Abstract
The activation of muscle-specific gene expression requires the coordinated action of muscle regulatory proteins and chromatin-remodeling enzymes. Microarray analysis performed in the presence or absence of a dominant-negative BRG1 ATPase demonstrated that approximately one-third of MyoD-induced genes were highly dependent on SWI/SNF enzymes. To understand the mechanism of activation, we performed chromatin immunoprecipitations analyzing the myogenin promoter. We found that H4 hyperacetylation preceded Brg1 binding in a MyoD-dependent manner but that MyoD binding occurred subsequent to H4 modification and Brg1 interaction. In the absence of functional SWI/SNF enzymes, muscle regulatory proteins did not bind to the myogenin promoter, thereby providing evidence for SWI/SNF-dependent activator binding. We observed that the homeodomain factor Pbx1, which cooperates with MyoD to stimulate myogenin expression, is constitutively bound to the myogenin promoter in a SWI/SNF-independent manner, suggesting a two-step mechanism in which MyoD initially interacts indirectly with the myogenin promoter and attracts chromatin-remodeling enzymes, which then facilitate direct binding by MyoD and other regulatory proteins.
In eukaryotes, activation of gene expression involves the ordered assembly of transcriptional regulators, chromatin-modifying enzymes, RNA polymerase II, and associated general transcription factors onto cis-acting elements that are embedded in chromatin. Chromatin-remodeling enzymes play an integral role in gene activation by perturbing chromatin structure and making specific loci permissive for transcription. Molecular analysis of multiple gene activation events suggests that the temporal recruitment of transcription factors and chromatin-remodeling enzymes is gene specific and dictated by the interplay between specific activators and local chromatin structure (1, 52, 55).
Two classes of enzymes have been shown to remodel chromatin structure either by catalyzing covalent modifications of histones or by hydrolyzing ATP to mobilize nucleosomes. Among the latter class of enzymes are the SWI/SNF chromatin-remodeling complexes. A distinguishing feature of this family is the presence of a bromodomain in the ATPase subunit, which promotes interaction with acetylated histones and links the activities of the two classes of chromatin remodelers in the regulation of gene expression (22). SWI/SNF enzymes physically interact with histone acetyltransferases (HATs), histone deacetylases (HDACs), and methyltransferases, showing the potential for coordination of chromatin-remodeling activities (reviewed in reference 53).
Mammalian SWI/SNF chromatin-remodeling enzymes are multisubunit complexes that contain either the Brg1 or Brm ATPase subunits and can activate or repress expression of a subset of genes (39, 53). They function in cell cycle control, and some of the subunits are tumor suppressors (49). Diverse SWI/SNF complexes exist that are distinguished by the particular ATPase, the presence of unique subunits, and tissue-specific isoforms of common subunits (60, 61). The Brg1- and Brm-containing complexes are similar biochemically but display different physiological characteristics. In mice, disruption of Brg1 is early embryonic lethal while disruption of Brm has a mild effect on proliferation (6, 48). Moreover, the two ATPase subunits can be associated with different promoters (25, 38).
Mammalian SWI/SNF enzymes have been shown to facilitate the binding of TBP and other factors involved in polymerase II (Pol II) preinitiation complex formation and to promote transcriptional elongation both in vitro and in vivo (5, 11, 24, 33, 52, 55). Multiple models to explain targeting of SWI/SNF enzymes to specific regulatory sequences exist: interactions with RNA polymerase II holoenzyme (62), binding of bromodomains to acetylated histones (22), and recruitment by sequence-specific transcriptional activators (12). In yeast, interaction with activators is critical for SWI/SNF function (47), and in mammalian cells, SWI/SNF components interact with numerous activators, at least some of which likely target SWI/SNF to specific promoters (8, 11, 18, 26, 30, 32, 35, 46).
During the differentiation of skeletal muscle, the myogenic basic helix-loop-helix family of regulatory factors (MRFs) heterodimerize with ubiquitously present E proteins and bind to 6-bp elements called E boxes. MRFs interact with members of the myocyte enhancer family (MEF2) of proteins, which bind a conserved A/T-rich sequence in the regulatory regions of muscle-specific genes, to synergistically activate downstream muscle gene expression (42). Although each MRF can bind to the E box with similar affinities, myogenin promotes myogenesis less efficiently than Myf5 in mouse embryos and is less effective than MyoD in activating endogenous muscle-specific genes when introduced into fibroblasts (3, 19). MyoD-mediated gene activation is associated with chromatin remodeling in the regulatory regions of muscle-specific genes and depends on a cysteine-histidine-rich region and a carboxy-terminal region (16, 19). The carboxy-terminal alpha-helical region of MyoD that is distinct from that of myogenin specifies the ability to initiate muscle-specific gene expression (3).
During embryogenesis and skeletal muscle regeneration, it is the induction of MyoD and/or other MRF proteins that is critical for commitment to the skeletal muscle lineage, such as occurs in primary cell cultures and in activated satellite cells (44, 58). To model events controlling myogenic differentiation via the induction of MyoD, we have utilized the well-established model of MyoD-induced transdifferentiation of fibroblast cells, first used to identify MyoD as the regulator of myogenic differentiation (14).
We previously used this system to establish a role for SWI/SNF chromatin-remodeling enzymes in MyoD-mediated activation of two muscle-specific genes and correlated activation of myogenin transcription with changes in myogenin promoter chromatin structure (16). We later extended our results to show that several muscle-specific genes were also inhibited by dominant-negative SWI/SNF enzymes but that cell cycle control and expression of key cell cycle regulators, such as p21, cyclin D3, and Rb, were unaffected during muscle differentiation induced by the different MRFs (17, 50).
To more specifically describe the role that SWI/SNF chromatin-remodeling enzymes play in muscle differentiation, we performed a microarray analysis of cells differentiated by MyoD in the presence or absence of dominant-negative BRG1 and found that a subset of genes activated by MyoD require SWI/SNF enzymes. We demonstrate that MyoD induces histone H4 acetylation and localization of Brg1 at the myogenin promoter; however, stable MyoD binding to the promoter occurs only after chromatin modifications. Expression of dominant-negative BRG1 interferes with MyoD binding to its cognate E box on the myogenin promoter but does not affect acetylation of histone H4. This raises a paradox: interaction of SWI/SNF and acetylation of histones at the myogenin promoter require MyoD, but MyoD does not stably bind to the promoter in the absence of functional SWI/SNF enzyme. To address this, we demonstrate that the Pbx-1 homeodomain factor, which cooperates with MyoD to stimulate myogenin transcription, is constitutively bound to the myogenin promoter in a SWI/SNF-independent manner. This suggests a novel mechanism by which MyoD interacts with the promoter indirectly via Pbx-1 and recruits chromatin-remodeling enzymes, which then facilitate the binding of MyoD and other regulators. Demonstration of physical interactions between Brg1 and MyoD and Brg1 and Pbx support this conclusion. Models describing the role of SWI/SNF enzymes in the activation of the myogenin locus that address these and other recently published data (54) are discussed.
MATERIALS AND METHODS
Cell culture.
The B22 cell line inducibly expressing dominant-negative BRG1 (15) was infected with retrovirus expressing MyoD- or MyoD-related regulators as previously described (14, 16, 17, 50). Briefly, this protocol involves culturing cells for 3 days in the presence of tetracycline (dominant-negative BRG1 repressed) or in the absence of tetracycline (dominant-negative BRG1 expressed) and passaging the cells so that 24 h later the cells are at about 50% confluence. The cells were infected with the retrovirus and incubated for 30 h. A low-serum differentiation medium was then added to induce myogenic differentiation. The time at which the differentiation medium was added is referred to as time zero. Samples were collected at the times indicated (hours) for analysis. Control samples were mock infected but still subjected to the differentiation protocol and are labeled “M” or “mock” for mock differentiated. Since the dominant-negative allele was derived from the human gene (28), capital letters are used throughout this report when describing the protein produced from this allele. Endogenous Brg1 in mouse-derived cell lines and the total amount of protein in mouse cells expressing the dominant-negative human allele are referred to as “Brg1.”
Microarray analysis.
For microarray analysis, RNA was prepared with the RNeasy kit from QIAGEN, and cDNA was generated as described previously (2). MyoD target genes were identified by comparing cells infected with MyoD-producing retrovirus (n = 3) to control cells (n = 3). Data points identified as unreliable by the scanner software were discarded. Data were normalized using the Lowess algorithm in the GeneSpring 6.0 analysis package (Silicon Genetics, Redwood City, CA). Normalized data were then transformed into log2 space. A heterocedastic t test was performed on each gene. The false discovery rate was estimated using Storey's q value (57). The resulting q values were used in conjunction with the magnitude severalfold change to identify significant genes at the thresholds described in the text. Brg1-dependent genes were identified in a similar fashion by comparing cells expressing a dominant-negative BRG1 (n = 3) to control cells (n = 3). Estimation of false-discovery rate (q value) for the Brg1 analysis was limited to the 94 genes identified as upregulated by MyoD.
RNA analysis.
For reverse transcription (RT)-PCR, RNA was isolated and reverse transcribed as previously described (17). The cDNA was amplified with AmpliTaq Gold (Applied Biosystems) with 200 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, and 0.1 μg of each primer as described previously (17). MyoD and myogenin were amplified for 20 cycles with previously described primers (59). Hprt was amplified for 20 cycles with previously described primers (17). Amplification of p21 was for 20 cycles with (5′-ACACACAGAGAGAGGGCTAGG-3′) and (5′-AGATCCACAGCGATATCCAGAC-3′). Flag-BRG1 was amplified for 23 cycles with a primer to the BRG1 coding region (5′-GTACAAGGACAGCAGCAGTGGA-3′) and primer to the Flag coding region (5′-TTTGTCATCGTCGTCCTTGTAGTC-3′). [32P]dATP incorporation was detected with PhosphorImager (Molecular Dynamics), and quantification was performed using ImageQuant software.
Antibodies, protein extracts, Western analysis, and immunoprecipitations (IP).
