Abstract
CENP-A is an evolutionarily conserved, centromere-specific variant of histone H3 that is thought to play a central role in directing kinetochore assembly and in centromere function. Here, we have analyzed the consequences of disrupting the CENP-A gene in the chicken DT40 cell line. In CENP-A-depleted cells, kinetochore protein assembly is impaired, as indicated by mislocalization of the inner kinetochore proteins CENP-I, CENP-H, and CENP-C as well as the outer components Nuf2/Hec1, Mad2, and CENP-E. However, BubR1 and the inner centromere protein INCENP are efficiently recruited to kinetochores. Following CENP-A depletion, chromosomes are deficient in proper congression on the mitotic spindle and there is a transient delay in prometaphase. CENP-A-depleted cells further proceed through anaphase and cytokinesis with unequal chromosome segregation, suggesting that some kinetochore function remains following substantial depletion of CENP-A. We furthermore demonstrate that CENP-A-depleted cells exhibit a specific defect in maintaining kinetochore localization of the checkpoint protein BubR1 under conditions of checkpoint activation. Our data thus point to a specific role for CENP-A in assembly of kinetochores competent in the maintenance of mitotic checkpoint signaling.
The kinetochores of eukaryotic chromosomes are specialized protein structures that assemble on centromeric DNA and direct accurate chromosome segregation at cell division. In human somatic cells, this process occurs with less than one error once every 105 divisions. However, the frequency of errors is increased in some cancers (31). In order to ensure high levels of precision, the kinetochore mediates three functions: spindle attachment, a check that the attachment is complete, and regulated cohesion of the sister chromatids.
Kinetochore proteins were first detected and subsequently identified by immunological reactivity in mammalian cells (16, 43). Since then, a wide range of efforts to understand the molecular architecture of this essential structure have extended our knowledge of kinetochore components in organisms from yeasts to humans. More than 60 kinetochore proteins have now been identified in budding yeast, many of which are conserved in metazoans (34, 40). The vertebrate kinetochore has a distinct, well-characterized, trilaminar structure, and some kinetochore functions have now been related to these visible structural domains (3, 9, 48). Thus, it is now known that the outer kinetochore contains microtubule-binding proteins and motor proteins, like CENP-E (10) or the dynein/dynactin complex, which orchestrate chromosome movement along the spindle (28). It is also the binding site for mitotic checkpoint components, such as the Mad and Bub proteins, which are involved in the monitoring of proper chromosome alignment before anaphase entry (44). The outer kinetochore domain also contains the Ndc80(Hec1)/Nuf2 complex, which plays a role in regulation of stable kinetochore-microtubule attachment and binding of the Mad checkpoint proteins (11, 12, 27, 39, 41).
The domain located between the two sister kinetochores of duplicated chromatids, the pairing domain (15), is the site of localization for proteins involved in cohesion (cohesin complex) and mitotic regulation (the passenger complex INCENP/AuroraB/Survivin/Borealin) (1, 22, 42).
The inner kinetochore is the site of proteins that maintain their specific localization throughout the cell cycle, and thus it is thought that this structure plays a key role in recruiting and possibly coordinating the functions of the rest of the kinetochore. In vertebrates, six proteins have thus far been localized to the inner kinetochore: CENP-A, -B, -C, -H, and -I and Mis12 (16, 23, 45, 53). Homologues, although with weak sequence identity in some cases, have now been identified in Saccharomyces cerevisiae (except for CENP-B) and Schizosaccharomyces pombe, suggesting that a conserved molecular core underlies eukaryotic kinetochore assembly (40, 47, 60). In budding yeast, kinetochore assembly is primarily driven by centromere-specific sequences (40). In humans and presumably other metazoans, centromere assembly is driven by a wide variety of sequences, as demonstrated by the existence of neo-centromeres, but occurs most readily on alphoid DNA, as proven by the ability of cloned alphoid DNA to seed the formation of artificial chromosomes. Once established, however, centromere identity has been proposed to be determined by epigenetic mechanisms (33).
The inner kinetochore component CENP-A is an essential evolutionarily conserved centromere-specific histone H3 variant found only at active centromere (54). CENP-A-containing nucleosomes can be assembled in vitro and are more compact and conformationally more rigid than the corresponding H3-H4 heterotetramers (4, 61). In vivo, CENP-A nucleosomes are interspersed with H3 nucleosomes on extended chromatin fibers. Three-dimensional analysis of metaphase chromosomes by deconvolution microscopy has shown that CENP-A chromatin is organized as a cylindrical structure devoid of H3 nucleosomes and predominantly localized beneath the kinetochore (6). Immunoelectron microscopy data have indicated that CENP-A is localized both within the inner kinetochore plate and in the chromatin immediately subjacent to it (C. A. Cooke and W. C Earnshaw, unpublished data). Those structural data fit with the hypothesis that CENP-A-containing nucleosome could provide the mark for the kinetochore assembly (54).
This hypothesis is further corroborated by the “kinetochore-null” phenotype observed after RNA interference (RNAi) depletion of CENP-A in Caenorhabditis elegans embryos: chromosome segregation is completely deficient, and this is followed by random distribution of DNA in the daughter cells (46). Inactivation or depletion of CENP-A leads to mislocalization of other outer or inner kinetochore components in worms, Drosophila, and mammals (5, 23, 29, 46), thus confirming the primary role of CENP-A in kinetochore assembly. However, knockdown of CENP-A by RNAi in human cells does not impair kinetochore localization of the inner component Mis12, suggesting that, in vertebrates, kinetochore assembly might not follow a simple linear pathway (23). In addition, knockdown of CENP-A by RNAi does not completely impair chromosome segregation but only leads to missegregation defects. It remained, however, unclear whether this phenotype could result from partial depletion of CENP-A, since no cell death was associated with the RNAi depletion of this essential gene (29, 46).
In order to further elucidate the part played by CENP-A in vertebrate kinetochore assembly and function, we have performed a conditional knockout of the CENP-A gene in the chicken DT40 cell line. This cell line allows combination of genetical tools and cytological analysis (7, 20, 21, 27, 30, 45, 52). Our results indicate that CENP-A knockout in DT40 cells results in missegregation after a mitotic delay and furthermore point to a specific involvement of CENP-A in spindle checkpoint signaling.
MATERIALS AND METHODS
Cell culture and transfection.
The chicken lymphoma B-cell line was cultured and transfected as previously described (7). G418 (Sigma) was used at a final concentration of 2 mg/ml, hygromycin (Roche) at a final concentration of 1.5 mg/ml, zeocin (Invitrogen) at a final concentration of 1 mg/ml, and histidinol at a final concentration of 1 mg/ml for stable transfectants. To suppress expression of the tetracycline-repressible CENP-A transgene, doxycycline (Dox) (Clontech) was used at a final concentration of 100 ng/ml. For microtubule drug experiments, cells were treated with 0.5 μg/ml of nocodazole (Sigma) or 10 μM of paclitaxel (Taxol; Sigma) for the period of time indicated in the text.
Construction of targeting and expression vectors.
To generate a tetracycline-repressible construct for GgCENP-A, GgCENP-A cDNA (49) was cloned into the EcoRI and XbaI sites of pUHD 10.3 (24). In order to distinguish between the GgCENP-A transgene and the endogenous GgCENP-A transcript, a silent mutation (GCG→GCC) creating an EagI restriction site was introduced by site-directed mutagenesis at codon 89 of the GgCENP-A cDNA.
To construct the neomycin-resistant targeting vector, a 2.9-kb fragment upstream of the start codon and a 4.8-kb fragment downstream of the stop codon were generated by long-range PCR on DT40 genomic DNA and subcloned into pBluescript. A neomycin-resistant cassette driven by the chicken β-actin promoter was inserted between the left and right arms. For the hygromycin-resistant targeting vector, a hygromycin-resistant cassette driven by the β-actin promoter was inserted between a left 1.9-kb fragment and a right 2-kb fragment generated by long-range PCR. The hygromycin-resistant targeting construct was designed to disrupt exons 1 to 3 of the open reading frame, including the start codon.
To generate the expression construct for the Mad2-green fluorescent protein (GFP) fusion gene, chicken Mad2 cDNA (T. Fukagawa, unpublished data) was inserted into the pEGFP-N1 plasmid (Clontech).
RT-PCR screening of DT40 cells expressing tetracycline-repressible CENP-A.
