Abstract
Mutations in genes mexR and nalC have previously been shown to drive overexpression of the MexAB-OprM multidrug efflux system in Pseudomonas aeruginosa. A transposon insertion multidrug-resistant mutant of P. aeruginosa overproducing MexAB-OprM was disrupted in yet a third gene, PA3574, encoding a probable repressor of the TetR/AcrR family that we have dubbed NalD. Clinical strains overexpressing MexAB-OprM but lacking mutations in mexR or nalC were also shown to carry mutations in nalD. Moreover, the cloned nalD gene reduced the multidrug resistance and MexAB-OprM expression of the transposon mutant and clinical isolates, highlighting the significance of the nalD mutations vis-à-vis MexAB-OprM overexpression in these isolates.
Pseudomonas aeruginosa is an opportunistic human pathogen characterized by an innate resistance to multiple classes of antimicrobials (11), attributable in part to a family of broadly specific, so-called multidrug efflux systems (23, 24) that work synergistically with low outer membrane permeability (9, 19) to limit antimicrobial accumulation in this organism. Several multidrug efflux systems in P. aeruginosa have been described to date (23), although the major system contributing to intrinsic multidrug resistance is encoded by the mexAB-oprM operon (10, 18, 26). MexAB-OprM exports a wide variety of antimicrobials, including most classes of antibiotics, biocides, dyes, detergents, organic solvents (i.e., aromatic hydrocarbons; reviewed in reference 23]), and homoserine lactones associated with quorum sensing (8, 22). The last play a role in cell density-dependent expression of a number of virulence factors in P. aeruginosa, and thus, the activity of this efflux system can influence virulence (30). Indeed, a recent study suggests that the MexAB-OprM efflux system of P. aeruginosa promotes the release of a molecule(s) ultimately important for the virulence of this organism (12). The observation that MexAB-OprM hyperexpression in nalB strains impairs fitness and virulence (30) also suggests that this efflux system has a physiological role in P. aeruginosa independent of antimicrobial efflux and resistance. Consistent with this, mutants hyperexpressing MexAB-OprM were readily selected in vivo in a rat model of acute P. aeruginosa pneumonia in the absence of any antibiotic selection (16). The specific nature of the selective in vivo growth advantage provided by this efflux system is, however, unknown.
Hyperproduction of MexAB-OprM has been documented in lab and clinical multidrug-resistant isolates carrying lesions in the mexR gene (4, 15, 20, 28, 35, 37) (so-called nalB mutants [21]), encoding a repressor of mexAB-oprM expression (27, 35). MexAB-OprM hyperexpression also occurs independently of mutations in mexR or the mexR and mexAB-oprM promoter regions (35, 37). These so-called nalC mutants (5, 20, 35) carry a mutation in a recently identified gene (PA3721, also known as nalC) that encodes a TetR family repressor of an adjacent two-gene operon, PA3720-PA3719 (5). It is, in fact, the increased expression of PA3719 that results from disruption of the nalC repressor gene that promotes mexAB-oprM hyperexpression (5), apparently as a result of a direct interaction between the PA3719 gene product and the MexR repressor (S. Fraud, unpublished data). Intriguingly, MexR levels are greatly increased in nalC strains (L. Cao, R. Srikumar, and K. Poole, unpublished data), suggesting that MexR repressor activity is modulated in such mutants, perhaps in response to the increase in PA3720-PA3719 expression. In the present report mutations in yet a third gene, PA3574 (nalD), are shown to enhance mexAB-oprM expression, producing a multidrug-resistant phenotype in lab and clinical isolates of P. aeruginosa.