Commercial antibodies utilized were phosphatidylinositol 3-kinase (PI 3-kinase) (06-496; Upstate), Pbx1 (sc-889; Santa Cruz), Mef2 (sc-313; recognizes the Mef2A, -C, and -D isoforms; Santa Cruz), and MyoD (sc-304; Santa Cruz). MyoD chromatin immunoprecipitation (ChIP) experiments were confirmed using an affinity-purified rabbit antibody generated against a fusion protein between glutathione S-transferase (GST) and full-length MyoD. Polyclonal rabbit antisera raised against GST fused to a unique portion of BRG1 (15) was used for all experiments except for the coimmunoprecipitation studies in Fig. 8D and 9A, which utilized affinity-purified antibody isolated from rat antisera that was generated against the same GST-BRG1 fusion protein. Flag-tagged proteins were detected using rabbit antisera against a peptide encoding the Flag epitope or M2 Flag antibody (Sigma).
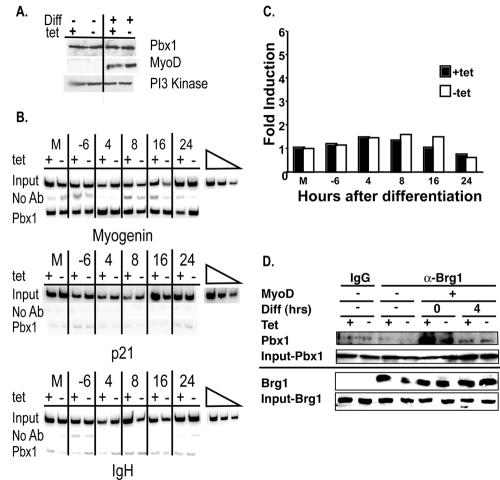
FIG. 8.
Pbx1 mediates targeting of Brg1 to the myogenin promoter. (A) Pbx1 protein levels are unaffected by differentiation (Diff.) or by the expression of dominant-negative BRG1. Protein extracts from mock-differentiated cells (−) or cells differentiated with MyoD (+) in the presence or absence of tetracycline were run on an SDS-polyacrylamide gel and probed with Pbx1, MyoD, or PI 3-kinase antibodies. (B) Pbx1 association with the myogenin promoter, the p21 promoter, and the IgH enhancer as measured by ChIP during a time course of differentiation induced by MyoD. M indicates samples that were mock differentiated for 24 h. The linearity of the PCRs was demonstrated by a twofold titration of input DNAs using the mock-differentiated, plus-tetracycline (+tet) sample. PCR amplification of 1% of the input DNA is shown. (C) Quantification of Pbx1 association with the myogenin promoter. Band intensities in each lane were normalized to input. Induction relative to the plus-tetracycline, mock-differentiated sample is shown. The data reflect the average of values from two independent experiments. (D) Endogenous Pbx1 and Brg1 coimmunoprecipitate from MyoD-differentiated but not mock-differentiated cells. Nuclear extracts from mock (−) or MyoD-differentiated (+) cells were immunoprecipitated with Brg1 antibody or purified IgG as indicated, and the immunoprecipitated material was run on an SDS-polyacrylamide gel, transferred to a membrane, and probed for the presence of Pbx1 and Brg1. The levels of Pbx1 and Brg1 present in 10% of the input for each sample are shown.
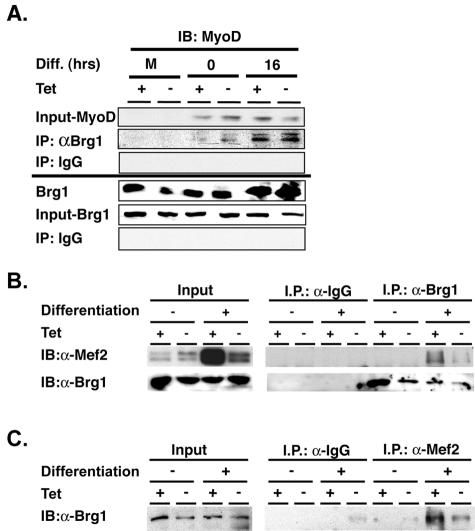
FIG. 9.
Brg1 interacts with MyoD and Mef2 in differentiating cells. Extracts were prepared from mock-differentiated cells or cells differentiated with MyoD in the presence or absence of tetracycline. (A) Immunoprecipitation from nuclear extracts from cells that were mock differentiated (M) or MyoD differentiated was performed using purified IgG or antibody against Brg1 at the times indicated. Samples were run on an SDS-polyacrylamide gel and transferred to a membrane for Western blotting with MyoD and Brg1 antibodies. Ten percent of the input for each sample is shown. The samples shown in Fig. 8D and 9A were from the same time course; the Brg1 input and Brg1 IP bands for the mock and 0-h time point are the same data that were presented in Fig. 8D. (B and C) Nuclear extracts were immunoprecipitated from cells mock (−) or MyoD (+) differentiated in the presence or absence of tetracycline for 36 h using purified IgG or antibody against Brg1 or Mef2 and probed for the presence of Mef2 or Brg1. Confirmation of Mef2 immunoprecipitation by the Mef2 antibody could not be obtained because the Mef2 band was obscured by the antibody heavy chains.
Isolation of protein and Western analyses were previously described (15). For coimmunoprecipitations in Fig. 9B, and C, cells were washed three times with phosphate-buffered saline and lysed with hypotonic buffer (10 mM Tris-HCl, pH 7.5, 10 mM KCl, 3 mM MgCl2, 10 mM NaF, 2 mM sodium orthovanadate, 0.5 mM dithiothreitol, 0.2 mM phenylmethylsulfonyl fluoride, 3 μg/ml cytochalasin B, 5 μg/ml leupeptin, 2 μg/ml pepstatin, and 2 μg/ml aprotonin). Cell lysates were incubated at 4°C for 30 min, homogenized, and centrifuged at 3,000 × g for 10 min. Nuclei were then lysed with IP buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 200 mM KCl, 0.5 mM EDTA, 1 mM dithiothreitol, 50 mM NaF, 2 mM sodium orthovanadate, 1 mM β-glycerophosphate, 0.5% NP-40, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml pepstatin, and 2 μg/ml aprotonin). Nuclear extracts were incubated with 200 μg/ml DNase I and 10 μg/ml RNase A for 30 min at 26°C and then centrifuged at 15,000 × g for 15 min. The supernatant (250 μl) was rocked with 2 μg of antibody for 12 h at 4°C, followed by the addition of protein A Sepharose (Amersham) and an additional incubation for 6 h. The beads were washed three times in lysis buffer and eluted with 2% sodium dodecyl sulfate (SDS) sample buffer. The coimmunoprecipitation experiments in Fig. 8D and 9A utilized a previously published protocol (43). The coimmunoprecipitation of Flag and MyoD (data not shown) utilized a different previously published protocol (15).
ChIPs.
ChIPs were performed using the antibodies listed above as described previously (52), except that immune complexes were eluted with 0.1 M NaHCO3 and 1% SDS, and following reversal of cross-links, the DNA was purified by proteinase K digestion followed by phenol-chloroform extraction and ethanol precipitation. The purified DNA was dissolved in 50 μl Tris-EDTA, and 2 μl was used for PCR. For acetylated H4 ChIPs, the dissolved DNA was diluted 20-fold before PCR. Inputs were 0.5% to 1% of chromatin before immunoprecipitation. PCRs were performed with QIAGEN HotStart Taq master mix with 2 μCi [α-32P]dATP for 32 cycles. PCR products were run on polyacrylamide gels and exposed to a PhosphorImager. Band intensities were quantified using the ImageQuant program. Primers to the myogenin regulatory region (34), the immunoglobulin H (IgH) enhancer (2), and the β-actin promoter (52) were described. Primers to the p21 promoter region were 5′-GTTGGTCTCCATCGGAATAG-3′ and 5′-GCCACATACATCTATGAACA-3′.
Restriction enzyme accessibility assay.
Restriction enzyme accessibility experiments were performed as described previously (16) through purification of the digested genomic DNA. To visualize the cleaved DNA via PCR, a modified ligation-mediated PCR protocol was used. One microgram of digested DNA was ligated to 1 μl of 100 mM adaptor as described in reference 7 using Ligation kit version 2 (Takara). PCR amplification was performed with QIAGEN HotStart Taq master mix under the following conditions: 94°C for 15 min, followed by 26 cycles of 94°C for 30 s and then 65°C for 60 s, followed by 72°C for 60 s. PCR products were resolved in a 1.2% Tris-acetate-EDTA-agarose gel and stained with Sybr Green I. Primers used were the sense primer LM-PCR1, as described in reference 7, and the antisense primer used for myogenin ChIP (34). Quantification was performed by densitometry using NIH image 1.62 software.
RESULTS
A subset of MyoD-regulated genes are highly dependent on the activity of the SWI/SNF complex.
Our previous studies demonstrated that SWI/SNF enzymes are necessary for MyoD to activate muscle gene transcription but not for MyoD to stimulate the expression of several cell cycle-regulated genes. To broadly assess the requirement of SWI/SNF enzymes for MyoD-mediated gene expression, we used spotted cDNA expression arrays with approximately 5,400 tiled features representing 4698 UniGene clusters (UniGene build no. 128, September 2003). We used NIH 3T3 murine fibroblasts that possess a tetracycline-suppressible, dominant-negative BRG1 allele (15, 28) and compared cells transduced with MyoD to control cells. Following 24 h in differentiation medium, MyoD increased the expression of 94 genes and decreased that of 70 genes (q < 0.10; change in expression greater than twofold) (Tables 1 and 2; see also Table S1 in the supplemental material). These 94 genes (represented by 96 array features) were analyzed for their dependence on a functional Brg1-based SWI/SNF complex. In the presence of dominant-negative BRG1, 29 genes did not achieve full activation by MyoD, as determined by statistical criteria (q < 0.05) and a twofold or more decrease in expression level (Table 1; see also Table S1 in the supplemental material). Some of the genes regulated by MyoD that were not dependent on SWI/SNF activity, such as pRb, cyclin D3, and p21 (17) (Table 2), are expressed prior to MyoD induction, whereas others, such as the beta and gamma subunits of the nicotinic acetylcholine receptor, are not expressed in fibroblasts and are induced by MyoD even in the presence of dominant-negative BRG1 (Table 2). We had previously documented that MyoD could activate cell cycle-regulated genes in the absence of functional SWI/SNF enzymes; however, the ability of MyoD to induce the expression of some previously silent loci in the absence of SWI/SNF function was not previously recognized. Of the 70 genes repressed by MyoD, only five were derepressed more than twofold in the presence of dominant-negative BRG1, suggesting that SWI/SNF enzymes play a limited role in MyoD-mediated gene repression (see Table S1 in the supplemental material).