Cells heterozygous for the disruption of CENP-A locus and transfected with the Tet-repressible transgene (CENP-A+/−tetCENP-A) were screened for expression and regulation of the transgene by reverse transcription-PCR (RT-PCR). Zeocin-resistant clones were grown for 48 h with (+) or without (−) doxycycline and harvested for RNA extraction. Total RNAs were reverse transcribed with a specific reverse primer located in exon 4 of GgCENP-A (5′-TGCAGGTCTTTGGGGTACAGTGTGACG-3′) using Superscript reverse transcriptase (Gibco-BRL). Amplification was carried out with the ampliTaq Gold (Perkin-Elmer) under conditions recommended by the manufacturer and using a specific forward primer located at the boundaries between exons 2 and 3 of GgCENP-A (5′-CGCGTGGTGCGGGAGATCTGCTT-3′). RT-PCR (176 bp) products were then digested with EagI, and expression and regulation of the transgene were diagnosed by the appearance of restriction fragments of 85 bp and 91 bp in the −Dox conditions.
PCR screening of DT40 cells targeted for the second CENP-A allele.
CENP-A+/−tetCENP-A clones transfected with the hygromycin targeting construct were screened for a second allele targeting event by PCR. Cells from 150 μl of cell culture were centrifuged in PCR microplates, washed in phosphate-buffered saline (PBS), and lysed in 30 μl of lysis buffer (1× PCR buffer, 0.5% Tween 20, 100 μg/ml proteinase K) for 45 min at 56°C. A further 60 μl of 1× PCR buffer was added to each well and mixed. Lysate (6 μl) was used as a template in each PCR performed with the Expand Long Template PCR system (Roche Applied Science) according to the instructions provided by the manufacturer. The forward primer (5′-ATTTCTGGCGCCGCCGGACGAACTAAACC-3′) was located in the hygromycin resistance cassette, and the reverse primer (5′-CCCAACAGTGACCTATAGAGACCTCCCTCCACC-3′) was designed on the basis of the GgCENP-A genomic sequence (49) and was located downstream of the 3′ arm of the targeting construct.
Immunoblotting.
Immunoblotting analysis was performed on DT40 nuclear extracts as previously described (49).
Cell cycle analysis.
Cell cycle analysis was carried out as described previously (21). Cells were analyzed on a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA) using CellQuest software.
FACS analysis of phosphohistone H3.
Cells (1.5 × 106 to 2 × 106) were harvested, washed in PBS supplemented with 1% bovine serum albumin (1% BSA-PBS) and fixed in 70% ethanol at −20°C overnight. Fixed cells were resuspended in 0.25% Triton X-100-PBS for 15 min on ice, washed in 1% BSA-PBS, and incubated for 1 h at room temperature with anti-phosphohistone H3 antibody (7.5 μg/ml; Upstate Biotechnology). After two rinses in 1% BSA-PBS, cells were labeled with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit immunoglobulin G (IgG) (1/15; Jackson Immunoresearch Laboratories, Inc). Stained cells were washed twice, resuspended in propidium iodide-RNase staining buffer (BD PharMingen), and subsequently analyzed by fluorescence-activated cell sorting (FACS) on a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA) using CellQuest software.
TUNEL assay.
Cells (1.5 × 106 to 2 × 106) were harvested, washed in 1% BSA-PBS, and fixed in 2% paraformaldehyde in PBS for 15 min at 4°C. Cells were washed once, resuspended in 70% ethanol, and stored overnight at −20°C. Fixed cells were washed in 1% BSA-PBS, and a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) reaction was performed as described by the manufacturer (in situ cell death detection kit, fluorescein; Roche). Stained cells were washed twice, resuspended in propidium iodide-RNase staining buffer (BD PharMingen), and subsequently analyzed on a FACSCalibur flow cytometer (Becton Dickinson, Mountain View, CA) using CellQuest software.
Indirect immunofluorescence microscopy.
DT40 cells were attached on polylysine slides (BDH), fixed in 4% paraformaldehyde in cytoskeleton buffer {1.1 M Na2HPO4, 0.4 M KH2PO4, 137 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM EGTA, 5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 5.5 mM glucose, pH 6.1} for 5 min at 37°C, permeabilized in 0.15% Triton X-100 in cytoskeleton buffer, and rinsed in PBS. Alternatively (see Fig. 1D, 3D, and 4C), cells were fixed in cold methanol for 30 min. Antibody incubation was done in 1% BSA-PBS for 1 h at 37°C. The following antibodies were used: FITC-conjugated anti-α-tubulin monoclonal (1:50, Sigma), anti-phosphohistone H3 (1:200, Upstate Biotechnology), rabbit anti-BubR1 (1:500) (45), rabbit anti-INCENP (1:500) (17), rabbit anti-chicken CENP-A (1:50) (49), and rabbit anti-Xenopus CENP-E (1:500) (A. Abrieu, unpublished data). Primary antibodies were detected with (Alexafluor 594)-conjugated goat anti-rabbit IgG (1:200; Molecular Probes), and the DNA was counterstained with DAPI (4′,6-diamidino-2-phenylindole) (Sigma) at 0.1 μg/ml.
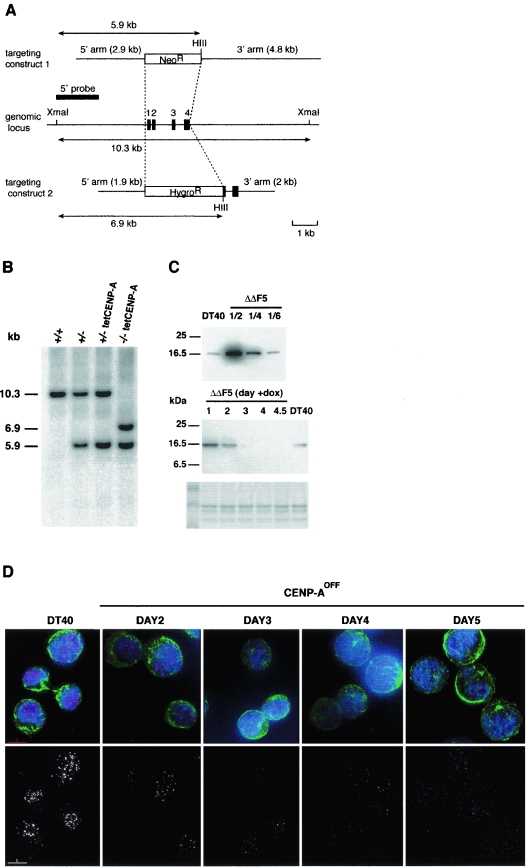
FIG. 1.
Generation of a conditional CENP-A−/− clone. (A) Schematic representation of the chicken CENP-A locus and the gene targeting constructs. Black boxes indicate the position of exons. HIII indicates HindIII sites. (B) Southern blot analysis of wild-type (+/+), heterozygous mutant (+/−), heterozygous mutant with random integration of the Tet-repressible CENP-A transgene (+/− tetCENP-A) and homozygous (−/−) mutant clones. DNA was digested with XmaI and HindIII and hybridized with the 5′ probe shown in panel A. First and second targeted events are diagnosed by the appearance of 5.9- and 6.9-kb restriction fragments replacing the cognate 10.3-kb fragment. (C) Western blot analysis of CENP-A expression. Nuclear extracts from DT40 cells or the CENP-A−/− cell line expressing the Tet-repressible transgene (ΔΔF5) were separated on a 15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel and analyzed by immunoblotting with affinity-purified rabbit anti-chicken CENP-A. Coomassie blue staining of histone proteins in the extracts was used as a loading control. (Upper panel) Serial dilutions of ΔΔF5 extracts were loaded to quantify overexpression level in the ΔΔF5 cell line. (Lower panel) Time course of CENP-A repression following addition of doxycycline in the ΔΔF5 cell line. (D) Immunofluorescence analysis of DT40 and ΔΔF5 cell line at the indicated time following addition of doxycycline. Cells were stained for CENP-A (red), α-tubulin (green), and DNA (blue). Images for each time point were acquired using the same acquisition parameters. Background signal is increased at late time points (days 4 and 5) due to nonspecific binding of the primary antibody (see main text).
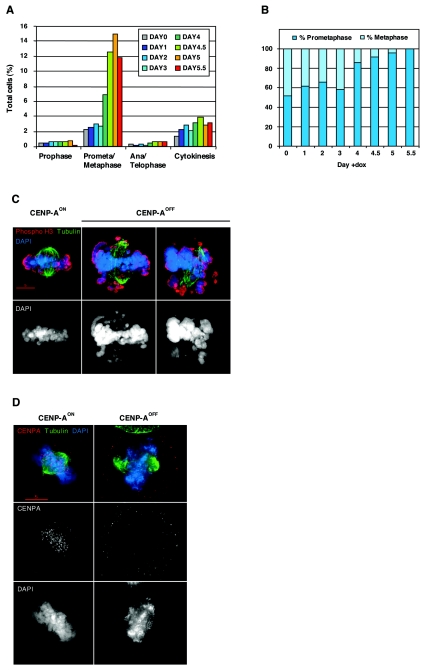
FIG. 3.