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacterial cells were cultivated in/on Luria broth and agar (L-agar) (5) with antibiotics, as necessary, at 37°C. Plasmid pDSK519 and its derivatives were maintained with 50 (in Escherichia coli), 500 (in P. aeruginosa K870 and its derivatives), or 2,000 (in P. aeruginosa clinical isolates) μg of kanamycin per ml, while plasmid pUT-mini-Tn5-tet was maintained in E. coli with either ampicillin (100 μg/ml) or tetracycline (10 μg/ml). Plasmid pK18MobSacB was maintained in E. coli with 30 to 50 μg per ml of kanamycin.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | φ80d lacZΔM15 endA1 recA1 hsdR17 (rK− mK+) supE44 thi-1 gyrA96 relA1 F− Δ(lacZYA-argF)U169 | 2 |
| S17-1 | thi pro hsdR recA Tra+ | 31 |
| P. aeruginosa | ||
| K767 | PAO1 prototroph | 21 |
| K870 | Smr derivative of K767 | 27 |
| K2346 | K870 nalD::mini-Tn5-tet | This study |
| K2347 | K2346 ΔPA3719 | This study |
| 2085 | Clinical nalB isolate | 20 |
| 2151 | Clinical nalB isolate | 20 |
| 1250 | Clinical nalC isolate | 20 |
| 1738 | Clinical nalC isolate | 20 |
| 1217 | Clinical nalD isolate | 20 |
| 1562 | Clinical nalD isolate | 20 |
| 1113 | Clinical nalD isolate | 20 |
| WL24 | Clinical nalD isolate | 20 |
| Plasmids | ||
| pDSK519 | Broad-host-range cloning vector; IncQ Kmr | 17 |
| pMLS003 | pDSK519::nalD | This study |
| pK18MobSacB | Broad-host-range gene replacement vector; sacB Kmr | 31 |
| pMLS004 | pK18MobSacB::ΔPA3719 | This study |
| pUT::mini-Tn5-tet | mini-Tn5-tet delivery vector: Apr Tcr | 6 |
| pEX18Tc | Broad-host-range gene replacement vector; sacB Tcr | 13 |
| pLC8 | pEX18Tc::ΔPA3719 | 5 |
DNA manipulations.
Standard protocols were used for restriction endonuclease digestions, ligations, transformation, plasmid isolation, and agarose gel electrophoresis, as described by Sambrook and Russell (29). Genomic DNA of P. aeruginosa was extracted by following the protocol of Barcak et al. (3). E. coli cells were made competent using the CaCl2 method (29) or that of Inoue et al. (14). Electroporation of pDSK519 and pMLS003 into clinical P. aeruginosa isolates was carried out as described previously (32). Chromosomal DNA flanking the mini-Tn5-tet element in putative nalD insertion mutants was sequenced using primer mini-Tn5-Right (5′-GCTTGCTCAATCAATCACC-3′). Oligonucleotide synthesis and nucleotide sequencing was carried out by Cortec DNA Services Inc., Kingston, Ontario, Canada. Once the flanking DNA sequences were obtained, disrupted genes were identified by BLASTN (http://www.ncbi.nlm.nih.gov/BLAST/) searches of the available P. aeruginosa genome sequence (http://www.pseudomonas.com).
Transposon mutagenesis.
P. aeruginosa strain K870, a streptomycin-resistant derivative of the wild-type P. aeruginosa PAO1 strain K767 was mutagenized with mini-Tn5-tet as described previously (5). Multidrug-resistant mutants overexpressing MexAB-OprM were identified initially by their characteristic resistance profile (5) and later using immunoblotting with an anti-MexB antiserum (see below). The mini-Tn5-tet element and flanking chromosomal DNA from selected mutants was obtained following PstI digestion of isolated chromosomal DNA and cloning of a mini-Tn5-tet-carrying PstI fragment as described before (5).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting.
Putative nalD mini-Tn5-tet mutants were screened for MexAB-OprM hyperexpression using a MexB-specific antiserum following Western immunoblotting of electrophoretically separated cell envelopes (34). Immunodetection of MexR (7) was also carried out in these mutants following electrophoresis of soluble (membrane-free) cell extracts as described previously (1).
Antimicrobial susceptibility testing.
The antimicrobial susceptibility of P. aeruginosa strains was assessed in microtiter trays using a twofold-serial-dilution technique as described previously (33, 20).
Cloning nalD (PA3574).
