one flask at a final concentration of 5 μg/ml. Twenty-five-ml aliquots were taken after 1 h and 24 h of growth. RNA was isolated as described (19). Probes for Northern blot analysis were fragments obtained by PCR and derived from ORFs of ERG2 (orf19.74), ERG7 (orf19.1570), ERG11 (orf19.922), and ERG25 (orf19.3732). The 2.8-kb ACT1 fragment was excised from plasmid pBR322-ACTIN (kindly provided by Beatrice Magee, University of Minnesota) by digestion with EcoRI and HindIII.
Protein expression, purification, and EMSA.
For protein expression and purification and electrophoretic mobility shift assay (EMSA), a DNA fragment encoding the DBD of Upc2p (amino acids 1 to 148) was amplified by PCR using the oligonucleotides CGGGATCCATGATGATGACAGTGAAACA and GGAATTCTTAAATCACCGGCTGAGTTTTGA. The fragment was digested with BamHI and EcoRI and ligated to the bacterial expression vector pGEX-F (17) cut with the same enzymes. The DBD was expressed in Escherichia coli as a glutathione S-transferase fusion and purified, and the glutathione S-transferase moiety was removed by thrombin cleavage as described previously (17). The following oligonucleotides were used as probes (Table 2): ERG2 SRE A, TCGAATTCGGATAAGT and TCGAACTTATCCGAAT; ERG2 SRE B, TCGACGTTATCCGAAT and TCGAATTCGGATAACG; ERG2 SRE C, TCGATGTCGTATAAAA and TCGATTTTATACGACA; ERG2 Mut C, TCGATGTCAGATAAAA and TCGATTTTATCTGACA (mutations are underlined); and ERG2 SRE (from S. cerevisiae), TCGACCTCGTATAAGC and TCGAGCTTATACGAGG.
TABLE 2.
Known and putative SREs found in various promotersa
| Promoter | Sequence | Position (bp) relative to ATG | Source or reference |
|---|---|---|---|
| S. cerevisiae | |||
| ERG2 | CTCGTATAAGC | −383 | 39 |
| UPC2 | TTCGTAAACGA | −587 | |
| CTCGTTTACGA | −582 | ||
| CTCGTTTAGAG | −356 | ||
| C. albicans | |||
| ERG2 | TTCGGATAAGT | −326 | SRE A, this study |
| TTCGGATAACG | −233 | SRE B, this study | |
| GTCGTATAAAA | −213 | SRE C, this study | |
| ATCGTATCACC | −441 | ||
| ERG7 | AACGTATTGTC | −314 | |
| TACGTTTAATC | −309 | ||
| GTCGTGTACAA | −213 | ||
| CACGTGAATCC | −164 | ||
| ATCGTTTAAAA | −84 | ||
| ERG11 | CACGTACAATC | −517 | |
| GTCGTATAGAT | −484 | ||
| CTCGTTTAGAG | −456 | ||
| GTCGTATATTC | −228 | ||
| ERG25 | GTCGTTTAGAA | −462 | |
| GTCGTATAACT | −427 | ||
| CACGTCTTCTT | −401 | ||
| CACGTGAATCC | −164 | ||
| ATCGTTTAAAA | −84 | ||
| UPC2 | ATCGTTAAACA | −179 |
Sequences of known (S. cerevisiae) and putative SREs are listed along with their position relative to the initiator codon. The search criterion was based (i) on sequences found within 600 bp upstream of the ATG and (ii) on nucleotides that match the SRE found in the S. cerevisiae ERG2 promoter (these nucleotides are shown in bold).
Binding reactions (4% glycerol, 4 mM Tris-HCl, pH 8.0, 40 mM NaCl, 4 mM MgCl2, 10 μM ZnSO4, 0.5 μg sheared salmon sperm DNA, approximately 60 ng of each radiolabeled probe, 0.2 μl protein extract, 0.2% bromophenol blue) were carried out for 20 min at room temperature before electrophoresis on a 0.5× Tris-borate-EDTA, 4% acrylamide gel that was prerun for 2 h at 120 V.
RESULTS
A C. albicans homologue of the S. cerevisiae UPC2 gene.