TABLE 1.
Selected MyoD-induced genes affected twofold or more by dominant-negative BRG1
| GenBank no. | UniGene no. | Name of gene product | Fold change
|
|
|---|---|---|---|---|
| MyoD+/BRG1+a | Dom. Neg. BRG1b | |||
| NM_009394 | Mm.1716 | Troponin C2, fast | 307.3 | −10.1 |
| NM_011620 | Mm.14546 | Troponin T3, skeletal, fast | 225.7 | −8.4 |
| NM_011619 | Mm.247470 | Troponin T2, cardiac | 219.3 | −2.4 |
| NM_016754 | Mm.14526 | Myosin light chain, phosphorylatable, fast skeletal muscle | 209.9 | −5.8 |
| X15784 | Mm.16528 | Myogenin | 186.3 | −10.7 |
| M38129 | Mm.340090 | Myosin, heavy polypeptide 3, skeletal muscle, embryonic | 90.7 | −19.1 |
| X15784 | Mm.16528 | Myogenin | 83.4 | −7.3 |
| NM_009405 | Mm.39469 | Troponin 1, skeletal, fast 2 | 71.2 | −7.4 |
| AL385643 | Mm.269621 | Myosin binding protein H | 19.8 | −10.4 |
| Al324268 | Mm.2375 | Creatine kinase, muscle | 16.6 | −8.3 |
| M19436 | Mm.247636 | Myosin, light polypeptide 4, alkali; atrial, embryonic | 12.9 | −4.1 |
| Al324023 | Mm.1000 | Myosin, light polypeptide 1, alkali; atrial, embryonic | 9.6 | −3.6 |
| NM_010518 | Mm.309617 | Insulin-like growth factor binding protein 5 | 9.1 | −2.0 |
| Al324248 | Mm.35134 | ATPase, Ca2+ transporting, cardiac muscle, fast twitch 1 | 8.6 | −4.1 |
| Al414541 | Mm.24059 | Schwannomin-interacting protein 1 | 6 | −2.2 |
| Al661474 | Mm.7342 | PDZ and LIM domain 3 | 4.3 | −2.2 |
| Al325234 | Mm.251322 | Enolase 3, beta muscle | 7.4 | −2.5 |
| NM_009668 | Mm.4383 | Bridging integrator 1 | 3.8 | −2.3 |
| Al604795 | Mm.220982 | Dysferlin | 3.3 | −2.1 |
| Al326773 | Mm.39968 | Histidine-rich calcium binding protein | 2.9 | −2.5 |
| Al447277 | Mm.295105 | (PTPRF), interacting protein (liprin), alpha 4 | 2.9 | −2.1 |
| M12866 | Mm.214950 | Actin, alpha 1, skeletal muscle | 2.9 | −2.2 |
| Al430815 | Mm.29475 | CK2-interacting protein 1 | 2.8 | −2 |
| Al325457 | Mm.22513 | Kinesin family member C3 | 2.8 | −2.3 |
| Al385590 | Mm.275654 | Glycogen synthase 3, brain | 2.3 | −2.8 |
| Al326236 | Mm.31646 | Actin-like 6 | −2.2 | −2.1 |
Induction of gene expression after 24 h of differentiation with MyoD in the absence of dominant-negative BRG1.
Fold change in induction after 24 h of differentiation with MyoD when dominant-negative BRG1 is expressed.
TABLE 2.
Selected MyoD-induced genes affected twofold or less by dominant-negative BRG1
| GenBank no. | UniGene no. | Name of gene product | Fold change
|
|
|---|---|---|---|---|
| MyoD+/BRG1+a | Dom. Neg. BRG1b | |||
| NM_010866 | Mm.1526 | Myogenic differentiation 1c | 42.7 | 1.1 |
| M30514 | Mm.2810 | Cholinergic receptor, nicotinic, gamma polypeptide | 19.6 | −1 |
| Al427434 | Mm.256342 | Kinesin family member 5C | 12.3 | 1.8 |
| Al385656 | Mm.4583 | Cholinergic receptor, nicotinic, alpha polypeptide 1 (muscle) | 10 | −1.9 |
| M14537 | Mm.86425 | Cholinergic receptor, nicotinic, beta polypeptide 1 (muscle) | 8.6 | −1.2 |
| NM_013597.2 | Mm.132788 | Myocyte enhancer factor 2A | 7.5 | −1.7 |
| Al323806 | Mm.195663 | Cyclin-dependent kinase inhibitor 1A (p21) | 7.2 | −1.5 |
| Al894122 | Mm.4261 | Kangai 1 (suppression of tumorigenicity 6, prostate) | 7.1 | −1.2 |
| Al326893 | Mm.280029 | Hairy and enhancer of split 6 (Drosophila) | 5.5 | 1.0 |
| Al323835 | Mm.273862 | Purinergic receptor (family A, group 5) | 4.9 | −1.0 |
| NM_009029 | Mm.273862 | Purinergic receptor (family A, group 5) | 4.8 | −1.2 |
| Al324186 | Mm.297976 | Glypican 1 | 4.7 | −1.4 |
| NM_011817 | Mm.281298 | Growth arrest and DNA-damage-inducible 45 gamma | 4.2 | 1.1 |
| Al451932 | Mm.4081 | Runt-related transcription factor 1 | 4.0 | −1.8 |
| Al450263 | Mm.333762 | Lectin, galactose binding, soluble 4 | 3.8 | 1.3 |
| Al415710 | Mm.25559 | Serine/threonine kinase 17b (apoptosis-inducing) | 3.5 | −1.9 |
| Al426448 | Mm.28683 | Transferrin receptor | 3.4 | −1.5 |
| Al528676 | Mm.347398 | B-cell leukemia/lymphoma 6 | 3.1 | −1.8 |
| NM_011484 | Mm.273174 | Signal transducing adaptor molecule (SH3 domain and ITAM motif) | 2.9 | −1.2 |
| Al428484 | Mm.30841 | Calcium channel, voltage-dependent, alpha2/delta subunit 1 | 2.9 | −1.2 |
| NM_007483 | Mm.687 | ras homolog gene family, member B | 2.9 | −1.6 |
| NM_010722 | Mm.7362 | Lamin B2 | 2.8 | 1.2 |
| Al326964 | Mm.3862 | Insulin-like growth factor 2 | 2.8 | −1.9 |
| Al450264 | Mm.180750 | Prion protein dublet | 2.8 | −1.7 |
| AB025099 | Mm.30262 | Kruppel-like factor 5 | 2.8 | −1.5 |
| Al326148 | Mm.289832 | Proteasome (prosome, macropain) 26S subunit, ATPase 3 | 2.7 | −1.3 |
| Al429452 | Mm.280805 | Huntingtin-interacting protein 1 | 2.6 | 1.1 |
| Al451071 | Mm.25594 | Protein kinase, cAMP dependent regulatory, type II beta | 2.5 | 1.3 |
| Al425917 | Mm.29495 | CUG triplet repeat, RNA binding protein 1 | 2.5 | −1.0 |
| Al326978 | Mm.280103 | ATPase, Na+/K+ transporting, alpha 1 polypeptide | 2.5 | −1.2 |
| Al573376 | Mm.22673 | Fc receptor, IgE, high-effinity I, gamma polypeptide | 2.5 | −1.3 |
| Al414367 | Mm.65906 | Pre-B-cell leukemia transcription factor-interacting protein 1 | 2.5 | 1.2 |
| Al447937 | Mm.12863 | Heparan sulfate 2-O-sulfotransferase 1 | 2.5 | −1.2 |
| Al324262 | Mm.294083 | Annexin A11 | 2.5 | −1.4 |
| Al449015 | Mm.206218 | Histone deacetylase 11 | 2.4 | 1.0 |
| Al324952 | Mm.289131 | 3-Hydroxy-3-methylglutaryl-coenzyme A synthase 2 | 2.4 | −1.1 |
| Al323871 | Mm.246520 | Cyclin D3 | 2.3 | −1.1 |
| Al894225 | Mm.19016 | Drebrin 1 | 2.3 | −1.1 |
| Al449069 | Mm.6529 | Dystrophia myotonica kinase, B15 | 2.3 | −1.3 |
| U22445 | Mm.177194 | Thymoma viral proto-oncogene 2 | 2.2 | −1.5 |
| Al428800 | Mm.206779 | Cytoplasmic polyadenylation element binding protein 3 | 2.2 | −1.3 |
| Al325922 | Mm.276826 | Cofilin 2, muscle | 2.2 | 1.3 |
Induction of gene expression after 24 h of differentiation with MyoD in the absence of dominant-negative BRG1.
Fold change in induction after 24 h of differentiation with MyoD when dominant-negative BRG1 is expressed.
Exogenously expressed from retroviral vector.
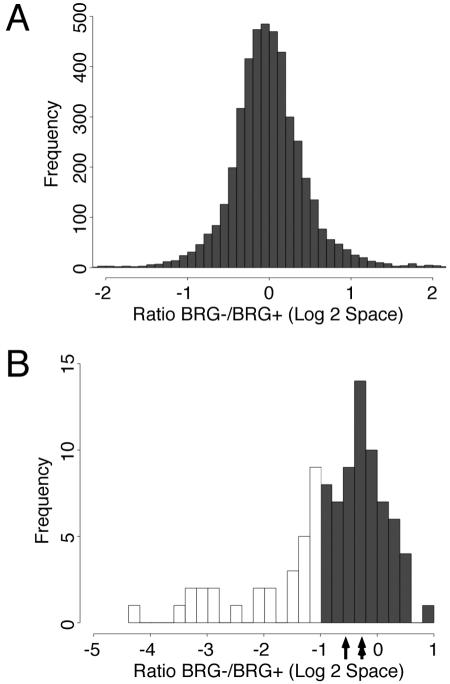
Despite the fact that only a subset of MyoD-regulated genes are highly dependent on SWI/SNF enzymes based on a severalfold change and statistical criteria, the expression of most of the MyoD-regulated genes was reduced in the absence of an active SWI/SNF complex. Graphing the log of the ratio of gene expression in the presence or absence of dominant-negative BRG1 revealed a distribution centered on zero for all the genes in the array (median ratio in log2 space is −0.02) (Fig. 1A), indicating that SWI/SNF activity does not globally alter gene expression. In contrast, limiting the analysis to the 94 genes regulated by MyoD shifted the center of the distribution below zero (median ratio in log2 space is −0.56), indicating that MyoD-regulated genes require SWI/SNF activity to achieve their full level of expression more than the typical gene spotted on the array (Fig. 1B). This is not solely due to the contribution of the highly SWI/SNF-dependent MyoD targets, because limiting the analysis to the MyoD target genes that were not identified as Brg1 dependent by the statistical criteria also provides a skewed histogram, with a median ratio of −0.23, as opposed to the median near 0 for all genes (Fig. 1B, double-headed arrow). The subset of genes that is highly dependent on SWI/SNF for MyoD activation is represented in the asymmetric tail of genes with negative ratios (29 genes demonstrate more than a twofold decrease in expression in the presence of dominant-negative BRG1; see Fig. 1B, unfilled bars). Therefore, the array data reveal a modest global dependence of MyoD-regulated genes on SWI/SNF enzymes and a more profound dependence for approximately one-third of the MyoD-regulated genes tested.