The nalD (PA3574) gene was amplified from P. aeruginosa K870 chromosomal DNA using primers PA3574-F (5′-AAAAGCTTAAGCTTGCAGCATAACACCGAAGAC-3′; tandem HindIII sites are underlined) and PA3574-R (5′-AAGGATCCGGATCCCAGGTACTCGAGGCGATC-3′; tandem BamHI sites are underlined) in a PCR mixture formulated as described previously (33) but with 1 U Vent DNA polymerase (NEB) and no dimethyl sulfoxide. Amplification of PA3574 was achieved by incubation for 45 s at 95°C, followed by 25 cycles of 45 s at 95°C, 45 s at 54°C, and 60 s at 72°C, with a final 7-min elongation at 72°C. The PA3574 PCR product was subsequently purified using the QIAGEN PCR purification kit, digested with HindIII and BamHI, and cloned into pBluescript (Stratagene). Nucleotide sequencing with universal primers confirmed the absence of mutations in pBluescript-borne PA3574. PA3574 was subsequently excised from pBluescript on a SalI-BamHI fragment and cloned into pDSK519 to yield pMLS003, in which PA3574 expression was driven by the resident lac promoter of pDSK519.
Construction of a ΔPA3719 mutant.
A ΔPA3719 derivative of nalD::mini-Tn5-tet strain K2346 was constructed using the previously described PA3719 deletion vector pLC8, from which the ΔPA3719 fragment was excised using EcoRI and HindIII and cloned into pK18MobSacB. The resultant vector, pMLS004, was introduced into E. coli S17-1 and subsequently mobilized into K2346 via conjugal transfer as described previously (25). K2346 transconjugants harboring pMLS004 in the chromosome were selected on 15 μg/ml chloramphenicol (to counterselect E. coli S17-1) and 1,000 μg/ml kanamycin and subsequently streaked onto L-agar containing sucrose (10% [wt/vol]; to screen for bacteria in which pK18MobSacB has been lost but the intended deletion possibly retained) (33). Sucrose-resistant colonies were then screened for deletion of nalD via colony PCR using Taq polymerase and the previously described primers 3719/20F and 3719/20R (5). Briefly, individual colonies were resuspended in 30 μl of distilled H2O, heated at 95 to 100°C for 5 min, and stored on ice, and then 10 μl was added to a 100-μl PCR mixture containing Q solution (QIAGEN). Reaction mixtures were heated to 95°C for 5 min, followed by 25 cycles of 45 s at 97°C, 45 s at 54°C, and 1 min at 72°C, followed by 7 min at 72°C.
Plasmid mobilization.
Plasmid pMLS003 was mobilized from E. coli DH5α into the P. aeruginosa PA3574::miniTn5-tet (i.e., NalD−) strain K2346 using a previously described triparental-mating procedure (36), with plasmid-carrying P. aeruginosa selected on L-agar containing 500 μg/ml kanamycin and 0.5 μg/ml imipenem (to counterselect E. coli).
RT-PCR.
RNA isolation from overnight cultures of P. aeruginosa K870 and K2346 and subsequent reverse transcription-PCR (RT-PCR) to assess the expression of rpsL and PA3573 was carried out as described previously (33) using primer pairs rpsLF and rpsLR (33) and PA3573F (5′-GATTTCTACCTGCCGAGC-3′) and PA3573R (5′-GCATCAACTGGGAAAACG-3′). The PCR portion was carried out for 18 and 19 cycles (rpsL) or 29 and 30 cycles (PA3573). In instances where expression of mexAB-oprM was assessed using real-time RT-PCR, the protocol of Llanes et al. (20) was employed.
PCR amplification of nalD from clinical isolates.
The nalD gene of clinical P. aeruginosa isolates was amplified via PCR according to a previously described protocol (20) using primers NalD1 (5′-GCGGCTAAAATCGGTACACT-3′) and NalD2 (5′-ACGTCCAGGTGGATCTTGG-3′) and an annealing temperature of 61°C. Both strands of the 789-bp nalD product were subsequently sequenced using NalD1, NalD2, and the internal primers NalDSeq1 (5′-TCAACGAGATGCTCAACC-3′) and NalDSeq2 (5′-CTGGTTGAGCATCTCGTTGA-3′).
RESULTS AND DISCUSSION
Identification of the nalD gene.
