Abstract
Background
Melanoma brain metastases (MBM) continue to be a significant clinical problem with limited treatment options. Highly invasive melanoma cells migrate along the vasculature and perivascular cells may contribute to residual disease and recurrence. PTEN loss and hyperactivation of AKT occur in MBM; however, a role for PTEN/AKT in perivascular invasion has not been described.
Methods
We used in vivo intracranial injections of murine melanoma and bulk RNA sequencing of melanoma cells co-cultured with brain endothelial cells (brECs) to investigate brain colonisation and perivascular invasion.
Results
We found that PTEN-null melanoma cells were highly efficient at colonising the perivascular niche relative to PTEN-expressing counterparts. PTEN re-expression (PTEN-RE) in melanoma cells significantly reduced brain colonisation and migration along the vasculature. We hypothesised this phenotype was mediated through vascular-induced TGFβ secretion, which drives AKT phosphorylation. Disabling TGFβ signalling in melanoma cells reduced colonisation and perivascular invasion; however, the introduction of constitutively active myristolated-AKT (myrAKT) restored overall tumour size but not perivascular invasion.
Conclusions
PTEN loss facilitates perivascular brain colonisation and invasion of melanoma. TGFβ-AKT signalling partially contributes to this phenotype, but further studies are needed to determine the complementary mechanisms that enable melanoma cells to both survive and spread along the brain vasculature.
Subject terms: Melanoma, Cancer microenvironment, CNS cancer, Metastasis
Background
Melanoma has an unusually high frequency of metastasise to the brain; among stage IV patients, 40–50% will develop clinically detectable intracranial disease, while on autopsy, brain metastases can be detected in over 70% of patients [1]. In addition, MBM carries a poor prognosis with high morbidity and mortality [2, 3]. Despite the findings that systemic therapies, particularly targeted therapies consisting of BRAF/MEK inhibitors and immunotherapy with checkpoint blockade, have drastically improved patient outcomes, these treatments have less efficacy in the brain. For example, results from the COMBI-MB phase 2 trial for dabrafenib plus trametinib demonstrated intracranial responses in patients with BRAF-mutant melanoma and brain metastases; however, the median duration of intracranial control was 6.5 months compared to extracranial responses of 10.2 months. Tumour progression was also higher in patients with intracranial or intracranial/extracranial disease (47% and 25%, respectively) compared to patients with only extracranial disease progression (9%) [4]. For immunotherapy combining nivolumab plus ipilimumab, there was comparable efficacy between extracranial and asymptomatic MBM, but lower efficacy in symptomatic MBM as described in CheckMate-204 [5]. Furthermore, the addition of locoregional therapy (stereotactic radiosurgery, conventional radiotherapy, and surgery) can further improve outcomes when combined with systemic therapies [6]. However, while locoregional therapies are effective at treating MBM, they cannot prevent the reoccurrence of new intracranial lesions. In addition, multiple lesions in the brain are associated with worse survival in several studies [7–9]. Thus, understanding the mechanisms of melanoma dispersal and survival within the brain is an important clinical challenge.
Melanoma, with its neural-crest lineage origins and high plasticity, is suggested to revert to developmental programmes that recapitulate invasion along the vasculature [10, 11]. Indeed, MBM are strongly associated with an angiotropic/ perivascular invasive phenotype in human samples [12–15]. In cutaneous melanoma, angiotropism (also called vascular co-option) is associated with poor survival and an increased risk of metastasis [16, 17]. In addition, in murine models, melanoma cells proliferate along the microvasculature and only occasionally induce angiogenesis after large macrometastases have formed; melanoma cells can also be found within a perivascular niche during dormancy [18]. Targeting the molecular mechanisms that facilitate cancer cell:endothelial cell (EC) interactions in the brain could therefore be applied as a therapeutic modality [12, 18–21].
Despite pre-clinical and clinical descriptions of perivascular invasion in MBM, few molecular mechanisms have been elucidated [22]. One mechanism of vascular co-option that has been studied in lung and breast cancer metastasis to the brain includes the upregulation of serpins to protect from FasL-mediated cell killing alongside an upregulation of the adhesion molecule L1CAM [23]. It has also been shown that PTEN is often lowly expressed or mutated across a variety of brain metastatic cancers [24]. In melanoma, PTEN is one of the most commonly mutated members of the PI3K pathway (14% of TCGA melanoma tumours) and often co-occurs with BRAF mutations [25, 26]. In murine models, PTEN loss cooperates with BRAF to promote tumorgenicity [27]. In addition, AKT1 overexpression combined with PTEN silencing leads to MBM in 80% of tumour-bearing mice [28]. These findings align with human studies describing loss of PTEN expression in association with decreased overall survival and time to brain metastasis formation in stage IIB/C BRAF-mutant melanoma [29]. In addition, higher levels of activated AKT and lower PTEN expression is found in MBM compared to extracranial (lung and liver) metastases [30–32].
Given previous evidence that loss of PTEN likely plays a post-extravasation role in brain metastases [24], we used intracranial injection models to examine brain colonisation and perivascular invasion in the post-colonisation stages of metastases. We found that PTEN-null melanoma cells were highly invasive along the brain perivascular niche compared to PTEN-expressing counterparts. Bulk RNA sequencing of melanoma cells differing in PTEN status co-cultured with brECs revealed upregulation of TGFβ and PI3K activation in PTEN-null melanoma cells. Finally, we used gain/loss of function studies to demonstrate that PTEN is an important brake on both brain colonisation and perivascular invasion, whereas AKT has a more prominent role for overall tumour size rather than perivascular motility.
Methods
Cell culture
D4M.3A and SM1WT1 cells were cultured in DMEM/F-12 (#11330032, Gibco, USA) supplemented with 5% foetal bovine serum (FBS) (#16000-044, Gibco). D4M.3A were acquired from Drs. Constance Brinkerhoff and David Mullins [33] and SM1WT1 cells were a gift from Dr. Tobias Bald. Brain endothelial cells (b.End3) and mouse embryonic fibroblasts (MEFs) were acquired from ATCC and cultured in 4.5 g/L D-glucose DMEM (#11965092, Gibco) supplemented with 10% FBS. Mouse dermal ECs (MDEC) were isolated by our lab [34] and cultured in 1 g/L D-glucose DMEM (#11885-084, Gibco) supplemented with 10% FBS, 10% Nu-Serum IV (BD, USA), 5 ng/mL bFGF (Peprotech, USA), 10 ng/mL VEGFA (Peprotech), and 20 USP units/mL heparin (Sigma, USA). All media were supplemented with antimycotic/antibiotic (Gibco) and plasmocin (InvivoGen, USA). Cell lines were periodically tested for mycoplasma. ECs and MEFs were cultured on 0.5% gelatin-coated plates. For co-culture experiments, ECs and MEFs were allowed to adhere for 2 days prior to seeding with melanoma cells. FACS was carried out using the ARIA Fusion Cell Sorter to re-separate melanoma cells from ECs or MEFs. For TGFβ treatments, cells were incubated in low serum (0.5% FBS in DMEM/F-12) with 10 ng/mL TGFβ2 (Peprotech).
Generation of cells with expression of PTEN or myrAKT
For expression of PTEN or myrAKT, target genes were cloned into the pLV-mCherry vector (#36084, Addgene, USA) to obtain the pLV-Pten-p2A-mCherry or pLV-myrAKT-p2A-mCherry plasmid, respectively. The mouse Pten gene was amplified using primers (forward: 5’-gcgtctagagccaccATGACAGCCATCATCAAAGAGATCG-3’; reverse: 5’-gcgggatccaggaccggggttttcttccacgtctcctgcttgctttaacagagagaagttcgtggcctcgagaccggtGACTTTTGTAATTTGTGAATGCTGA-3’) and mouse myrAKT gene was amplified using primers (forward: 5’-gcgacgcgtgccaccATGGGGAGCAGCAAG-3’; reverse: 5’-gcgggatccaggaccggggttttcttccacgtctcctgcttgctttaacagagagaagttcgtggcctcgagaccggtAGCGTAGTCTGGGACGTCGTA-3’). Plasmids (pLV-Pten-p2A-mCherry or pLV-myrAKT-p2A-mCherry, psPAX2, pMD2.6) were co-transfected into HEK293T cells using lipofectamine 3000 (L3000001, Thermo Fisher, USA). D4M.3A cells were infected with virus and FACS selected for mCherry+ cells.
Generation of CRISPR knockout (KO) cell lines
To generate D4M.3A TGFβR2 KO and SM1WT1 PTEN KO cells, target guides were designed using the Broad Institute Genetic Perturbation Platform’s sgRNA design tool (https://portals.broadinstitute.org/gppx/crispick/public). The following sgRNA sequences were used: AAAAAGTCCGCGATTACGTC and AAAACGGCTCGATCGGTGAT for Non-targeting; CCTCCAATTCAGGACCCACG and GGTTTGATAAGTTCTAGCTG for PTEN; and ACCTGCAGGAGTACCTCACG for TGFβR2. These guides were cloned into the lentiCRISPRV2 plasmid (#52961, Addgene) as described previously [35]. Cloned lentiCRISPRV2 was co-transfected into HEK293T cells with VSVG and delta8.9 plasmids using X-tremeGene HP DNA transfection reagent (Sigma). D4M.3A and SM1WT1 cells were incubated with lentivirus and 8 µg/mL polybrene. Two days post-infection, cells were puromycin-selected (2 or 6 µg/mL, respectively) prior to western confirmation of KO. Limiting dilution assays were used to select for clonal populations.