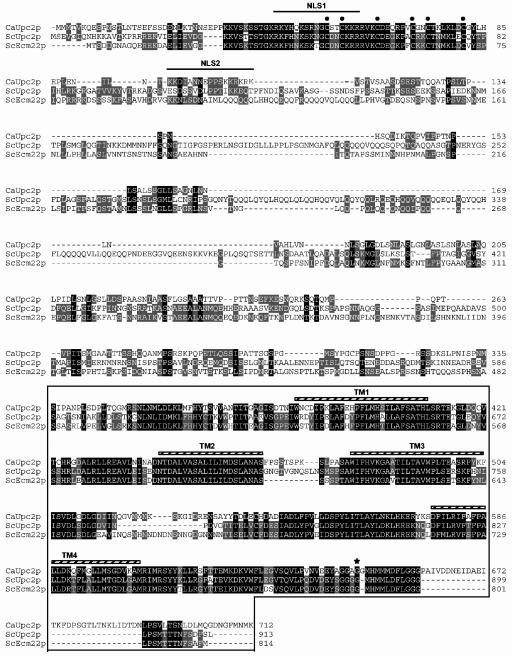
The Saccharomyces cerevisiae genome contains 54 ORFs that encode putative zinc cluster proteins, forming one of the largest known families of uniquely fungal transcriptional regulators (5). The recent sequencing of the diploid C. albicans genome (22) reveals at least 77 putative members in this family, many of which show strong homologies to characterized zinc cluster proteins in budding yeast (S. Znaidi, B. Turcotte, A. Nantel, and M. Raymond, unpublished results). A BLAST genome search identified a C. albicans homologue of the S. cerevisiae proteins Upc2p and Ecm22p. The UPC2 ORF (orf19.391) is predicted to encode a protein of 712 amino acids exhibiting high homology to the S. cerevisiae Upc2p and Ecm22p (Fig. 1). The cysteine-rich N terminus predicts a zinc finger DBD, the hallmark of this family of transcriptional regulators. The N-terminal DBD of the C. albicans Upc2p and the putative C-terminal activation domain are highly related to their S. cerevisiae counterparts.
FIG. 1.
Homology between the gene product of the C. albicans ORF orf19.391 and S. cerevisiae Upc2p and Ecm22p. Deduced amino acid sequences of C. albicans ORF orf19.391 (CaUpc2p) with S. cerevisiae Upc2p (ScUpc2p) and Ecm22p (ScEcm22p) are compared. Amino acid numbering is shown on the right part of the figure. ORF orf19.391 was translated using the C. albicans genetic code. Two putative bipartite nuclear localization signals, NLS1 and NLS2 (predicted by PSORT II), are indicated by a line. Cysteines of the zinc cluster domains of the various Upc2p proteins are represented by dots. The box at the bottom of the figure corresponds to a highly conserved region among the three proteins which includes four transmembrane segments (TM1 to -4; striped bars) predicted by TMAP. The position of the amino acid change (Gly to Asp) in the S. cerevisiae upc2-1 mutant (11) is shown by an asterisk.
The UPC2 gene has multiple alleles in SGY-243.
In order to characterize the UPC2 gene product, we attempted to construct a strain carrying deletions of both UPC2 alleles in an SGY-243 background (Table 1). After deleting two alleles with a KO cassette based on the pMB-7 plasmid described by Fonzi and Irwin (14), Southern blot analysis revealed the presence of at least one extra copy of the UPC2 gene, suggesting the presence of multiple alleles, the occurrence of gene conversion, or chromosome duplication. Consequently, we constructed three additional internal KO cassette plasmids (Fig. 2A). We used a strain carrying a single deletion of UPC2 to perform three more transformation rounds. Southern blot analysis revealed the presence of four UPC2 alleles in this particular genetic background (Fig. 2B). The complete removal of the wild-type UPC2 allele was confirmed by the disappearance of the 6.9-kb fragment (Fig. 2B).
Deletion of UPC2 renders C. albicans susceptible to drugs.