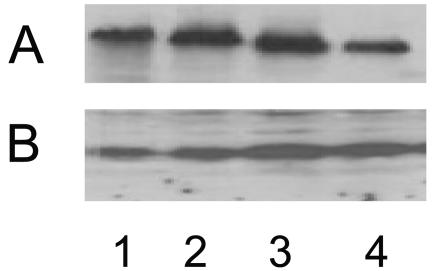
To identify potentially novel genes controlling mexAB-oprM expression in P. aeruginosa, strain K870 was subjected to mini-Tn5-tet transposon insertion mutagenesis and mutants showing a multidrug resistance phenotype characteristic of MexAB-OprM overexpression were selected. One such mutant, K2346, showed increased resistance to chloramphenicol, carbenicillin, nalidixic acid, and novobiocin at levels below that of previously described nalB (mexR) mutants but very similar to that of nalC mutants (35) (Table 2). Western immunoblotting confirmed, too, a modest increase in MexB expression in K2346 (Fig. 1, lane 2; cf. lane 1), consistent with enhanced expression of the MexAB-OprM multidrug efflux system in this mutant. Subsequent recovery of the mini-Tn5-tet element from K2346 and sequencing of flanking P. aeruginosa chromosomal DNA identified PA3574, encoding a probable repressor of the TetR/AcrR family (http://www.pseudomonas.com), as the disrupted gene in the mutant. Introduction of the cloned, wild-type PA3574 gene on plasmid pMLS003 into K2346 reduced MexAB-OprM expression (Fig. 1, lane 4; cf. lane 3) and multidrug resistance (Table 2), confirming that the PA3574 disruption was, indeed, responsible for the elevated MexAB-OprM expression and attendant multidrug resistance of strain K2346. This gene has been dubbed nalD to reflect its connection to other genes and/or mutations (i.e., nalB and nalC) that are similarly associated with enhanced MexAB-OprM production and because such a designation has already been used to refer to MexAB-OprM-overproducing mutants lacking nalB (mexR) and nalC mutations (5, 20).
TABLE 2.
Antimicrobial susceptibility of nalD P. aeruginosa
| Strain | nalD statusa | MIC (μg/ml)b
|
mexA expressionc | ||||||
|---|---|---|---|---|---|---|---|---|---|
| CAM | NAL | TET | NOV | CAR | TIC | ATM | |||
| K870 | WT | 16 | 64 | 8 | 512 | 128 | |||
| K2346 | Null | 64 | 256 | 256 | 1,024 | 512 | |||
| K2346(pDSK519)d | Null | 64 | 256 | 256 | 1,024 | 512 | |||
| K2346(pMLS003)d | WT | 8 | 64 | 64 | 256 | 64 | |||
| K2347 | Null | 64 | 256 | 256 | 1,024 | 512 | |||
| PAO1 | WT | 32 | 64 | 32 | 16 | 8 | 1.00 | ||
| 1113(pDSK519)e | ΔT410-G433 | 64 | 256 | 64 | 64 | 32 | 1.69 | ||
| 1113(pMLS003)e | WT | 32 | 32 | 16 | 8 | 2 | 0.34 | ||
| WL24(pDSK519)e | ? | 128 | >1,028 | 128 | 64 | 16 | 2.41 | ||
| WL24(pMLS003)e | WT | 32 | >1,028f | 32 | 16 | 2 | 0.25 | ||
The indicated strains expressed wild type (WT), nalD::mini-Tn5-tet-disrupted (Null), or mutant nalD genes (specific mutations are highlighted as base changes in the gene itself). Mutations in nalD were verified following PCR amplification of the gene and nucleotide sequencing of the PCR product obtained. ?, the nalD gene could not be amplified by PCR, suggesting a possible deletion of the gene in this strain.
CAM, chloramphenicol; NAL, nalidixic acid; TET, tetracycline; NOV, novobiocin; CAR, carbenicillin; TIC, ticarcillin; ATM, aztreonam.
mexA expression (as a measure of mexAB-oprM expression) was quantitated using real-time RT- PCR and normalized to expression levels seen in wild-type strain PAO1.
pDSK519 is the vector without nalD; pMLS003 is pDSK519::nalD.
Clinical isolate.
The apparent lack of an impact of the cloned nalD gene on the nalidixic acid MIC likely reflects the presence of another determinant(s) of resistance to this agent in strain WL24, which may be masking any impact of mexAB-oprM expression on resistance.
FIG. 1.
Immunoblot showing expression of MexB (A) and MexR (B) in P. aeruginosa strains K870 (NalD+; lane 1), K2346 (NalD−; lane 2), K2346/pDSK519 (NalD−; lane 3), and K2346/pMLS003 (NalD+; lane 4).