FIG. 1.
(A) Histogram of the expression ratio in cells expressing dominant-negative BRG1 (BRG1−) to nonexpressing cells (BRG1+) for the 4,282 array features that reported reliable data. The median value in log2 space is −0.02. (B) Histogram of the BRG1−/BRG1+ expression ratio for the 94 genes upregulated by MyoD. Hollow boxes indicate the 29 genes identified as strongly BRG1 dependent at the twofold change (q < 0.05) threshold. The single-headed arrow indicates the median ratio for all 94 MyoD-dependent genes (−0.56). The double-headed arrow indicates the median for the 65 genes not identified as strongly BRG1 dependent (−0.23).
To better document the role of SWI/SNF enzymes during the induction of the subset of muscle marker genes that are highly dependent on these chromatin remodelers for MyoD-mediated gene activation, we elected to focus on the myogenin gene. Previously, we demonstrated that myogenin activation by MyoD or other members of the MyoD family of muscle regulatory proteins failed in the presence of dominant-negative SWI/SNF enzymes and showed that SWI/SNF-dependent myogenin activation correlated with a SWI/SNF-dependent increase in promoter accessibility at the endogenous myogenin locus (16, 17, 50). We therefore sought to build on this base of knowledge by temporally examining the interplay of SWI/SNF enzymes and myogenic transcription factors during myogenin activation.
Kinetics of myogenin activation during MyoD-mediated differentiation.
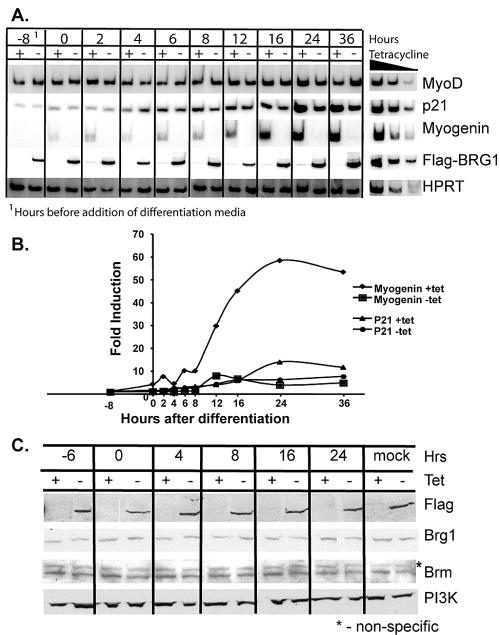
We first performed a temporal analysis of gene expression. We infected cells with a MyoD-expressing retrovirus for 30 h, induced differentiation by adding a low-serum differentiation medium at time zero, and took samples for analysis at the indicated time points. RT-PCR analysis showed that MyoD was expressed 8 h before the addition of differentiation medium and remained constant throughout differentiation in the presence and absence of tetracycline (Fig. 2A). A slight increase in the amount of myogenin mRNA was apparent in the hours following addition of the differentiation medium, with a significant increase in mRNA levels after 8 h of differentiation. Myogenin expression was inhibited by dominant-negative BRG1 at all time points (Fig. 2A and B). Induction of p21 cyclin-dependent kinase inhibitor occurred 4 h after differentiation and continued to increase but was less than twofold affected by dominant-negative BRG1, in agreement with previous work (17, 50) and with the microarray results (Table 2).
FIG. 2.
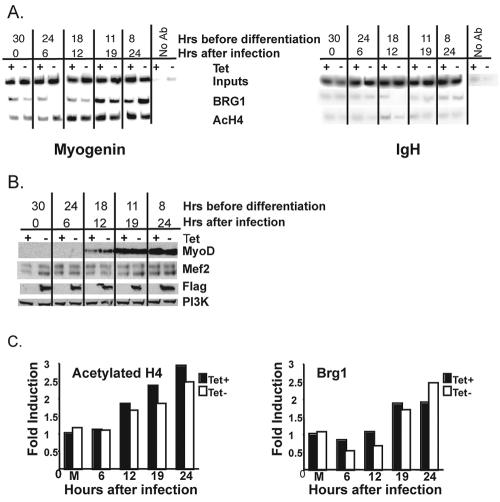
(A) Time course of myogenin, p21, and ectopic MyoD expression during differentiation. Cells expressing or not expressing dominant-negative flag-tagged BRG1 were infected with retrovirus containing MyoD. Thirty hours later, differentiation was initiated by replacement of the medium with a low-serum differentiation medium (time zero). mRNA levels of each gene were examined at the indicated time points by RT-PCR. The −8 time point is 8 h prior to addition of differentiation medium. A titration of twofold dilutions of cDNA shows the linearity of the PCRs. The 36-h time point, plus-tetracycline (Tet) sample, was used for all titrations except for Flag-tagged dominant-negative BRG1, which was amplified with the 36-h, minus-tetracycline sample. (B) Quantification of mRNA levels observed in panel A. Fold induction was defined as the ratio of myogenin or p21 levels in a given sample relative to the levels of Hprt in the same sample and standardized to the −8-h time point. (C) Time course of dominant-negative BRG1, total Brg1, and Brm protein levels during differentiation. A Western blot was performed with protein extracts isolated at the indicated time points. “Mock” refers to the samples that were not infected with the MyoD-encoding retrovirus but were instead mock infected, subjected to the same differentiation protocol, and harvested 24 h after the addition of differentiation medium.
The levels of Brg1 and Brm as well as the levels of Flag-tagged dominant-negative BRG1 remained constant during MyoD-mediated differentiation (Fig. 2C). Interestingly, overall expression of Brg1 did not change when dominant-negative BRG1 was present, demonstrating that induction of dominant-negative BRG1 did not result in overexpression of total Brg1. This effectively eliminates the possibility that expression of dominant-negative BRG1 results in nonspecific effects on transcription and promoter localization due to overexpression. It also suggests that a compensating mechanism exists for regulating Brg1 levels in cells. Tight regulation and compensation for levels of the SWI/SNF enzyme subunits Brm, Baf57, and Ini1 have previously been demonstrated (9, 20, 48).
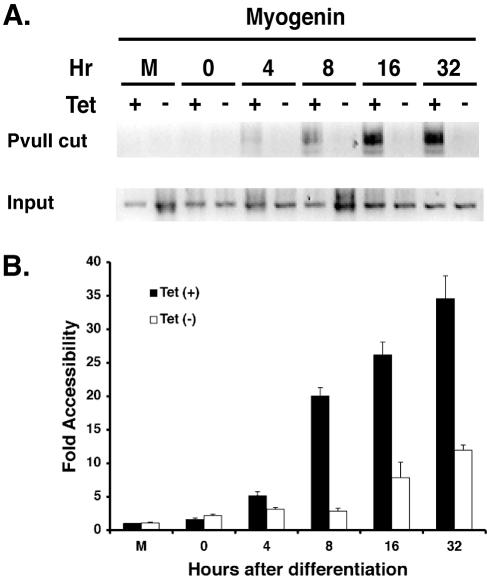
We then examined changes in nuclease accessibility at the endogenous myogenin promoter. Previously, we reported a differentiation-dependent increase in restriction enzyme accessibility at a PvuII site 370 bp upstream of the transcription initiation site and showed that the change in accessibility required SWI/SNF chromatin remodeling activity (16). We monitored the change in accessibility at this site as a function of time of differentiation, using a modified protocol in which a linker DNA was ligated to the purified, digested genomic DNA fragments followed by PCR amplification to permit visualization of the cleaved DNA (see Materials and Methods). We observed a small but noticeable increase in accessibility at 4 h postdifferentiation and a continued increase in accessibility as differentiation proceeded (Fig. 3). Thus, the change in promoter accessibility precedes the onset of myogenin mRNA accumulation.
FIG. 3.
Restriction enzyme accessibility increases at the endogenous myogenin promoter as a function of MyoD-induced differentiation and requires functional Brg1 based-SWI/SNF enzymes. Nuclei were isolated from cells that were differentiated in the presence or absence of tetracycline at the indicated time points or from cells that were mock differentiated (M) in the presence or absence of tetracycline. The mock-differentiated cells were not infected with the MyoD-encoding retrovirus but were subjected to the differentiation protocol for 32 h. (A) A modified LM-PCR protocol (see Materials and Methods) was utilized to visualize cleaved genomic DNA isolated from nuclei digested with PvuII, which cleaves the myogenin promoter at −370 relative to the start site of transcription. The PCR product was visualized by Sybr Green I staining, and an inverse image is shown. To monitor the input DNA, 10% of the amount of purified, cleaved DNA that was used for ligation-mediated PCR was used to amplify the sequences between −143 and −5 of the myogenin promoter, which contain no PvuII site. (B) Quantification of the change in nuclease accessibility at the myogenin promoter. The relative values for each time point were normalized to input and graphed relative to the value obtained for cleavage in the mock-differentiated, plus-tetracycline [Tet(+)] sample, which was arbitrarily set at 1.0. Each value is the mean ± standard deviation from three independent experiments.
