Summary
Precise, on-target CRISPR-Cas9 genome editing has been shown in Schistosoma mansoni, involving both non-homology end joining and homology-directed repair pathways. Here, we present a multiplexed CRISPR-Cas9 protocol for large transgene integration into the S. mansoni genome. We describe steps for deploying multiplexed ribonucleoprotein complexes (RNPs) and donor DNA preparation. We then detail procedures for introducing RNPs into schistosome eggs by square-wave electroporation in the presence of a 5′ phosphorothioate-modified double-stranded donor transgene.
For complete details on the use and execution of this protocol, please refer to Ittiprasert et al. (2023).1
Subject areas: Model Organisms, Molecular Biology, Gene Expression, CRISPR
Graphical abstract

Highlights
-
•
Simplify DNA donor preparation for optimal yield and its stability
-
•
Ribonucleoprotein complex of Cas9 enzyme and gRNA assembly
-
•
Co-electroporation of multiple RNPs and dsDNA donor into the cells
-
•
Instructions on genotyping for transgene integration following transfection of eggs
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Precise, on-target CRISPR-Cas9 genome editing has been shown in Schistosoma mansoni, involving both non-homology end joining and homology-directed repair pathways. Here, we present a multiplexed CRISPR-Cas9 protocol for large transgene integration into the S. mansoni genome. We describe steps for deploying multiplexed ribonucleoprotein complexes (RNPs) and donor DNA preparation. We then detail procedures for introducing RNPs into schistosome eggs by square wave electroporation in the presence of a 5′ phosphorothioate-modified double-stranded donor transgene.
Before you begin
Schistosome eggs
Timing: 1 day
Where electroporation is employed to introduce the CRISPR/Cas-material into eggs of S. mansoni, it is necessary to isolate healthy and well developing schistosome eggs. Eggs recover from the liver lobes of S. mansoni-infected mice2 no later than 1 day. The S. mansoni-infected mice provide by the Schistosomiasis Resource Center (SRC), Biomedical Research Institute (BRI), without cost beyond shipping charges to registered researchers with BEI Resources, https://www.beiresources.org/Catalog.aspx?f_instockflag=In+Stock%23∼%23Temporarily+Out+of+Stock&q=schistosoma (see detail in key resources table).
-
1.
Ensure that the eggs are free of contaminating host tissues, cells and other debris, and maintain the eggs in DMEM medium containing 20% heat-inactivated fetal bovine serum at 37°C, 5% CO2.1,3
-
2.
For transfection of schistosome eggs with CRISPR materials delivered using square wave electroporation, suspend the eggs in Opti-MEM as the electroporation medium at a concentration of 50,000 eggs/mL.
Note: The mice from SRC are housed at the Animal Research Facility of George Washington University, which is accredited by the American Association for Accreditation of Laboratory Animal Care (AAALAC no. 000347).
Plasmid construct coding for the donor transgene/repair template
Timing: 2 days
To prepare the donor repair template, a custom designed plasmid, which can be synthesized, constructed and nucleotide sequence must be proved by Sanger direct sequencing. The plasmid’s cargo includes the nucleotides of the homology arms, an appropriate gene promoter, a transgene, e.g., the EGFP reporter, and a cognate terminator sequence, and serves as the PCR template for dsDNA donor synthesis using a high fidelity Taq DNA polymerase.
-
3.
Generate dsDNA donor template using primers including five nucleotides at the 5′ end with phosphorothioate (P) modification.
-
4.
Purify PCR amplicon by column purification kit, for example, NucleoSpin Gel and PCR Clean up (Takara) and Monarch PCR and DNA Clean up Kit (NEB).
-
5.
Assess the concentrated and purified dsDNA donor by spectrophotometry (NanoDrop, Thermo Fisher). DNA donor length and integrity confirm by visualizing following using ethidium-stained agarose gel (1%) electrophoresis.