As stated above, a S. cerevisiae Δupc2 strain is hypersensitive to the antifungal drug ketoconazole (4). We wished to test if a C. albicans strain lacking the UPC2 gene product also demonstrated a similar phenotype. Wild-type (SGY-243) and deletion strains were serially diluted and spotted on rich medium plates supplemented with ketoconazole and fluconazole (Fig. 3A). All strains in this spotting assay lacked the URA3 auxotrophic marker. The wild-type strain or a strain deleted of one UPC2 allele showed similar growth in the absence of drugs, while removal of two or more UPC2 alleles resulted in a moderate growth advantage. Deletion of one allele of UPC2 did not impair growth on plates containing ketoconazole or fluconazole. Interestingly, strains lacking two, three, or all copies of UPC2 had severely impaired growth on ketoconazole. On fluconazole plates, marked growth defects were observed only upon removal of three or four UPC2 alleles. We also tested the deletion strains' ability to grow on fluphenazine, a calmodulin inhibitor. When normalized to the control plate, a slight susceptibility to fluphenazine was observed only with the strain lacking the four UPC2 alleles.
FIG. 3.
Phenotypes of strains carrying deletions of the UPC2 gene. Strains were grown overnight in YPD supplemented with uridine. Cells were serially diluted and spotted on plates as described in Material and Methods. (A) Wild-type strain (WT) and strains carrying one to four deletions of the UPC2 gene (Table 1) were spotted on YPD-uridine plates without drugs or YPD-uridine plates containing ketoconazole, fluconazole, or fluphenazine, as indicated on the figure. Cells were grown for 24 h (upper left panel) or 48 h (upper right and lower panels). (B) Spotting experiments were carried out with two independent integrants (Table 1). A wild-type strain with an integrated pCaEXP plasmid, WT + pCaEXP (strains CSM2-2 and CSM2-3); a UPC2 deletion strain with an integrated pCaEXP plasmid, Δupc2 + pCaEXP (strains CSM13-1 and CSM13-2); and a UPC2 deletion strain with an integrated UPC2 overexpression vector, Δupc2 + pCaEXP-UPC2 (strains CSM15-1 and CSM15-2) were spotted on minimal medium plates (lacking methionine, cysteine, and uridine) without drugs or containing ketoconazole, fluconazole, or fluphenazine, as indicated on the figure. Cells were grown for 24 h (upper left panel), 48 h (upper right and lower left panels), or 72 h (lower right panel). (C) Strains used in Fig. 3B were grown for 24 h in the presence of oxygen or for 48 h in the absence of oxygen on minimal plates (lacking methionine, cysteine, and uridine).
Overexpression of UPC2 confers resistance to drugs.
The observed drug phenotype was reversed by overexpressing the UPC2 gene in a Δupc2 strain, demonstrating that this gene is at least partially responsible for conferring drug resistance (Fig. 3B). The UPC2 ORF was placed under the control of the MET3 promoter in the overexpression vector pCaEXP-MCS and then integrated into the Δupc2 strain at the RP10 locus (8). The empty vector was also integrated at the same locus into the wild-type and deletion strains to serve as controls. Two separate integrants from each transformation were spotted to ensure that there was no variability between transformants that may have been caused by a spontaneous mutation. In the absence of drugs, all strains grew at relatively the same rate (Fig. 3B). Furthermore, the deletion strain demonstrated severely impaired growth on minimal media in the presence of either ketoconazole or fluconazole, as observed on rich medium (Fig. 3A). Strikingly, overexpression of the UPC2 gene product in the deletion strain not only rescued its phenotype but also conferred resistance to ketoconazole, fluconazole, and fluphenazine that even surpasses that of the wild-type strain.
A strain lacking UPC2 has impaired growth under anaerobic conditions.
In S. cerevisiae, Upc2p regulates genes in the oxygen-dependent ergosterol biosynthetic pathway. Under hypoxic conditions, UPC2 is upregulated and plays a not-well-understood yet critical role in anaerobic sterol uptake (1). We show that the UPC2 homologue is also important in C. albicans when grown under anaerobic conditions. In this spotting assay, strains used in Fig. 3B were grown aerobically overnight before being serially diluted on minimal medium plates and incubated under aerobic or anaerobic conditions. Under anaerobic conditions, growth of strains lacking the UPC2 gene was severely impaired when compared to the wild-type strain (Fig. 3C), while overexpression of UPC2 lead to stronger growth than the wild-type strain (Fig. 3C).