Histone acetylation and recruitment of SWI/SNF to muscle-specific promoters occurs in response to MyoD-mediated differentiation.
We previously showed that chromatin remodeling is inhibited by dominant-negative BRG1 in MyoD-differentiated cells (16). Upon differentiation, hyperacetylation of histones surrounding the MyoD and Mef2 binding sites of several muscle-specific genes has been reported (34, 37). To determine whether dominant-negative BRG1 inhibits acetylation of histone H4, we differentiated cells with MyoD in the presence or absence of tetracycline, cultured the cells in differentiation media for 36 h, and then performed ChIP analysis. Figure 4 shows that muscle differentiation resulted in histone H4 hyperacetylation at the myogenin promoter and that H4 hyperacetylation was not inhibited by dominant-negative BRG1 in cells differentiated in the absence of tetracycline. This indicates that hyperacetylation of histone H4 occurs independently of SWI/SNF chromatin remodeling. We also looked at the acetylation status of the p21 promoter, because p21 expression is up-regulated during muscle differentiation but is not dependent on functional SWI/SNF chromatin-remodeling enzymes (17). We found a high level of histone acetylation at the p21 promoter in both mock-differentiated and differentiated cells compared to results with the silent IgH enhancer (Fig. 4B and C). This substantiates our previous conjecture that in fibroblasts, the promoters of genes that are constitutively expressed at low levels or that are regulated in a cell cycle-dependent manner may not require extensive chromatin remodeling.
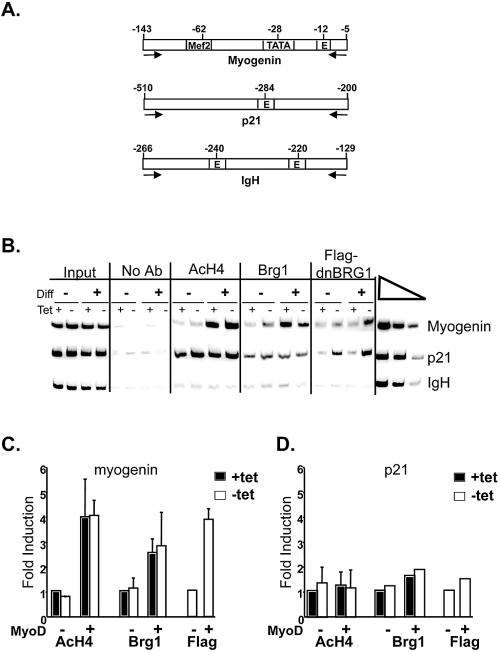
FIG. 4.
Brg1 and hyperacetylated H4 are associated with the myogenin promoter. (A) A schematic diagram indicating the regions of the myogenin and p21 promoters and the IgH enhancer that were amplified. Arrows indicate the location and direction of primers used for amplification. The approximate locations of transcription factor binding sites are indicated. (B) Chromatin immunoprecipitations were performed with antisera against BRG1, Flag, or tetra-acetylated histone H4 (AcH4) or with no antibody (No Ab) on mock-differentiated (−) or MyoD-differentiated (+) samples that had been cultured in the presence or absence of tetracycline (tet) and that were harvested for analysis 36 h after the onset of differentiation. PCR amplification of 1% of the input DNA is shown on the left. A twofold titration of input DNA using the undifferentiated, plus-tetracycline sample was performed (far right) to show that the PCR was in the linear range. (C and D) Quantification of the levels of hyperacetylated H4, Brg1, and Flag-tagged dominant-negative BRG1 present on the myogenin and p21 promoters by ChIP analysis. Band intensities in each lane were normalized to input. Induction relative to the plus-tetracycline, mock-differentiated sample is shown for hyperacetylated H4 and Brg1. Induction relative to the minus-tetracycline, mock-differentiated sample is shown for Flag-tagged dominant-negative BRG1. The data reflect the means ± standard deviations from three to four independent experiments, except for the levels of Brg1 and Flag-tagged dominant-negative Brg1 on the p21 promoter, which reflect the average values from two independent experiments.
To show that Brg1 is directly acting at these regulatory regions, we performed ChIP analysis with a BRG1 antibody and an antibody to the Flag epitope to detect epitope-tagged dominant-negative BRG1. We found that Brg1 was recruited to the myogenin promoter upon differentiation in the presence and absence of tetracycline and that Flag-tagged dominant-negative BRG1 could also be localized to these regions in differentiated cells (Fig. 4B and C). Surprisingly, although p21 up-regulation during muscle differentiation is not appreciably affected by dominant-negative BRG1 (17, 50) (Fig. 2A and B and Table 2), Brg1 was localized to the promoter region both in mock-differentiated and differentiated cells (Fig. 4B). SWI/SNF enzymes have been reported to play a direct role in the regulation of p21 transcription in other cell types that are actively proliferating; therefore, the requirement for SWI/SNF enzymes may be cell type or cell cycle stage specific (23, 27). In fibroblasts, the local chromatin structure on the p21 promoter may not require extensive chromatin remodeling by SWI/SNF enzymes during muscle differentiation and/or there may be redundant mechanisms for achieving remodeling. Thus, SWI/SNF enzymes likely contribute to p21 expression but are not required as they are for many of the muscle-specific genes.
MRF and Mef2 association with muscle-specific promoters at an endpoint of muscle differentiation is inhibited by dominant-negative BRG1.
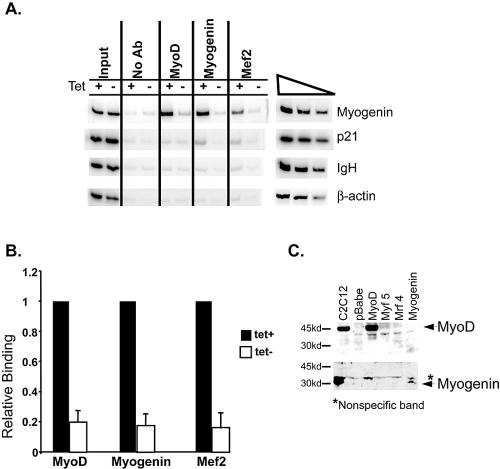
It is generally thought that transcriptional regulators play a critical role in targeting of chromatin-remodeling complexes to specific promoters. Both MyoD and Mef2 have been shown to interact with HATs and/or HDACs (41). We therefore conducted ChIP analysis of cells following differentiation in the presence or absence of dominant-negative BRG1 using antibodies to MyoD, myogenin, and Mef2 to localize these proteins on the myogenin promoter. Figure 5A and B shows that MyoD, myogenin, and Mef2 were associated with the myogenin promoter in differentiated cells with wild-type Brg1 activity; however, there was marked inhibition of binding by all three proteins when dominant-negative BRG1 was expressed. No binding of MyoD, myogenin, or Mef2 was observed on the β-actin promoter or the inactive IgH enhancer, which contains a consensus E box. Since MyoD and myogenin are structurally similar, we infected separate NIH 3T3 cell cultures with retroviruses encoding one of each of the MyoD family of myogenic regulators (50) to demonstrate that the MyoD and myogenin antibodies do not cross-react (Fig. 5C). This indicates that both MyoD and myogenin can occupy the myogenin promoter.
FIG. 5.
Binding of muscle regulatory proteins to the myogenin promoter is inhibited by dominant-negative BRG1. (A) Chromatin immunoprecipitations were performed with antisera against MyoD, myogenin, or Mef2 or with no antibody (No Ab) on MyoD-differentiated samples that had been cultured in the presence or absence of tetracycline (tet) and that were harvested for analysis 36 h after the onset of differentiation. PCR amplification of 0.5% of the input DNA is shown on the left. A twofold titration of input DNA using the undifferentiated, plus-tetracycline sample was performed (far right) to show that the PCR was in the linear range. (B) Quantification of the level of MyoD, myogenin, or Mef2 present on the myogenin promoter by ChIP analysis. Band intensities in each lane were normalized to input. The decrease in promoter association due to the expression of dominant-negative BRG1 is expressed relative to the differentiated, plus-tetracycline sample, which was set at 1.0. The data reflect the mean ± standard deviation from three independent experiments. (C) MyoD and myogenin antisera do not cross-react. Protein extracts from C2C12 myotubes or NIH 3T3 cells infected with MyoD, myogenin-, MRF4-, or Myf5-containing retrovirus or the empty retroviral vector (pBABE) were run on an SDS-polyacrylamide gel, blotted, and probed with either anti-MyoD or antimyogenin antiserum.
We have previously shown that ectopic expression of MyoD is not affected by dominant-negative BRG1 (16) (Fig. 1; see also Fig. 8A), while endogenous myogenin induction is profoundly inhibited and Mef2 induction is inhibited to a lesser extent (16, 17) (Fig. 2A and B; see also Fig. 9). Thus, the observed inhibition of myogenin and Mef2 binding in the presence of dominant-negative BRG1 is at least in part due to reduced levels of these proteins. However, this is not the case for the inhibition of MyoD binding caused by dominant-negative BRG1. The inhibition of MyoD association with the myogenin promoter suggests that SWI/SNF enzyme function is required to form a stable DNA binding complex within chromatin and does not support the idea that MyoD stably bound to chromatin directly targets Brg1-containing SWI/SNF enzymes to muscle-specific promoters.
Activation of p21 expression is critical for muscle differentiation and is promoted by MyoD (21, 45, 63). Although there are potential E boxes in the upstream region of p21, we did not detect significant levels of MyoD, myogenin, or Mef2 binding on the endogenous p21 upstream region at the end of the differentiation protocol by ChIP analysis (Fig. 5A), suggesting that MyoD activates p21 expression by an indirect mechanism and/or that our ChIP experiments do not detect an indirect association of MyoD with the p21 promoter through protein-protein interactions as previously was demonstrated for MyoD and CREB on the Rb promoter (36).
Kinetics of myogenin promoter interactions during MyoD-mediated differentiation.