Note: P bond substitutes a sulfur atom for a non-bridging oxygen in the phosphate backbone of the oligo. This modification renders the inter-nucleotide linkage resistant to nuclease degradation inside the cell. Hence, primers are introduced with P bonds at 5 nucleotides at their 5′-ends, which impede exonuclease degradation.
Stock solutions
Timing: 30 min
-
6.Prepare reagents for the formation of ribonucleoprotein complexes (RNP). Perform these steps on wet ice.
-
a.Dilute the Cas9 nuclease (Alt-R Streptococcus pyogenes Cas9 protein, Integrated DNA Technologies [IDT]) in phenol red-free Opti-MEM medium (Thermo Fisher Scientific), mix by pipetting to 1 μg/μL and aliquot in 0.5 mL tubes. Store at ‒80°C; avoid freeze/thaw cycles.
-
b.Resuspend synthetic guide RNA (sgRNA) solution in nuclease-free duplex buffer (IDT), mix by pipetting to 10 μg/μL, and store at ‒20°C. sgRNA is stable for at least 6 months; avoid freeze/thaw cycles.
-
a.
-
7.
Dilute the dsDNA donor in Opti-MEM at a concentration of 1 μg/μL; store aliquots at ‒20°C.
Designing the guide RNA
Timing: 30 min
The genome of S. mansoni is characterized by a high AT content (65%); therefore, it is critical to judiciously select the target binding sequence of the guide RNA (gRNA). To design the overlapping gRNAs for Cas9, the protospacer adjacent motif (PAM) site NGG can be used to introduce multiple, double strand breaks (DSB) due to the substrate sequence specificity of the Cas9 nuclease.
Note: In our experience, gRNAs low in GC-content, and specific for an AT-rich locus around the expected cleavage-site result in poor or only modest at best CRISPR/Cas-efficiencies in editing of schistosome eggs. Accordingly, we presume that it may be more efficient to deploy multiple and overlapping gRNAs at an AT rich target site to generate a staggered ended DSB.
-
8.To enhance efficiency of the targeted gene knock-out, choose two to three overlapping gRNAs specific for the target site.Note: Deploying the overlapping gRNAs in unison leads to the deletion of most of genes of interest.
-
a.To induce homology-directed repair (HDR) mediated integration of a transgene, choose gRNA sequences with PAM sites situated at 6–8 nt distance to other PAM to catalyze the deletion of large DNA fragments and to liberate staggered ended DSBs.Note: Bioinformatic tools including CHOPCHOP (https://chopchop.cbu.nib.no)4,5,6 and Breaking-Cas (https://bioinfogp.cnb.csic.es/tools/breakingcas/)7 provide assistance in the prediction of gRNAs with optimal on-target efficiency but with lack predicted off-target cleavage activity on the S. mansoni genome.
 CRITICAL: Using multiple gRNAs to generate large deletions of target DNA fragment, chose each gRNA with a similar CRISPR efficiency with the goal of preventing bias in DNA cleavage mediated by a more efficient gRNA. In addition, avoid gRNAs with predicted off-target site(s) proximal to the programmed target sites.
CRITICAL: Using multiple gRNAs to generate large deletions of target DNA fragment, chose each gRNA with a similar CRISPR efficiency with the goal of preventing bias in DNA cleavage mediated by a more efficient gRNA. In addition, avoid gRNAs with predicted off-target site(s) proximal to the programmed target sites.
-
a.
DNA donor template
Timing: 5–6 h
Non-homologous end joining (NHEJ), rather than HDR, is generally the dominant mechanism responsible for repairing DSBs in the genome of cells when a single gRNA combined without (or even with) single-stranded oligodeoxynucleotides (ssODNs) is provided as DNA donor template.8,9 However, enhancing the efficiency of DSB repair carried out by HDR to knock in (KI) a large transgene can be improved by engineering a staggered DSB while, in tandem, providing a suitably prepared donor DNA template.10 Here, we use an example from our recent report1 to illustrate the KI procedure using a linear dsDNA carrying a large reporter transgene flanked by the S. mansoni ubiquitin gene promoter and its terminator. Our example uses the Ubi-EGFP-Ubi (total length, ∼ 4.5 kb) donor DNA template for HDR into schistosome gene safe harbor 1 (GSH1), as detailed below.
