Abstract
The phosphoinositide phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] regulates the activity of many actin-binding proteins and as such is an important modulator of cytoskeleton organization during cell migration, for example. In migrating cells actin remodeling is tightly regulated and localized; therefore, how the PI(4,5)P2 level is spatially and temporally regulated is crucial to understanding how it controls cell migration. Here we show that the LIM protein Ajuba contributes to the cellular regulation of PI(4,5)P2 levels by interacting with and activating the enzymatic activity of the PI(4)P 5-kinase (PIPKIα), the predominant enzyme in the synthesis of PI(4,5)P2, in a migration stimulus-regulated manner. In migrating primary mouse embryonic fibroblasts (MEFs) from Ajuba−/− mice the level of PI(4,5)P2 was decreased with a corresponding increase in the level of the substrate PI(4)P. Reintroduction of Ajuba into these cells normalized PI(4,5)P2 levels. Localization of PI(4,5)P2 synthesis and PIPKIα in the leading lamellipodia and membrane ruffles, respectively, of migrating Ajuba−/− MEFs was impaired. In vitro, Ajuba dramatically activated the enzymatic activity of PIPKIα while inhibiting the activity of PIPKIIβ. Thus, in addition to its effects upon Rac activity Ajuba can also influence cell migration through regulation of PI(4,5)P2 synthesis through direct activation of PIPKIα enzyme activity.
The phosphoinositide phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] is an important regulator of cytoskeletal organization during diverse cellular functions such as vesicle trafficking, endocytosis, phagocytosis, focal adhesion formation, and cell migration. PI(4,5)P2 binds to and affects the function of many actin-binding and actin-remodeling proteins such as profilin, gelsolin, vinculin, talin, cofilin, α-actinin, WASP, Arp2/3, and the Rho family of GTPases (for reviews see references 2, 14, and 28). In general PI(4,5)P2 production is concentrated at the plasma membrane, and the total cell amount is maintained at a relatively constant level; however, localized dynamic changes have been observed in response to growth factor stimulation and cell adhesion to the extracellular matrix, at sites of phagocytosis, and in actin-rich protrusion and membrane ruffles (3, 6, 8). How the PI(4,5)P2 level is spatially and temporally regulated is therefore crucial to understanding the cellular responses that it controls.
The synthesis of PI(4,5)P2 is regulated by a family of enzymes, phosphatidylinositol phosphate kinases (PIPKs) (reviewed in reference 14). The PIPKs can be subdivided into three subfamilies, types I, II, and III, based on their substrate specificity and the position that they phosphorylate on the inositol ring. For type I and II there are α, β, and γ isoforms of each that have been identified and characterized biochemically. Recently a new PIPK type I homolog, PIPKH, has been identified, and it appears to localize and regulate other type I kinases but has little kinase activity itself (7). PIP kinases comprising type I utilize PI(4)P as substrate and phosphorylate the D5 position of the inositol ring, while the substrate for type II kinases is PI(5)P and the enzymes phosphorylate the D4 position of the inositol ring. Type III PIPK, PIPfyve, utilizes PI(3)P as substrate and produces PI(3,5)P2 (37). Type I and II kinases can also utilize PI(3)P but with much-reduced efficiency (41, 45). Since PI(4)P is the major phosphatidylinositol phosphate in cells and its concentration is relatively high, type I PIPKs are thought to play a more prominent role in the production of PI(4,5)P2.
The subcellular localization of these enzyme families also differs. Many of the early studies were done in cells overexpressing tagged forms of the enzymes; however, recently new antibodies have allowed for the localization of endogenous enzymes (13, 15, 26). In fibroblasts type Iα PIPK is found at membrane ruffles, diffusely throughout the cytosol, and within the nucleus. Type Iβ is primarily present on perinuclear cytosolic vesicular structures, and type Iγ is found at focal adhesions. Type II enzymes are present in the cytosol and nucleus and associated with the endoplasmic reticulum. Type III enzymes contain a FYVE motif that targets them to endomembranes, where the production of PI(3,5)P2 regulates membrane integrity (21, 36).
The generation of PI(4,5)P2 at specific subcellular sites of cytoskeletal rearrangement is most likely modulated by selective targeting and activation of specific PIPKs. For example, PIPKIγ is targeted to sites of focal adhesions through an interaction with the focal adhesion protein talin, and this association also results in an increase of the kinase activity of the enzyme (13, 26). The localized generation of PI(4,5)P2 by PIPKIγ at focal adhesions appears to be critical for the formation of focal complexes and their maturation to focal adhesions (13, 26). PIPKIα localizes to membranes in response to G-protein-coupled receptor activation (8) and to membrane ruffles in response to platelet-derived growth factor stimulation (15). Finally in B cells the BTK kinase associates with PIPKIα and, in response to B-cell receptor signaling, brings PIPKIα to membranes to generate local synthesis of PI(4,5)P2 (35).
The Rho family of G proteins are major regulators of actin cytoskeletal dynamics (17). RhoA activity regulates the formation of stress fibers and focal adhesions, while Rac1 regulates lamellipodium production and membrane ruffling. Regulated formation and turnover of these structures are important during cell migration. To effect actin assembly, Rho family proteins require PI(4,5)P2 synthesis (31). Rho, Rac, and another G protein, Arf6, have all been shown to regulate PIPK activity (20, 40, 43), and some PIPKs can associate with Rac and Rho, in vitro (34, 38, 39). Rac can also activate phosphatidylinositol 4-kinase, an enzyme that leads to the production of PI(4)P, the major cellular substrate for PIPKs (44). Recently we have shown that the focal adhesion LIM protein Ajuba affects cell migration by influencing the localization of p130Cas to nascent focal complexes and thus Rac1 activity through the efficient assembly of the p130Cas/Crk/DOCK180/ELMO Rac GEF (32).
Ajuba is a cytosolic protein and a member of the zyxin/Ajuba family of LIM proteins (4, 5), defined by the presence of three related tandem LIM domains at the C termini (LIM region) and unrelated, proline-rich N termini (PreLIM region). These proteins are components of integrin-mediated adhesive complexes in fibroblasts (5, 32) and cell-cell junction adhesive complexes in epithelial cells (19, 27) and interact with filamentous actin either indirectly or directly (11, 27). Thus, in addition to regulating cell migration, they contribute to the formation or strengthening of cell-cell junctions in part by linking adhesive receptors to the actin cytoskeleton (27). This family of LIM proteins also contains nuclear export signals, and the proteins shuttle in and out of the nucleus, suggesting that they could function to transduce signals from cell adhesive sites to the nucleus (1, 23, 29, 30). Indeed Ajuba was recently shown to interact with and affect the activity of the mitotic kinase Aurora A (18).