Upc2p increases ERG and UPC2 mRNA levels upon fluconazole treatment.
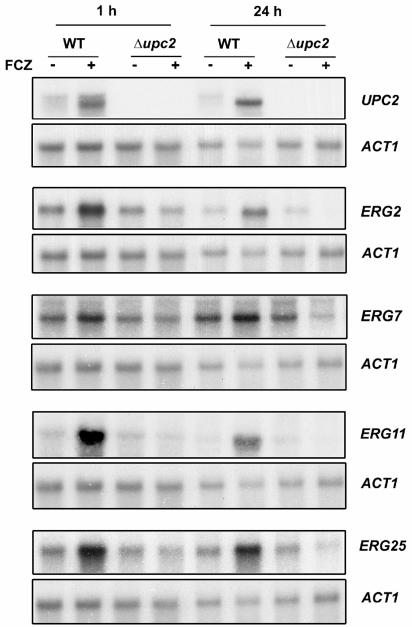
Given that Upc2p controls expression of the ergosterol biosynthetic genes in S. cerevisiae (39, 41), we wanted to determine if its Candida homologue plays a similar role. Wild-type and Δupc2 strains were treated or not with fluconazole (1 h, 24 h) and RNA was isolated for Northern blot analysis (Fig. 4). As expected, no UPC2 mRNA could be detected in the deletion strain. Interestingly, treatment with fluconazole (1 h, 24 h) increased UPC2 mRNA levels (Fig. 4, top panel). In the wild-type strain, antifungal treatment also increased mRNA levels of the ergosterol biosynthetic genes ERG2, ERG7, ERG11, and ERG25. The effect was sustained even after 24 h of exposure to fluconazole. Importantly, increases in mRNA levels were not observed for tested genes in cells lacking Upc2p, while basal levels remained unchanged. Probing with an actin control probe showed similar loading.
FIG. 4.
Northern blot analysis of UPC2 and ERG genes. Wild-type (WT) and Δupc2 strains were treated (+) or not (−) for 1 h or 24 h with 5 μg/ml fluconazole (FCZ), and RNA was isolated for Northern blot analysis. RNAs were hybridized with various probes as indicated on the right part of the figure.
Purified Upc2p DBD binds in vitro to DNA elements in the ERG2 promoter.
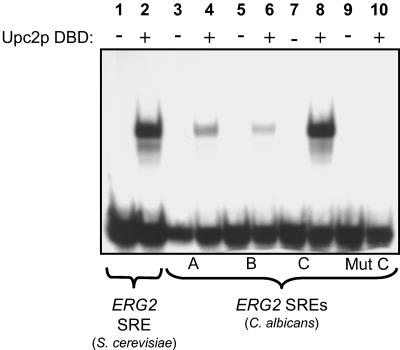
We wanted to obtain additional evidence that Upc2p is a transcriptional activator of ergosterol genes. In S. cerevisiae, Upc2p regulates ERG2 gene expression by binding directly to an SRE in its promoter (39). Upon examination of the C. albicans ERG2 promoter (up to 600 bp upstream of the coding region), we identified several similar elements with closely related sequences (Table 2). ERG2 SREs “A” and “B” contain the sequence TCGGATAA, while the ERG2 SRE “C” located closest to the ATG, differs (residue in boldface and underlined) slightly (TCGTATAA). (Boldface letters correspond to nucleotides that differ from the core sequence of the SRE “C” [see Table 2].) The purified DBD (amino acids 1 to 148) of the C. albicans Upc2p was used in an EMSA to test if it could bind to these elements (Fig. 5). Results show that the DBD of Upc2p binds to an SRE found in the promoter of S. cerevisiae ERG2 (Fig. 5, lanes 1 and 2). Probes in lanes 3 to 8 correspond to the putative SREs “A,” “B,” and “C” from the C. albicans ERG2 promoter. The DBD of Upc2p preferentially binds to SRE “C” that contains the sequence TCGTATAA, while reduced binding is observed with SREs “A” and “B” which contain the sequence TCGGATAA (Fig. 5, compare lanes 4, 6, and 8). Moreover, mutating the SRE “C” to TCAGATAA completely abolished binding of Upc2p (Fig. 5, lane 10). All together, these results suggest that Candida Upc2p controls the expression of ERG2 and related genes by binding to SREs found in their promoters.