Crystal violet (CV) assay
Cells were seeded at 2000 cells/well in a 96-well plate. On days 0–3, plates were stained with 0.5% CV for 20 min, washed 3× with tap water, dried, and solubilized with methanol. Optical density at 570 nm was measured with a plate reader.
EdU assay
Melanoma cells were seeded at 100,000 cells/well in a 6-well plate overnight and incubated with 10 µM EdU for 2 h prior to staining with the Click-It Plus EdU Alexa Fluor-647 kit (C10634, Thermo Scientific, USA) and analysed with the Accuri C6 flow cytometer and FlowJo.
Adhesion assay
Melanoma cells were seeded at 100,000 cells/well in a 6-well plate containing a confluent monolayer of brECs or MEFs. Cells were washed with PBS to remove unadhered cells, fixed with 4% paraformaldehyde, and DAPI stained. Images were taken using the Nikon Eclipse Ti-E inverted microscope/NIS-Elements software and adhered cells were counted in ImageJ.
Immunoblotting
Protein extraction and western blotting were carried out using standard methods. Primary antibodies included GAPDH (1:2500, #5174, Cell Signaling, USA), pAKT (1:1000, #4060, Cell Signaling), AKT (1:1000, #4691, Cell Signaling), PTEN (1:1000, #9188, Cell Signaling), TGFβR2 (1:500, #79424, Cell Signaling). Secondary antibody: HRP-conjugated peroxidase goat anti-rabbit IgG (1:10,000, #PI-1000, Vector Laboratories, USA).
Luminex
To detect protein levels of TGFβ, supernatant was collected from mono-cultured and co-cultured cells, concentrated 50X using Microsep Advance Centrifugal Filters (MCP010C41, Pall Laboratory, USA), and run on a Luminex panel containing active TGFβ1-3 (Millipore, USA).
Flow cytometry
Cells were fixed using 4% paraformaldehyde for 15 min at room temperature (RT) and permeabilized with 90% methanol on ice for 10 min. Primary antibody for pAKT (1:200, #4060, Cell Signaling) was incubated for 1 h at RT, followed by goat anti-rabbit IgG Alexa Fluor-488 secondary antibody (1:100, #A11034, Invitrogen, USA) for 30 min at RT. Cells were analysed using the Accuri C6 flow cytometer and FlowJo.
Real-time quantitative PCR
RNA extraction was performed using a Quick-RNA Microprep or Miniprep Kit (Zymo Research R1050 or R1055, Zymo Research, USA). cDNA was synthesised using iScript cDNA Synthesis Kit (1708891EDU, Bio-Rad, USA), and qPCR reactions were completed using QuantStudio 12K Flex Real-time PCR System. Primers used included: Tgfβ1 (forward: 5’-GATACGCCTGAGTGGCTGTCTT-3’; reverse: 5’-GCCCTGTATTCCGTCTCCTTG-3’); Tgfβ2 (forward: 5’-GACCTTCTCGTCTCGTCCCT-3’, reverse: 5’-TGGAGTTCAGTGTGTCAGGC-3’); Tgfβ3 (forward: 5’-GAGTGGCTGTTGAGGAGAGAGTC-3’, reverse: 5’-CATTGGGCTGAAAGGTGTGAC-3’); Vegfa (forward: 5’-GCAGCGACAAGGCAGACTAT-3’, reverse: 5’-AAATCCCAGAGCACAGACTCC-3’); Vegfb (forward: 5’-CCCAGAGCTGCCATCTAACAA-3’, reverse: 5’-GAGACAGCCAGCCAGAAGATG-3’); Fgf2 (forward: 5’-CGTCAAACTACAACTCCAAGCAG-3’, reverse: 5’-CAGCCGTCCATCTTCCTTCA-3’); Plgf (forward: 5’-CGACTCGTCCCTGCTGAATG-3’, reverse: 5’-ATCCGTGGCTGGCTTCTTTC-3’); Pdgfb (forward: 5’-TCCTTCCTCTCTGCTGCTACCT-3’, reverse: 5’-GCTCAGCATTTCATACAGTTCCTC-3’).
Bulk RNA sequencing and analysis
RNA was harvested with Quick-RNA Microprep (Zymo Research R1050) and sent to Novogene for bulk RNA sequencing and analysis. In brief, 1 µg RNA/sample was used as input material, library was prepped using NEB Next Ultra RNA Library prep for Illumina (NEB, USA), and purified PCR products (AMPure XP system) were assessed for quality on the Agilent Bioanlyzer 2100 system. Index-coded samples were clustered on a cBot Cluster Generation System using PE Cluster Kit cBot-HS (Illumina, USA). Raw reads were processed through fastp and clean reads were obtained by removing adapter, poly-N sequences, and reads with low quality. All downstream analyses were based on clean data with high quality.
Paired-end clean reads were aligned to the reference genome (NCBI/UCSC/Ensembl) using the Spliced Transcripts Alignment to a Reference (STAR v2.5) software. STAR used the method of Maximal Mappable Prefix (MMP) to generate a precise mapping result for junction reads. For principal component analysis, samples were clustered using expression level FPKM to see the correlation using hierarchical clustering distance method with the function of heatmap, SOM (Self-organisation mapping) and kmeans using silhouette coefficient to adapt the optimal classification with default parameter in R. Differential expression analysis was performed using the DESeq2 R package (2_1.6.3). The resulting P values were adjusted using Benjamini and Hochberg’s approach for controlling the false discovery rate (FDR). Genes with an adjusted P value < 0.05 found by DESeq2 were assigned as differentially expressed. For KEGG enrichment analysis, R package clusterProfiler was used to test the statistical enrichment of differential expression genes in KEGG pathways (http://www.genome.jp/kegg/). Gene length bias was corrected and adjusted P values < 0.05 were considered significant enrichment.
Additional analysis was performed by the UVA Bioinformatics Core. In brief, in-house developed programmes were used for adaptor identification, and any contamination of adaptor sequence was removed with cutadapt (https://cutadapt.readthedocs.io/en/stable/). Reads were mapped to the genome with STAR. DESeq2 was used to perform differential gene expression and low-expression genes (few replicates/low counts) were excluded. Differentially expressed genes were ranked based on log2fold change and FDR-corrected P values. The ranked file was used to perform pathway analysis using GSEA software. The enriched pathways were selected based on enrichment scores as well as normalised enrichment scores.
Animal studies
All experiments were performed in accordance with the University of Virginia guidelines for animal handling and care. Cdh5-CreERT2 mice were generated by Ralf Adams (Max Planck Institute for Molecular Biomedicine). Ai6 ZsGreen (#007906) and Pdgfrb-P2A-CreERT2 (#030201) were purchased from The Jackson Laboratory. Cdh5-CreERT2 and Ai6 ZsGreen mice were crossed to generate Cdh5-CreERT2; ZSGreenl/s/l mice (herein called ECiZSGreen). Pdgfrb-P2A-CreERT2 and Ai6 ZsGreen mice were crossed to generate Pdgfrb-P2A-CreERT2; ZSGreenl/s/l mice (herein called PDGFRβiZSGreen). All mice were on a C57BL/6 background. To reduce confounding factors, mice were fed a standard diet and co-housed with mice of the same cohort. Littermates were used when possible and males and females were randomised in equal proportions between control and experimental groups. Intraperitoneal injection of 75 mg/kg tamoxifen was performed three times over the course of 7 days in mice ages 4–10 weeks to induce expression of the Cre-controlled ZSGreen reporter; experiments were performed at least one week after induction. Stereotaxic intracranial injection or organotypic brain slice culture experiments were carried out with mice 7–16 weeks old. Studies were not blinded and animals were excluded from quantitative analysis when no bulk tumour was detected. A power analysis using preliminary data of bulk tumour size of PTEN-null vs. PTEN-RE mice was used to determine an estimated sample size of n = 6 mice for intracranial experiments (alpha probability value = 0.05 and power value = 0.8).
Stereotaxic intracranial injection
Using stereotaxic coordinates, 5000 mCherry-labelled melanoma cells/3 µL of HBSS was injected intracranially into the right striatum of induced ECiZSGreen or PDGFRβiZSGreen mice. For Empty vs. PTEN-RE experiment, 25,000 cells/3 µL of HBSS was used to allow for sufficient colonisation. Mice were euthanized and brains were harvested two weeks post-injection. Brain tissue was fixed in 4% paraformaldehyde overnight, followed by soaking in 30% sucrose for 48 h at 4 °C prior to embedding in O.C.T compound (Fisher Scientific, USA). A Leica microtome was used to cut 40 µm sections, which were mounted with Vectashield (H-1700-10, Vector Labs, USA). Images were taken using the Nikon Eclipse Ti-E inverted microscope/NIS-Elements software. Image analysis was performed using ImageJ; each data point represents the mean of n = 3 sections from one mouse. An area ROI was drawn around the mCherry+ bulk tumour, the number of PVTCs (colocalized mCherry and ZSGreen clusters outside the area ROI), and the farthest distance of a PVTC from the edge of the area ROI (invasive front) was measured. For analysis between two groups, the Mann–Whitney t-test was used because not all groups passed a normal distribution assumption. For the analysis of three groups, one-way ANOVA with Tukey’s multiple comparisons test was used for bulk tumour size and no. of PVTCs, which had a normal distribution for all groups, whereas the Kruskal–Wallis test was used for distance measurements as these groups did not have a normal distribution.