To further understand how Ajuba regulates actin dynamics, and cell migration specifically, we performed a yeast two-hybrid protein-protein interactive screen to identify potential Ajuba-interacting proteins implicated in cytoskeletal regulation. We found that Ajuba associates with the PIPK enzymes PIPKIα and PIPKIIβ and affects their subcellular localization and enzyme activity. Ajuba−/− primary MEFs have reduced levels of PI(4,5)P2 and a corresponding elevation of the substrate PI(4)P. PI(4,5)P2 levels are corrected upon reintroduction of Ajuba into these cells. These results suggest that in addition to regulating Rac activity Ajuba could also influence cell migration by regulating the levels of cellular PI(4,5)P2 through a direct interaction with and activation of PIPKIα.
MATERIALS AND METHODS
Yeast two-hybrid screen.
The PreLIM region of human Ajuba was subcloned into the pAS2 plasmid and sequenced. Parental AH109 yeast cells (Clontech) were transformed, and production of Gal4-PreLIM-Ajuba fusion protein in yeast was confirmed by Western blotting using purified Ajuba antibody. Competent yeast cells containing pAS2-PreLIM were then transformed with a human keratinocyte Matchmaker cDNA library (Clontech). Transformants (5 × 106) were screened. Twenty primary positives were identified. Colonies were cured of the bait plasmid by segregation, and the remaining library plasmid was rescued by transforming XL1-Blue bacteria. Retransformation of AH109 yeast cells with appropriate specificity controls was carried out. Of the 20 primary positives 5 persisted as true positives. All were sequenced, and two clones contained partial sequence for PIPKIIβ.
Plasmid construction, antibodies, and cell lines.
The cDNAs of PIPKIα and -β and PIPKIIα and -β isoforms were PCR amplified from a human keratinocyte cDNA library. PIPKIγ661 was from R. Anderson (University of Wisconsin) (26). All PCR products were subcloned into pCMV5-HA and sequenced. Myc-tagged Ajuba, zyxin, and LPP plasmids have been described elsewhere (16, 23). Antihemagglutinin (anti-HA) antibodies conjugated to agarose (Santa Cruz) or M2AG (Sigma), polyclonal Myc antiserum, and Ajuba antiserum have been described previously (16, 27). PIPKIα antiserum has been described in reference 15. Littermate Ajuba-null and wild-type (wt) MEFs were generated as described previously (32). Primary MEFs within the first 12 passages were transfected using the Nucleofector MEF2 kit (Amaxa, Inc., Gaithersburg, MD). HEK 293 cells were transfected using Trans-IT LT-1 (Mirrus, Madison, WI). All cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum.
Immunoprecipitation.
HEK 293 cells (4 × 105/well) were plated in a 12-well plate overnight before transfection. Cells were transfected and 24 h later collected and lysed with IP buffer (20 mM HEPES, pH 7.5, 120 mM NaCl, 5 mM NaF, 1 mM sodium orthovanadate, 0.5 mM EDTA, 1 mM dithiothreitol, 5% glycerol, 0.1% NP-40, protease inhibitors), and cytoplasmic extract was clarified by spinning at 12,000 rpm for 10 min and mixed with primary antibodies overnight. Beads were washed with IP buffer five times, and bound products were eluted with 20 μl of sodium dodecyl sulfate (SDS)-polyacrylamide gel loading buffer. Five microliters was loaded per lane and separated on a 10% SDS-polyacrylamide gel, under reducing conditions, and transferred to nitrocellulose membranes for Western blot analysis.
wt, Ajuba-null, and Ajuba-null MEFs rescued with Flag-tagged Ajuba were grown to confluency. To stimulate migration, half of the plates were scratch wounded multiple times such that 50% of cells were removed. Stimulated and unstimulated cells were harvested and lysed 1.5 to 2 h postwounding. Flag immunoprecipitation was performed as above. Bound material was eluted from the beads with 100 μg/ml Flag peptide. Eluted proteins from the same immunoprecipitate were analyzed by both Western blotting and in vitro lipid kinase assay.
Protein purification.
His-tagged PIPKIα and PIPKIIα were purified as described previously (45). PIPKIβ and PIPKIIβ were subcloned into a pBacPAK9 plasmid (Clontech) containing a His6 and Flag tag fragment between the BamHI and EcoRI sites. The kinases were expressed in Sf9 cells. For protein purification, crude kinase extracts were loaded onto 1 ml Talon metal affinity columns (Clontech) equilibrated with high-salt buffer (1× phosphate-buffered saline containing 650 mM NaCl). The column was washed thoroughly with 50 ml of high-salt buffer, and then proteins were eluted with a 30 ml linear gradient of 0 to 100 mM imidazole in high-salt buffer. Fractions containing purified kinase were combined and concentrated with Centricon-10 concentrators (Amicon) and dialyzed overnight against 50 mM Tris, pH 7.6, 1 mM EGTA, 10% glycerol (for PIPKIα, 150 mM NaCl was also present in the dialyzing buffer). The PreLIM region of Ajuba was expressed and purified from bacterial extracts on glutathione-agarose beads, as a glutathione S-transferase (GST)-PreLIM fusion protein. The PreLIM peptide was cleaved from the GST moiety with Prescission protease (Amersham), and the released peptide was dialyzed overnight.
Lamellipodium purification.
Lamellipodium purification was performed as described previously (9, 32). Briefly, MEFs were serum starved overnight and then plated in the upper chamber of wells containing a 3.0-μm porous Transwell filter (Fisher Co., Pittsburgh, PA) coated on both sides with fibronectin (5 mg/ml fibronectin [Sigma] in phosphate-buffered saline). Cells were allowed to attach and spread for 2 h. Lamellipodium formation was induced by the addition of serum to the bottom chamber. Cell bodies or lamellipodia were isolated after 60 to 90 min from the top or the bottom of the membrane, respectively, into lysis buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 1 mM NaF, 1 mM sodium orthovanadate, 0.1% NP-40, and protease inhibitors).