CENP-A-deficient cells are delayed in prometaphase. (A) Distribution of mitotic stages in CENP-AON (day 0) and CENP-AOFF cells during the depletion time course. Counting was performed on cells stained for tubulin, phosphohistone H3, and DNA. Five hundred to 2,000 cells, including at least 100 mitotic cells, were counted for each time point. (B) Ratio of prometaphase and metaphase cells in the CENP-AON (day 0) and CENP-AOFF populations during the depletion time course. Fifty prometaphase/metaphase cells with a bipolar spindle where both poles were localized in the same plane were counted for each time point. Equatorial chromosome alignment defined the metaphase stage. (C) CENP-AON and CENP-AOFF (doxycycline, day 4.5) were stained for tubulin (green), phosphohistone H3 (red), and DNA (blue). Metaphase cells as observed in control CENP-AON cells were rarely observed in CENP-AOFF cells. Instead, the frequency of cells with misaligned chromosomes increased. (D) CENP-AON and CENP-AOFF (doxycycline, day 4.5) were stained for tubulin (green), CENP-A (red), and DNA (blue). Kinetochore signals were no longer observed on CENP-A-deficient prometaphases.
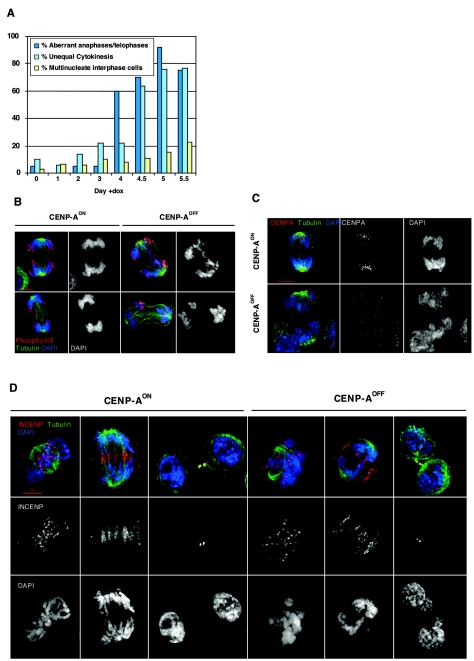
FIG. 4.
CENP-A-deficient cells exhibit severe chromosome segregation defects. (A) Quantification of anaphases/telophases with lagging chromosomes, unequal cytokinesis, or multinucleate interphase cells in CENP-AON (day 0) and CENP-AOFF cells. The percentage shown in each category represents the fraction of aberrant events out of the total number of events falling into the category. For each time point until day 5, the anaphases/telophases, cytokinesis, and interphases recorded were 20, 50, and 500, respectively. For day 5.5, the values were 8, 13, and 500, respectively. (B) CENP-AON and CENP-AOFF (doxycycline, day 4.5) anaphases (upper panel) and telophases (lower panel) were stained for tubulin (green), phosphohistone H3 (red), and DNA (blue). Few or multiple lagging chromosomes were frequently observed in CENP-AOFF cells. (C) CENP-AON and CENP-AOFF (doxycycline, day 4.5) anaphase cells stained for tubulin (green), CENP-A (red), and DNA (blue). Discrete CENP-A signals located at poles in CENP-AON cells were no longer visible in CENP-AOFF cells. (D) CENP-AON and CENP-AOFF (doxycycline, day 4.5) cells in prometaphase, anaphase, or cytokinesis stained for tubulin (green), INCENP (red), and DNA (blue). INCENP localizes at kinetochores in CENP-AOFF prometaphase cells and transfers to the midzone in missegregating CENP-AOFF cell anaphases. Cytokinesis in CENP-AOFF cells resulted in daughter cells with several nuclei of unequal size.
For Fig. 5, chromosome spreads were processed as described previously (14) and stained with antibodies to chicken CENP-C (1:1,000) (20), chicken CENP-H (1:2,000) (21), chicken CENP-I (1:2,000) (45) or chicken Hec1 (1/1,000) (27). For CENP-H and CENP-I antibodies, cells were fixed in cold methanol for 30 min.
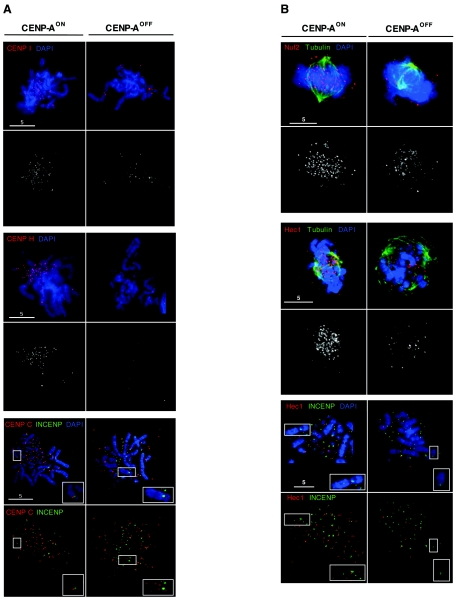
FIG. 5.
CENP-AOFF cells are depleted of the inner kinetochore proteins CENP-I, CENP-H, and CENP-C and of the Nuf2/Hec1 complex. (A) CENP-AON and CENP-AOFF metaphase spread (doxycycline, day 5) cells were stained for CENP-I (red, upper panel), CENP-H (red, middle panel), CENP-C (red, lower panel), and DNA (blue). Cells stained with CENP-C were also stained with INCENP (green, lower panel). While CENP-AON cells display strong kinetochore signals for CENP-I, CENP-H, and CENP-C, CENP-AOFF cells showed reduction of signal number and/or intensity for CENP-I and CENP-H. CENP-C signals in CENP-AOFF cells become diffuse and were no longer kinetochore localized on mitotic chromosomes. Inset panels show the inner centromere localization of INCENP and the absence of CENP-C staining for the chromosomeindicated by the box. (B) CENP-AON and CENP-AOFF prometaphase cells (doxycycline, day 5) were stained for tubulin (green), DNA (blue), and Nuf2 (red, upper panel) or Hec1 (red, middle panel). Nuf2 and Hec1 staining is strongly reduced in CENP-AOFF cells. CENP-AON and CENP-AOFF metaphase spreads (lower panel) were stained with Hec1 (Red), INCENP (green), and DNA (blue). Inset panels show that kinetochore localization of Hec1 is abolished in CENP-AOFF cells. For all immunofluorescence stainings, control slides stained with the anti-CENP-A antibody were included and no significant chromosomal signal could be detected in CENP-AOFF cells.
For whole-cell immunofluorescence in Fig. 5, cells were fixed in cold methanol for 30 min, washed in PBS, and treated three times for 5 min in TEEN buffer (1 mM triethanolamine-HCl, pH 8.5, 0.2 mM Na EDTA, 25 mM NaCl), 0.1% Triton X-100, 0.1% BSA prior to incubation with chicken Nuf2 antibody (1/1,000) (27) diluted in TEEN buffer, 0.1% Triton X-100, 0.1% BSA. Slides were washed three times for 5 min in KB buffer (10 mM Tris-HCl, pH 7.7, 0.15 M NaCl, 0.1% BSA) and incubated with Alexafluor 594-conjugated anti-rabbit IgG (1:200; Molecular Probes) diluted in KB buffer. After a further wash in KB buffer, DNA was counterstained with DAPI (Sigma; 0.1 μg/ml). For whole-cell immunostaining with Hec1 antibody, cells were fixed in 4% paraformaldehyde diluted in PBS prior to the methanol fixation.
Three-dimensional data sets were collected using a DeltaVision system (Applied Precision), deconvolved, and projected into a single plan.
RESULTS
Generation of DT40 cells conditionally deficient for CENP-A expression.