3D confocal images
Brain tissues from intracranially-injected mice were harvested as described above and sectioned at 100 µm. Floating brain sections were washed in PBST (PBS + 0.2% Triton X-100) and incubated in blocking solution (PBS + 0.3% Triton X-100 + 5% normal goat serum + 5% BSA) for 1 h with shaking at RT. Sections were incubated with CD31 (1:100, BD 550274) or PDGFRβ (1:100, #14-1402-81, eBioscience, USA) primary antibodies with orbital shaking overnight at 4 °C, washed with PBST, and then incubated with goat anti-rat IgG Alexa Fluor-647 (1:500, #A21247, Invitrogen) for 1.5 h shaking at RT. Sections were washed with PBST and mounted with Vectashield, imaged with the ZEISS LSM 900 with Airyscan 2 microscope and 3D images were generated using Imaris software.
Organotypic brain slice culture
Brains from induced ECiZSGreen mice were dissected into 250 µm slices in HBSS using a tissue slicer (Stoelting). Brain slices were then cultured on Millicell cell culture inserts (PICM0RG50, Millipore, USA) in culture media for 1 h at 37 °C and 5% CO2 prior to seeding of mCherry+ melanoma cells (5000 cells/2 µL HBSS). Melanoma cells were incubated on brain slices for 48 h before time-lapse imaging was taken using the ZEISS LSM 900 with Airyscan 2 microscope. Confocal z-stack images were taken over a 3 h period with 20 min intervals. Velocities were measured for individual migratory perivascular melanoma cells using the ImageJ cell tracker plugin.
Statistics
All statistical analyses were performed using GraphPad Prism software, and P values less than 0.05 were considered significant. Quantitative data represents the mean ± SD from at least n = 3 or more samples per data point. For analysis between two groups, the Mann–Whitney t-test was used because not all groups passed a normal distribution assumption. For the analysis of three groups, one-way ANOVA was used if the sample had a normal distribution, whereas the Kruskal–Wallis test was used for when there was not a normal distribution. Additional experimental details (number of animals or experimental replicates) are provided in the figure legends.
Results
Melanoma cells deficient in PTEN invade along blood vessels in the brain microenvironment
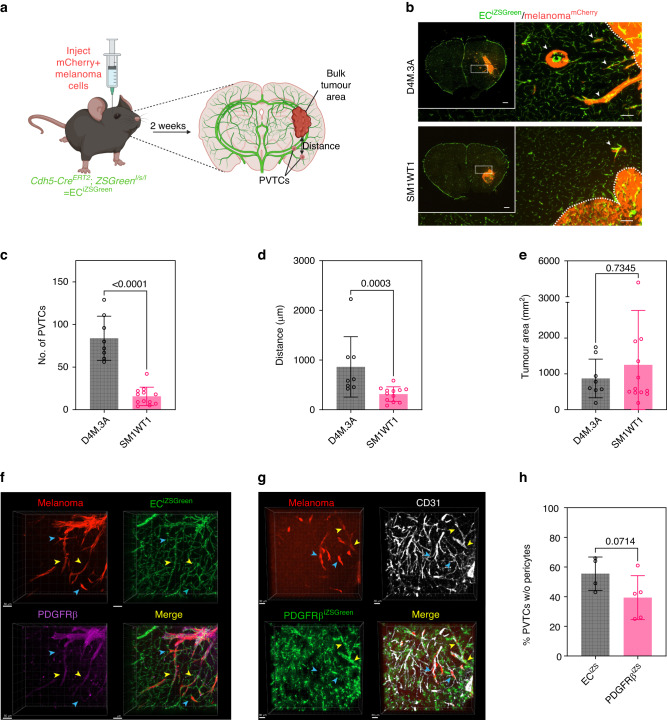
It was previously shown that angiotropism/perivascular invasion is a common feature in patients with MBM; however, the mechanisms that guide melanoma cells motility along the surface of the vasculature are not well-understood [12–15, 22]. Thus, to model the behaviour of melanoma cells in the brain post-colonisation, we used intracranial injections of two murine melanoma cell lines derived from genetically engineered mice differing in their Pten status (BRafCA,PtenloxP,Tyr::CreERT2 = D4M.3A and BRafCA,Tyr::CreERT2 = SM1WT1 [33, 36]). For these studies, we used mice where the endothelium is genetically labelled with ZSGreen by crossing Cdh5Cre mice with Ai6 reporter mice, whereas melanoma cells were labelled with mCherry [37–39]. The results showed that while both melanoma lines could readily colonise the brain, there was a striking difference in perivascular invasion; namely, D4M.3A cells showed a 5-fold increase in perivascular tumour clusters (PVTCs) compared to SM1WT1. Furthermore, these PVTCs travelled approximately 3-fold further when measured at a distance from the invasive front, whereas the bulk tumour area was not significantly different (Fig. 1a–e). To more closely examine the position of these PVTCs, we used 3D confocal imaging of 100 µm sections of D4M.3A intracranially injected brains with PDGFRβ staining to mark pericytes (Fig. 1f). Using this model, we found that approximately 50% of PVTCs directly contacted the vasculature without PDGFRβ+ pericytes being present. However, given that D4M.3A melanoma cells express their own PDGFRβ, we additionally quantified PVTCs with displaced pericytes using D4M.3A cells injected into genetically inducible PDGFRβiZSGreen mice and stained for CD31 to label the vasculature (Fig. 1g). In this case, we found approximately 40% of PVTCs without pericytes being present (Fig. 1h). Thus, these complementary models demonstrated that melanoma cells can undergo pericyte-like spreading and suggest that murine melanoma cell lines, differing in PTEN status, can be used as a model to further dissect the molecular mechanisms that mediate perivascular invasion in the brain in the post-colonisation phase of brain metastasis.
Fig. 1. Melanoma cells deficient in PTEN invade along blood vessels in the brain microenvironment.
a Schematic of intracranial injection experiments. b Representative images of brains from ECiZSGreen mice (vasculature labelled green) injected intracranially with mCherry+ D4M.3A cells or mCherry+ SM1WT1 cells. Scale bar = 100 µm and insert scale bar = 500 µm. Arrows point to perivascular tumour clusters (PVTCs). Quantification of c number of PVTCs, d max distance of PVTCs from the invasive front, and e bulk tumour area; D4M.3A, n = 8 mice and SM1WT1, n = 12 mice. P values were determined by the Mann–Whitney t-test. f 3D confocal imaging of a 100 µm brain section of mCherry+ D4M.3A cells intracranially injected into an ECiZSGreen mouse with PDGFRβ antibody staining; scale bar = 50 µm. g 3D confocal imaging of a 100 µm brain section of mCherry+ D4M.3A cells intracranially injected into a PDGFRβiZSGreen mouse with CD31 antibody staining; scale bar = 30 µm. Yellow arrows indicate PVTCs with interdigitating pericytes, whereas blue arrows indicate PVTCs that have displaced pericytes in (f) and (g). h Quantification of the percentage of PVTCs without pericytes found in either the ECiZSGreen or PDGFRβiZSGreen mouse models; ECiZSGreen, n = 4 mice and PDGFRβiZSGreen, n = 5 mice. P values were determined by the Mann–Whitney t-test.
Perivascular invasive melanoma upregulates transcriptional programmes important for adhesion, migration, and survival
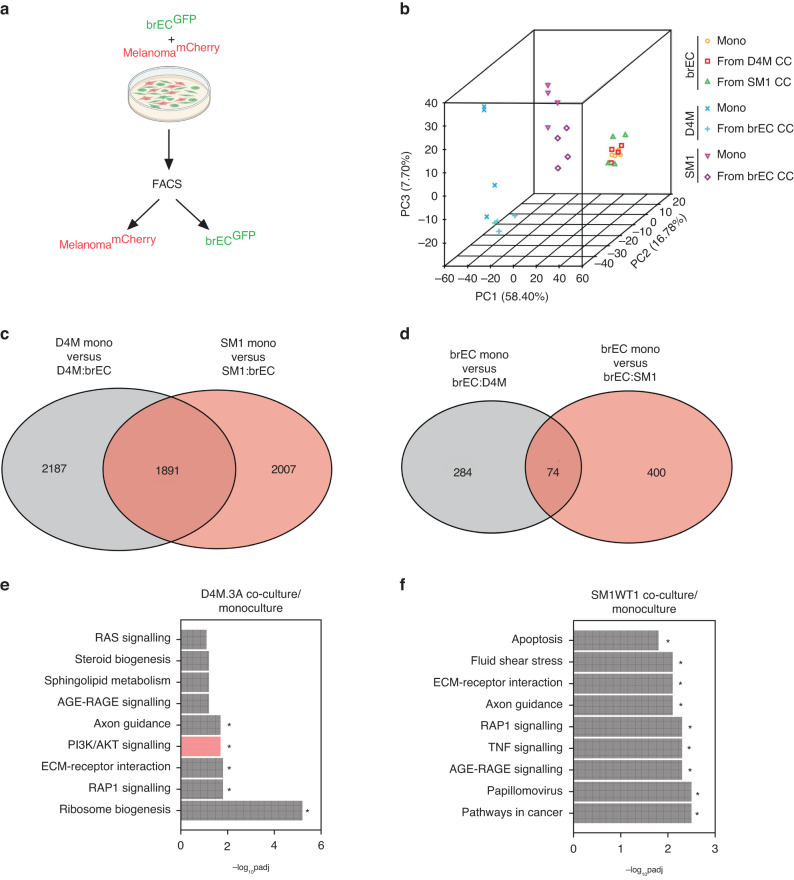
Next, we sought to identify pathways that might contribute to the perivascular invasive phenotype by these different melanoma cells. We co-cultured D4M.3A or SM1WT1 cells with murine brECs and used FACS to re-isolate melanoma cells from brECs (Fig. 2a). After bulk RNA sequencing, we found that all three cell lines clustered away from one another by principal component analysis (PCA) suggesting they have distinct transcriptomic identities (and as an indicator of cell purity after FACS). In addition, brECs before and after co-culture were almost indistinguishable from one another, whereas there was a clear separation between the melanoma mono-culture versus co-culture clusters (Fig. 2b). These data suggest that melanoma cells, after contacting brECs, undergo large-scale changes in gene expression, whereas EC gene expression is relatively stable. This was further supported by examining the number of differentially expressed genes found upon co-culture; melanoma cells differentially expressed 10× more genes than brECs after co-culture (~4000 vs 400). In addition, while 1891 differentially expressed genes were shared agnostic to melanoma cell line identity, there were an additional 2187 genes that were only differentially expressed in D4M.3A and 2007 genes differentially expressed only in SM1WT1 after co-culture with brECs. For brECs, 74 genes were differentially expressed regardless of melanoma identity, whereas 284 genes were changed only after D4M.3A co-culture and 400 genes after SM1WT1 co-culture (Fig. 2c, d). These data suggest there are both common and distinct features found between PTEN-null and PTEN-expressing melanoma cells upon contact with the brain endothelium.