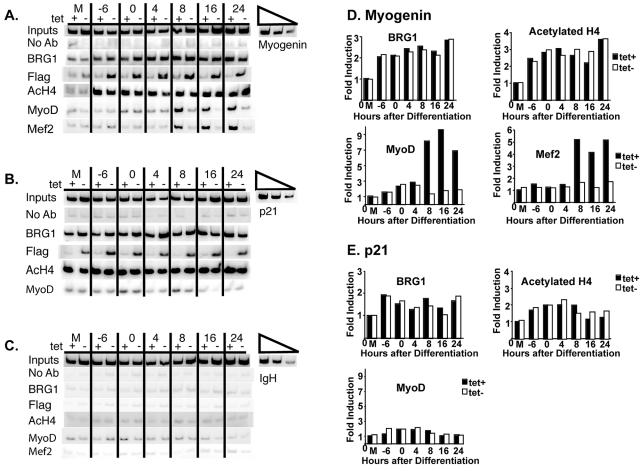
To determine how promoter interactions might influence the timing of myogenin expression, we performed ChIPs over the time course of differentiation (Fig. 6A). We found that Brg1 was recruited to the promoter 6 h before addition of differentiation medium and remained present throughout differentiation. Dominant-negative BRG1 was also present on the myogenin promoter, as seen by the ChIPs of Flag-tagged dominant-negative BRG1. Likewise, histone H4 on the myogenin promoter was hyperacetylated by 6 h prior to differentiation and was unaffected by dominant-negative BRG1. This indicates that chromatin-remodeling enzymes are associated with the myogenin promoter prior to significant gene expression and suggests that additional chromatin modifications may occur before activation of transcription. On the p21 promoter, recruitment of Brg1 and acetylation of histone H4 did not change significantly as a function of muscle differentiation, and neither was affected by expression of dominant-negative BRG1 (Fig. 6B). Neither Brg1 nor hyperacetylated H4 was present on the inactive IgH enhancer (Fig. 6C).
FIG. 6.
Brg1 and acetylated H4 associate with the myogenin promoter prior to stable binding of MyoD and Mef2. Shown is a time course of histone H4 acetylation, total Brg1, dominant-negative BRG1, MyoD, and Mef2 association with (A) the myogenin promoter, (B) the p21 promoter, and (C) the IgH enhancer as measured by ChIP during a time course of differentiation induced by MyoD in the presence or absence of tetracycline (tet). M indicates samples that were mock differentiated for 24 h. “No Ab” indicates ChIP reactions performed in the absence of antibody. Linearity of the PCRs was demonstrated by twofold titrations of input DNA using the mock-differentiated, plus-tetracycline sample. PCR amplification of 1% of the input DNA is shown. (D and E) Quantification of the levels of acetylated H4, Brg1, MyoD, or Mef2 present on the myogenin promoter (D) or on the p21 promoter (E) by ChIP analysis. Band intensities in each lane were normalized to input. Induction relative to the plus-tetracycline, mock-differentiated sample is shown. The data reflect the average value from two independent experiments.
ChIPs with the MyoD antibody showed that MyoD was bound at 8, 12, and 24 h after the addition of differentiation medium but not if the cells were differentiated in the presence of dominant-negative BRG1. Stable association of MyoD with the myogenin promoter was dependent on functional SWI/SNF enzymes and occurred just prior to the significant increase in myogenin gene expression after 8 h postdifferentiation (Fig. 2A and B and 6A). These results were confirmed with a different antibody against MyoD (data not shown). ChIP experiments to detect Mef2 binding on the promoter generated similar results (Fig. 6A). In contrast, amplification of the p21 promoter or IgH enhancer showed no or minimal interaction of MyoD with these sequences (Fig. 6B and C).
These results indicate that histone hyperacetylation and recruitment of Brg1 occur early during the differentiation process and well prior to stable binding of MyoD to the myogenin promoter. Bromodomains of chromatin-remodeling enzymes display high affinity for acetylated histones, and histone acetylation can facilitate the recruitment of SWI/SNF enzymes (22). To determine whether histone acetylation occurs prior to the recruitment of Brg1 to the myogenin promoter, we conducted ChIPs at earlier time points. Figure 7A shows that histone H4 hyperacetylation could first be detected 12 h after infection with the MyoD retrovirus or 18 h before addition of the differentiation medium at time zero, whereas recruitment of Brg1 occurred 19 h after retroviral infection or 11 h before addition of differentiation media. These results demonstrate that histone H4 acetylation coincides with the appearance of detectable levels of MyoD protein (Fig. 7B and C) and precedes the recruitment of Brg1, suggesting that histone acetylation facilitates the interaction of SWI/SNF complexes with the myogenin promoter. Thus, during MyoD-directed differentiation, the myogenin promoter becomes hyperacetylated on H4 prior to Brg1 recruitment. Although these initial events depend on MyoD, they precede stable binding of MyoD to the promoter in a SWI/SNF-dependent manner, raising the question of how the chromatin-remodeling enzymes become specifically localized to the promoter.
FIG. 7.
(A) Histone H4 hyperacetylation precedes the binding of Brg1 at the myogenin promoter. Shown is a time course of histone H4 hyperacetylation and Brg1 association with the myogenin promoter. Cells were grown in the presence or absence of tetracycline (Tet), infected or not with MyoD-encoding retrovirus, and harvested for ChIP at the indicated times prior to the addition of differentiation medium. Time points are also indicated as hours following retroviral infection. Association with the IgH enhancer is presented as a control. One percent of the input is shown. (B) Western analysis of MyoD and Mef2 protein present at the indicated times prior to addition of differentiation medium. Anti-Flag antiserum was used to document the presence of the Flag-tagged dominant-negative BRG1. PI 3-kinase (PI3K) is shown as a loading control. (C) Quantification of the levels of acetylated H4 and Brg1 present on the myogenin promoter by ChIP analysis. Band intensities in each lane were normalized to input. Induction of levels of association is presented relative to the plus-tetracycline, mock-differentiated sample. The data reflect the average value from two independent experiments.
Pbx1 is constitutively associated with the myogenin promoter and participates in the recruitment of chromatin-remodeling complexes.
Recent studies suggest that MyoD is recruited to the myogenin promoter through interaction of its cysteine/histidine and helix 3 regions with a homeodomain protein complex containing Pbx and Meis that is constitutively bound to the myogenin promoter (4). If targeting of chromatin-remodeling enzymes occurs via the Pbx-Meis site, one would predict that factor binding to this site would be independent of chromatin-remodeling activities. To address this question, we performed ChIPs with an antibody against Pbx1 during a time course of differentiation in the presence or absence of dominant-negative BRG1. Western analyses showed that Pbx1 levels did not change during MyoD-mediated muscle differentiation and were not affected by dominant-negative BRG1 (Fig. 8A). Figure 8B and C shows that, as previously reported, Pbx1 was constitutively associated with the myogenin promoter (4). Significantly, association of Pbx1 was not affected by dominant-negative BRG1; thus, Pbx1 potentially plays a role in the recruitment of Brg1 and other chromatin-remodeling activities to the myogenin promoter, perhaps via a direct interaction or via indirect recruitment through MyoD bound to the Pbx/Meis complex.
The previously published studies indicate that MyoD and Pbx1 interact at the myogenin promoter both in vivo and in vitro (4, 29). We therefore tested whether endogenous Brg1 and endogenous Pbx1 can physically interact before the onset of stable MyoD binding and the initiation of myogenin transcription. Figure 8D demonstrates that Brg1 from nuclear extracts prepared from differentiated cells coimmunoprecipitated with Pbx1 at the onset of differentiation and at 4 h postdifferentiation. The interaction was not observed in mock-differentiated cells and was not appreciably affected by the expression of dominant-negative BRG1 in the cells, indicating that the mutant BRG1 molecule also likely interacts with Pbx1. The results reveal that a specific Brg1-Pbx1 interaction occurs in the presence of MyoD, thereby supporting the idea that SWI/SNF enzymes are targeted by a MyoD-Pbx1 complex that is present on the promoter prior to the stable interaction of MyoD with the chromatin.
Brg1 interacts with MyoD and Mef2 in differentiated cells.
To further examine mechanisms for recruitment of SWI/SNF enzymes to the myogenin promoter, we investigated whether endogenous Brg1 could stably associate with MyoD and/or Mef2 in differentiating cells. Pulldown of Brg1 from differentiated cell nuclear extracts demonstrated that MyoD and Brg1 could be coimmunoprecipitated at the onset of differentiated as well as at later times and that MyoD was capable of interacting with the dominant-negative Brg1 protein as well (Fig. 9A). As an independent confirmation of this interaction, an antibody against the Flag epitope that marks the dominant-negative BRG1 was used for immunoprecipitation to show that the mutant BRG1 interacted with MyoD (data not shown). Others have also demonstrated that MyoD and Brg1 can be coimmunoprecipitated from extracts of differentiating cells (54).
Finally, we demonstrate that an antibody against BRG1 can coimmunoprecipitate Mef2 from MyoD-differentiated cells (Fig. 9B), while the converse experiment showed that an antibody against Mef2 could coimmunoprecipitate Brg1 (Fig. 9C). An interaction between Mef2 and Brg1 was also detected in the absence of tetracycline when dominant-negative BRG1 was expressed. The lighter band likely results from the inhibitory effect that dominant-negative BRG1 has on Mef2 expression (see input in Mef2 Western, Fig. 9B). Previously, we reported that expression of dominant-negative SWI/SNF complexes inhibited the expression of Mef2C RNA (17). These results demonstrate that endogenous SWI/SNF enzymes can associate with endogenous Mef2 in MyoD-differentiated cells.
Taken together, the results indicate that endogenous Brg1 interacts with both Pbx1 and MyoD at the onset of differentiation and with MyoD and Mef2 later during differentiation. The physical interactions between Brg1 and Pbx1 and between Brg1 and MyoD support the idea that MyoD is initially targeted to the myogenin promoter via the constitutively bound Pbx1. Because MyoD and Mef2 have been shown to physically and functionally interact (42), these data also suggest that Brg1-based SWI/SNF enzymes may be part of a higher-order complex containing both MyoD and Mef2 during the activation of muscle-specific genes. The data suggest that Brg1-based SWI/SNF enzymes are associated with the myogenin promoter throughout the differentiation process via protein-protein interactions with several regulatory factors.
DISCUSSION
Stable recruitment of MyoD and Mef2 transcriptional activators requires functional SWI/SNF enzymes.