PA3574 occurs immediately adjacent to a divergently transcribed gene encoding a putative exporter of the major facilitator superfamily, PA3573, which might be a target for PA3574 regulation. RT-PCR revealed, however, that disruption of PA3574 had no impact on PA3573 expression (data not shown), indicating that the PA3574 mutation in K2346 did not impact MexAB-OprM expression via influence on PA3573. In this it differs from a nalC mutant, where a mutation in another TetR/AcrR family repressor, nalC (also known as PA3721), increased expression of the adjacent, divergently transcribed two-gene operon PA3720-PA3719, with increased PA3719 alone responsible for MexAB-OprM hyperexpression (5). To see, however, whether the positive impact of a nalD mutation on MexAB-OprM expression similarly involved PA3719 alone the PA3719 gene was deleted from strain K2346 (producing K2347). No change in MexAB-OprM production or resistance was observed as a result of the PA3719 loss in K2347 (data not shown), indicating that yet another mechanism exists in P. aeruginosa for enhancing MexAB-OprM expression. Unlike nalC mutations, too, which are characterized by hyperproduction of a stable but apparently nonfunctional MexR repressor (Cao et al., unpublished), disruption of nalD had no, or at best a very modest, impact on MexR production (Fig. 1B).
nalD mutations in clinical isolates.
To assess the importance of mutations in nalD in clinical strains overproducing MexAB-OprM, the nalD gene of previously described MexAB-OprM-overproducing, multidrug-resistant clinical strains lacking mutations in mexR or nalC (i.e., strains 1217, 1562, 1113, and WL24 [20]) was amplified by PCR and sequenced. As controls, clinical isolates overproducing MexAB-OprM but with mutations in mexR (strains 2085 and 2151) or nalC (strains 1250 and 1738) were also examined. mexR strain 2151 harbored no mutations in PA3574, while the remaining mexR and nalC strains carried only silent mutations in this gene (strain 2085: C276T, TGCCys→TGTCys; T295C, TTGLeu→CTGLeu; C333T, ATCIle→ ATTIle; C540T, GACAsp→GATAsp; strain 1250: T450C, CGTArg→CGCArg; G477A, CCGPro→CCAPro; T555C, GATAsp→GACAsp; strain 1738: T555C, GATAsp→GACAsp). In contrast, strains 1217, 1562, and 1113 all carried mutations in nalD (strains 1217 and 1562 had a substitution leading to a Ser32Asn change in NalD; strain 1113 had a 24-bp deletion [Table 2]), with strain 1113 also harboring two silent mutations in this gene (C276T, TGCCys TGTCys; T295C, TTGLeu CTGLeu). The nalD gene could not be amplified from strain WL24, indicating its lack in this strain, possibly due to deletion. Nonetheless, introduction of the cloned, wild-type nalD gene into WL24 as well as 1113 (the high kanamycin MICs for strains 1217 and 1562 precluded introduction of the nalD vector pMLS003 into these isolates) reduced resistance levels and mexAB-oprM expression (Table 2), indicating that like the nalD::mini-Tn5-tet mutation of K2346, the nalD mutations of these clinical strains were responsible for elevated MexAB-OprM production and multidrug resistance. Despite our inability to assess complementation of the nalD mutant strains 1217 and 1562 with cloned nalD, the fact that these strains lack other mutations that might explain enhanced mexAB-oprM expression and that nalD mutations do provide for elevated mexAB-oprM expression and multidrug resistance in other mutants argue strongly that the nalD mutations in 1217 and 1562 do contribute to the resistance and efflux phenotypes of these isolates.
Conclusions.