Fig. 2. Perivascular invasive melanoma upregulates transcriptional programmes important for adhesion, migration, and survival.
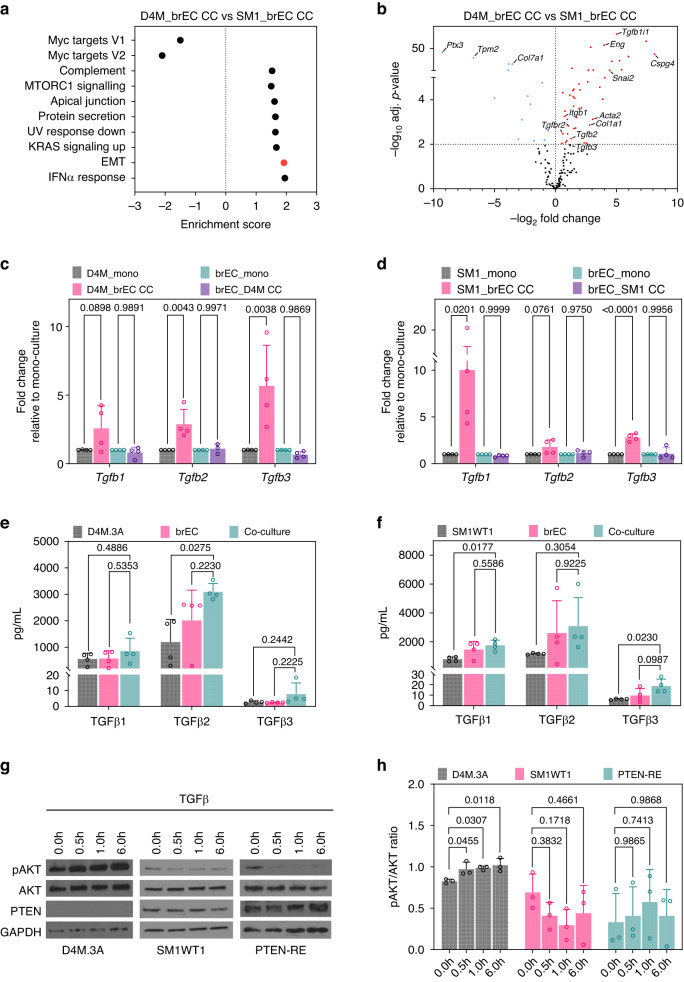
a Schematic of co-culture experiments. b PCA of bulk RNA sequencing in mono- vs. co-culture (CC) samples. c Venn diagram with the number of differentially expressed genes found between melanoma mono-cultures and co-cultures with brECs. d Venn diagram with the number of differentially expressed genes found between brEC mono-culture and co-cultures with melanoma. e KEGG enrichment comparing D4M.3A co-culture from mono-culture controls. f KEGG enrichment comparing SM1WT1 co-cultures from mono-culture controls. n = 4 samples per condition. An adjusted P value < 0.05 was considered significant.
Next, we used pathway enrichment analysis to carry out an unbiased screen of gene/pathway changes that occur in these different melanoma cells in contact with brECs. To examine potential functions of the differentially expressed genes, we examined KEGG enrichment scores and found pathways involved in cell adhesion/migration (axon guidance, ECM-receptor interaction, RAP1) to be commonly enriched in both PTEN-null and PTEN-intact melanoma cells upon co-culture. However, D4M.3A also upregulated pathways involved in high energetic states (PI3K/AKT signalling, ribosomal biogenesis), whereas SM1WT1 cells co-cultured with brECs were enriched for pathways indicative of cellular stress and inflammation (apoptosis, TNF signalling, AGE-RAGE signalling, fluid shear stress) (Fig. 2e, f). Taken together, these data suggest that melanoma cells of differing PTEN status differentially activate specific pathways once in contact with brECs. Of interest was the hyperactivation of PI3K/AKT, specific to D4M.3A, suggesting that brECs might produce factors that selectively activate this pathway.
PTEN re-expression reduces perivascular invasion along the brain vasculature in vivo
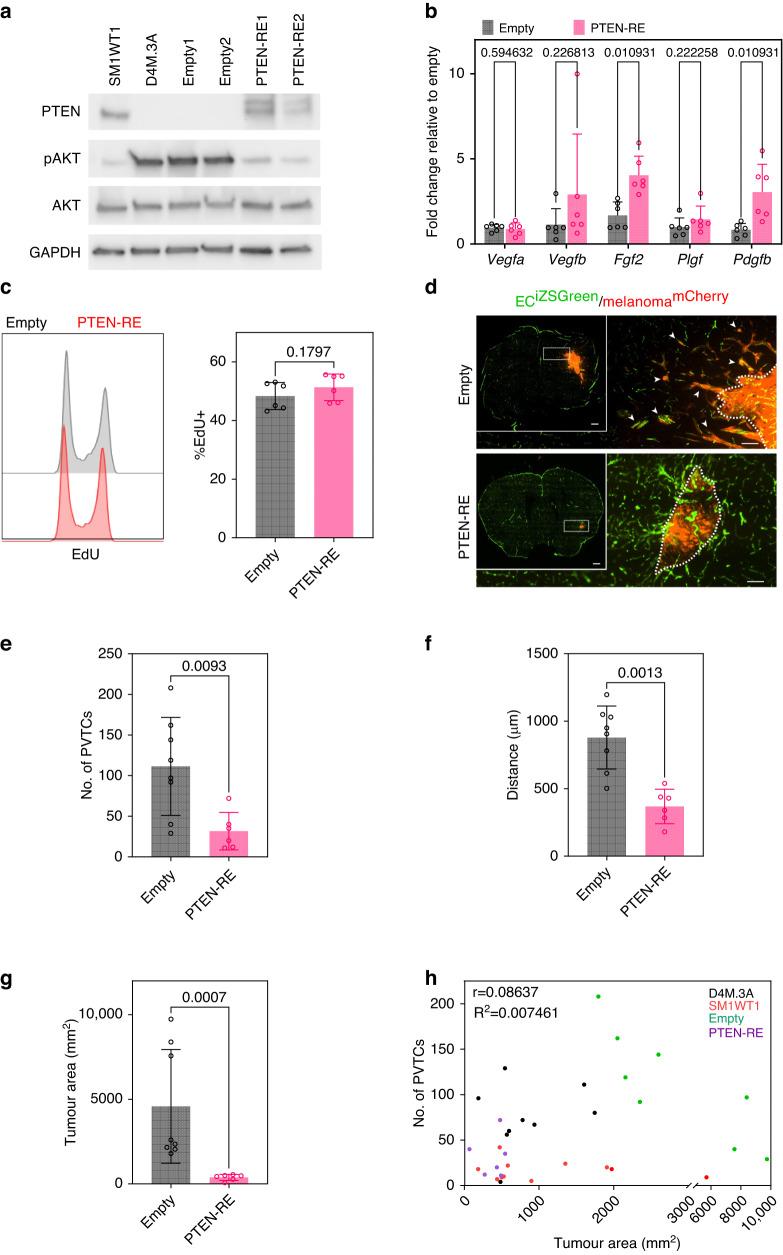
To further characterise the role of PTEN in perivascular survival and invasion while minimising intrinsic cell line variability, D4M.3A PTEN-RE (re-expression) cells were generated and compared to empty vector controls; as expected, PTEN-RE resulted in a reduction of pAKT in D4M.3A cells to levels comparable in PTEN-expressing SM1WT1 (Fig. 3a). Since vascular co-option and angiogenesis have been described as two alternative mechanisms of tumour vascularisation, we characterised the expression of several pro-angiogenic factors in our empty vector controls versus PTEN-RE melanoma cells [34]. We found that PTEN-RE cells expressed higher mRNAs for pro-angiogenic factors such as Fgf2 and Pdgfb, but not Vegfa/b or Plgf (Fig. 3b), suggesting that PTEN-RE cells may be predisposed to a more angiogenic phenotype. Furthermore, we characterised the rate of proliferation in vitro and found no significant differences when comparing empty vector and PTEN-RE lines (Fig. 3c). However, PTEN-RE lines demonstrated a significantly impaired ability to colonise the brain in vivo; indeed, no bulk tumours formed when 5000 cells were engrafted in the brain of 4/6 mice, suggesting that D4M.3A melanoma cells strongly depend on PTEN to establish in the brain microenvironment. To increase the likelihood of tumour establishment using PTEN-RE melanoma cells, the seeding density of injected cells was increased to 25,000. Here, we found that PTEN-RE reduced the number of PVTCs by 4-fold and reduced the distance travelled from the invasive front by 2.5-fold when compared to empty vector controls (Fig. 3d–f). These differences in the number of PVTCs and distance from the invasive front were similar to those found in D4M.3A versus SM1WT1 tumours. However, since PTEN-RE also had a 12-fold reduction in bulk tumour area (Fig. 3g) and the reduction in PVTCs observed in PTEN-RE mice could be a function of reduced overall colonisation, we carried out a Pearson’s correlation to compare overall tumour area versus number of PVTCs. The results showed a relatively weak association between tumour area and the ability to form PVTCs when multiple tumours were plotted, suggesting that the ability to form PVTCs is not solely a function of bulk tumour size (r = 0.08637) (Fig. 3h). Surprisingly, deleting PTEN in SM1WT1 cells did not promote perivascular invasion in vivo, suggesting that while reconstituting PTEN function in PTEN-null melanoma impairs perivascular invasion, loss of PTEN alone is not a sufficient driver of this mode of invasion in the brain (Supplementary Fig. S1). It is possible, therefore, that PTEN loss cooperates with additional factors expressed by D4M.3A cells to drive perivascular invasion in the brain.