We found that Brg1 was localized to the myogenin promoter during differentiation. Stable association of muscle-specific activators with the myogenin promoter was inhibited by dominant-negative BRG1, indicating that functional SWI/SNF enzymes are necessary for activator binding. Although in vitro studies using reconstituted templates and purified factors long ago demonstrated that SWI/SNF enzymes have the potential to facilitate activator binding to chromatin (13, 31, 60), there is only one other direct example of this occurring in mammalian cells. Ma et al. recently demonstrated that ectopic expression of BRG1 in BRG1-deficient cells stimulated MMP2 transcription and increased the binding of Sp1 and AP2 to the MMP2 promoter (35). Temporal analysis of protein binding events at other mammalian promoters and enhancers has revealed that the order is gene specific and that SWI/SNF chromatin-remodeling enzymes are generally recruited during the later stages of the activation process (1, 40, 52, 55, 56). For example, induction of the beta interferon gene by viral infection results in ordered binding of an enhancesome complex to a nucleosome-free region, recruitment of the GCN5 histone acetyltransferase, association of the Pol II/CBP complex, and subsequent recruitment of SWI/SNF enzymes. Chromatin remodeling by SWI/SNF enzymes promotes association of TBP with the TATA box and transcription initiation (1, 33). During enterocyte differentiation, activation of α1 antitrypsin transcription starts with association of the activator, HNF-1, TBP, and TFIIB with the promoter followed by Pol II, TFIID components, TFIIH, and mediator and then by recruitment of the activator, HNF-4α, CBP/PCAF, and BRM containing SWI/SNF enzymes (55). We recently demonstrated that during adipocyte differentiation, activation of the PPARγ nuclear hormone receptor promoter involves recruitment of SWI/SNF enzymes to the promoter after binding of the C/EBPβ activator and that SWI/SNF-promoter interactions facilitated or stabilized the binding of Pol II-associated general transcription factors (52). In these and other cases, localization of SWI/SNF enzymes and chromatin remodeling at the promoter occurred after activator binding, implying in most of these cases that SWI/SNF enzymes are needed to complete PIC formation and/or function. Indeed, early in vitro experiments indicated that SWI/SNF-mediated remodeling of nucleosomes could permit TBP/TFIIA binding to nucleosome particles (24). SWI/SNF enzymes are also required, both in vitro and in vivo, to promote transcriptional elongation of the hsp70 gene (5, 11). Thus, our data reveal an additional role for SWI/SNF chromatin-remodeling enzymes in cells and show that SWI/SNF enzymes are needed at different steps during activation of different genes.
Requirement for MyoD to initiate chromatin remodeling at the myogenin promoter: a model for SWI/SNF recruitment to the myogenin promoter.
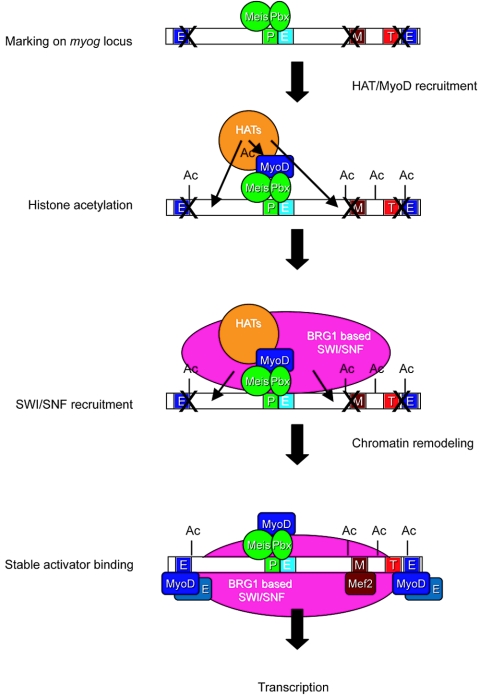
The simplest mechanism for recruitment of chromatin-remodeling enzymes to muscle-specific promoters is recruitment by DNA-bound MyoD. In our study, there is an apparent paradox: expression of MyoD was necessary for early histone acetylation and SWI/SNF recruitment, yet ChIP assays showed that MyoD formed a stable, DNA-bound complex only after these changes occurred. A model to explain this apparent paradox is that MyoD initially associates with the myogenin promoter indirectly via interactions with Pbx1/Meis proteins at a Pbx binding site next to a noncanonical E box previously identified at −123/97 of the myogenin promoter. The initiation of myogenin expression in differentiating cells was recently shown to require the interaction of MyoD with a DNA-bound complex containing the Pbx homeodomain protein in the absence of a canonical MyoD binding site, and protein interactions between MyoD and the Pbx complex were shown to be necessary for the initial association of MyoD with the myogenin promoter (4). The interaction of MyoD with these factors might induce a conformational change in the Pbx/Meis proteins that permits targeting of Brg1 in differentiating cells. Alternatively, others have demonstrated that Pbx proteins can interact with HDACs and act to repress transcription, and then, upon specific cell signaling, Pbx can become associated with HATs and promote transcription (51). A similar switch potentially could occur upon muscle differentiation, with the Pbx protein facilitating transcription via interaction with HATs and/or SWI/SNF enzymes. Indeed, coimmunoprecipitation of endogenous Brg1 and endogenous Pbx1 (Fig. 8D) provides support for models involving targeting of SWI/SNF components by Pbx/Meis proteins in differentiating cells.
However, given the existing data indicating that both SWI/SNF and HAT enzymes can also interact with MyoD, we propose that recruitment of chromatin-remodeling enzymes occurs early during differentiation via interaction with MyoD bound to the promoter indirectly through the Pbx/Meis proteins. Because the interaction of MyoD is indirect in this scenario, it would not be easily cross-linked at early time points in our ChIP assays. MyoD is known to interact with p300 and P/CAF (reviewed in reference 41); targeting these HATs to the myogenin promoter would result in acetylation of histone tails, which would help promote the association of SWI/SNF complexes through interactions with the bromodomain present on the ATPase subunit. The ability of MyoD and Brg1 to stably associate (Fig. 9) (54), combined with the ability of Brg1 to interact with Pbx (Fig. 8D), would further promote the association of Brg1-based SWI/SNF enzymes with the myogenin promoter. Thus, SWI/SNF association with the promoter can be facilitated by its interactions with acetylated chromatin, with MyoD, and with Pbx. Subsequent chromatin remodeling by SWI/SNF would then open the canonical E boxes and the Mef2 site for factor binding, resulting in stably bound activator complex at the myogenin promoter that can be detected by ChIP assay. A schematized version of these events is presented in Fig. 10.
FIG. 10.
A cartoon model of the order of events occurring during activation of the endogenous myogenin locus in MyoD-differentiated cells. Bold “X” marks placed over transcription factor binding sites indicate that the sites are inaccessible to factor binding. “P” indicates a Pbx binding site, “M” indicates a Mef2 binding site, “T” indicates the TATA box, and “E” indicates E boxes, which bind MyoD. The nonconsensus E box adjacent to the Pbx1 binding site is indicated in light blue to distinguish it from consensus binding sites elsewhere.
This model is generally consistent with the recent findings of Simone et al. (54). Using differentiating C2C12 myoblasts, they demonstrated that MyoD is recruited to the myogenin promoter and induces histone acetylation. Chromatin remodeling at the promoter required the subsequent recruitment and activity of SWI/SNF enzymes, which were dependent on an active p38 kinase. In that study, MyoD was cross-linked to the myogenin promoter prior to chromatin remodeling, which could suggest that MyoD is directly bound to the promoter but alternatively could indicate that a tighter association exists between MyoD and the Pbx complex in C2C12 myoblasts than exists in the MyoD-differentiated fibroblasts, thereby permitting MyoD to be visualized at the promoter by ChIP at the beginning of the differentiation process. Both possibilities support a model of early recruitment of MyoD to the myogenin promoter prior to acetylation and SWI/SNF recruitment, though the loss of MyoD binding in the presence of dominant-negative BRG1 (Fig. 5 and 6) and the contribution of Pbx complexes during myogenin activation in C2C12 cells (4) argue for the latter possibility. Further experiments will be needed to evaluate the role of each of the potential interactions occurring at the myogenin promoter during the activation of gene expression.
MyoD-induced genes show differential requirements for SWI/SNF enzymes.
The expression of the vast majority of genes measured by the expression array was not altered by the inhibition of SWI/SNF activity, whereas most genes induced by MyoD were modestly affected by SWI/SNF inhibition and a subset of MyoD-regulated genes were highly dependent on SWI/SNF (see Fig. 1). Our data demonstrate that chromatin modification and SWI/SNF activity are necessary for MyoD to form a stable interaction with DNA at the myogenin promoter, suggesting that chromatin remodeling may be necessary for MyoD binding at the subset of genes that are highly dependent on SWI/SNF activity. We suggest that the interaction between MyoD and Pbx is necessary to initially target MyoD to the myogenin promoter. It is interesting that there is a significant overlap between the MyoD-regulated genes that are highly dependent on SWI/SNF and the subset of genes that require the domains of MyoD that interact with Pbx for full activation (unpublished data), suggesting that these domains of MyoD might be necessary for targeting the protein to multiple different promoters as an initial step in gene activation.
Summary.
During the establishment of new cell lineages, changes in chromatin structure become apparent as previously silent genes are activated. How activators initially gain access to their binding sites within condensed chromatin structure has been a topic of intense investigation. In some cases, such as during differentiation of liver cells, the HNF3 homeodomain protein can bind to nucleosomal DNA and disrupt nucleosomal structure in the absence of ATP-dependent chromatin-remodeling enzymes (10). In other cases, gene-specific activators recruit chromatin-remodeling enzymes to specific promoters (12). Our results suggest that during muscle differentiation, muscle-specific activators may both recruit and require chromatin-remodeling activities for stable binding to the regulatory regions of muscle-specific genes.