Lipid kinase assay.
Lipid kinase activity assays were carried out for 10 min (unless indicated otherwise) at 37°C in 50-μl reaction volumes containing 50 mM Tris, pH 7.6, 10 mM MgCl2, 0.5 mM EGTA, 20 μM PIP substrate (prepared in isotonic KCl solution), and 50 μM [32P]ATP (5 μCi/reaction). The reactions were stopped by the addition of 100 μl of 1 N HCl, and the lipid products were extracted using 200 μl of chloroform-methanol (1:1). The organic phase was washed with 100 μl of methanol-1 N HCl (1:1). The organic phase was then spotted on thin-layer chromatography (TLC) plates and run as described previously (45). After autoradiography, the spots on the TLC plates corresponding to products were scraped into vials and counted using Cerenkov radioactivity in a Beckman scintillation counter.
Determination of inositol phospholipid levels.
MEFs were incubated in phosphate-free Dulbecco's modified Eagle's medium (Invitrogen) containing 400 μCi of [32P]orthophosphate (ICN) for 2 h at 37°C. For phosphoinositide isolation from lamellipodia or cell bodies, MEFs were labeled while adhering to the filter in the upper chamber of the Transwell (see above). Lipids were extracted by an addition of 0.5 ml methanol:chloroform:8% HClO4 (20:10:1). Phosphoinositide deacylation and separation on an anion-exchange column were done as described before (24).
Live cell imaging.
Live cell imaging was performed on cells grown on the glass coverslips as described previously (42). Following transfection, cells were grown to 100% confluence, scratch wounded, and placed in a heated chamber (Delta T from Bioptechs, Butler, PA) containing L-15 medium plus 10% fetal calf serum. Images were taken using an upright BX61WI microscope (Olympus), a xenon illumination source (Lambda LS; Sutter Instruments, San Rafael, CA), and a water-immersion lens (0.9 numerical aperture) with a cooled charge-coupled device camera (Coolsnap HQ; Roper Scientific) and processed using Metamorph software (Universal Imaging, Media, PA).
Interfering RNA (RNAi) knockdown of PIPKIα.
Lentiviruses expressing PIPKIα-specific small interfering RNA (siRNA) or control scrambled siRNA were generated (33). These viruses also contain an internal ribosome entry site-green fluorescent protein (GFP) cassette. The PIPKIα target sequence was amino acids 71 to 77 (AAGGTGCCATCCAGTTAGGCA). wt MEFs were infected, and GFP expression was determined. Plates were lysed, and PIPKIα protein levels were determined by Western blotting.
RESULTS
Ajuba associates with PIPKIα and PIPKIIβ in a migration stimulus-regulated manner.
To further understand how Ajuba influences actin cytoskeleton dynamics, particularly in migrating cells, we performed a yeast two-hybrid protein-protein interactive screen to identify candidate interacting proteins with these capabilities. Since skin keratinocytes express high levels of Ajuba and keratinocytes from Ajuba−/− mice are defective in cell-cell adhesion (27), we screened a human keratinocyte library with the N-terminal PreLIM region of Ajuba (directing association with F-actin [27]). In this screen the PIP kinase PIPKIIβ was one such candidate protein identified.
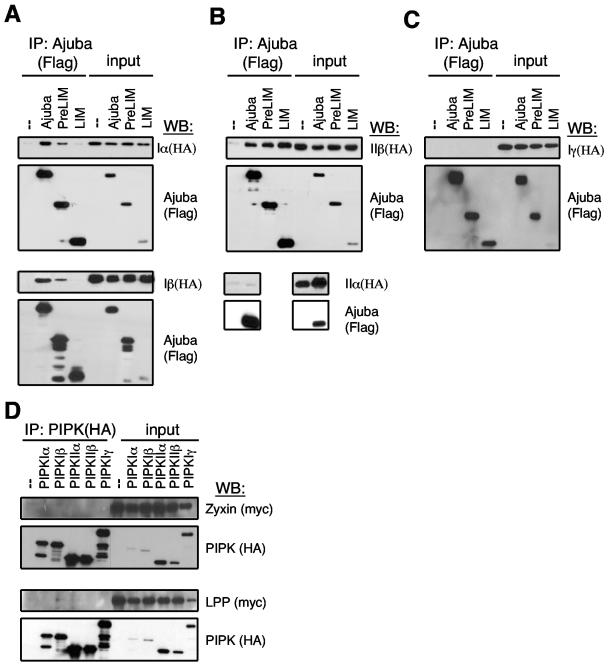
To confirm that the interaction between Ajuba and PIPKIIβ occurred in mammalian cells, we carried out coimmunoprecipitation experiments. We also determined whether other PIP kinase enzymes interacted with Ajuba and if there was specificity among Ajuba family members for interacting with PIPKs. HEK 293 cells were transiently transfected with various HA-tagged PIPKs and Flag-tagged full-length Ajuba, the N-terminal PreLIM region of Ajuba, or the C-terminal LIM region of Ajuba. From cell extracts the Ajuba isoforms were immunoprecipitated with an anti-Flag antibody, and the presence of PIPK in the immunoprecipitate was determined by Western blotting with anti-HA antibodies. In this analysis Ajuba was found to interact with PIPKIIβ, PIPKIα, and PIPKIβ but not PIPKIIα or PIPKIγ (Fig. 1A to C). For PIPKIα and PIPKIβ the PreLIM region of Ajuba directed its association (the LIM region did not interact) (Fig. 1A); however, for PIPKIIβ both the PreLIM and LIM regions were able to associate with the enzyme (Fig. 1B). Therefore, while Ajuba interacted with more than one member of the PIPK enzyme family, there was specificity, as closely related PIPKIIα and PIPKIγ enzymes did not interact.
FIG. 1.