Fig. 3. Pten re-expression reduces perivascular invasion along the brain vasculature in vivo.
a Protein expression of PTEN, pAKT, and AKT in SM1WT1, D4M.3A, D4M.3A Empty clones and D4M.3A PTEN-RE clones; n = 3. b mRNA expression of pro-angiogenic genes in Empty vs. PTEN-RE, n = 6. c In vitro proliferation of D4M.3A Empty and PTEN-RE clones by Click-It EdU, n = 6. d Representative images of brains from ECiZSGreen mice (vasculature labelled green) injected intracranially with mCherry+ Empty or mCherry+ PTEN-RE cells. Scale bar = 100 µm and insert scale bar = 500 µm. Arrows point to perivascular tumour clusters (PVTCs). Quantification of e number of PVTCs, f max distance of PVTCs from invasive front, and g bulk tumour area; Empty, n = 8 mice and PTEN-RE, n = 6 mice. h Pearson correlation of tumour area vs no. of PVTCs of D4M.3A, SM1WT1, Empty, and PTEN-RE injected mice; n = 34 mice in total. P values were determined using a Mann–Whitney t-test.
Pten re-expression reduces melanoma adhesion to brECs and inhibits migration on ex vivo brain slices
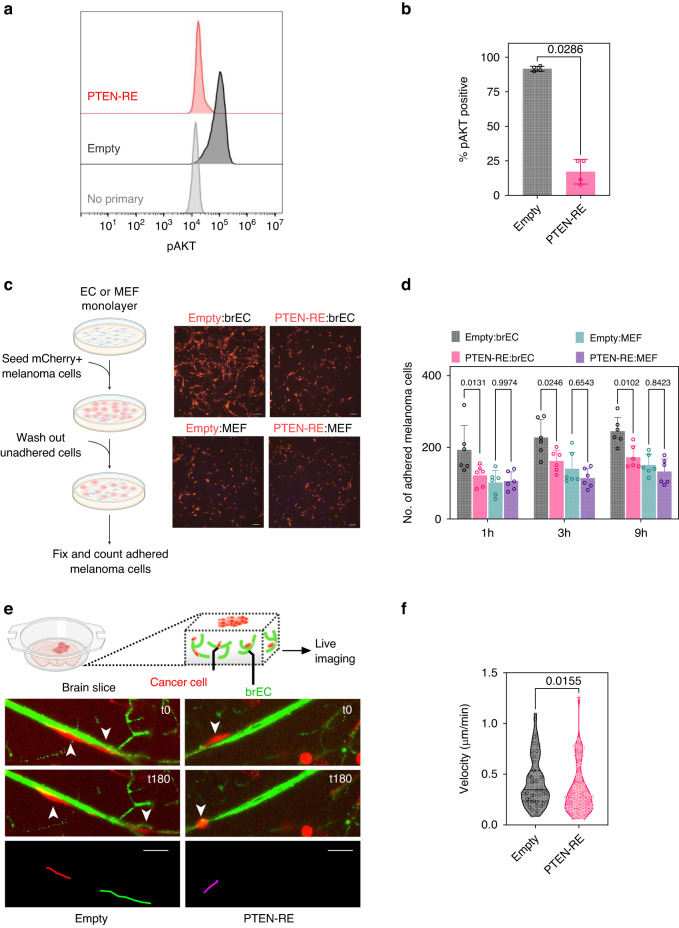
Next, we functionally characterised Empty versus PTEN-RE melanoma cells co-cultured with brECs in vitro. We found that, as expected, PTEN-RE cells have a significantly lower expression of pAKT measured using FACS (Fig. 4a, b). Interestingly, PTEN-RE cells also showed a 30% reduction in adhesion to brECs, suggesting a role for melanoma PTEN in mediating adhesion to the vasculature. Furthermore, since no difference in adhesion was observed between Empty and PTEN-RE cells plated on mouse embryonic fibroblasts (MEFs), this is likely a vascular-selective rather than global loss of adhesion (Fig. 4c, d). Since these were monolayers of ECs that do not resemble the 3D tube-like structures of brain vessels in vivo, we used an ex vivo brain slice culture model to carry out real-time tracking of individual melanoma cells along the vasculature. The results showed that PTEN-RE cells migrated 20% slower along the vasculature when compared to empty vector controls (Fig. 4e, f). Taken together, impaired initial adhesion to the vasculature and reduced migration upon PTEN-RE may account for the fewer numbers of PVTCs observed in our in vivo intracranial model of MBM perivascular invasion.
Fig. 4. Pten re-expression reduces melanoma adhesion to brECs and inhibits migration on ex vivo brain slices.
a Representative histogram of pAKT intensity as measured by flow cytometry in D4M.3A Empty vector or PTEN-RE melanoma cells after co-culture with brECs. No primary refers to the no primary antibody control. b Quantification of protein levels from flow analysis of pAKT in Empty or PTEN-RE cells co-cultured with brECs; n = 4. c Experimental schematic and representative image of mCherry+ Empty or mCherry+ PTEN-RE cells adhered to a monolayer of either brECs or MEFs. Scale bar = 100 µm. d Quantification of number of melanoma cells adhered to brECs or MEFs; n = 6. e Experimental schematic and representative live-imaging migration path of mCherry+ Empty or mCherry+ PTEN-RE cells on an ex vivo brain slice from ECiZSGreen mice (vasculature labelled green). Scale bar = 50 µm. f Quantification of average velocity of individual cells in a 20-min interval over a 180-min timeframe; Empty, n = 79 cells and PTEN-RE, n = 60 cells. P values were determined by Mann–Whitney t-test for (b) and (f) and two-way ANOVA with Tukey’s multiple comparisons for (d).
TGFβ is upregulated in brEC:melanoma co-cultures
To identify potential vascular-induced factors that may contribute to pAKT signalling when melanoma cells are juxtaposed to the brain vasculature, we returned to our bulk RNA sequencing data using melanoma cells co-cultured with brECs. Using Gene Set Enrichment Scores (GSEA) from D4M.3A cells co-cultured with brECs versus SM1WT1 cells co-cultured with brECs, we found an enrichment of several pathways including complement, MTORC1 signalling, apical junction, protein secretion, UV response down, KRAS signalling up, epithelial-mesenchymal transition (EMT), and interferon (IFNα) response (Fig. 5a). Since EMT is a well-known transcriptional programme that is typically activated in highly motile cell types, we next examined candidate EMT gene expression under these same two conditions and found that D4M.3A cells, when co-cultured with brECs, showed elevated expression of several TGFβ pathway effectors, such as Eng and Tgfb1i1, as well as enrichment of TGFβ-regulated genes including Acta2, Itgb1, and Col1a1 when compared to SM1WT1 (Fig. 5b).
Fig. 5. TGFβ is upregulated in brEC:melanoma co-cultures.
a GSEA pathway analysis of D4M.3A co-cultured cells compared to SM1WT1 co-cultured cells; n = 4. b Volcano plot of selected EMT genes comparing D4M.3A co-cultured cells to SM1WT1 co-cultured cells; n = 4. TGFβ transcript levels of c D4M.3A:brECs co-cultures and d SM1WT1:brEC co-cultures; n = 4. TGFβ protein levels detected by Luminex in the supernatant of e D4M.3A:brECs co-cultures or f SM1WT1:brECs co-cultures; n = 4. g Western blot of D4M.3A, SM1WT1, or PTEN-RE cells treated with a time course of TGFβ and h quantification of pAKT/AKT ratio; n = 3. P values were determined by one-way ANOVA with Tukey’s multiple comparisons for each isoform in (c)–(f) and one-way ANOVA with Dunnett’s multiple comparisons test in (h).
Of particular interest was the expression of Tgfb ligands since Tgfb is a well-known driver of EMT [40]. Thus, we used qPCR to validate the bulk RNA sequencing data focusing on Tgfb ligands. We found an approximately 5-fold increase in Tgfb2 and Tgfb3 transcripts in D4M.3A cells upon contact with the vasculature, whereas Tgfb1 and Tgfb3 were elevated in SM1WT1 (Fig. 5c, d). At the protein level, there was an approximately three-fold increase in TGFβ2 protein in the supernatant of co-cultured D4M.3A cells and a 2- to 3-fold increase in TGFβ1 and TGFβ3 protein concentrations of co-cultured SM1WT1 cells compared to mono-culture controls (Fig. 5e, f). The upregulation of Tgfb appears to be vascular-selective because it was similarly found upon co-culture with an additional EC line (MDEC) but not upon co-culture with MEFs (Supplementary Fig. S2a, b). In addition, Tgfb transcripts were not upregulated upon exposure to brEC conditioned media (CM) or brEC extracellular matrix (ECM); these data indicate that upregulation of Tgfb in melanoma cells is predominantly mediated by direct contact with ECs, though not necessarily specific to brEC (Supplementary Fig. S2c, d).