Conditional disruption of genes encoding essential centromeric proteins has been successfully engineered in DT40 cells by the sequential disruption of the genomic loci and introduction of a rescuing cDNA construct under the control of a Tet-repressible promoter (57). We undertook this approach with the centromeric histone H3 variant CENP-A. We had previously characterized the chicken CENP-A cDNA and genomic locus (49). We generated a CENP-A deletion construct in which the entire CENP-A coding sequence was replaced with a neomycin resistance cassette (Fig. 1A). We recovered one targeted clone following transfection of this construct into DT40 cells and analysis of 23 neomycin-resistant clones. Correct genomic targeting was confirmed by Southern blot analysis with 5′ and 3′ probes (Fig. 1B and data not shown). This clone was cotransfected with a chicken CENP-A cDNA construct under the control of a tetracycline-repressible promoter and a Tet-repressible transactivator containing a zeocin (Zeo) resistance cassette (21, 24).
In order to target the second CENP-A allele, we generated a second CENP-A deletion construct containing the hygromycin resistance cassette and shorter 5′ and 3′ arms (Fig. 1A). This allowed us to design a PCR strategy for screening of targeted hygromycin-resistant clones (see Materials and Methods). Four independent lines of DT40 cells targeted for the first CENP-A allele and expressing the tetracycline-regulated CENP-A transgene (CENP-A+/−tetCENP-A) were transfected with the second CENP-A disruption construct. For each cell line, we screened about 100 neomycin- and hygromycin-resistant clones, and for one cell line, we isolated three clones in which the remaining CENP-A allele was targeted (CENP-A−/−tetCENP-A). Southern blot hybridization confirmed the targeting event (Fig. 1B). One of these CENP-A conditionally null clones, ΔΔF5, was chosen for further analysis.
Western blotting with anti-GgCENP-A antibody showed that the level of CENP-A expression from the Tet-repressible transgene in the ΔΔF5 cell line was ≈6 times higher than that of endogenous CENP-A level in the DT40 cell line (Fig. 1C). Following addition of doxycycline, CENP-A protein level decreased gradually and became undetectable under our blotting conditions after 4 days of treatment (Fig. 1C). Progressive decrease of CENP-A protein level was also observed by indirect immunofluorescence. CENP-A spots were reduced over time in number and/or intensity, and by day 4 the typical strong punctate nuclear pattern was no longer detectable (Fig. 1D). Fluorescent signal could still be detected at late time points (days 4 and 5). Since CENP-A was undetectable in nuclear extracts by Western blotting at these time points (Fig. 1C), this signal was most likely due to nonspecific binding of the anti-GgCENP-A antibody. In agreement with this interpretation, this signal did not localize with chromosomes (data not shown and see Fig. 3D and 4C).
Analysis of the depletion phenotype described hereafter was performed at day 4.5 or day 5 of the depletion time course.
CENP-A-deficient cells are delayed in mitosis but undergo apoptotic cell death in G1.
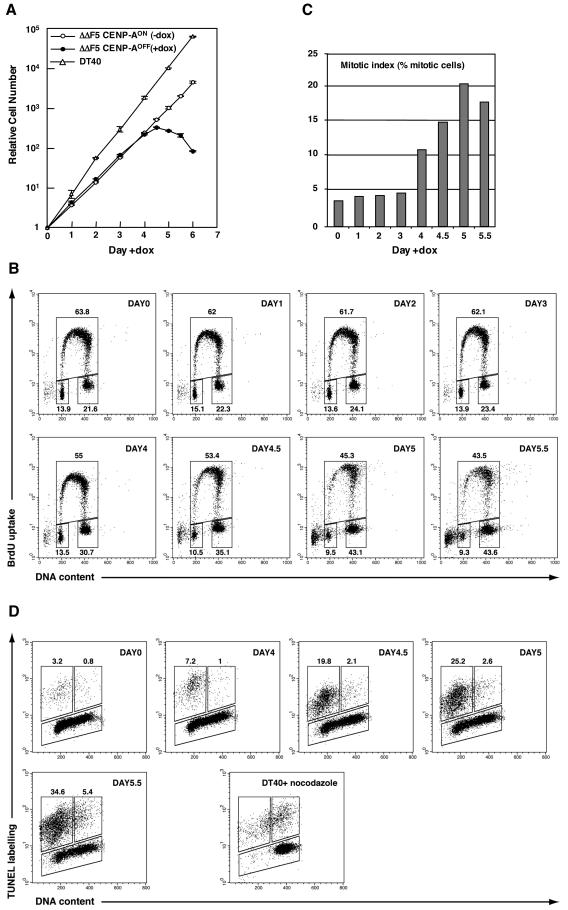
Clone ΔΔF5 grown in the absence of doxycycline (CENP-AON) has a doubling time of 12.5 h compared to 8.8 h for DT40 cells (Fig. 2A). However, the fractions of ΔΔF5 cells in the different phases of the cell cycle were essentially the same as for DT40 cells, indicating no major delay in any of the cell cycle stages (data not shown). ΔΔF5 cells grown with doxycycline (CENP-AOFF) stopped proliferating after 7 to 8 cell cycles postaddition of the drug (day 4), and extensive cell death was visible in the culture by day 4.5 (Fig. 2A). Cell cycle analysis performed during the depletion time course revealed an accumulation of cells in G2/M from day 4 (up to 43.6% at day 5.5; Fig. 2B). Concomitantly, the mitotic index increased (Fig. 2C). By combining our cell cycle data (Fig. 2B) and the mitotic index (Fig. 2C), we could estimate that the fraction of G2 cells did not vary significantly between day 0 and day 5 (17.9% and 22.4%, respectively) and that the increase in mitotic cells could account for the increase in G2/M cells. We therefore conclude that CENP-AOFF cells accumulate in mitosis. However, the mitotic index reached a plateau value of ≈20% at days 5 and 5.5, and cell cycle analysis revealed that CENP-AOFF cells were still cycling even when extensive cell death had occurred in the culture (43.5% in S phase at day 5.5). These data strongly suggest that the accumulation of the CENP-AOFF cells in mitosis results from a transient delay rather than an absolute mitotic arrest.
FIG. 2.
Cell cycle analysis of CENP-AOFF cells. (A) Growth curve of parental DT40 and ΔΔF5 cells in the presence or absence of doxycycline. (B) Cell cycle distribution of ΔΔF5 cells after addition of doxycycline as measured by bromodeoxyuridine (BrdU) incorporation and DNA content in flow cytometry analysis. Cells were pulse-labeled with BrdU and stained with FITC-anti-BrdU to detect BrdU incorporation (vertical axis, logarithmic scale) and propidium iodide to detect total DNA (horizontal axis, linear scale). The lower-left gate identifies G1 cells, the upper gate identifies cells incorporating BrdU (S phase), and the lower-right gate represents G2/M cells. The numbers show the percentage of cells falling in each gate, excluding the apoptotic sub-G1 cells. (C) Mitotic index of ΔΔF5 cells following addition of doxycycline. Fraction of mitotic cells was determined by flow cytometry analysis of phosphohistone H3 staining. (D) Flow cytometric analysis of apoptosis and cell cycle in the ΔΔF5 cell line at the indicated time following addition of doxycycline. DNA strand breaks were labeled by TUNEL reaction using fluorescein-conjugated dUTP (vertical axis, logarithmic scale), and cells were stained with propidium iodide to determine the DNA content (horizontal axis, linear scale). The upper-left gate identifies positive apoptotic cells with a 2N DNA content, whereas the upper-right gate identifies positive apoptotic cells with a 4N DNA content. DT40 cells treated with nocodazole for 16 h were used as a control for apoptosis occurring with a 4N DNA content.
DNA content analysis of CENP-AOFF cells during the repression time course also showed that sub-G1 cells accumulate at later time points (day 4 to day 5.5), reminiscent of DNA fragmentation occurring during apoptotic cell death (Fig. 2B). We confirmed the occurrence of extensive apoptosis following the loss of CENP-A by TUNEL assay (Fig. 2D). Furthermore, we showed by dual analysis of TUNEL labeling and DNA content that the apoptotic cell death of CENP-AOFF cells occurred mainly in the G1 phase of the cell cycle.
CENP-A-deficient cells are delayed in prometaphase and go through mitosis with severe chromosome segregation defects.