Mapping interaction of Ajuba and related LIM proteins with different PIPK enzymes. A to C. HEK 293 cells were cotransfected with control empty vector (−), Flag-Ajuba, Flag-PreLIM, or Flag-LIM regions of Ajuba and HA-tagged PIPK type Iα and Iβ (A), PIPK type IIβ and IIα (B), or PIPK type Iγ (C). Ajuba isoforms were immunoprecipitated with Flag antibodies, and bound products were Western blotted with HA antibodies (i.e., PIPK, upper panels) and Flag antibodies (i.e., Ajuba, lower panels). A fraction (5%) of each cell lysate was Western blotted for each protein (input). D. HEK 293 cells were cotransfected with HA-tagged PIPK enzymes and Myc-zyxin (upper two panels) or Myc-LPP (lower two panels). PIPKs were immunoprecipitated with HA antibodies (left), and bound products were Western blotted with Myc antibodies (i.e., LIM protein) and HA antibodies (i.e., PIPK enzyme). A fraction (5%) of each cell lysate was Western blotted for each protein (input).
To determine whether there was specificity within the Ajuba family of LIM proteins for interacting with PIPKs, we tested whether the related LIM proteins zyxin and LPP interacted with the various PIPKs. HEK 293 cells were transiently transfected with HA-tagged PIPKs and Myc-tagged zyxin or LPP. PIPKs were immunoprecipitated with anti-HA antibody, and bound products were Western blotted for the presence of zyxin and LPP with anti-Myc antibody. Neither zyxin nor LPP interacted with any of the PIPKs (Fig. 1D). Thus, Ajuba's interaction with PIPKIα, PIPKIβ, and PIPKIIβ was specific among related LIM protein family members.
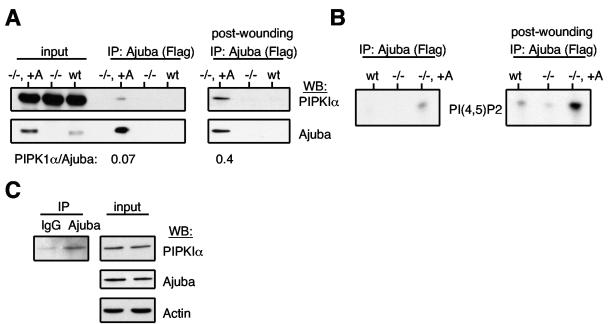
To determine whether PIPKs associated with endogenous Ajuba in cells, we first immunoprecipitated Ajuba from wt MEFs, Ajuba-null MEFs (negative control), and Ajuba-null MEFs that had been rescued with Flag-tagged Ajuba. Ajuba-rescued null MEFs expressed approximately 5- to 10-fold more Ajuba than what was present in wt MEFs (Fig. 2A). Cells were either growing at steady state or stimulated to migrate by performance of multiple scratch-wound procedures in a confluent plate of cells, such that approximately 50% of cells were removed (32). After 1.5 to 2 hours, a time at which cells had begun to migrate into the wounds, cells were lysed and Ajuba was immunoprecipitated. An aliquot of each immunoprecipitated product was subjected to both in vitro PIP kinase assays (Fig. 2B) and Western blotting for the presence of PIPKIα (Fig. 2A).
FIG. 2.
PIPKIα associates with Ajuba in cells, and migration stimuli increase this interaction. Ajuba (anti-Flag) was immunoprecipitated from Ajuba-null MEFs (−/−), wild-type MEFs (wt), and Ajuba-null MEFs rescued with Flag-Ajuba (−/−, +A). An aliquot of the immunoprecipitated products was Western blotted for endogenous PIPKIα (upper panels) or Ajuba (anti-Ajuba antibodies, lower panels). A fraction (5%) of each lysate was also Western blotted for each protein (input). The left set of panels are immunoprecipitates from cells growing at steady state. The right set of panels are immunoprecipitates from cells stimulated to migrate by scratch wounding. The ratio of PIPKIα to Ajuba in the Ajuba immunoprecipitate under each condition, as determined by densitometry, is listed below the respective lanes. B. The remainder of the same Ajuba immunoprecipitate as in panel A was subjected to in vitro PIP kinase reaction, in the presence of [32P]ATP and PI(4)P as substrate. The left panel is an immunoprecipitate from cells growing at steady state. The right panel is an immunoprecipitate from cells stimulated to migrate by scratch wounding. C. wt MEFs were scratch wounded, as above, and lysed. Control IgG or Ajuba antiserum was added, and immunoprecipitates were Western blotted for the presence of PIPKIα (left panel). The right panels are input controls for the levels of PIPKIα, Ajuba, and actin in the respective cell lysates.
PIPKIα Western blots of Ajuba immunoprecipitates from cells growing at steady state detected a small amount of enzyme associated with Ajuba (Fig. 2A, left panel). When cells had been stimulated to migrate, the amount of PIPKIα associated with Ajuba greatly increased (Fig. 2A, right panel). The ratio of PIPKIα to Ajuba in Ajuba immunoprecipitates was 0.07 in cells at steady state and increased to 0.4 in cells stimulated to migrate (Fig. 2A).
PIP kinase assays were performed in the presence of [32P]ATP and either PI(4)P (PIPK type I substrate; Fig. 2B) or PI(5)P (PIPK type II substrate; data not shown) as substrate. The reaction products were separated on TLC plates, scraped, deacylated, and identified by anion-exchange high-pressure liquid chromatography (HPLC) against known standards. Compared to Ajuba null-cells, Ajuba immunoprecipitated from Ajuba-rescued Ajuba-null cells produced a small amount of PI(4,5)P2 when incubated with PI(4)P (Fig. 2B, left panel). When cells were incubated with PI(5)P as substrate, there was little, if any, PI(4,5)P2 synthesized (data not shown). When PIP kinase assays were performed on Ajuba immunoprecipitates from cells stimulated to migrate, a significant increase in PIPK type I activity was detected (Fig. 2B, right panel). Again little PIPK type II activity was detected in stimulated cells (data not shown). We did not detect any PI(3,4)P2 or PI(3,5)P2 produced, indicating that Ajuba did not interact with PI3K, another phosphatidylinositol phosphate kinase capable of utilizing the same substrates.
Finally, to determine whether Ajuba and PIPKIα associate in cells expressing endogenous levels of each, we wounded wt MEFs, added Ajuba antiserum or control nonspecific immunoglobulin G (IgG) to total cell extracts, and then Western blotted immunoprecipitated products for the presence of PIPKIα. Although control IgG immunoprecipitate contained a small amount of PIPKIα, there was a significant increase in the amount of PIPKIα present in the Ajuba immunoprecipitate (Fig. 2C), indicating that PIPKIα and Ajuba can associate in wt MEFs stimulated to migrate (i.e., wounded).