Since previous studies have shown that TGFβ can phosphorylate AKT in divergent cell types and promote protein synthesis, survival, and invasion [40–43], we treated melanoma cells of differing PTEN status with TGFβ and assayed pAKT by western blotting. The results showed that only melanoma cells lacking PTEN were able to upregulate pAKT in response to TGFβ and this effect was diminished by PTEN-RE (Fig. 5g, h). Taken together, these data suggest that TGFβ is induced in the brain tumour perivascular niche, which can then lead to activation of pAKT in PTEN-null melanoma cells.
TGFβ signalling contributes to brain colonisation and perivascular invasion
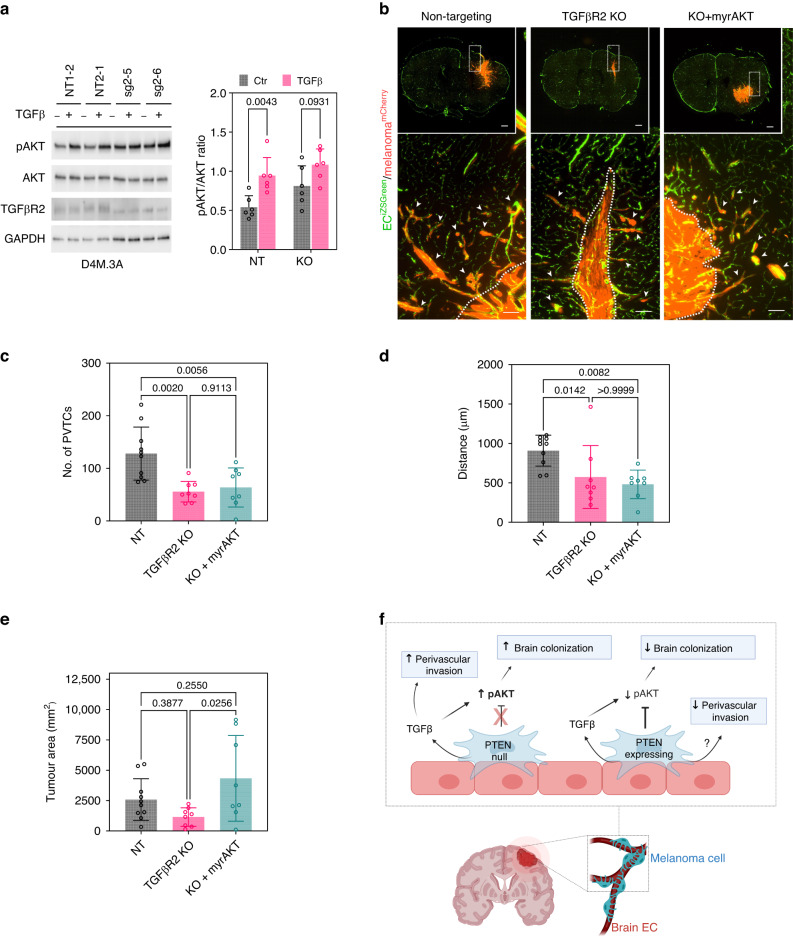
Finally, to determine whether TGFβ-dependent pAKT signalling is a driver of brain colonisation and perivascular invasion, we deleted TGFβR2 (TGFβR2 KO) in PTEN-null melanoma cells. TGFβ signalling requires heterodimerization with TGFβR1 to propagate intracellular signalling; thus, deleting TGFβR2 results in a completely disabled TGFβ signalling cascade downstream of all three TGFβ isoforms [40]. Although the stimulation of pAKT was modest since D4M.3A cells lack PTEN and therefore have high constitutive pAKT levels, after deleting the receptor, TGFβR2 KO cells no longer upregulated pAKT when challenged with TGFβ (Fig. 6a). Using the intracranial model, we found an approximate two-fold reduction in the numbers of PVTCs and distance travelled from the invasive front in TGFβR2 KO cells compared to NT cells. To test whether this TGFβ-dependent loss of perivascular invasion was mediated through AKT, we introduced myrAKT to allow for constitutively active AKT in TGFβR2 KO cells. Interestingly, myrAKT did not restore the number of PVTCs or distance travelled from the invasive front. However, the addition of myrAKT rescued the overall brain colonisation of TGFβR2 knockout cells to levels comparable to NT controls (Fig. 6b–e). Taken together, these data suggest that in PTEN-null melanoma, a TGFβ-AKT axis promotes melanoma brain colonisation, while AKT-independent mechanisms involving perivascular niche TGFβ signalling contribute to perivascular invasion (Fig. 6f).
Fig. 6. TGFβ signalling contributes to brain colonisation and perivascular invasion.
a Protein levels of pAKT upon treatment with TGFβ in NT vs TGFβR2 KO cells with quantification of pAKT/AKT ratio; n = 3. b Representative images of brains from ECiZSGreen mice (vasculature labelled green) injected intracranially with mCherry+ NT cells, mCherry+ TGFβR2 KO cells, or mCherry+ TGFβR2 KO cells with myrAKT rescue. Scale bar = 100 µm and insert scale bar = 500 µm. Arrows point to perivascular tumour clusters (PVTCs). Quantification of c number of PVTCs, d max distance of PVTCs from invasive front, and e bulk tumour area; NT, n = 10 mice; TGFβR2 KO, n = 8 mice; and KO+myrAKT, n = 8 mice. f A model of brain colonisation and perivascular invasion in PTEN-null vs. PTEN-expressing melanoma cells in contact with brECs. PTEN loss cooperates with perivascular TGFβ to mediate pAKT-dependent colonisation, whereas PTEN and TGFβ mediate perivascular invasion through AKT-independent mechanisms. P values were determined by Mann–Whitney for (a), one-way ANOVA with Tukey’s multiple comparison test for (c) and (e), and a Kruskal–Wallis test for (d).
Discussion
Cancer cell retention within a perivascular niche post-extravasation was previously described as an important requirement for metastatic colonisation [12, 18, 20]. This may be of particular importance for MBM, which are commonly found as either solitary cells or clusters of cells juxtaposed to the vasculature in human samples [12–15]; these perivascular melanoma cells are typically highly motile and invasive [15, 18, 44–46]. However, only a few molecular mechanisms that control perivascular invasion in MBM are known [22]. In our work, we have used gain/loss of function studies in a murine intracranial model to investigate how vascular-derived brain microenvironment factors may cooperate with tumour genetic alterations in established brain metastases. Herein, we have found that ectopic PTEN expression in PTEN-null melanoma robustly reduces melanoma brain colonisation and perivascular invasion. In addition, PTEN-null melanoma cells are enriched in the activation of survival pathways compared to PTEN-expressing counterparts when co-cultured with brECs; furthermore, PTEN-RE cells showed a vascular-selective loss of adhesion and migration along brECs. These data suggest that PTEN expression in melanoma cells impacts how they interact with and invade upon ECs in the brain perivascular niche.
PTEN is a commonly lost tumour suppressor gene in many cancers; notably, PTEN loss is associated with MBM, resistance to therapeutics, and worse overall survival [29, 32, 47]. In addition to genomic alterations, PTEN expression can be downregulated by epigenetic and post-transcriptional modifiers (e.g., methylation or miRNAs). For example, in the brain it was previously found that astrocytes secrete exosomal miRNA to induce PTEN loss [24]. While our studies demonstrate that loss of PTEN alone is probably not sufficient to promote perivascular invasion, the importance of PTEN loss in MBM warranted investigating how PTEN-null melanoma cells acquire additional features that enable them to invade along the vasculature. Given that previous studies in melanoma have shown that PTEN silencing and AKT activation have distinct outcomes that can synergise to promote MBM [28], we were interested in vascular-mediated factors that might activate AKT in the context of PTEN loss. TGFβ was a good candidate given its ability to induce EMT and upregulate AKT-mTOR signalling to promote increased cell size and invasion [41, 48]. In our studies, we found that melanoma:EC contact promotes upregulation of TGFβ regardless of PTEN status; however, there was a differential response to TGFβ-induced activation of pAKT depending on PTEN status in MBM.
PTEN-null melanoma upregulated pAKT in the presence of TGFβ, and disabling melanoma response to TGFβ by knocking out TGFβR2 reduced brain colonisation and perivascular invasion. However, we found that myrAKT primarily contributed to tumour size and not numbers of or migration of PVTCs. This was surprising given recent reports describing the importance of AKT activation in co-opting the vasculature in early MBM [49]; however, later stages of perivascular invasion may utilise different mechanisms and suggest the possibility of AKT-independent mechanisms that can be explored. For example, one possible area of investigation includes the AKT-independent activation of mTOR [50]. In addition, while it is postulated that PTEN functions in melanogenesis through its lipid phosphatase activities and inhibition of PI3K signalling, PTEN may also activate important adhesion components such as focal adhesion kinase (FAK) through its protein phosphatase activity [51–53]. Additional factors that could function in this context could be small GTPase proteins such as RHO, RAC, and CDC42, which have also been implicated in glioblastoma invasion, and can cross-talk with PI3K and TGFβ [40, 54, 55]. Finally, serpins that can inhibit plasminogen activator promote survival and vascular co-option in brain metastases [23]. While serpine1 (PAI-1) was not one of the upregulated isoforms described in the breast and lung cancer brain metastases models by Valiente et al., this serpin can selectively inhibit plasminogen activator and is a well-known downstream factor of TGFβ that may also be modified by PTEN [39, 56], thus making it another intriguing mechanism for further investigation.