Supplementary Material
Acknowledgments
This work was funded by an American Heart Association Scientist Development Grant and by a grant from the Medical Foundation through support from the June Rockwell Levy Foundation and the Charles A. King Trust to I.L.D., by NIH grants AR045113 to S.J.T. and GM56244 to A.N.I., and by a Scholar Award from the Leukemia and Lymphoma Society to A.N.I. C.A.B. was supported by the Chromosome Metabolism and Cancer Training Grant (NIH T32 CA09657).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Bergstrom, D. A., B. H. Penn, A. Strand, R. L. Perry, M. A. Rudnicki, and S. J. Tapscott. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell 9:587-600. [DOI] [PubMed] [Google Scholar]
- 3.Bergstrom, D. A., and S. J. Tapscott. 2001. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol. Cell. Biol. 21:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkes, C., D. A. Bergstrom, B. H. Penn, K. J. Sever, S. Knoepfler, and S. J. Tapscott. 2004. Pbx and Meis mark genes for activation by MyoD indicating a role for homeodomain genes in establishing myogenic potential. Mol. Cell 14:465-477. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. A., A. N. Imbalzano, and R. E. Kingston. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 10:1479-1490. [DOI] [PubMed] [Google Scholar]
- 6.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 7.Carey, M., and S. T. Smale. 2000. Transcriptional regulation in eukaryotes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 8.Cheng, S. W., K. P. Davies, E. Yung, R. J. Beltran, J. Yu, and G. V. Kalpana. 1999. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat. Genet. 22:102-105. [DOI] [PubMed] [Google Scholar]
- 9.Chi, T. H., M. Wan, K. Zhao, I. Taniuchi, L. Chen, D. R. Littman, and G. R. Crabtree. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418:195-199. [DOI] [PubMed] [Google Scholar]
- 10.Cirillo, L. A., F. R. Lin, I. Cuesta, D. Friedman, M. Jarnik, and K. S. Zaret. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9:279-289. [DOI] [PubMed] [Google Scholar]
- 11.Corey, L. L., C. S. Weirich, I. J. Benjamin, and R. E. Kingston. 2003. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 17:1392-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cosma, M. P. 2002. Ordered recruitment: gene-specific mechanism of transcription activation. Mol. Cell 10:227-236. [DOI] [PubMed] [Google Scholar]
- 13.Côté, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 14.Davis, R., H. Weintraub, and A. Lassar. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51:987-1000. [DOI] [PubMed] [Google Scholar]
- 15.de la Serna, I. L., K. A. Carlson, D. A. Hill, C. J. Guidi, R. O. Stephenson, S. Sif, R. E. Kingston, and A. N. Imbalzano. 2000. Mammalian SWI-SNF complexes contribute to activation of the hsp70 gene. Mol. Cell. Biol. 20:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Serna, I. L., K. A. Carlson, and A. N. Imbalzano. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187-190. [DOI] [PubMed] [Google Scholar]
- 17.de la Serna, I. L., K. Roy, K. A. Carlson, and A. N. Imbalzano. 2001. MyoD can induce cell cycle arrest but not muscle differentiation in the presence of dominant negative SWI/SNF chromatin remodeling enzymes. J. Biol. Chem. 276:41486-41491. [DOI] [PubMed] [Google Scholar]
- 18.Fryer, C. J., and T. K. Archer. 1998. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature 393:88-91. [DOI] [PubMed] [Google Scholar]
- 19.Gerber, A. N., T. R. Klesert, D. A. Bergstrom, and S. J. Tapscott. 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 11:436-450. [DOI] [PubMed] [Google Scholar]
- 20.Guidi, C. J., T. M. Veal, S. N. Jones, and A. N. Imbalzano. 2004. Transcriptional compensation for loss of an allele of the Ini1 tumor suppressor. J. Biol. Chem. 279:4180-4185. [DOI] [PubMed] [Google Scholar]
- 21.Guo, K., J. Wang, V. Andres, R. C. Smith, and K. Walsh. 1995. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol. Cell. Biol. 15:3823-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 23.Hendricks, K. B., F. Shanahan, and E. Lees. 2004. Role for BRG1 in cell cycle control and tumor suppression. Mol. Cell. Biol. 24:362-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 25.Kadam, S., and B. M. Emerson. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377-389. [DOI] [PubMed] [Google Scholar]
- 26.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang, H., K. Cui, and K. Zhao. 2004. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol. Cell. Biol. 24:1188-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khavari, P. A., C. L. Peterson, J. W. Tamkun, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 29.Knoepfler, P. S., D. A. Bergstrom, T. Uetsuki, I. Dac-Korytko, Y. H. Sun, W. E. Wright, S. J. Tapscott, and M. P. Kamps. 1999. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx-Meis1/Prep1. Nucleic Acids Res. 27:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kowenz-Leutz, E., and A. Leutz. 1999. A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4:735-743. [DOI] [PubMed] [Google Scholar]
- 31.Kwon, H., A. N. Imbalzano, P. A. Khavari, R. E. Kingston, and M. R. Green. 1994. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature 370:477-481. [DOI] [PubMed] [Google Scholar]
- 32.Liu, H., H. Kang, R. Liu, X. Chen, and K. Zhao. 2002. Maximal induction of a subset of interferon target genes requires the chromatin-remodeling activity of the BAF complex. Mol. Cell. Biol. 22:6471-6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 34.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 35.Ma, Z., M. J. Chang, R. Shah, J. Adamski, X. Zhao, and E. N. Benveniste. 2004. Brg-1 is required for maximal transcription of the human matrix metalloproteinase-2 gene. J. Biol. Chem. 279:46326-46334. [DOI] [PubMed] [Google Scholar]
- 36.Magenta, A., C. Cenciarelli, F. De Santa, P. Fuschi, F. Martelli, M. Caruso, and A. Felsani. 2003. MyoD stimulates RB promoter activity via the CREB/p300 nuclear transduction pathway. Mol. Cell. Biol. 23:2893-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mal, A., and M. L. Harter. 2003. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl. Acad. Sci. USA 100:1735-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall, T. W., K. A. Link, C. E. Petre-Draviam, and K. E. Knudsen. 2003. Differential requirement of SWI/SNF for androgen receptor activity. J. Biol. Chem. 278:30605-30613. [DOI] [PubMed] [Google Scholar]
- 39.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 40.Martens, J. H., M. Verlaan, E. Kalkhoven, and A. Zantema. 2003. Cascade of distinct histone modifications during collagenase gene activation. Mol. Cell. Biol. 23:1808-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. Signaling chromatin to make muscle. Curr. Opin. Cell Biol. 14:763-772. [DOI] [PubMed] [Google Scholar]
- 42.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 43.Nerlov, C., and E. B. Ziff. 1995. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. EMBO J. 14:4318-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parker, M. H., P. Seale, and M. A. Rudnicki. 2003. Looking back to the embryo: defining transcriptional networks in adult myogenesis. Nat. Rev. Genet. 4:497-507. [DOI] [PubMed] [Google Scholar]
- 45.Parker, S. B., G. Eichele, P. Zhang, A. Rawls, A. T. Sands, A. Bradley, E. N. Olson, J. W. Harper, and S. J. Elledge. 1995. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science 267:1024-1027. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen, T. A., E. Kowenz-Leutz, A. Leutz, and C. Nerlov. 2001. Cooperation between C/EBPalpha TBP/TFIIB and SWI/SNF recruiting domains is required for adipocyte differentiation. Genes Dev. 15:3208-3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prochasson, P., K. E. Neely, A. H. Hassan, B. Li, and J. L. Workman. 2003. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell 12:983-990. [DOI] [PubMed] [Google Scholar]
- 48.Reyes, J. C., J. Barra, C. Muchardt, A. Camus, C. Babinet, and M. Yaniv. 1998. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). EMBO J. 17:6979-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts, C. W., and S. H. Orkin. 2004. The SWI/SNF complex-chromatin and cancer. Nat. Rev. Cancer 4:133-142. [DOI] [PubMed] [Google Scholar]
- 50.Roy, K., I. L. de la Serna, and A. N. Imbalzano. 2002. The myogenic basic helix-loop-helix family of transcription factors shows similar requirements for SWI/SNF chromatin remodeling enzymes during muscle differentiation in culture. J. Biol. Chem. 277:33818-33824. [DOI] [PubMed] [Google Scholar]
- 51.Saleh, M., I. Rambaldi, X. J. Yang, and M. S. Featherstone. 2000. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20:8623-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salma, N., H. Xiao, E. Mueller, and A. N. Imbalzano. 2004. Temporal recruitment of transcription factors and SWI/SNF chromatin remodeling enzymes during adipogenic induction of the PPARγ nuclear hormone receptor. Mol. Cell. Biol. 24:4651-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sif, S. 2004. ATP-dependent nucleosome remodeling complexes: Enzymes tailored to deal with chromatin. J. Cell Biochem. 91:1087-1098. [DOI] [PubMed] [Google Scholar]
- 54.Simone, C., S. V. Forcales, D. A. Hill, A. N. Imbalzano, L. Latella, and P. L. Puri. 2004. p38 pathway targets SWI-SNF chromatin remodeling complex to muscle specific loci. Nat. Genet. 36:738-743. [DOI] [PubMed] [Google Scholar]
- 55.Soutoglou, E., and I. Talianidis. 2002. Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901-1904. [DOI] [PubMed] [Google Scholar]
- 56.Spilianakis, C., A. Kretsovali, T. Agalioti, T. Makatounakis, D. Thanos, and J. Papamatheakis. 2003. CIITA regulates transcription onset via Ser5-phosphorylation of RNA Pol II. EMBO J. 22:5125-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storey, J. D., and R. Tibshirani. 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100:9440-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tajbakhsh, S. 2003. Stem cells to tissue: molecular, cellular and anatomical heterogeneity in skeletal muscle. Curr. Opin. Genet. Dev. 13:413-422. [DOI] [PubMed] [Google Scholar]
- 59.Valdez, M. R., J. A. Richardson, W. H. Klein, and E. N. Olson. 2000. Failure of Myf5 to support myogenic differentiation without myogenin, MyoD, and MRF4. Dev. Biol. 219:287-298. [DOI] [PubMed] [Google Scholar]
- 60.Wang, W., J. Côte, Y. Xue, S. Zhou, P. A. Khavari, S. R. Biggar, C. Muchardt, G. V. Kalpana, S. P. Goff, M. Yaniv, J. L. Workman, and G. R. Crabtree. 1996. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 15:5370-5382. [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, C. J., D. M. Chao, A. N. Imbalzano, G. R. Schnitzler, R. E. Kingston, and R. A. Young. 1996. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell 84:235-244. [DOI] [PubMed] [Google Scholar]
- 63.Zhang, P., C. Wong, D. Liu, M. Finegold, J. W. Harper, and S. J. Elledge. 1999. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 13:213-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.