FIG. 5.
The DBD of Upc2p binds in vitro to SREs. EMSA was performed with various probes (as indicated at the bottom of the figure) in the presence (+) or absence (−) of the purified DBD of Upc2p. Core sequences of the probes are given in Table 2. Complete sequences of the oligonucleotides used in EMSA can be found in Materials and Methods.
DISCUSSION
Many studies have focused on processes that are responsible for increased drug resistance in C. albicans and other pathogenic fungi. For example, increased drug efflux and alteration of the ergosterol biosynthetic pathway constitute two well-known mechanisms used by yeast cells to escape drug toxicity (35, 40). However, less is known about the transcriptional regulators that are involved in mediating PDR. The zinc cluster protein Fcr1p complements a Δpdr1 Δpdr3 strain in S. cerevisiae, but appears to act as a negative regulator of multidrug resistance in C. albicans (38). Tac1p, another zinc cluster protein, regulates the expression of the ABC transporters CDR1 and CDR2 (10). Moreover, the bZip transcription factor Cap1p and Fcr3p are also involved in PDR, although their targets are not well defined (6, 43, 45).
In S. cerevisiae, we have previously shown that deletion of UPC2 results in hypersensitivity to the antifungal ketoconazole while deletion of the highly related gene ECM22 results in sensitivity to the translation inhibitor cycloheximide (4). Upc2p and Ecm22p, which are both members of the zinc cluster family of transcriptional regulators (5), have overlapping roles, including activation of sterol biosynthetic genes (36, 39). We have identified only one homologue of S. cerevisiae Upc2p/Ecm22p in the C. albicans genome (Fig. 1) (data not shown).
Our study in C. albicans shows that a Δupc2 strain is hypersensitive to the antifungals ketoconazole and fluconazole (Fig. 3A). Conversely, overexpression of Upc2p in the KO strain results in increased resistance to these drugs as well as to fluphenazine when compared to a wild-type strain (Fig. 3B). Moreover, the Δupc2 strain shows growth defects under anaerobic conditions (Fig. 3C), while overexpression of UPC2 leads to growth superior to that of the wild-type strain. In S. cerevisiae, Upc2p is involved in sterol uptake under anaerobic conditions (41) while the upc2-1 mutant is hyperactive even under aerobic conditions (11, 41). However, we did not observe reduced growth of a double Δupc2 Δecm22 KO strain (BY4742 background) under anaerobic conditions (S. MacPherson and B. Turcotte, unpublished results). This phenotypic difference may be due to the fact that these two fungal species proliferate in quite different environments.
Northern blot analysis has shown that mRNA levels of the ERG genes tested are increased upon exposure to fluconazole (Fig. 4). Strikingly, this effect is abolished in a Δupc2 strain while basal levels of ERG mRNAs remained unchanged. Perturbation of the sterol metabolism has been associated with altered drug sensitivity (20, 21, 34, 44). In keeping with our results, other studies have shown that exposure to the antifungal itraconazole increases expression of ergosterol biosynthetic genes (13, 18). Thus, the azole susceptibility of a Δupc2 strain could be explained by its inability to increase ergosterol synthesis.