We further examined the mitotic progression of CENP-A-deficient cells by immunofluorescence analysis. The distribution of the different mitotic stages revealed a gradual increase in prometaphase cells over time (Fig. 3A and 3B). While prometaphase represents ≈1.2% of the total number of cells at day 0, this value reached 14.3% at day 5, indicative of an ≈12-fold increase. In contrast, the number of metaphase cells, where chromosomes have reached alignment at the spindle equator, decreased over time, and virtually no metaphase cell could be detected at day 5.5 (Fig. 3B). As shown in Fig. 3C, we could observe both prometaphases where chromosomes had congressed to the spindle equator with only a few nonaligned chromosomes and those with a more dramatic phenotype with all chromosomes randomly distributed over the mitotic spindle. We cannot exclude that CENP-A levels below our detection threshold remain on centromeres of correctly aligned chromosomes, but we did not detect CENP-A signals on any of the prometaphase cells (Fig. 3D). We conclude that CENP-A-deficient DT40 cells display defects in proper chromosome congression. The prometaphase spindle of CENP-AOFF cells (n = 30) was 39% longer than that of control cells (n = 20), suggesting a deficiency in pulling forces originating from kinetochore fibers.
The percentage of anaphase and cytokinesis cells did not decrease significantly over the time-course of depletion (Fig. 3A), thus confirming the transient nature of the prometaphase delay. However, close cytological examination of those mitotic figures revealed aberrant behaviors whose frequency increased over time (Fig. 4A). At day 5, 92% of anaphases were abnormal. These included lagging chromosomes in the middle of two well-separating DNA masses or an even more disorganized phenotype with many lagging chromosomes, no clear concentration at the spindle pole, and obvious unequal separation of DNA masses (Fig. 4B, 4C, and 4D). No CENP-A signal could be detected on centromeres in those anaphase cells (Fig. 4C). We found that in DT40, as in C. elegans (46), localization of the passenger protein INCENP was not impaired in CENP-A-deficient cells (Fig. 4D), and we used INCENP labeling and observation of its transfer to the spindle midzone as a marker to confirm that the aberrantly segregating cells were indeed in anaphase. As the frequency of missegregating anaphases increased, we also observed an increase in cytokinesis with multinucleated daughter cells (Fig. 4A and 4D). The micronuclei were of various sizes, indicating that the nuclear envelope had apparently reformed around “orphan chromosomes” that did not move normally to the poles during anaphase. It was also obvious in some cases that the two daughter cells had unequal DNA content. The number of multinucleated interphase cells also increased in the culture over time but ultimately did not exceed 22%. Taken together, our data indicate that CENP-A-deficient cells go through mitosis with severe chromosome segregation defects. Our TUNEL assay results strongly suggest that this missegregation phenotype leads to an apoptotic cell death in the subsequent G1 phase of the cell cycle.
CENP-A-deficient cells are depleted for CENP-I, CENP-H, CENP-C, and the Nuf2/Hec1 complex.
We had previously determined that some interdependency relationships exist for the recruitment of kinetochore proteins in DT40 cells: we have shown that the inner centromere components CENP-I and CENP-H are mutually necessary for their binding to the kinetochore and are both required for the recruitment of CENP-C (21, 45). Also the kinetochore-bound fraction of the transient mitotic Nuf2/Hec1 complex was significantly reduced in cells depleted for CENP-I or CENP-H (27). Furthermore, we had previously shown that depletion of any of these centromere proteins does not affect CENP-A kinetochore localization. We used immunofluorescence analysis with specific antibodies for chicken CENP-I, -H, and -C, Nuf2, and Hec1 to examine whether their kinetochore localization was affected in CENP-AOFF cells.
We found that kinetochore localization of the inner kinetochore components CENP-I, CENP-H, and CENP-C was severely impaired in CENP-A-depleted cells: analysis of metaphase chromosomes showed that the typical kinetochore staining observed for control cells was lost upon depletion of CENP-A (Fig. 5A). We noticed, however, that a few chromosomes occasionally still retained some kinetochore staining for CENP-I and CENP-H. Depletion of CENP-A resulted in diffuse CENP-C signals, and double-immunofluorescence staining with antibodies against CENP-C and INCENP, which unambiguously labels the inner centromere, showed that kinetochore localization of CENP-C was lost for the vast majority of chromosomes (Fig. 5A, lower panel).
We also showed that the mitotic kinetochore association of the Nuf2/Hec1 complex was affected in CENP-A-depleted cells (Fig. 5B): prometaphase CENP-AOFF cells no longer displayed the punctate centromeric signal (Fig. 5B, upper and middle panels), and double-immunofluorescence analysis of metaphase spreads with INCENP and Hec1 confirmed that the kinetochore signal was abolished by the depletion of CENP-A (Fig. 5B, lower panel).
Taken together, our results indicate that the inner kinetochore components CENP-I, CENP-H, and CENP-C as well as the Nuf2/Hec1 complex in the outer kinetochore fail to be normally recruited at kinetochores of DT40 CENP-A-depleted cells.
Binding of BubR1 is not stably maintained in CENP-A-deficient prometaphase cells, while recruitment of CENP-E and Mad2 is severely affected.
In many eukaryotes, cell division is under the control of a spindle assembly checkpoint that prevents anaphase onset until all chromosomes have achieved proper alignment on the mitotic spindle (44). The ability of CENP-A-deficient cells to enter anaphase despite the failure to complete chromosome biorientation suggested that those cells eventually override the spindle checkpoint. In order to gain some insight into the checkpoint status of CENP-A-deficient cells, we therefore examined the behavior of the mitotic checkpoint machinery using antibodies against BubR1, CENP-E, and Mad2.
All three proteins are key components of vertebrate mitotic checkpoint signaling. BubR1 and Mad2 are directly involved in the generation of the wait-anaphase signal (62, 64) and the kinesin-like motor protein CENP-E plays a role in recruitment and activation of the BubR1 kinase (38, 59). Their kinetochore localization depends upon microtubule-kinetochore interactions as they accumulate at unattached kinetochores in prometaphase cells and subsequent chromosome biorientation on the metaphase plate correlates with a significant decrease of the BubR1 and CENP-E signals and loss of Mad2 localization (26).
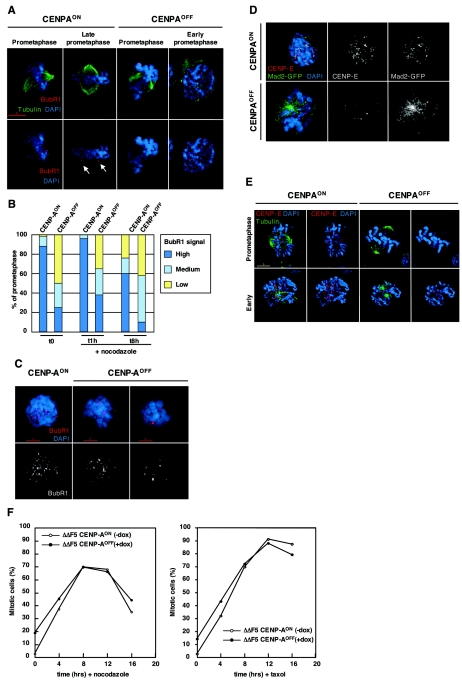
When CENP-AON cells were stained with BubR1 antibody, we observed a strong kinetochore staining in prometaphase which was significantly reduced upon chromosome alignment in late prometaphase (Fig. 6A). At the population level, 88% of CENP-AON prometaphase cells exhibited high staining (Fig. 6B). In contrast, in CENP-AOFF cells, we observed that 75% of prometaphase cells exhibited either medium or low signals (few or very few kinetochores remained positively stained, respectively; Fig. 6B). The vast majority of those prometaphase cells had bipolar spindles and displayed many unaligned chromosomes (Fig. 6A, compare the prometaphase stage harboring unaligned chromosomes in CENP-AON cells versus that in CENP-AOFF cells; CENP-AON, left panel, and CENP-AOFF, left panel). Strikingly, when we scored BubR1 signals only in early prometaphase of CENP-AOFF cells (soon after nuclear envelope breakdown), we noticed that those cells displayed a kinetochore staining as strong as control cells (n = 14, Fig. 6A). This indicated that BubR1 could be efficiently recruited to kinetochores in CENP-AOFF cells but did not maintain its kinetochore localization despite the failure of proper chromosome alignment.
FIG. 6.