These analyses indicated that PIPKIα associated with Ajuba in cells and that a migration stimulus significantly increased the amount of PIPKIα associated with Ajuba, signifying that the association between Ajuba and PIPKI was regulated, specifically by a migration stimulus.
Ajuba-null MEFs have decreased levels of intracellular PI(4,5)P2.
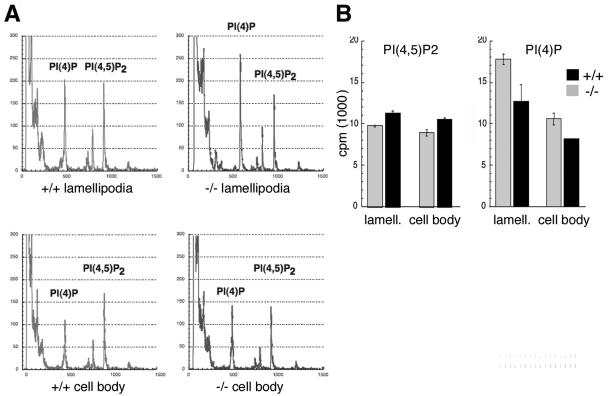
During cell migration lamellipodia are produced at the leading edge, in the direction of migration. Lamellipodium production results from local actin assembly, and thus, lamellipodia are enriched for proteins and second messenger molecules important for the formation and disassembly of the actin cytoskeleton (31). In light of Ajuba's association with PIPK enzymes and defective migration of Ajuba−/− cells (32), we asked whether there were differences in phosphatidylinositide levels in Ajuba−/− MEFs in response to migration stimuli. To address this, we employed two approaches. In the first approach we made use of a recently described assay to isolate lamellipodia (pseudopodia) from the remaining cell body (9, 32). Serum-starved early-passage Ajuba−/− MEFs and control wt littermate MEFs were incubated with inorganic 32Pi, and then cells were seeded into the upper chamber of a well containing fibronectin-coated filters (both sides) with 3-μm pores. Cell migration was stimulated by the addition of medium containing 10% fetal calf serum to the lower chamber. After 90 min lamellipodia were isolated from the bottom of the filters while the cell body fraction remained on the upper surface. Total phosphoinositol lipids were extracted from each fraction, deacylated, and then separated by anion-exchange HPLC against known standards (Fig. 3A). Since Ajuba−/− MEFs produce less lamellipodia than do wt MEFs (32), all samples were first normalized by protein levels prior to loading on HPLC. Compared to wt MEFs, both lamellipodia and cell body fractions from Ajuba−/− cells had decreased levels of PI(4,5)P2 and increased levels of PI(4)P (Fig. 3A and B). The difference between cells for the two products appeared to be somewhat greater in lamellipodia than in the cell bodies (Fig. 3A and B). Compared to wt MEFs, the level of PI(4,5)P2 in Ajuba−/− lamellipodia was reduced on average by 14%, whereas the level of substrate PI(4)P was 40% higher (Fig. 3B). In the cell body of Ajuba−/− MEFs the PI(4,5)P2 level was reduced 10%, while the level of PI(4)P substrate was increased 25% (Fig. 3B).
FIG. 3.
Ajuba-null MEFs have decreased levels of PI(4,5)P2 and increased levels of PI(4)P. A. Wild-type (+/+) or Ajuba-null (−/−) MEFs were incubated with 32Pi. Cellular lipids were isolated from lamellipodia and cell bodies of migrating cells, as described in Materials and Methods. Representative HPLC chromatograms of lamellipodium (upper panels) and cell body (lower panels) fractions are shown. B. From multiple experiments, incorporated 32P counts were quantified at each peak and presented as the mean and standard deviation about the mean. Gray columns, Ajuba−/−; black columns, wild type (+/+).
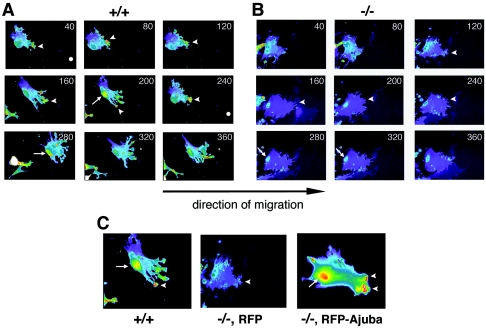
In the second approach PI(4,5)P2 levels in live, migrating Ajuba−/− and control wt MEFs were measured. Cells were transiently transfected with a plasmid expressing the PH domain of phospholipase Cδ (PLCδ) fused to GFP. The GFP-PLCδ PH domain specifically interacts with PI(4,5)P2 in cells (i.e., a PIP2 sensor) (12). To ensure that cells across experimental groups expressed equivalent amounts of GFP-PLCδ PH, red fluorescent protein (RFP) was cotransfected with GFP-PLCδ PH (1:1 ratio) and only cells with equivalent levels of RFP were compared. Following transfection, cells were induced to migrate by making scratch wounds on a plate of confluent cells and fluorescent video microscopy of live migrating cells at the wound edge was performed (Fig. 4A and B; see also movies S1 and S2 in the supplemental material). The most striking difference between Ajuba−/− and wt cells was the reduction in GFP-PLCδ PH fluorescence in Ajuba−/− MEFs. In wt MEFs GFP-PLCδ PH accumulated throughout the extended lamellipodia in the direction of migration (Fig. 4A; see also movie S1 in the supplemental material). The highest concentration of probe was detected in the membrane ruffles formed at the leading edge of the lamellipodia (Fig. 4A, arrowheads). During migration, cells maintained a gradient of PIP2 from the lamellipodia facing the wound to the back of the cell opposite the extending lamellipodia. GFP-PLCδ PH also accumulated at the back of the cell preceding retraction of the trailing tail (Fig. 4A, arrow). This tail accumulation of GFP-PLCδ PH was not due to the bulging of the backside of a cell, as the RFP intensity (indicative of cell volume) did not increase over the same time period. In contrast to wt MEFs, Ajuba−/− MEFs did not migrate effectively into the wound, did not produce broad lamellipodia, and displayed reduced membrane ruffle formation (32; also data not shown). Localization of GFP-PLCδ PH to the leading lamellipodia was reduced and was evident in membrane ruffles only transiently and at low levels (Fig. 4B, arrowheads; see also movie S2 in the supplemental material). Compared to the gradual gradient of GFP-PLCδ PH from the leading edge to the rear of the cell in wt MEFs, in Ajuba−/− MEFs the GFP-PLCδ PH gradient was not maintained, and we did not detect any accumulation of GFP-PLCδ PH in the tail region (Fig. 4B, arrows; see also movie S2 in the supplemental material).