More direct evidence for Upc2p in regulating ERG gene transcription was provided by EMSA analysis. The purified DBD of Upc2p bound in vitro to SREs derived from the S. cerevisiae and C. albicans ERG2 promoters (Fig. 5). Of two related DNA elements, TCGTATAA was preferred over the related sequence TCGGATAA. In S. cerevisiae, Upc2p and Ecm22p also recognize the core sequence TCGTATAA (39), an observation that can be explained by the strong homology of the DBDs of these proteins with their Candida counterpart (Fig. 1). Interestingly, the preferred SRE site is also found in the ERG7, ERG11, and the ERG25 promoters, whose azole-induced expression is also dependent on Upc2p (Table 2). Moreover, similar sites are also found in the promoters of other genes of the ergosterol pathway (data not shown). UPC2 mRNA levels are increased upon antifungal drug treatment (Fig. 4), suggesting an autoregulatory mechanism. Again, this parallels S. cerevisiae where UPC2 promoter activity is increased under anaerobic conditions (1). Azole induction of UPC2 expression may be explained by the presence of a SRE in the promoter of this gene (Table 2), although regulation by another transcription factor is also possible.
While this work was under review, Silver et al. reported similar observations with a strain different from the one used in our study (37). Silver et al. also observed increased sensitivity of a Δupc2 strain to azoles. Moreover, the Δupc2 strain was sensitive to various other drugs, including terbinafine, fenpropimorph, lovastatin, and calcofluor white. Removal of UPC2 resulted in decreased ergosterol levels and cholesterol uptake (37). Northern blot analysis showed Upc2p-dependent increase of UPC2, ERG2, and ERG11 mRNA levels upon fluconazole treatment, in agreement with our observations. Thus, two independent studies link drug resistance and sterol metabolism to the transcription factor Upc2p in C. albicans.
The sequencing of the C. albicans genome reveals the presence of 77 putative ORFs encoding zinc cluster proteins, many of which have very close homologues in budding yeast. Based on what we know of this family of transcriptional regulators in S. cerevisiae, it is highly likely that these putative zinc cluster proteins play important roles, encompassing a wide spectrum of cellular functions. Importantly, many may be implicated in PDR as observed in S. cerevisiae (4). Clearly, further characterization of the Upc2 protein, as well as assignment of roles to other putative regulators, will be required to better comprehend the mechanism of drug resistance in this pathogenic species. As stated above, zinc cluster proteins are characterized by the presence of a highly conserved Zn(II)2Cys6 zinc finger required for DNA recognition. Interestingly, this motif is only found in fungi. Given the emergence of fungi that show resistance to cytotoxic compounds, zinc cluster proteins may constitute new targets for antifungal drugs. For instance, a drug disrupting the zinc finger of Upc2p could be effective when used in combination with other compounds such as azoles.
Acknowledgments
We would like to thank P. E. Sudbery for providing plasmid pCaEXP and Beatrice Magee for the actin plasmid. We are grateful to Pfizer Canada for the gift of fluconazole. We thank Sadri Znaidi for help with the sequence comparison. We also thank Karen Hellauer for technical assistance and Marc Larochelle for critical review of the manuscript.
This work was supported by grants from the Canadian Institutes of Health Research to B.T. (grant 204575) and M.R. (grant 15679). M.R. is supported by a scholarship from the Fonds de la Recherche en Santé du Québec (FRSQ), B.A. by a studentship from FRSQ, and X.D.D. by a postdoctoral fellowship from FRSQ.