-
9.dsDNA donor design and synthesis:
-
a.Generate dsDNA donor templates using primers including 5 nucleotides at the 5′end with 5′-phosphorothioate (P) modifications.Note: The DNA sequence complexity and/or hairpin structure of the long dsDNA donor is critically associated with HDR efficiency. Using 5′-end-modifications with P bonds strongly enhances HDR and favors the efficiency of single-copy donor integration through retention of monomeric donor conformation. In turn, this facilitates targeted integration of the repair template (transgene) to heal the programmed Cas9-catalyzed chromosomal break.11
-
b.Synthesized-5′-phosphorothioate-modified (≥5 nt) primers by standard desalting of 20–24 nt complementary to the desired insert, e.g., homology arms flanking promoter-enhanced green fluorescent protein gene-terminator. The 100 μM primer stock solution may be aliquoted and stored at ‒20°C. A 10 μM working solution is used to execute PCR.
-
c.Perform PCRs to amplify the desired transgene donor from plasmid DNA as a template using a high-fidelity Taq DNA polymerase. The PCR master mix must be freshly prepared.
-
d.Agarose gel electrophoresis is used to size obtained amplicons and to confirm amplification of a single fragment. If the amplicon profile seen in the gel differs from the expected product, the annealing temperature of the PCR step must be optimized to resolve this issue.
-
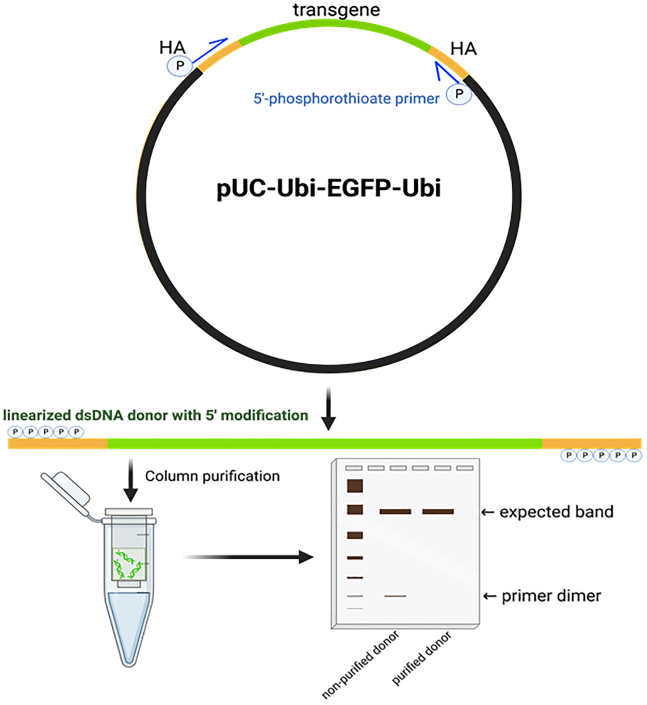
e.Purification of the DNA donor from the PCR amplicon is accomplished using a spin-column kit, for example, NucleoSpin Gel and PCR Clean-up column (Takara) or Monarch PCR and DNA Clean up Kit (NEB). This column kit removes excess primer and/or primer dimers. The long dsDNA donor can be eluted in deionized, nuclease-free water or Opti-MEM at 70°C (Figure 1).
-
f.Purified dsDNA donor may be stored at ‒20°C, thawed on wet ice and warmed to 37°C before transfection (optional).Note: Dimers may not be visible on the agarose gel. It is helpful to use a primer analysis tool such as the OligoAnalyzer Tool from IDT and OligoEvaluator by Sigma-Aldrich to predict self-dimer or hetero-dimer formation to avoid using primers that exhibit these characteristics. If primer-dimer formation remains a concern, the PCR amplicon can be cleaned by paramagnetic beads (AMPure XP) to size-exclude shorter products (e.g., < 200 bp). This is critical purification reagent for size selected-amplicon purification. The column kit is not recommended.