Mutations in at least three different genes (mexR, nalC, and nalD) can provide for increased expression of MexAB-OprM, highlighting the complexity of mexAB-oprM regulation in P. aeruginosa. While the increase in PA3719 seen in nalC strains provides for elevated mexAB-oprM expression, owing to an apparent impact on MexR repressor activity (Cao et al., unpublished), mutations in nalD appear to work independently of PA3719, indicating that yet a second pathway exists in P. aeruginosa by which mexAB-oprM expression is influenced. Whether this relates to different environmental or cell-associated signals capable of impacting mexAB-oprM expression (i.e., a variety of conditions require MexAB-OprM export activity) is at present unknown. Intended DNA array studies may, however, provide insights vis-à-vis the gene(s) that is the immediate target(s) for the putative NalD repressor by identifying genes that are coregulated with mexAB-oprM in nalD mutants. This will also, hopefully, address the issue of the intended function(s) of this broadly specific antimicrobial efflux system, since the function (if known) of genes coregulated with mexAB-oprM may provide clues as to MexAB-OprM function in P. aeruginosa. Clearly, MexAB-OprM exports multiple substrates and its expression is associated with multiple phenotypes (increased antimicrobial resistance, reduced fitness, improved in vivo survival, changes in virulence), suggesting that it has multiple roles in P. aeruginosa and is not limited to antimicrobial export and resistance.
Acknowledgments
This work was supported by operating grants from the Canadian Cystic Fibrosis Foundation (CCFF) to K.P. and the French Cystic Fibrosis Association (Vaincre la mucoviscidose) to P.P. L.C. holds an Ontario Graduate Scholarship.
REFERENCES
- 1.Adewoye, L., A. Sutherland, R. Srikumar, and K. Poole. 2002. The MexR repressor of the mexAB-oprM multidrug efflux operon in Pseudomonas aeruginosa: characterization of mutations compromising activity. J. Bacteriol. 184:4308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Short protocols in molecular biology, 2nd ed. John Wiley & Sons, Inc., New York, N.Y.
- 3.Barcak, G. J., M. S. Chandler, R. J. Redfield, and J. F. Tomb. 1991. Genetic systems in Haemophilus influenzae. Methods Enzymol. 204:321-342. [DOI] [PubMed] [Google Scholar]
- 4.Boutoille, D., S. Corvec, N. Caroff, C. Giraudeau, E. Espaze, J. Caillon, P. Plesiat, and A. Reynaud. 2004. Detection of an IS21 insertion sequence in the mexR gene of Pseudomonas aeruginosa increasing β-lactam resistance. FEMS Microbiol. Lett. 230:143-146. [DOI] [PubMed] [Google Scholar]
- 5.Cao, L., R. Srikumar, and K. Poole. 2004. MexAB-OprM hyperexpression in NalC type multidrug resistant Pseudomonas aeruginosa: identification and characterization of the nalC gene encoding a repressor of PA3720-PA3719. Mol. Microbiol. 53:1423-1436. [DOI] [PubMed] [Google Scholar]
- 6.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6567-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, K., L. Adewoye, and K. Poole. 2001. MexR repressor of the mexAB-oprM multidrug efflux operon of Pseudomonas aeruginosa: identification of MexR binding sites in the mexA-mexR intergenic region. J. Bacteriol. 183:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, K., L. Passador, R. Srikumar, E. Tsang, J. Nezezon, and K. Poole. 1998. Influence of the MexAB-OprM multidrug efflux system on quorum-sensing in Pseudomonas aeruginosa. J. Bacteriol. 180:5443-5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germ, M., E. Yoshihara, H. Yoneyama, and T. Nakae. 1999. Interplay between the efflux pump and the outer membrane permeability barrier in fluorescent dye accumulation in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 261:452-455. [DOI] [PubMed] [Google Scholar]
- 10.Gotoh, N., H. Tsujimoto, K. Poole, J.-I. Yamagishi, and T. Nishino. 1995. The outer membrane protein OprM of Pseudomonas aeruginosa is encoded by oprK of the mexA-mexB-oprK multidrug resistance operon. Antimicrob. Agents Chemother. 39:2567-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock, R. E. W., and D. P. Speert. 2000. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Res. Updat. 3:247-255. [DOI] [PubMed] [Google Scholar]
- 12.Hirakata, Y., R. Srikumar, K. Poole, N. Gotoh, T. Suematsu, S. Kohno, S. Kamihira, R. E. Hancock, and D. P. Speert. 2002. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J. Exp. Med. 196:109-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoang, T. T., R. R. Karkhoff-Schweizer, A. J. Kutchma, and H. P. Schweizer. 1998. A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77-86. [DOI] [PubMed] [Google Scholar]