FIG. 4.
Migrating Ajuba-null MEFs have decreased levels of PI(4,5)P2, as detected by the PI(4,5)P2 sensor GFP-PLCδ PH. Wild-type (+/+) MEFs (A) or Ajuba-null (−/−) MEFs (B) were transfected with GFP-PLCδ PH, and scratch wounds were made. Migration into the wound is towards the right. Time-lapse frames (minutes) acquired during migration show the fluorescence intensity of the GFP-PLCδ PH probe. Pseudocolors reflect the fluorescent intensity from low (blue) to high (red). Arrowheads identify membrane ruffles at the leading edge. Arrows identify the retracting tail. C. Wild-type MEFs (+/+) were transfected with GFP-PLCδ PH probe (left panel), Ajuba-null MEFs (−/−) were cotransfected with the GFP-PLCδ PH probe and RFP (middle panel), and Ajuba-null MEFs were cotransfected with GFP-PLCδ PH probe and RFP-Ajuba (right panel). Migration was stimulated by scratch wounding cells, and cell migration into the wound (towards the right) was imaged. Arrowheads identify membrane ruffles at the leading edge. Arrows identify the retracting tail.
To ensure that the abnormal GFP-PLCδ PH staining profile in migrating Ajuba−/− MEFs was due to the absence of Ajuba, Ajuba−/− MEFs were transiently transfected with GFP-PLCδ PH and RFP-Ajuba or as a control GFP-PLCδ PH and RFP alone. RFP-Ajuba rescued the motility defect of Ajuba−/− cells, as expected (32). Coexpression of Ajuba and GFP-PLCδ PH in Ajuba−/− cells resulted in a GFP-PLCδ PH staining profile similar to that of wt cells, including enrichment in leading-edge lamellipodia, accumulation in membrane ruffles (Fig. 4C, arrowheads; see also movie S3 in the supplemental material), and accumulation in the tail preceding retraction (Fig. 4C, arrow; see also movie S3 in the supplemental material).
In summary, by two different methods, Ajuba−/− MEFs were found to contain reduced levels of PI(4,5)P2 and increased levels of PI(4)P compared to wt MEFs, particularly in the lamellipodia and membrane ruffles at the leading edge of migrating cells. Reintroduction of Ajuba into these cells rescued not only the migration defect but also the level of PI(4,5)P2. This reduction in PI(4,5)P2 level and corresponding increase in PI(4)P level in Ajuba−/− MEFs suggested that synthesis of PI(4,5)P2 could be regulated by Ajuba in these cells.
Ajuba influences the membrane localization of PIPKIα and enhances PIPKIα enzymatic activity while inhibiting PIPKIIβ activity in vitro.
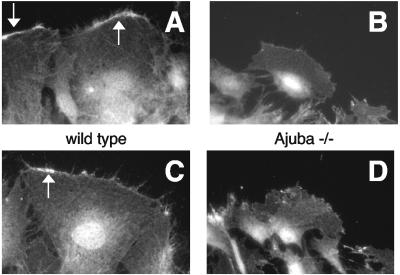
Ajuba could regulate PI(4,5)P2 synthesis by influencing the localization of PIPK enzymes, PIPK enzyme activity, or both. To determine if Ajuba influenced enzyme localization, wt and Ajuba-null MEFs were induced to migrate, by scratch wounding, and after a period of time fixed, and indirect immunofluorescence with antiserum that detects endogenous PIPKIα (15) was performed. In wild-type MEFs PIPKIα was localized to the leading-edge membrane ruffles and nucleus and diffusely throughout the cytoplasm, as expected (Fig. 5A and C) (15). However, in Ajuba-null MEFs there was little PIPKIα detected at the leading edge (Fig. 5B and D). It should be noted that migrating Ajuba-null cells produced less membrane ruffles at the leading edge than did migrating wild-type cells. In Ajuba-null cells PIPKIα was still detected in the nucleus and cytosol, however. When migrating Flag-Ajuba-rescued Ajuba-null cells were costained for PIPKIα (anti-PIPKIα antibody) and Ajuba (anti-Flag antibody), we did not detect significant colocalization of these two proteins (data not shown).
FIG. 5.
PIPKIα localization to the leading-edge membrane ruffles is decreased in migrating Ajuba-null MEFs. Wild-type (A and C) or Ajuba−/− (B and D) MEFs were stimulated to migrate by scratch wounding and then fixed, and an immunofluorescence assay with an antibody that detects endogenous PIPKIα was performed. The direction of migration is towards the top of the page. Arrows identify membrane ruffles at the leading edge.
These results suggested the possibility that Ajuba is required for the leading-edge membrane ruffle localization of PIPKIα. In addition, the diminution of leading-edge membrane ruffling in migrating Ajuba-null cells could also be contributing to the decreased PIPKIα staining observed at this site.
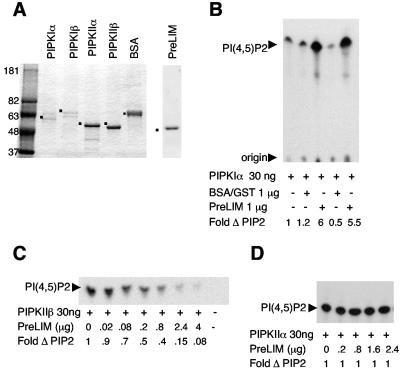
To determine whether Ajuba's interaction with PIPKs influenced their enzymatic activity, purified PIPKIα, PIPKIIβ, and PIPKIIα were mixed with the purified PreLIM region of Ajuba. His-tagged PIPKs and GST-PreLIM Ajuba were expressed and purified from Escherichia coli as described previously (16, 45). The purity of the isolated proteins is shown in Fig. 6A. In the presence of substrate and [32P]ATP all purified enzymes were active (data not shown). The PreLIM region of Ajuba was used rather than full-length Ajuba, as expression of GST-Ajuba was not stable in bacteria, and purification resulted in multiple degradation products, thereby decreasing the effective concentration of binding domain in the sample.