-
a.
Figure 1.
Schematic diagram of the long double stranded DNA donor
Donor contains 5′phosphorothioate modification at the first 5 nt of both DNA strands. Plasmid DNA encoding gene cassette of an ubiquitin promoter driven EGFP with its terminator (green line) flanked on both sides by homology arms (HA) (yellow lines). A pair of 5′-phosphorothioated (P) primers (blue arrows) is used to amplify the dsDNA. PCR products are purified using a DNA column purification kit and inspected by agarose gel electrophoresis comparing with non-purified PCR (primer dimer contamination) amplicon. Purified DNA from the PCR, not from agarose gel, serves as the donor template for HDR.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Streptococcus pyogenes Alt-R HiFi Cas9 nuclease | IDT | Cat no. 1081058 |
| Opti-MEM | Thermo Fisher Scientific | Cat no. 31985062 |
| DMEM | Thermo Fisher Scientific | Cat no. 11320033 |
| Fetal bovine serum | Gibco | Cat no. A5256801 |
| Antibiotic antimycotic | Gibco | Cat no. 15240062 |
| Phusion high-fidelity DNA polymerase | New England Biolabs | Cat no. M0530 |
| NucBlue Live ReadyProbes reagent | Thermo Fisher Scientific | R37605 |
| Deoxynucleotide set, 100 mM | MilliporeSigma | Cat no. DNTP100A |
| Critical commercial assays | ||
| AmPure XP bead-based reagent | Beckman Coulter | A63881 |
| Experimental models: Organisms/strains | ||
| Mouse liver tissues from Schistosoma mansoni-infected mice | Biomedical Research Institute, Rockville, MD | https://www.afbr-bri.org/schistosomiasis/ |
| Oligonucleotides | ||
| Custom guide RNA | IDT | N/A |
| HA-donor forward primer | This study | 5′-PS bond-AGATTGCTAGAAATTTATGAAAG-3′ |
| HA-donor reverse primer | This study | 5′-PS bond-ACTGCCGAATTTATAATATTTGG-3′ |
| Control-F | This study | TGTTATCGTCCGTCGCTTCA |
| Control-R | This study | GCGTTCAAACATTGCCCACT |
| 5′ KI-F | This study | AGGGTTTTGGTTCGCAGGAT |
| 5′ KI-R | This study | CGGAGACAATCTGGAAAGGTCA |
| 3′ KI-F | This study | GTCCGCGTAATCGTCGTTACTA |
| 3′ KI-R | This study | GTGGTTCCATACTATGCAGTTTCC |
| Recombinant DNA | ||
| pUC-Ubi-EGFP-Ubi | This study | N/A |
| Software and algorithms | ||
| CHOPCHOP | http://chopchop.cbu.uib.no/ | |
| Breaking-Cas | https://bioinfogp.cnb.csic.es/tools/breakingcas/ | |
| Other | ||
| NanoDrop One | Thermo Fisher Scientific | Cat no. ND-ONE-W |
| DNAzol ES | Molecular Research Center, Inc. | Cat no. DN128 |
| Electroporation cuvette | Cole-Parmer, BTX | Cat no. 45-0126 |
| Electro Square Porator | Cole-Parmer, BTX | Model ECM830 |
Materials and equipment
PCR reaction master mix for long dsDNA donor preparation
| Reagent | Amount |
|---|---|
| 5× Phusion High Fidelity buffer | 10 μL |
| 10 mM dNTPs | 1 μL |
| 10 μM HA-donor forward primer | 2.5 μL |
| 10 μM HA-donor reverse primer | 2.5 μL |
| pUC-Ubi-EGFP-Ubi plasmid DNA | variable |
| DMSO | 1.5 μL |
| Phusion DNA polymerase | 0.5 μL |
| Nuclease-free deionized water | to 50 μL |
| Total | 50 μL |
[Store Stock solution at 20°C, expiration date suggested by manufacture, the PCR reaction master mix must be freshly prepared]
PCR cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Initial denaturation | 98°C | 30 s | 1 |
| Amplification | 98°C | 10 s | 30 cycles |
| 55°C | 30 s | ||
| 72°C | 3 min | ||
| Final extension | 72°C | 10 min | 1 |