REFERENCES
- 1.Abramova, N. E., B. D. Cohen, O. Sertil, R. Kapoor, K. J. A. Davies, and C. V. Lowry. 2001. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157:1169-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams, A., D. E. Gottschling, and T. Stearns. 1997. Methods in yeast genetics, vol. 1. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 3.Akache, B., S. MacPherson, M. A. Sylvain, and B. Turcotte. 2004. Complex interplay among regulators of drug resistance genes in Saccharomyces cerevisiae. J. Biol. Chem. 279:27855-27860. [DOI] [PubMed] [Google Scholar]
- 4.Akache, B., and B. Turcotte. 2002. New regulators of drug sensitivity in the family of yeast zinc cluster proteins. J. Biol. Chem. 277:21254-21260. [DOI] [PubMed] [Google Scholar]
- 5.Akache, B., K. Q. Wu, and B. Turcotte. 2001. Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res. 29:2181-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alarco, A.-M., and M. Raymond. 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181:700-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer, B. E., H. Wolfger, and K. Kuchler. 1999. Inventory and function of yeast ABC proteins: about sex, stress, pleiotropic drug and heavy metal resistance. Biochim. Biophys. Acta—Biomembranes 1461:217-236. [DOI] [PubMed] [Google Scholar]
- 8.Care, R. S., J. Trevethick, K. M. Binley, and P. E. Sudbery. 1999. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol. Microbiol. 34:792-798. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, B. D., O. Sertil, N. E. Abramova, K. J. A. Davies, and C. V. Lowry. 2001. Induction and repression of DAN1 and the family of anaerobic mannoprotein genes in Saccharomyces cerevisiae occurs through a complex array of regulatory sites. Nucleic Acids Res. 29:799-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coste, A. T., M. Karababa, F. Ischer, J. Bille, and D. Sanglard. 2004. TAC1, Transcriptional Activator of CDR Genes, is a new transcription factor involved in the regulation of Candida albicans ABC transporters CDR1 and CDR2. Eukaryot. Cell 3:1639-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crowley, J. H., F. W. Leak, Jr., K. V. Shianna, S. Tove, and L. W. Parks. 1998. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180:4177-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui, Z., T. Shiraki, D. Hirata, and T. Miyakawa. 1998. Yeast gene YRR1, which is required for resistance to 4-nitroquinoline N-oxide, mediates transcriptional activation of the multidrug resistance transporter gene SNQ2. Mol. Microbiol. 29:1307-1315. [DOI] [PubMed] [Google Scholar]
- 13.De Backer, M. D., T. Ilyina, X.-J. Ma, S. Vandoninck, W. H. M. L. Luyten, and H. Vanden Bossche. 2001. Genomic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gow, N. A. R., P. W. Robbins, J. W. Lester, A. J. P. Brown, W. A. Fonzi, T. Chapman, and O. S. Kinsman. 1994. A hyphal-specific chitin synthase gene (CHS2) is not essential for growth, dimorphism, or virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 91:6216-6220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hellauer, K., B. Akache, S. MacPherson, E. Sirard, and B. Turcotte. 2002. Zinc cluster protein Rdr1p is a transcriptional repressor of the PDR5 gene encoding a multidrug transporter. J. Biol. Chem. 277:17671-17676. [DOI] [PubMed] [Google Scholar]
- 17.Hellauer, K., M.-H. Rochon, and B. Turcotte. 1996. A novel DNA binding motif for yeast zinc cluster proteins: the Leu3p and Pdr3p transcriptional activators recognize everted repeats. Mol. Cell. Biol. 16:6096-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henry, K. W., J. T. Nickels, and T. D. Edlind. 2000. Upregulation of ERG genes in Candida species by azoles and other sterol biosynthesis inhibitors. Antimicrob. Agents Chemother. 44:2693-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 20.Jensen-Pergakes, K. L., M. A. Kennedy, N. D. Lees, R. Barbuch, C. Koegel, and M. Bard. 1998. Sequencing, disruption, and characterization of the Candida albicans sterol methyltransferase (ERG6) gene: drug susceptibility studies in erg6 mutants. Antimicrob. Agents Chemother. 42:1160-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia, N., B. Arthington-Skaggs, W. Lee, C. A. Pierson, N. D. Lees, J. Eckstein, R. Barbuch, and M. Bard. 2002. Candida albicans sterol C-14 reductase, encoded by the ERG24 gene, as a potential antifungal target site. Antimicrob. Agents Chemother. 46:947-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelly, R., S. M. Miller, M. B. Kurtz, and D. R. Kirsch. 1987. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol. Cell. Biol. 7:199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolaczkowski, M., and A. Goffeau. 1997. Active efflux by multidrug transporters as one of the strategies to evade chemotherapy and novel practical implications of yeast pleiotropic drug resistance. Pharmacol. Ther. 76:219-242. [DOI] [PubMed] [Google Scholar]