FIG. 6.
Ajuba activates PIPKIα activity but inhibits PIPKIIβ activity, in vitro. A. Coomassie blue-stained SDS-polyacrylamide gel electrophoresis of an aliquot of purified PIPKs and PreLIM Ajuba peptide. For comparison 1 μg of BSA was run in lane 6. Dots identify enzymes or PreLIM peptide. Molecular standards in kDa are run in lane 1. B. In vitro kinase activity of PIPKIα in the presence of a 30-fold excess of PreLIM Ajuba peptide or control BSA or GST. Fold increase in enzyme activity is presented below each lane (control, no additions, lane 1 = 1). C. In vitro kinase activity of PIPKIIβ in the presence of fixed amounts of PIPKIIβ and increasing amounts of PreLIM Ajuba peptide. Fold decrease in enzyme activity is presented below each lane (control, no additions, lane 1 = 1). D. In vitro kinase activity of PIPKIIα in the presence of fixed amounts of PIPKIIα and increasing amounts of PreLIM Ajuba peptide. Fold change in enzyme activity is presented below each lane (control, no additions, lane 1 = 1).
In the presence of fixed amounts of PIPKIα enzyme and PI(4)P substrate the addition of increasing amounts of Ajuba PreLIM peptide to the kinase reaction resulted in enzyme activation [i.e., increased PI(4,5)P2 production] (data not shown). When the amounts of PreLIM peptide and PI(4)P substrate were fixed, increasing amounts of added PIPKIα also resulted in increased PI(4,5)P2 synthesis (data not shown). A maximum sixfold stimulation of the PIPKIα kinase activity was observed in the presence of a 30-fold molar excess of Ajuba PreLIM peptide (Fig. 6B). This stimulation was specific to Ajuba PreLIM peptide, as the addition of an equal amount of unrelated protein, such as bovine serum albumin (BSA) or GST, had no effect (Fig. 6B).
Interestingly, Ajuba PreLIM peptide had the opposite effect on the kinase activity of PIPKIIβ. In the presence of fixed amounts of PIPKIIβ and PI(5)P substrate the addition of increasing amounts of Ajuba PreLIM peptide inhibited the production of PI(4,5)P2 (Fig. 6C). At the same 30-fold molar excess of PreLIM peptide PIPKIIβ activity was inhibited four- to fivefold (Fig. 6C). Importantly the addition of Ajuba PreLIM peptide to PIPKIIα, an enzyme that did not interact with Ajuba, had no effect on its kinase activity (Fig. 6D). Thus, Ajuba profoundly increased the activity of PIPKIα while inhibiting the activity of PIPKIIβ, in vitro.
PIPKIα is required for fibroblast migration.
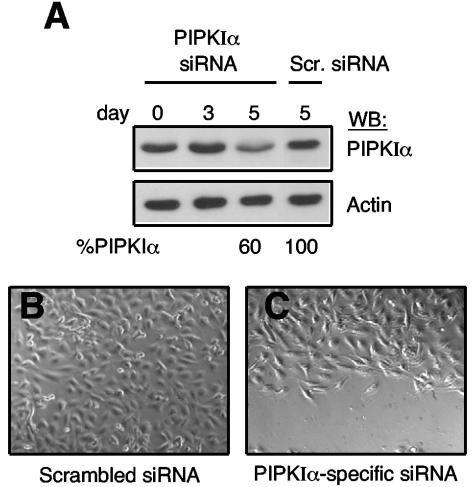
Since Ajuba can influence the activity of PIPKIα and Ajuba-null cells have decreased amounts of PI(4,5)P2 and are inhibited in their cell migration, we determined whether PIPKIα was required for the migration of wt MEFs. To do so, PIPKIα levels were reduced using an RNAi strategy. Primary wt MEFs were infected with retroviruses expressing an siRNA against PIPKIα or as a control a scrambled siRNA. The time to significant reduction in PIPKIα protein level following RNAi treatment was relatively long. The level of PIPKIα did not begin to significantly decrease until day 5 (Fig. 7A). In control scrambled RNAi-treated MEFs we did not observe any decrease in PIPKIα level at day 5 (Fig. 7A). The level of the focal adhesion protein vinculin was not changed in PIPKIα siRNA or control scrambled siRNA-treated cells (data not shown). Interestingly at 7 or more days following RNAi infection the PIPKIα levels further dropped and cells began to die. Similar results were observed in PIPKIα siRNA-treated HEK293 cells. Cell viability appeared to depend upon the degree of PIPKIα protein reduction. High-level PIPKIα knockdown resulted in cell death, whereas cells with partial knockdown remained viable for longer periods. Thus, for migration assays, we analyzed MEFs with approximately a 50 to 60% decrease in PIPKIα protein level (e.g., day 5). When control scrambled siRNA-treated MEFs and PIPKIα siRNA-treated MEFs were induced to migrate, by scratch wounding, cells containing reduced levels of PIPKIα were inhibited in their migration (Fig. 7B and C). Scrambled siRNA-treated wt MEFs migrated at the same rate as untreated wt MEFs (data not shown). This result suggested that PIPKIα is important for an appropriate cell migration response; however, some caution is necessary, as PIPKIα knocked-down cells began to die 2 days after these migration assays were performed.
FIG. 7.
PIPKIα is required for fibroblast migration. (A) wt MEFs were infected with lentiviruses expressing a PIPKIα-specific siRNA or scrambled control (Scr.), and on the indicated days postinfection cell lysates were immunoblotted for PIPKIα and actin. The amount of PIPKIα present on day 5, relative to day 0, is listed below the day 5 lanes. (B and C) Confluent plates of wt MEFs 5 days after infection with control scrambled siRNA (B) or PIPKIα-specific siRNA (C) were scratch wounded, and 10 h later differential interference contrast images of wound closure were obtained. The direction of migration is towards the bottom of the page.