Localization of the checkpoint proteins BubR1, CENP-E, and Mad2 is affected in CENP-A-depleted cells. (A) CENP-AON and CENP-AOFF cells (doxycycline, day 4.5) were stained for tubulin (green), BubR1 (red), and DNA (blue). Unaligned chromosomes in CENP-AON prometaphase cells have strong BubR1 signals. In late prometaphase, signal intensity decreases upon chromosome alignment while out-of-the-plate chromosomes retain strong staining, as indicated by arrows. CENP-AOFF cells only retain a few kinetochores staining on misaligned chromosomes. CENP-AOFF cells still exhibit strong staining in early prometaphase. (B) Quantitation of CENP-AON and CENP-AOFF (doxycycline, day 4.5) prometaphase cells with high (many kinetochores), medium (few kinetochores), or weak (very few kinetochores) BubR1 signals. Cells were either nontreated (t = 0) or received nocodazole for 1 h or 8 h. Fifty prometaphase cells were counted in each case. (C) CENP-AON and CENP-AOFF (doxycycline, day 4.5) were treated with nocodazole (1 h) and stained for BubR1 (red) and DNA (blue). CENP-AON prometaphase cells retain a strong BubR1 staining when treated with the spindle drug, while most of CENP-AOFF cells had either few (CENP-AOFF, left panel) or very few (CENP-AOFF, right panel) kinetochores stained. (D) Nocodazole-treated (2 h) CENP-AON and CENP-AOFF cells stably expressing a Mad2-GFP construct were stained for CENP-E (red) and DNA (blue). Kinetochore localization of CENP-E and Mad2-GFP was lost in nocodazole-arrested prometaphase cells. (E) CENP-AON and CENP-AOFF cells (doxycycline, day 4.5) were stained for tubulin (green), CENP-E (red), and DNA (blue). CENP-E signals were hardly detectable in prometaphase cells, whereas some signals could be detected in early prometaphase. (F) Mitotic index of CENP-AON and CENP-AOFF (doxycycline day 4.5) during a 16-h time course of nocodazole or paclitaxel treatment. The fraction of mitotic cells was determined by flow cytometry analysis of phosphohistone H3 staining. For all immunofluorescence stainings, control slides stained with the anti-CENP-A antibody were included and no significant chromosomal signal could be detected in CENP-AOFF cells.
Depletion of BubR1 from kinetochores normally depends on the establishment of kinetochore-microtubule attachment and tension between attached sister kinetochores (51). In order to determine whether depletion of the BubR1 kinetochore signals in CENP-AOFF cells could be due to microtubule-kinetochore interactions, we examined the behavior of BubR1 after blocking microtubule attachment with spindle drugs. CENP-AON and CENP-AOFF cells were treated for one hour with nocodazole, a microtubule-depolymerizing agent. As described for mammalian cells (56), CENP-AON prometaphase DT40 cells had prominent BubR1 staining (Fig. 6B and 6C, high signal) after nocodazole treatment (96% of the cell population). In CENP-AOFF-treated cells, only 38% of prometaphase cells were as strongly stained as control cells whereas the majority of prometaphase cells had either a medium or low staining (Fig. 6B). We thus concluded that the depletion of BubR1 from CENP-AOFF kinetochores cannot be reversed by spindle-damaging agents and is therefore not dependent on kinetochore-microtubule attachment. After a longer nocodazole treatment (8 h), only 10% of CENP-AOFF prometaphase cells retained a strong staining compared to 60% in CENP-AON cells (Fig. 6B), confirming that CENP-A-deficient cells are defective in maintenance of the checkpoint protein BubR1 under conditions of spindle checkpoint activation.
Immunostaining with CENP-E antibodies and analysis of a CENP-A-deficient cell line that stably expressed Mad2-GFP revealed that depletion of CENP-A severely affected kinetochore localization of those checkpoint proteins. The prominent kinetochore staining pattern observed in CENP-AON nocodazole-arrested prometaphase cells disappeared in CENP-AOFF cells (Fig. 6D). We noticed, however, that early prometaphase cells exhibited some level of kinetochore binding of CENP-E (Fig. 6E, n = 10), whereas no signal could be detected in prometaphase cells where bipolar spindle had already formed.
In conclusion, our data thus indicate that CENP-AOFF cells exhibit defects in kinetochore localization of the checkpoint proteins BubR1, CENP-E, and Mad2. We show that depletion of CENP-A affects the kinetochore recruitment of CENP-E and Mad2 and that, although CENP-A-deficient cells are able to recruit the checkpoint signaling component BubR1, they are deficient in maintaining its kinetochore localization in a process that appears to be independent of kinetochore-microtubule interactions.
CENP-A-depleted cells retain their ability to activate the spindle checkpoint in response to spindle drugs.
The ability of CENP-A-depleted cells to exit mitosis without normal chromosome segregation as well as their defects in localizing the checkpoint proteins BubR1, CENP-E, and Mad2 led us to test whether those cells were still able to activate the spindle checkpoint in response to spindle-damaging drugs. We thus challenged CENP-AOFF cells with nocodazole, which depolymerizes microtubules, or paclitaxel, which abolishes tension at attached kinetochores (58). Quantitative flow cytometry analysis with anti-phosphohistone H3, a mitotic marker, revealed that CENP-AOFF cultures accumulated mitotic cells similarly to control cultures over a 16-h time course of nocodazole or paclitaxel treatment (Fig. 6F). Thus, the loss of CENP-A did not impair the overall activation of the checkpoint signaling pathway in response to spindle-damaging agents.
DISCUSSION
Conditional knockout of the centromeric histone protein CENP-A in the vertebrate DT40 cell line led to a gradual decrease in CENP-A protein levels, consistent with CENP-A being a long-lived protein (50). CENP-A was not detectable by immunoblotting after 7 to 8 generations, and the typical CENP-A distribution, as observed by indirect immunofluorescence microscopy, was no longer visible on interphase or mitotic cells from day 4 of the repression time course. This correlated with a cessation of cell proliferation at day 4 and apoptotic cell death at day 4.5, thus confirming that CENP-A is essential for viability (29).
Our phenotypic analysis of CENP-A-depleted DT40 cells first revealed a congression defect at mitosis, as indicated by the accumulation of cells in prometaphase, with condensed chromosomes scattered around an extended bipolar spindle. This suggests that CENP-A-depleted kinetochores are deficient in forming the microtubule-kinetochore interactions necessary for generating chromosome biorientation as well as for establishing a mechanically stable mitotic spindle. However, despite this defect in chromosome congression, CENP-A-depleted cells were only transiently delayed in prometaphase and subsequently completed chromosome segregation and cytokinesis, albeit with severe missegregation defects. This missegregation phenotype corroborates previous observations when depleting CENP-A either by genetic knockout in mice (29), or by RNAi in Drosophila or human cells (5, 23). Use of the DT40 genetic system allowed us to further demonstrate that vertebrate CENP-A knockout cells die in the G1 stage of the cell cycle. Anti-CENP-A antibody injection in human cells or Drosophila embryos has also been previously shown to result in interphase arrest (5, 18). In DT40 CENP-A-depleted cells, we observed apoptosis in G1 without any accumulation of cells in that stage, suggesting that apoptosis is likely to be due to the massive missegregation that results from CENP-A depletion.
Proper kinetochore assembly did not occur in DT40 CENP-A-depleted cells. CENP-A depletion impairs the localization of the inner kinetochore components CENP-C, CENP-H, and CENP-I as well as the outer kinetochore components Nuf2/Hec1, CENP-E, and Mad2. We had previously shown that depletion of CENP-I or CENP-H in DT40 cells affects the kinetochore recruitment of the Nuf2/Hec1 complex, which in turn is involved in the recruitment of Mad2 (27). Thus, our data confirm that CENP-A is an upstream component of a hierarchical kinetochore assembly pathway (19).
Mislocalization of the inner kinetochore components in DT40 CENP-A-depleted cells is in agreement with what has been observed in embryos from the CENP-A knockout mouse (29) or after depleting CENP-A by RNAi in human cells (23). However, localization of the outer component Mad2 was found to be unaffected after RNAi depletion of human CENP-A (23). This discrepancy could reflect differences in functional interactions among kinetochore components between humans and chickens. However, since human CENP-I and Nuf2/Hec1 are involved in efficient kinetochore targeting of Mad2 (11, 37, 39), it is possible that the observed Mad2 targeting after RNAi depletion of CENP-A could be due to residual levels of kinetochore-bound CENP-A. Indeed, levels of CENP-A depletion are likely to be different between the two experimental systems, since in the RNAi experiments, human cells were examined after three generations whereas we performed the analysis of the DT40 cells at least eight generations after inducible CENP-A expression was turned off.
Examining kinetochore assembly in DT40 CENP-AOFF cells, we also found that CENP-A depletion does not affect the initial recruitment of the outer kinetochore protein BubR1 nor the localization of the inner centromere component INCENP. We also noticed that after nocodazole treatment of CENP-AOFF cells, BubR1 staining adopted a typical crescent morphology (Fig. 6C), reflecting the expansion of the kinetochore outer domain (26). We conclude from those data that CENP-A depletion does not impair the entire kinetochore structure. Indeed, our phenotypic analysis revealed that chromosome segregation was not completely abrogated after depletion of CENP-A in vertebrate cells. At time points where CENP-A was undetectable by immunofluorescence, we could observe some chromosome alignment in prometaphase cells as well as anaphase A chromatid migration to the poles, suggesting that functional interactions may still occur between microtubules and chromosomes. Taken together, our results suggest that some kinetochore function remains even following substantial depletion of the centromeric histone variant.