| 12°C | ∞ |
PCR master mix for transgene investigation1
| Reagent | Amount |
|---|---|
| 2× GoTaq G2 DNA polymerase mix | 10 μL |
| 10 μM forward primer (5′-KI-F or 3′-KI-R or control-F) | 0.5 μL |
| 10 μM reverse primer (5′-KI-R or 3′-KI-R or control-R) | 0.5 μL |
| Genomic DNA | variable |
| Nuclease-free deionized water | to 20 μL |
| Total | 20 μL |
[Store Stock solution at 20°C, expiration date suggested by manufacture, the PCR reaction master mix must be freshly prepared]
PCR cycling conditions
| Steps | Temperature | Time | Cycles |
|---|---|---|---|
| Initial denaturation | 95°C | 2 min | 1 |
| Amplification | 94°C | 15 s | 40 cycles |
| 58°C | 30 s | ||
| 72°C | 1 min | ||
| Final extension | 72°C | 10 min | 1 |
| 12°C | ∞ |
To optimize the PCR thermal cycling conditions, we recommended performing initial annealing with a temperature gradient, 52°C–59°C.
Step-by-step methodological details of Cas9 mediated genome editing
Genome editing by Cas9 mediated by multiple gRNAs
Timing: <30 min
Ribonucleoprotein complexes (RNPs) of each synthetic gRNA (sgRNA) and Cas9 nuclease must be prepared individually. The assembly of the RNP complex is performed at ambient temperature. Thereafter, retain the RNPs on wet ice until transfection by electroporation (Figure 2).
-
1.
Prepare dsDNA donor at a concentration of 1 μg/μL in Opti-MEM.
-
2.
Prepare each sgRNA at 1 μg/μL from a stock solution at 10 μg/μL in Opti-MEM. In this protocol the S. mansoni GSH1 was targeted by three sgRNAs. Therefore, gRNAs in the example shown here are designated sgRNA#1, sgRNA#2 and sgRNA#3.
-
3.
Prepare the Cas9 working solution by diluting to a concentration of 1 μg/μL in Opti-MEM.
-
4.Transfer and mix 5 μL of Opti-MEM, 10 μL of sgRNA#1 (or sgRNA#2 or sgRNA#3) and 10 μL of Cas9 into a new tube by gently pipetting (do not vortex).
-
a.Incubate at ambient temperature for 15 min.
-
b.Subsequently, keep RNP on wet ice until used for electroporation.
-
a.
-
5.
Using a pipette, dispense 200 μL of schistosome eggs (50,000 eggs/mL of Opti-MEM, as described above) into a 4 mm gap electroporation cuvette and add 10 μL of 1 μg/μL of the dsDNA donor, 25 μL of each RNP. Total volume in the cuvette will be ∼285 μL–300 μL.
-
6.
Immediately proceed with square wave electroporation of a single pulse of 125 V for 20 ms (msec).
-
7.
After electroporation, maintain the schistosome eggs in DMEM supplemented with 20% FBS, 2% antibiotic-antimycotic (penicillin-streptomycin-amphotericin B) at 37°C, 5% CO2. Change the culture medium daily (this is necessary) until the experiment terminates.
Note: We recommend preparing a fresh working concentration of gRNA in Opti-MEM. Promptly use the RNPs for electroporation after the preparation and assembly of the RNP complex. The electroporation cuvette can be used at 23°C–28°C. A 1:1 ratio sgRNA:Cas9 at 10 μg of each reagent may be optimal, based on our studies using a range of 5 μg–20 μg of gRNA and Cas9. Although we have not seen a positive correlation between enhanced efficiency of editing and the increasing concentrations of the RNP reagents, we have not titrated the concentration of the dsDNA donor with respect to efficiency of HDR. We have observed, however, that less donor DNA resulted into a comparably lower efficiency of HDR.