One technical limitation of our studies was that TGFβR2 may not be entirely deleted in the melanoma lines we generated resulting in a lowly expressed truncated protein with residual function; thus, it is possible that we did not eliminate the complete response to TGFβ signalling in melanoma cells. Another limitation of our studies includes the possibility that the mechanism(s) investigated in this context are not specific to the brain microenvironment; moreover, our intracranial model does not allow for the study of early metastatic processes prior to extravasation into the brain microenvironment. In addition, we found that PTEN-null melanoma also upregulates TGFβ upon contact with dermal ECs, suggesting there may be high levels of TGFβ within the perivascular niche of primary tumours found in the dermis, where vascular co-option may also be present. While TGFβ-mediated vascular co-option in primary melanoma has yet to be explored, there are studies that have demonstrated the importance of TGFβ signalling during vascular co-option at extracranial sites; in particular, TGFβ1-RUNX1 signalling mediates vascular co-option in colorectal cancer liver metastases. In this context, cancer cells with vascular co-option potential expressed high levels of TGFβR2 and RUNX1 and were able to induce upregulation of TGFβ1 secretion by hepatocytes [57, 58]. Thus, TGFβ can contribute to vascular co-option in various cancers, although how it participates during cellular cross-talk between cancer cells and the vasculature may vary based on additional microenvironmental cues.
In summary, we have described a new role for PTEN and TGFβ in perivascular invasion by MBM, though further studies are needed to dissect how these complex signalling networks ultimately drive adhesion to and movement along the vasculature. For example, despite lack of expression of L1CAM, one of the most well-studied adhesion molecules in vessel co-option of various cancers [20], D4M.3A cells are still highly proficient at perivascular invasion suggesting that alternative pathways to co-opt the vasculature exist (unpublished observation). Through a better understanding of how melanoma:EC interactions drive perivascular spreading in the brain, one could uncover new treatment modalities that prevent MBM progression and recurrence.
Supplementary information
Acknowledgements
We would like to thank the UVA Flow Cytometry Core, particularly Alex Wendling and Taylor Harper, for their assistance with Luminex and FACS experiments. The UVA Bioinformatics Core, including Pankaj Kumar, assisted with bulk RNA sequencing analysis. The UVA Biospecimen and Tissue Research Facility provided use of equipment for cryosections (Leica microtome). We would also like to acknowledge Drs Kimberly Stegmaier and Brian Crompton, who provided the CRISPR plasmids and protocols. Sonia Patel and Yuvraj Sethi assisted with genotyping mice. BioRender was used to design all schematics.
Author contributions
SW and ACD conceptualised the study and wrote the manuscript. SW carried out the experiments. CPR and YZ assisted with in vivo and in vitro functional studies. SA assisted with additional experiments.
Funding
SW is supported by the National Institute of Health/National Cancer Institute (NIH/NCI) Ruth L. Kirschstein NRSA for Individual Predoctoral Fellows Award (5F30CA268842-02) and previously received support from the NIH/NCI T32 CA009109 and NIH/NIGMS T32 GM007267 training grants, a trainee award from the UVA Comprehensive Cancer Center, and a medical student research award from the Melanoma Research Foundation. CPR is supported by the NIH/NCI training grant T32 CA009109. ACD is supported by grants from the National Institutes of Health/National Cancer Institute (2RO1 CA177875 and RO1 CA2558451), the Melanoma Research Alliance (ID612638), and funds from the Emily Couric Cancer Center at the University of Virginia. Portions of this research were supported by the NCI Cancer Center Support Grant 5P30CA044579 and by the UVA Genome Analysis and Technology Core (RRID:SCR_018883). Additional support was provided by the University of Virginia Flow Cytometry Core (RRID:SCR_018883). While the authors were supported by the NIH, the content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Data availability
Bioinformatics data will be made available immediately upon publication. Data can be accessed from the NCBI GEO database under accession code GSE247232 or the NCBI SRA database under the accession code PRJNA1032415.
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
This project is considered non-human studies research and no IRB review was needed. All animal studies were performed in accordance with the University of Virginia guidelines for animal handling and care.
Consent for publication
All authors have been provided with a copy of the manuscript prior to submission and consent to publication.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02530-5.
References
- 1.Becco P, Gallo S, Poletto S, Frascione MPM, Crotto L, Zaccagna A, et al. Melanoma brain metastases in the era of target therapies: an overview. Cancers (Basel) 2020;12:1640. doi: 10.3390/cancers12061640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutzmer R, Vordermark D, Hassel JC, Krex D, Wendl C, Schadendorf D, et al. Melanoma brain metastases – interdisciplinary management recommendations 2020. Cancer Treat Rev. 2020;89:102083. doi: 10.1016/j.ctrv.2020.102083. [DOI] [PubMed] [Google Scholar]
- 3.Berghoff AS, Preusser M. Targeted therapies for melanoma brain metastases. Curr Treat Options Neurol. 2017;19:13. doi: 10.1007/s11940-017-0449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies MA, Saiag P, Robert C, Grob JJ, Flaherty KT, Arance A, et al. Dabrafenib plus trametinib in patients with BRAF(V600)-mutant melanoma brain metastases (COMBI-MB): a multicentre, multicohort, open-label, phase 2 trial. Lancet Oncol. 2017;18:863–73. doi: 10.1016/S1470-2045(17)30429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tawbi HA, Forsyth PA, Hodi FS, Algazi AP, Hamid O, Lao CD, et al. Long-term outcomes of patients with active melanoma brain metastases treated with combination nivolumab plus ipilimumab (CheckMate 204): final results of an open-label, multicentre, phase 2 study. Lancet Oncol. 2021;22:1692–704. doi: 10.1016/S1470-2045(21)00545-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Franklin C, Mohr P, Bluhm L, Grimmelmann I, Gutzmer R, Meier F, et al. Impact of radiotherapy and sequencing of systemic therapy on survival outcomes in melanoma patients with previously untreated brain metastasis: a multicenter DeCOG study on 450 patients from the prospective skin cancer registry ADOREG. J Immunother Cancer. 2022;10:e004509. doi: 10.1136/jitc-2022-004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang P, Boehling NS, Koay EJ, Bucheit AD, Jakob JA, Settle SH, et al. Melanoma brain metastases harboring BRAF (V600K) or NRAS mutations are associated with an increased local failure rate following conventional therapy. J Neurooncol. 2018;137:67–75. doi: 10.1007/s11060-017-2695-2. [DOI] [PubMed] [Google Scholar]
- 8.Kavouridis VK, Harary M, Hulsbergen AFC, Lo YT, Reardon DA, Aizer AA, et al. Survival and prognostic factors in surgically treated brain metastases. J Neurooncol. 2019;143:359–67. doi: 10.1007/s11060-019-03171-6. [DOI] [PubMed] [Google Scholar]
- 9.Berger A, Bernstein K, Alzate JD, Mullen R, Silverman JS, Sulman EP, et al. Significant survival improvements for patients with melanoma brain metastases: can we reach cure in the current era? J Neurooncol. 2022;158:471–80. doi: 10.1007/s11060-022-04036-1. [DOI] [PubMed] [Google Scholar]
- 10.Bailey CM, Morrison JA, Kulesa PM. Melanoma revives an embryonic migration program to promote plasticity and invasion. Pigment Cell Melanoma Res. 2012;25:573–83. doi: 10.1111/j.1755-148X.2012.01025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lugassy C, Kleinman HK, Vermeulen PB, Barnhill RL. Angiotropism, pericytic mimicry and extravascular migratory metastasis: an embryogenesis-derived program of tumor spread. Angiogenesis. 2020;23:27–41. doi: 10.1007/s10456-019-09695-9. [DOI] [PubMed] [Google Scholar]
- 12.Carbonell WS, Ansorge O, Sibson N, Muschel R. The vascular basement membrane as “soil” in brain metastasis. PLoS One. 2009;4:e5857. doi: 10.1371/journal.pone.0005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodewald AK, Rushing EJ, Kirschenbaum D, Mangana J, Mittmann C, Moch H, et al. Eight autopsy cases of melanoma brain metastases showing angiotropism and pericytic mimicry. Implications for extravascular migratory metastasis. J Cutan Pathol. 2019;46:570–8. doi: 10.1111/cup.13465. [DOI] [PubMed] [Google Scholar]
- 14.Siam L, Bleckmann A, Chaung HN, Mohr A, Klemm F, Barrantes-Freer A, et al. The metastatic infiltration at the metastasis/brain parenchyma-interface is very heterogeneous and has a significant impact on survival in a prospective study. Oncotarget. 2015;6:29254–67. doi: 10.18632/oncotarget.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berghoff AS, Rajky O, Winkler F, Bartsch R, Furtner J, Hainfellner JA, et al. Invasion patterns in brain metastases of solid cancers. Neuro Oncol. 2013;15:1664–72. doi: 10.1093/neuonc/not112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnhill R, Dy K, Lugassy C. Angiotropism in cutaneous melanoma: a prognostic factor strongly predicting risk for metastasis. J Invest Dermatol. 2002;119:705–6. doi: 10.1046/j.1523-1747.2002.01871.x. [DOI] [PubMed] [Google Scholar]