Although, as for any conditional knockout, we cannot formally exclude that minute amounts of undetectable CENP-A could result in partial kinetochore function, our data suggest that CENP-A may not be the only mark for kinetochore assembly in somatic cells. This raises the interesting possibility that CENP-A could be required at a given developmental stage to specify the epigenetic mark of centromeric chromatin but might later become partly dispensable for kinetochore assembly in somatic cells. Such a mechanism would be consistent with the observation that CENP-A depletion in somatic cells leads to missegregation, whereas in C. elegans embryos, where the nature of the epigenetic mark may not be as well established, it results in a “kinetochore-null” phenotype.
Importantly, the fact the CENP-A-depleted cells undergo a transient mitotic delay before proceeding through mitosis with chromosome missegregation reveals that those cells are able to activate the mitotic checkpoint but that the checkpoint response is subsequently overridden. We find that CENP-A depletion affects kinetochore localization of the checkpoint components Mad2, BubR1, and CENP-E. Indeed, Mad2 protein fails to target unattached kinetochore in CENP-A-depleted cells, whereas BubR1 can be efficiently recruited but fails to maintain its kinetochore localization under conditions of checkpoint activation. Recruitment of CENP-E was severely diminished by depletion of CENP-A, although we could observe some initial recruitment in early prometaphase cells. Our data thus may suggest that CENP-A-depleted cells are deficient in generating a MAD2-dependent signal from unattached kinetochores, but that checkpoint activation can occur through a BubR1-dependent pathway. We hypothesize that subsequent defective maintenance in kinetochore localization of BubR1 and/or of its activator CENP-E further results in premature inactivation of the signaling pathway and progression through mitosis.
It is worth noting that CENP-A-depleted cells could activate and efficiently maintain the checkpoint upon treatment with spindle-damaging agents. This ability to maintain a prolonged arrest when treated with spindle poisons despite only a transient mitotic arrest in the absence of drugs is reminiscent of what has been observed when depleting the kinetochore components CENP-I and CENP-E (37, 55, 59). As previously suggested by those authors, we could explain this phenotype if a certain threshold level of inhibitory signal generated by unattached kinetochores is necessary for a cell to sustain a mitotic arrest. Levels below this threshold would be unable to sustain checkpoint activation. An argument supporting this hypothesis is given by immunodepletion experiments in Xenopus, where it could be shown that readdition of 20% of the normal level of kinase-competent BubR1 to BubR1-depleted extracts did not restore checkpoint activation, suggesting that normal levels of BubR1 may be required for checkpoint function (38). In our case, the premature depletion of kinetochore-bound BubR1 and/or CENP-E in normal mitotic progression of CENP-AOFF cells would result in generation of a weakened inhibitory signal from kinetochores that are unattached and/or not under tension, and this might not be strong enough to sustain a strong mitotic block. In the presence of spindle-damaging agents, the summation of these inhibitory signals from all kinetochores would then reach the threshold necessary for checkpoint maintenance.
A further question raised by our results is how mitotic checkpoint activation, which is independent of CENP-A for its initiation, becomes CENP-A dependent for its maintenance.
Phenotypic analysis following depletion or inhibition of the individual components of the passenger protein complex INCENP/Aurora B/Survivin has established a role for Aurora B and Survivin in mitotic checkpoint maintenance and has revealed a phenotype reminiscent of CENP-A depletion, with major missegregation defects and defects in kinetochore recruitment of Mad2, BubR1, and CENP-E (2, 8, 13, 25, 32, 36). It has been shown that Ser7 of human CENP-A is specifically phosphorylated by Aurora A and Aurora B at mitosis (35, 63). It is thus tempting to speculate that phosphorylation of the centromeric histone by the Aurora B kinase could be a key step for a CENP-A-dependent mechanism of checkpoint maintenance. Although Ser7 is not strictly conserved in chickens, a conserved serine residue (Ser8) is present in the N-terminal domain in a similar basic and proline-rich environment (49). Complementation experiments in the DT40 CENP-A conditionally null cell line with mutants of this residue may shed further light on the possible role of CENP-A in the checkpoint-signaling pathway.
Acknowledgments
We are grateful to A. Abrieu for providing the anti-Xenopus CENP-E antibody and Gabriele Reinkensmeier for technical assistance with FACS analysis. We thank François-Xavier Barre, Chris-Tyler Smith, and Ed Southern for discussions and comments.
This work was supported by grants from Cancer Research UK, BBSRC, and the Wellcome Trust. V.R. was supported by fellowships from European Molecular Biology Organization and Ligue Nationale contre le Cancer, and W.C.E. is a Wellcome Trust Principal Research Fellow. The FACS machine was provided by the Wellcome trust.
REFERENCES
- 1.Adams, R. R., M. Carmena, and W. C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49-54. [DOI] [PubMed] [Google Scholar]
- 2.Adams, R. R., H. Maiato, W. C. Earnshaw, and M. Carmena. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biggins, S., and C. E. Walczak. 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13:R449-R460. [DOI] [PubMed] [Google Scholar]
- 4.Black, B. E., D. R. Foltz, S. Chakravarthy, K. Luger, V. L. Woods, Jr., and D. W. Cleveland. 2004. Structural determinants for generating centromeric chromatin. Nature 430:578-582. [DOI] [PubMed] [Google Scholar]
- 5.Blower, M. D., and G. H. Karpen. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3:730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blower, M. D., B. A. Sullivan, and G. H. Karpen. 2002. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell 2:319-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buerstedde, J. M., and S. Takeda. 1991. Increased ratio of targeted to random integration after transfection of chicken B cell lines. Cell 67:179-188. [DOI] [PubMed] [Google Scholar]
- 8.Carvalho, A., M. Carmena, C. Sambade, W. C. Earnshaw, and S. P. Wheatley. 2003. Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J. Cell Sci. 116:2987-2998. [DOI] [PubMed] [Google Scholar]
- 9.Cleveland, D. W., Y. Mao, and K. F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407-421. [DOI] [PubMed] [Google Scholar]
- 10.Cooke, C. A., B. Schaar, T. J. Yen, and W. C. Earnshaw. 1997. Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 106:446-455. [DOI] [PubMed] [Google Scholar]
- 11.DeLuca, J. G., B. J. Howell, J. C. Canman, J. M. Hickey, G. Fang, and E. D. Salmon. 2003. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 13:2103-2109. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca, J. G., B. Moree, J. M. Hickey, J. V. Kilmartin, and E. D. Salmon. 2002. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J. Cell Biol. 159:549-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ditchfield, C., V. L. Johnson, A. Tighe, R. Ellston, C. Haworth, T. Johnson, A. Mortlock, N. Keen, and S. S. Taylor. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Earnshaw, W. C., H. Ratrie, and G. Stetten. 1989. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma 98:1-12. [DOI] [PubMed] [Google Scholar]
- 15.Earnshaw, W. C., and J. B. Rattner. 1989. A map of the centromere (primary constriction) in vertebrate chromosomes at metaphase. Prog. Clin. Biol. Res. 318:33-42. [PubMed] [Google Scholar]
- 16.Earnshaw, W. C., and N. Rothfield. 1985. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91:313-321. [DOI] [PubMed] [Google Scholar]
- 17.Eckley, D. M., A. M. Ainsztein, A. M. Mackay, I. G. Goldberg, and W. C. Earnshaw. 1997. Chromosomal proteins and cytokinesis: patterns of cleavage furrow formation and inner centromere protein positioning in mitotic heterokaryons and mid-anaphase cells. J. Cell Biol. 136:1169-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueroa, J., R. Saffrich, W. Ansorge, and M. Valdivia. 1998. Microinjection of antibodies to centromere protein CENP-A arrests cells in interphase but does not prevent mitosis. Chromosoma 107:397-405. [DOI] [PubMed] [Google Scholar]