CRITICAL: Prepare each RNP complex individually. Attempting to assemble the complexes of all the gRNAs and the Cas9 nuclease in a same tube may result in inconsistent complex assembly of the RNP and its component Cas9 and individual gRNAs. Given that schistosome eggs settle rapidly to the bottom of the cuvette, ensure that the eggs are homogeneously suspended in the medium during electroporation by frequent, gentle agitation by hand.
Alternative: Tracking the entry of RNPs into the schistosome egg and its cells may be monitored using red fluorescent protein (RFP)-tagged Cas9 nuclease without negatively impacting CRISPR/Cas efficiency (Figure 3). Fluorescence from the tagged Cas9 is apparent up to 48 h post electroporation.
Figure 2.
Multiple RNP preparation and co-electroporation
(A) Individual, freshly prepared RNP complexes, donor, and parasite eggs in Opti-MEM, which are held on wet ice until electroporation.
(B) Electroporation proceeds at 23°C–28°C. It is not necessary to pre-chill the cuvette on ice.
Figure 3.
RNPs entry into parasite eggs
Delivery of RNPs into schistosome eggs containing a fully developed larva tracked by RFP-tagged Cas9 enzymes (IDT). Only RNP-electroporated eggs showed positive red fluorescence (lower panels). Vital cells have been stained using by live cell staining, Hoechst 33342 (blue). The RNP delivery into larval cells was visualized using the ReadyProbes Cell viability imaging kit, Blue (Invitrogen). Control treatment group eggs did not show the red fluorescent protein (RFP) signal.
Analysis for transgene integration
Timing: 1–2 days
The genotyping for transgene integration can be performed at day 7 following transfection of the eggs. Our experience revealed that the P-modified dsDNA donor is stable in cells of the cultured schistosome eggs for as long as 5–7 days if not incorporated by HDR. We have reported EGFP expression (Figure 4, left panel) in the developing embryo within the eggshell. The transgene fluorescence displayed in these eggs could only be differentiated at day 5 post and onwards after electroporation from signals from control group eggs transfected only with the donor repair template (i.e., without RNPs). The EGFP signal from these control group eggs was no longer detected by day 10.1 Hence, we recommend proceeding to genome sequence/genotyping analysis at or later than 10 days after transfection to avoid false positive and cross-reactive PCR from residual donor DNA repair template (Figure 4, right panel).
-
8.
Retrieve the eggs from the culture for analysis. Wash eggs 3 times in 1× PBS, then extract genomic DNA from the eggs using DNAzol ES, following the manufacturer’s protocol.
-
9.
To avoid false positive PCRs due to mosaicism, locate one of the primers used for genotyping outside of the homology arms (at least 50 nt away).
Note: This should prevent or at least minimize amplification from transiently retained (episomal) donor DNA molecules. We used Primer-Blast for primer design (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome) to predict precise knock-in at the S. mansoni GSH1 target site. In silico, none of the primers cross amplify with the wild type GSH1 locus and/or elsewhere in the schistosome genome using primer pairs specificity checking parameters with database settings of ‘nr’ and exclude ‘Schistosomatidae’.
-
10.
Undertake PCR as above using 5′ KI or 3′ KI specific primer pairs and genomic DNA of the eggs as DNA template. Also perform a positive control PCR assay for the integrity of the genomic DNAs using samples of the gDNA from each treatment and control group. The PCR master mix must be freshly prepared.
Figure 4.
Fluorescent reporter gene knocked-in into schistosome GSH1
EGFP expression driven by the ubiquitin promoter following Cas9 mediated transgene knock-in, programmed using multiple, overlapping gRNAs. Green fluorescent protein signal seen in viable eggs (green arrows) but not in non-viable eggs (which exhibit only auto fluorescence [AF] as the white arrow indicates) beyond day 5 post electroporation (top panel as published in Ittiprasert et al., 20231 as Figure 4B). Genotyping for transgene integration using PCR with primers specific for both the 5′KI and 3′KI integrations at the programmed target site. The PCR control shows positive bands in all DNA samples (bottom panel).