- 17.Wilmott J, Haydu L, Bagot M, Zhang Y, Jakrot V, McCarthy S, et al. Angiotropism is an independent predictor of microscopic satellites in primary cutaneous melanoma. Histopathology. 2012;61:889–98. doi: 10.1111/j.1365-2559.2012.04279.x. [DOI] [PubMed] [Google Scholar]
- 18.Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16:116–22. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 19.Kuczynski EA, Vermeulen PB, Pezzella F, Kerbel RS, Reynolds AR. Vessel co-option in cancer. Nat Rev Clin Oncol. 2019;16:469–93. doi: 10.1038/s41571-019-0181-9. [DOI] [PubMed] [Google Scholar]
- 20.Er EE, Valiente M, Ganesh K, Zou Y, Agrawal S, Hu J, et al. Pericyte-like spreading by disseminated cancer cells activates YAP and MRTF for metastatic colonization. Nat Cell Biol. 2018;20:966–78. doi: 10.1038/s41556-018-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seano G. Targeting the perivascular niche in brain tumors. Curr Opin Oncol. 2018;30:54–60. doi: 10.1097/CCO.0000000000000417. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Wang S, Dudley AC. Models and molecular mechanisms of blood vessel co-option by cancer cells. Angiogenesis. 2020;23:17–25. doi: 10.1007/s10456-019-09684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH, Lee DJ, et al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell. 2014;156:1002–16. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature. 2015;527:100–4. doi: 10.1038/nature15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Dutton-Regester K, Brown KM, Hayward NK. The genomic landscape of cutaneous melanoma. Pigment Cell Melanoma Res. 2016;29:266–83. doi: 10.1111/pcmr.12459. [DOI] [PubMed] [Google Scholar]
- 26.Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. The Genetic Evolution of Melanoma from Precursor Lesions. N Engl J Med. 2015;373:1926–36. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 27.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–52. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho JH, Robinson JP, Arave RA, Burnett WJ, Kircher DA, Chen G, et al. AKT1 activation promotes development of melanoma metastases. Cell Rep. 2015;13:898–905. doi: 10.1016/j.celrep.2015.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bucheit AD, Chen G, Siroy A, Tetzlaff M, Broaddus R, Milton D, et al. Complete loss of PTEN protein expression correlates with shorter time to brain metastasis and survival in stage IIIB/C melanoma patients with BRAFV600 mutations. Clin Cancer Res. 2014;20:5527–36. doi: 10.1158/1078-0432.CCR-14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niessner H, Forschner A, Klumpp B, Honegger JB, Witte M, Bornemann A, et al. Targeting hyperactivation of the AKT survival pathway to overcome therapy resistance of melanoma brain metastases. Cancer Med. 2013;2:76–85. doi: 10.1002/cam4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies MA, Stemke-Hale K, Lin E, Tellez C, Deng W, Gopal YN, et al. Integrated molecular and clinical analysis of AKT activation in metastatic melanoma. Clin Cancer Res. 2009;15:7538–46. doi: 10.1158/1078-0432.CCR-09-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanaya N, Kitamura Y, Lopez Vazquez M, Franco A, Chen KS, van Schaik TA, et al. Gene-edited and -engineered stem cell platform drives immunotherapy for brain metastatic melanomas. Sci Transl Med. 2023;15:eade8732. doi: 10.1126/scitranslmed.ade8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins MH, Steinberg SM, Alexander MP, Fisher JL, Ernstoff MS, Turk MJ, et al. Multiple murine BRaf(V600E) melanoma cell lines with sensitivity to PLX4032. Pigment Cell Melanoma Res. 2014;27:495–501. doi: 10.1111/pcmr.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudley AC, Khan ZA, Shih SC, Kang SY, Zwaans BM, Bischoff J, et al. Calcification of multipotent prostate tumor endothelium. Cancer Cell. 2008;14:201–11. doi: 10.1016/j.ccr.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knight DA, Ngiow SF, Li M, Parmenter T, Mok S, Cass A, et al. Host immunity contributes to the anti-melanoma activity of BRAF inhibitors. J Clin Invest. 2013;123:1371–81. doi: 10.1172/JCI66236. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Kim DJ, Anandh S, Null JL, Przanowski P, Bhatnagar S, Kumar P, et al. Priming a vascular-selective cytokine response permits CD8(+) T-cell entry into tumors. Nat Commun. 2023;14:2122. doi: 10.1038/s41467-023-37807-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCann JV, Liu A, Musante L, Erdbrugger U, Lannigan J, Dudley AC. A miRNA signature in endothelial cell-derived extracellular vesicles in tumor-bearing mice. Sci Rep. 2019;9:16743. doi: 10.1038/s41598-019-52466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCann JV, Xiao L, Kim DJ, Khan OF, Kowalski PS, Anderson DG, et al. Endothelial miR-30c suppresses tumor growth via inhibition of TGF-beta-induced Serpine1. J Clin Invest. 2019;129:1654–70. doi: 10.1172/JCI123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamouille S, Derynck R. Emergence of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin axis in transforming growth factor-beta-induced epithelial-mesenchymal transition. Cells Tissues Organs. 2011;193:8–22. doi: 10.1159/000320172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–51. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horowitz JC, Lee DY, Waghray M, Keshamouni VG, Thomas PE, Zhang H, et al. Activation of the pro-survival phosphatidylinositol 3-kinase/AKT pathway by transforming growth factor-beta1 in mesenchymal cells is mediated by p38 MAPK-dependent induction of an autocrine growth factor. J Biol Chem. 2004;279:1359–67. doi: 10.1074/jbc.M306248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi JY, Shin I, Arteaga CL. Type I transforming growth factor beta receptor binds to and activates phosphatidylinositol 3-kinase. J Biol Chem. 2005;280:10870–6. doi: 10.1074/jbc.M413223200. [DOI] [PubMed] [Google Scholar]
- 44.Bentolila NY, Barnhill RL, Lugassy C, Bentolila LA. Intravital imaging of human melanoma cells in the mouse ear skin by two-photon excitation microscopy. Methods Mol Biol. 2018;1755:223–32. doi: 10.1007/978-1-4939-7724-6_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bentolila LA, Prakash R, Mihic-Probst D, Wadehra M, Kleinman HK, Carmichael TS, et al. Imaging of angiotropism/vascular co-option in a murine model of brain melanoma: implications for melanoma progression along extravascular pathways. Sci Rep. 2016;6:23834. doi: 10.1038/srep23834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Riedstra CP, Wang S, Patel S, Bald T, Kumar P, et al. Melanoma cells appropriate pericyte:endothelial cell crosstalk during perivascular invasion in the brain. bioRxiv.2022.07.15.500231 [Preprint].
- 47.Cabrita R, Mitra S, Sanna A, Ekedahl H, Lovgren K, Olsson H, et al. The role of PTEN loss in immune escape, melanoma prognosis and therapy response. Cancers (Basel) 2020;12:742. doi: 10.3390/cancers12030742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.James CC, Zeitz MJ, Calhoun PJ, Lamouille S, Smyth JW. Altered translation initiation of Gja1 limits gap junction formation during epithelial-mesenchymal transition. Mol Biol Cell. 2018;29:797–808. doi: 10.1091/mbc.E17-06-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tehranian C, Fankhauser L, Harter PN, Ratcliffe CDH, Zeiner PS, Messmer JM, et al. The PI3K/Akt/mTOR pathway as a preventive target in melanoma brain metastasis. Neuro Oncol. 2022;24:213–25. doi: 10.1093/neuonc/noab159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva JM, Bulman C, McMahon M. BRAFV600E cooperates with PI3K signaling, independent of AKT, to regulate melanoma cell proliferation. Mol Cancer Res. 2014;12:447–63. doi: 10.1158/1541-7786.MCR-13-0224-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ertay A, Ewing RM, Wang Y. Synthetic lethal approaches to target cancers with loss of PTEN function. Genes Dis. 2023;10:2511–27. doi: 10.1016/j.gendis.2022.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–7. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 53.Tibarewal P, Zilidis G, Spinelli L, Schurch N, Maccario H, Gray A, et al. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Sci Signal. 2012;5:ra18. doi: 10.1126/scisignal.2002138. [DOI] [PubMed] [Google Scholar]
- 54.Caspani EM, Crossley PH, Redondo-Garcia C, Martinez S. Glioblastoma: a pathogenic crosstalk between tumor cells and pericytes. PLoS One. 2014;9:e101402. doi: 10.1371/journal.pone.0101402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwiatkowska A, Symons M. Signaling determinants of glioma cell invasion. Adv Exp Med Biol. 2020;1202:129–49. doi: 10.1007/978-3-030-30651-9_7. [DOI] [PubMed] [Google Scholar]
- 56.Brodaczewska K, Majewska A, Filipiak-Duliban A, Kieda C. Pten knockout affects drug resistance differently in melanoma and kidney cancer. Pharm Rep. 2023;75:1187–99. doi: 10.1007/s43440-023-00523-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rada M, Kapelanski-Lamoureux A, Petrillo S, Tabaries S, Siegel P, Reynolds AR, et al. Runt related transcription factor-1 plays a central role in vessel co-option of colorectal cancer liver metastases. Commun Biol. 2021;4:950. doi: 10.1038/s42003-021-02481-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Zuo T, McVicar A, Yang HL, Li YP, Chen W. Runx1 is a key regulator of articular cartilage homeostasis by orchestrating YAP, TGFbeta, and Wnt signaling in articular cartilage formation and osteoarthritis. Bone Res. 2022;10:63. doi: 10.1038/s41413-022-00231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bioinformatics data will be made available immediately upon publication. Data can be accessed from the NCBI GEO database under accession code GSE247232 or the NCBI SRA database under the accession code PRJNA1032415.