- 25.Le Crom, S., F. Devaux, P. Marc, X. Zhang, W. S. Moye-Rowley, and C. Jacq. 2002. New insights into the pleiotropic drug resistance network from genome-wide characterization of the YRR1 transcription factor regulation system. Mol. Cell. Biol. 22:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupetti, A., R. Danesi, M. Campa, M. Del Tacca, and S. Kelly. 2002. Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 27.Mamnun, Y. M., R. Pandjaitan, Y. Mahe, A. Delahodde, and K. Kuchler. 2002. The yeast zinc finger regulators Pdr1p and Pdr3p control pleiotropic drug resistance (PDR) as homo- and heterodimers in vivo. Mol. Microbiol. 46:1429-1440. [DOI] [PubMed] [Google Scholar]
- 28.Morschhauser, J. 2002. The genetic basis of fluconazole resistance development in Candida albicans. Biochim. Biophys. Acta—Mol. Basis Dis. 1587:240-248. [DOI] [PubMed] [Google Scholar]
- 29.Moye-Rowley, W. S. 2003. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog. Nucleic Acid Res. Mol. Biol. 73:251-279. [DOI] [PubMed] [Google Scholar]
- 30.Parks, L. W., and W. M. Casey. 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49:95-116. [DOI] [PubMed] [Google Scholar]
- 31.Perea, S., J. L. López-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perea, S., J. L. López-Ribot, B. L. Wickes, W. R. Kirkpatrick, O. P. Dib, S. P. Bachmann, S. M. Keller, M. Martinez, and T. F. Patterson. 2002. Molecular mechanisms of fluconazole resistance in Candida dubliniensis isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 46:1695-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinjon, E., G. P. Moran, C. J. Jackson, S. L. Kelly, D. Sanglard, D. C. Coleman, and D. J. Sullivan. 2003. Molecular mechanisms of itraconazole resistance in Candida dubliniensis. Antimicrob. Agents Chemother. 47:2424-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanglard, D., F. Ischer, T. Parkinson, D. Falconer, and J. Bille. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanglard, D., and F. C. Odds. 2002. Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2:73-85. [DOI] [PubMed] [Google Scholar]
- 36.Shianna, K. V., W. D. Dotson, S. Tove, and L. W. Parks. 2001. Identification of a UPC2 homolog in Saccharomyces cerevisiae and its involvement in aerobic sterol uptake. J. Bacteriol. 183:830-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silver, P. M., S. G. Oliver, and T. C. White. 2004. Role of Candida albicans transcription factor Upc2p in drug resistance and sterol metabolism. Eukaryot. Cell 3:1391-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talibi, D., and M. Raymond. 1999. Isolation of a putative Candida albicans transcriptional regulator involved in pleiotropic drug resistance by functional complementation of a pdr1 pdr3 mutation in Saccharomyces cerevisiae. J. Bacteriol. 181:231-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vik, Å., and J. Rine. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:6395-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilcox, L. J., D. A. Balderes, B. Wharton, A. H. Tinkelenberg, G. Rao, and S. L. Sturley. 2002. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J. Biol. Chem. 277:32466-32472. [DOI] [PubMed] [Google Scholar]
- 42.Wolfger, H., Y. M. Mamnun, and K. Kuchler. 2001. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res. Microbiol. 152:375-389. [DOI] [PubMed] [Google Scholar]
- 43.Yang, X. S., D. Talibi, S. Weber, G. Poisson, and M. Raymond. 2001. Functional isolation of the Candida albicans FCR3 gene encoding a bZip transcription factor homologous to Saccharomyces cerevisiae Yap3p. Yeast 18:1217-1225. [DOI] [PubMed] [Google Scholar]
- 44.Young, L. Y., C. M. Hull, and J. Heitman. 2003. Disruption of ergosterol biosynthesis confers resistance to amphotericin B in Candida lusitaniae. Antimicrob. Agents Chemother. 47:2717-2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, X., M. De Micheli, S. T. Coleman, D. Sanglard, and W. S. Moye-Rowley. 2000. Analysis of the oxidative stress regulation of the Candida albicans transcription factor, Cap1p. Mol. Microbiol. 36:618-629. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, X. T., Z. F. Cui, T. Miyakawa, and W. S. Moye-Rowley. 2001. Cross-talk between transcriptional regulators of multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 276:8812-8819. [DOI] [PubMed] [Google Scholar]