Expected outcomes
CRISPR/Cas9-mediated editing of a target locus using multiple RNPs and overlapping gRNAs in tandem was used to improve precise transgene integration into the AT-rich S. mansoni genome. Large gene deletions, as well as the transgene integration of a fluorescence (EGFP) reporter, could be accomplished with higher efficiency compared to a single gRNA approach. The stability of the dsDNA donor was significantly increased by 5′-phosphorothioate modification with homology arms of <1 kb in length. In this example, homology arms of 600 bp enabled HDR insertion of a 3 kb transgene into a predicted schistosome genome safe harbor termed GSH1. Moreover, HDR efficiency also was improved.1 The optimized conditions to deliver CRISPR materials by electroporation into the schistosome egg were sufficiently gentle such that growth and development of the embryo within the treated egg proceeds apparently in the same way as the control, wild type eggs. This method is advantageous for functional studies when using CRISPR/Cas approaches, including for transgene gain-of-function functional genomics. We anticipate that this protocol not only will benefit research by other labs studying schistosomes but will be modifiable for other species including other helminths with AT-rich genomes.
Limitations
It can be challenging to integrate large dsDNA donors (>3 kb) containing a transgene flanked by homology arms) into the genome. To minimize this hurdle, select/design the DNA repair donor without low DNA complexity, repetitive sequence(s) and/or self-hairpin forming loop(s). For the design of the donor sequence, non-frameshift-based processing can be considered to eliminate the above problems.
Troubleshooting
Problem 1
As soon as possible following electroporation, culture the eggs in 20% FBS, DMEM.
Related to this, the presence of non-viable parasite eggs, which can occur following from the egg preparation, confounds the investigation because larval development after electroporation does not proceed normally.
Potential solution
-
•
Use freshly prepared eggs (< 2 days in culture).
-
•
Change egg culture medium regularly.
Problem 2
No transgene insertion.
Potential solution
-
•
Ensure the integrity of the donor by visualizing following agarose gel electrophoresis. If degradation is observed (a smear rather than a single band of the correct size), prepare a fresh aliquot of the donor template.
-
•
Contamination by primer dimers or partial donor degradation inhibits HDR efficiency.
-
•
Optimize the assembly of the RNP complexes and the synthesis of the donor DNA template as suggested in this method. Excess quantities of non-parasite material including the Cas9 nuclease and donor repair template DNA can be toxic. By contrast, insufficient concentration of CRISPR/Cas reagents may result in sub-optimal outcome, including inefficient programmed target site cleavage and subsequent HDR.
Problem 3
Background EGFP signals from residual/non-integrated dsDNA donor in transcript reporter transgene expression study.
Potential solution
-
•
Ensure to set up an experimental group, where eggs are only transfected with the dsDNA donor (no RNPs). Information from this group is used for normalization.
-
•
For the 5′-phosphothioate modified-donor, stability within the parasite cells up to 5 days has been shown. To eliminate/minimize the bias of EGFP expression from non-integrated donor, the downstream experiments for both genotyping and EGFP expression should be undertaken only at day ≥5 after transfection.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Paul J. Brindley (pbrindley@gwu.edu).
Technical contact
Technical questions about this protocol should be directed to the technical contact, Wannaporn Ittiprasert (wannaporni@gwu.edu).
Materials availability
All materials are available upon reasonable request.
Data and code availability
This report did not generate or analyze any data sets.