- 14.Inoue, H., H. Nojima, and H. Okayama. 1991. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 15.Jalal, S., and B. Wretlind. 1998. Mechanisms of quinolone resistance in clinical strains of Pseudomonas aeruginosa. Microbiol. Drug Resist. 4:257-261. [DOI] [PubMed] [Google Scholar]
- 16.Join-Lambert, O. F., M. Michea-Hamzehpour, T. Kohler, F. Chau, F. Faurisson, S. Dautrey, C. Vissuzaine, C. Carbon, and J. C. Pechere. 2001. Differential selection of multidrug efflux mutants by trovafloxacin and ciprofloxacin in an experimental model of Pseudomonas aeruginosa acute pneumonia in rats. Antimicrob. Agents Chemother. 45:571-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 18.Li, X.-Z., H. Nikaido, and K. Poole. 1995. Role of MexA-MexB-OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, X.-Z., L. Zhang, and K. Poole. 2000. Interplay between the MexAB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 45:433-436. [DOI] [PubMed] [Google Scholar]
- 20.Llanes, C., D. Hocquet, C. Vogne, D. Benali-Baitich, C. Neuwirth, and P. Plesiat. 2004. Clinical strains of Pseudomonas aeruginosa overproducing MexAB-OprM and MexXY efflux pumps simultaneously. Antimicrob. Agents Chemother. 48:1797-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda, N., and S. Ohya. 1992. Cross-resistance to meropenem, cephems, and quinolones in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 36:1847-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pearson, J. P., C. Van Delden, and B. H. Iglewski. 1999. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J. Bacteriol. 181:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole, K. 2004. Efflux-mediated multiresistance in Gram-negative bacteria. Clin. Microbiol. Infect. 10:12-26. [DOI] [PubMed] [Google Scholar]
- 24.Poole, K. 2004. Efflux pumps, p. 635-674. In J.-L. Ramos (ed.), Pseudomonas, vol. I. Genomics, life style and molecular architecture. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 25.Poole, K., D. E. Heinrichs, and S. Neshat. 1993. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol. Microbiol. 10:529-544. [DOI] [PubMed] [Google Scholar]
- 26.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole, K., K. Tetro, Q. Zhao, S. Neshat, D. Heinrichs, and N. Bianco. 1996. Expression of the multidrug resistance operon mexA-mexB-oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob. Agents Chemother. 40:2021-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito, K., H. Yoneyama, and T. Nakae. 1999. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 179:67-72. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sanchez, P., J. F. Linares, B. Ruiz-Diez, E. Campanario, A. Navas, F. Baquero, and J. L. Martinez. 2002. Fitness of in vitro selected Pseudomonas aeruginosa nalB and nfxB multidrug resistant mutants. J. Antimicrob. Chemother. 50:657-664. [DOI] [PubMed] [Google Scholar]
- 31.Simon, R., U. Priefer, and A. Puehler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 32.Smith, A. W., and B. H. Iglewski. 1989. Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res. 17:10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobel, M. L., G. A. McKay, and K. Poole. 2003. Contribution of the MexXY multidrug transporter to aminoglycoside resistance in Pseudomonas aeruginosa clinical isolates. Antimicrob. Agents Chemother. 47:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srikumar, R., T. Kon, N. Gotoh, and K. Poole. 1998. Expression of Pseudomonas aeruginosa multidrug efflux pumps MexA-MexB-OprM and MexC-MexD-OprJ in a multidrug-sensitive Escherichia coli strain. Antimicrob. Agents Chemother. 42:65-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srikumar, R., C. J. Paul, and K. Poole. 2000. Influence of mutations in the mexR repressor gene on expression of the MexA-MexB-OprM multidrug efflux system of Pseudomonas aeruginosa. J. Bacteriol. 182:1410-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, Q., X.-Z. Li, A. Mistry, R. Srikumar, L. Zhang, O. Lomovskaya, and K. Poole. 1998. Influence of the TonB energy-coupling protein on efflux-mediated multidrug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 42:2225-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ziha-Zarifi, I., C. Llanes, T. Koehler, J.-C. Pechere, and P. Plesiat. 1999. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA-MexB-OprM. Antimicrob. Agents Chemother. 43:287-291. [DOI] [PMC free article] [PubMed] [Google Scholar]