- 19.Fukagawa, T. 2004. Assembly of kinetochores in vertebrate cells. Exp. Cell Res. 296:21-27. [DOI] [PubMed] [Google Scholar]
- 20.Fukagawa, T., and W. R. A. Brown. 1997. Efficient conditional mutation of the vertebrate CENP-C gene. Hum. Mol. Genet. 6:2301-2308. [DOI] [PubMed] [Google Scholar]
- 21.Fukagawa, T., Y. Mikami, A. Nishihashi, V. Regnier, T. Haraguchi, Y. Hiraoka, N. Sugata, K. Todokoro, W. Brown, and T. Ikemura. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603-4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gassmann, R., A. Carvalho, A. J. Henzing, S. Ruchaud, D. F. Hudson, R. Honda, E. A. Nigg, D. L. Gerloff, and W. C. Earnshaw. 2004. Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J. Cell Biol. 166:179-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hauf, S., R. W. Cole, S. LaTerra, C. Zimmer, G. Schnapp, R. Walter, A. Heckel, J. van Meel, C. L. Rieder, and J. M. Peters. 2003. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161:281-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman, D. B., C. G. Pearson, T. J. Yen, B. J. Howell, and E. D. Salmon. 2001. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 12:1995-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hori, T., T. Haraguchi, Y. Hiraoka, H. Kimura, and T. Fukagawa. 2003. Dynamic behavior of Nuf2-Hec1 complex that localizes to the centrosome and centromere and is essential for mitotic progression in vertebrate cells. J. Cell Sci. 116:3347-3362. [DOI] [PubMed] [Google Scholar]
- 28.Howard, J., and A. A. Hyman. 2003. Dynamics and mechanics of the microtubule plus end. Nature 422:753-758. [DOI] [PubMed] [Google Scholar]
- 29.Howman, E. V., K. J. Fowler, A. J. Newson, S. Redward, A. C. MacDonald, P. Kalitsis, and K. H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA 97:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson, D. F., P. Vagnarelli, R. Gassmann, and W. C. Earnshaw. 2003. Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev. Cell 5:323-336. [DOI] [PubMed] [Google Scholar]
- 31.Jallepalli, P. V., and C. Lengauer. 2001. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer 1:109-117. [DOI] [PubMed] [Google Scholar]
- 32.Kallio, M. J., M. L. McCleland, P. T. Stukenberg, and G. J. Gorbsky. 2002. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr. Biol. 12:900-905. [DOI] [PubMed] [Google Scholar]
- 33.Karpen, G. H., and R. C. Allshire. 1997. The case for epigenetic effects on centromere identity and function. Trends Genet. 13:489-496. [DOI] [PubMed] [Google Scholar]
- 34.Kitagawa, K., and P. Hieter. 2001. Evolutionary conservation between budding yeast and human kinetochores. Nat. Rev. Mol. Cell Biol. 2:678-687. [DOI] [PubMed] [Google Scholar]
- 35.Kunitoku, N., T. Sasayama, T. Marumoto, D. Zhang, S. Honda, O. Kobayashi, K. Hatakeyama, Y. Ushio, H. Saya, and T. Hirota. 2003. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev. Cell 5:853-864. [DOI] [PubMed] [Google Scholar]
- 36.Lens, S. M., R. M. Wolthuis, R. Klompmaker, J. Kauw, R. Agami, T. Brummelkamp, G. Kops, and R. H. Medema. 2003. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 22:2934-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, S. T., J. C. Hittle, S. A. Jablonski, M. S. Campbell, K. Yoda, and T. J. Yen. 2003. Human CENP-I specifies localization of CENP-F, MAD1 and MAD2 to kinetochores and is essential for mitosis. Nat. Cell Biol. 5:341-345. [DOI] [PubMed] [Google Scholar]
- 38.Mao, Y., A. Abrieu, and D. W. Cleveland. 2003. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell 114:87-98. [DOI] [PubMed] [Google Scholar]
- 39.Martin-Lluesma, S., V. M. Stucke, and E. A. Nigg. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297:2267-2270. [DOI] [PubMed] [Google Scholar]
- 40.McAinsh, A. D., J. D. Tytell, and P. K. Sorger. 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19:519-539. [DOI] [PubMed] [Google Scholar]
- 41.McCleland, M. L., R. D. Gardner, M. J. Kallio, J. R. Daum, G. J. Gorbsky, D. J. Burke, and P. T. Stukenberg. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meluh, P. B., and A. V. Strunnikov. 2002. Beyond the ABCs of CKC and SCC. Do centromeres orchestrate sister chromatid cohesion or vice versa? Eur. J. Biochem. 269:2300-2314. [DOI] [PubMed] [Google Scholar]
- 43.Moroi, Y., C. Peebles, M. J. Fritzler, J. Steigerwald, and E. M. Tan. 1980. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc. Natl. Acad. Sci. USA 77:1627-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signalling. Nat. Rev. Mol. Cell Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 45.Nishihashi, A., T. Haraguchi, Y. Hiraoka, T. Ikemura, V. Regnier, H. Dodson, W. C. Earnshaw, and T. Fukagawa. 2002. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell 2:463-476. [DOI] [PubMed] [Google Scholar]
- 46.Oegema, K., A. Desai, S. Rybina, M. Kirkham, and A. A. Hyman. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pidoux, A. L., W. Richardson, and R. C. Allshire. 2003. Sim4: a novel fission yeast kinetochore protein required for centromeric silencing and chromosome segregation. J. Cell Biol. 161:295-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pluta, A. F., A. M. Mackay, A. M. Ainsztein, I. G. Goldberg, and W. C. Earnshaw. 1995. Centromere—hub of chromosomal activities. Science 270:1591-1594. [DOI] [PubMed] [Google Scholar]
- 49.Regnier, V., J. Novelli, T. Fukagawa, P. Vagnarelli, and W. Brown. 2003. Characterization of chicken CENP-A and comparative sequence analysis of vertebrate centromere-specific histone H3-like proteins. Gene 316:39-46. [DOI] [PubMed] [Google Scholar]
- 50.Shelby, R. D., K. Monier, and K. F. Sullivan. 2000. Chromatin assembly at kinetochores is uncoupled from DNA replication. J. Cell Biol. 151:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skoufias, D. A., P. R. Andreassen, F. B. Lacroix, L. Wilson, and R. L. Margolis. 2001. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA 98:4492-4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sonoda, E., T. Matsusaka, C. Morrison, P. Vagnarelli, O. Hoshi, T. Ushiki, K. Nojima, T. Fukagawa, I. C. Waizenegger, J. M. Peters, W. C. Earnshaw, and S. Takeda. 2001. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev. Cell 1:759-770. [DOI] [PubMed] [Google Scholar]
- 53.Sugata, N., E. Munekata, and K. Todokoro. 1999. Characterization of a novel kinetochore protein, CENP-H. J. Biol. Chem. 274:27343-27346. [DOI] [PubMed] [Google Scholar]
- 54.Sullivan, K. F. 2001. A solid foundation: functional specialization of centromeric chromatin. Curr. Opin. Genet. Dev. 11:182-188. [DOI] [PubMed] [Google Scholar]
- 55.Tanudji, M., J. Shoemaker, L. L′Italien, L. Russell, G. Chin, and X. M. Schebye. 2004. Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell 15:3771-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor, S. S., D. Hussein, Y. Wang, S. Elderkin, and C. J. Morrow. 2001. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J. Cell Sci. 114:4385-4395. [DOI] [PubMed] [Google Scholar]
- 57.Wang, J., Y. Takagaki, and J. L. Manley. 1996. Targeted disruption of an essential vertebrate gene: ASF/SF2 is required for cell viability. Genes Dev. 10:2588-2599. [DOI] [PubMed] [Google Scholar]
- 58.Waters, J. C., R. H. Chen, A. W. Murray, and E. D. Salmon. 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141:1181-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weaver, B. A., Z. Q. Bonday, F. R. Putkey, G. J. Kops, A. D. Silk, and D. W. Cleveland. 2003. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 162:551-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Westermann, S., I. M. Cheeseman, S. Anderson, J. R. Yates III, D. G. Drubin, and G. Barnes. 2003. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 163:215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoda, K., S. Ando, S. Morishita, K. Houmura, K. Hashimoto, K. Takeyasu, and T. Okazaki. 2000. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc. Natl. Acad. Sci. USA 97:7266-7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu, H. 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14:706-714. [DOI] [PubMed] [Google Scholar]
- 63.Zeitlin, S. G., R. D. Shelby, and K. F. Sullivan. 2001. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J. Cell Biol. 155:1147-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou, J., J. Yao, and H. C. Joshi. 2002. Attachment and tension in the spindle assembly checkpoint. J. Cell Sci. 115:3547-3555. [DOI] [PubMed] [Google Scholar]