Acknowledgments
We thank the members of Biomedical Research Institute, Shady Grove, MD for support with S. mansoni-infected mice through NIH-NIAID contract HHSN272201700014I. The S. mansoni genome safe harbor coordinates were kindly shared by C. Grunau, Interactions Hôtes Pathogènes Environnement, University of Perpignan via Domitia, Perpignan, France. This work was funded in part by the Wellcome Trust grant 107475/Z/15/Z (Flatworm Functional Genomics Initiative, PI, Karl F. Hoffmann) and by NIH award CA164719 (PI, P. Brindley). For open access, the authors applied a CC by public copyright license to any author-accepted manuscript version arising from this submission. Zeiss 710 confocal microscope service for analysis of fluorescence spectra to differentiate the autofluorescence from the true EGFP was provided by the Nanofabrication and Imaging Center, George Washington University (NIH award number RR025565, PI, A. Popratiloff). Graphical abstract and schematic figures were created with BioRender.com with agreement number GK25IQ86RA.
Author contributions
Writing, W.I. and P.B.; methodology, W.I., M.M.M., and V.H.M.; supervision, P.B.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Wannaporn Ittiprasert, Email: wannaporni@gwu.edu.
Paul J. Brindley, Email: pbrindley@gwu.edu.
References
- 1.Ittiprasert W., Moescheid M.F., Chaparro C., Mann V.H., Quack T., Rodpai R., Miller A., Wisitpongpun P., Buakaew W., Mentink-Kane M., et al. Targeted insertion and reporter transgene activity at a gene safe harbor of the human blood fluke, Schistosoma mansoni. Cell Rep. Methods. 2023;3 doi: 10.1016/j.crmeth.2023.100535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalton J.P., Day S.R., Drew A.C., Brindley P.J. A method for the isolation of schistosome eggs and miracidia free of contaminating host tissues. Parasitology. 1997;115:29–32. doi: 10.1017/s0031182097001091. [DOI] [PubMed] [Google Scholar]
- 3.Rinaldi G., Eckert S.E., Tsai I.J., Suttiprapa S., Kines K.J., Tort J.F., Mann V.H., Turner D.J., Berriman M., Brindley P.J. Germline transgenesis and insertional mutagenesis in Schistosoma mansoni mediated by murine leukemia virus. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labun K., Montague T.G., Gagnon J.A., Thyme S.B., Valen E. CHOPCHOP v2: a web tool for the next generation of CRISPR genome engineering. Nucleic Acids Res. 2016;44:W272–W276. doi: 10.1093/nar/gkw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montague T.G., Cruz J.M., Gagnon J.A., Church G.M., Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oliveros J.C., Franch M., Tabas-Madrid D., San-León D., Montoliu L., Cubas P., Pazos F. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016;44:W267–W271. doi: 10.1093/nar/gkw407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ittiprasert W., Chatupheeraphat C., Mann V.H., Li W., Miller A., Ogunbayo T., Tran K., Alrefaei Y.N., Mentink-Kane M., Brindley P.J. RNA-Guided AsCas12a- and SpCas9-Catalyzed Knockout and Homology Directed Repair of the Omega-1 Locus of the Human Blood Fluke, Schistosoma mansoni. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms23020631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ittiprasert W., Mann V.H., Karinshak S.E., Coghlan A., Rinaldi G., Sankaranarayanan G., Chaidee A., Tanno T., Kumkhaek C., Prangtaworn P., et al. Programmed genome editing of the omega-1 ribonuclease of the blood fluke, Schistosoma mansoni. Elife. 2019;8 doi: 10.7554/eLife.41337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang D.E., Lee J.Y., Lee J.H., Koo O.J., Bae H.S., Jung M.H., Bae J.H., Hwang W.S., Chang Y.J., Lee Y.H., et al. Multiple sgRNAs with overlapping sequences enhance CRISPR/Cas9-mediated knock-in efficiency. Exp. Mol. Med. 2018;50:1–9. doi: 10.1038/s12276-018-0037-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutierrez-Triana J.A., Tavhelidse T., Thumberger T., Thomas I., Wittbrodt B., Kellner T., Anlas K., Tsingos E., Wittbrodt J. Efficient single-copy HDR by 5' modified long dsDNA donors. Elife. 2018;7 doi: 10.7554/eLife.39468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This report did not generate or analyze any data sets.