DISCUSSION
Here we show that the LIM protein Ajuba specifically regulates cellular PI(4,5)P2 levels by directly interacting with and activating PIPKIα to increase synthesis of PI(4,5)P2 from PI(4)P. Migration stimulus significantly increased the amount of PIPKIα associated with Ajuba, indicating that the interaction is regulated. In the absence of Ajuba protein migrating MEFs are defective in production of PI(4,5)P2 and have a corresponding increase in PI(4)P substrate. Reintroduction of Ajuba into Ajuba−/− MEFs corrects both PI(4,5)P2 levels and cell migration. In the absence of Ajuba PIPKIα does not localize appropriately to membrane ruffles at the leading edge of migrating cells and the level of PI(4,5)P2 in lamellipodia is also decreased. In vitro, Ajuba was found to be a potent activator of PIPKIα enzyme activity while inhibiting PIPKIIβ activity.
Ajuba−/− MEFS are inhibited in their migration and have been shown to be defective in activating Rac1 in response to migratory cues (32). Rho family GTPases, including Rac1, have been shown to bind to PIPKs, including PIPKIα, and influence their activity. In many situations PIPKIα appears to be a downstream effector of Rac1 signaling (8, 20, 40). Alternatively, the association of PIPKIα with active Rac1 can influence enzyme recruitment to membranes. Although PIPKIα and Rac1 activities cooperate to regulate membrane ruffling, for example, each protein also has independent functions in the production of membrane ruffles during migration (15). Since Ajuba positively influences Rac1 activity during migration, it is likely that Ajuba-dependent Rac1 activation also contributes to the activation of PIPKIα and thus PI(4,5)P2 production during migration. However, we have shown that Ajuba directly interacts with PIPKIα and that this association is increased following migration stimuli and can affect PIPKIα enzyme activity independently of Rac1, suggesting that Ajuba may also regulate PI(4,5)P2 levels and migration by directly activating PIPKIα. PIPKIα is required for proper cell migration, as siRNA-mediated knockdown of PIPKIα in wild-type MEFs inhibited their migration. Future work is directed at distinguishing the contribution of Ajuba's effect upon Rac1 to PIPKIα activity from Ajuba's direct effects upon PIPKIα. Regardless, both these effects of Ajuba contribute to the production of PI(4,5)P2 and thus enhanced migratory responses through modulation of the actin cytoskeleton. Since PIPKIα can also be a downstream effector of another G protein, Arf6, in membrane ruffle formation (20), this raises the possibility that Ajuba might also influence Arf6 activity.
Ajuba appeared to be necessary for the proper localization of PIPKIα to membrane ruffles at the leading edge of migrating cells. Despite this alteration in enzyme localization and the ability to coimmunoprecipitate Ajuba and PIPKIα, we were not able to colocalize these two proteins by immunofluorescence analysis of fixed and live migrating cells. This could be due to a number of possibilities. In cells Ajuba is present at focal adhesions and diffusely in the cytosol and shuttles in and out of the nucleus. It is not present at the leading edge of migrating cells or in membrane ruffles (where PIPKIα localizes). Therefore, Ajuba and PIPKIα may be interacting in other cellular compartments and Ajuba's role is to deliver the enzyme to sites of membrane ruffling, much like the proposed role for Ajuba in delivering p130Cas to nascent focal complexes in migrating cells (32). In support of this possibility we have recently observed Ajuba on motile cytosolic vesicles, and these appear to shuttle to and from the leading edge of migrating cells (S. Pratt and G. Longmore, unpublished). Furthermore, Ajuba's recruitment to adhesive complexes in fibroblasts and epithelial cells is regulated (S. Pratt and G. Longmore, unpublished; 27). The LIM region of Ajuba directs its localization to adhesive complexes in both cell types, while the PreLIM region directs Ajuba's association with PIPKIα and activates its enzyme activity. Accumulating evidence indicates that the LIM region and PreLIM region of this family of LIM proteins have distinct target interactions, suggesting that a possible role for these proteins is to couple different intracellular signaling pathways or functions in response to a stimulus. For example, Ajuba contributes to the function of adherens junctions in epithelial cells by coupling epithelial cell-cell junctions to the actin cytoskeleton through a LIM region interaction with α-catenin bound to E-cadherin and a PreLIM interaction with F-actin (27) or the recruitment of p130Cas to focal adhesions through its interaction with Ajuba's PreLIM region while the LIM region of Ajuba targets this protein complex to nascent adhesive complexes in migrating cells (32).
The decreased PIPKIα localization to the leading edge of Ajuba-null cells may also reflect the decrease in membrane ruffling apparent in Ajuba-null cells. Only the PIPKIα PIPK isoform localizes to membrane ruffles, and expression of a kinase-dead mutant of PIPKIα blocks membrane ruffling (15). When overexpressed PIPKIα did not stimulate membrane ruffles, however, in the presence of constitutively activated Rac, membrane ruffles were produced. These authors proposed that PIPKIα regulates localized production of PI(4,5)P2 while actual membrane ruffle formation requires Rac activation. Ajuba may provide a link between these two pathways to regulate membrane ruffling.
Although Ajuba and related LIM proteins are present at focal adhesions (5, 32), Ajuba did not interact with PIPKIγ (the type I PIPK isoform present at focal adhesions). Talin, another focal adhesion protein, was shown to localize PIPKIγ to focal adhesions and activate its enzymatic activity at these sites, thereby regulating focal adhesion formation (13, 26). The subcellular distribution of the zyxin/Ajuba family of LIM proteins is largely overlapping, and yet related LPP and zyxin did not interact with the PIPKs, suggesting some structural feature unique to Ajuba vis-à-vis an association with PIPK. The N-terminal PreLIM regions of this protein family are the most divergent, and with respect to Ajuba, PIPKIα, and PIPKIβ this was the region that directed the association.
The zyxin/Ajuba LIM proteins also contain nuclear export sequences in their PreLIM regions, and multiple studies have demonstrated that they shuttle in and out of the nucleus (23, 29, 30), and nuclear accumulation of Ajuba in embryonal carcinoma cells has been shown to affect cell proliferation and cell fate decisions, through pathways that are presently undetermined (23). Phosphoinositides are also present in the nucleus, and their composition varies during the cell cycle (10, 22). Endogenous PIPKIα has been shown to localize to the nucleus in a speckled pattern, where it is thought to function in mRNA processing (14). Whether Ajuba also interacts with PIPKIα in the nucleus and could influence phosphoinositide regulation in the nucleus remains to be determined.
How an association with Ajuba can have different effects upon two related PIPK enzymes is unclear. Ajuba profoundly increases the activity of PIPKIα while inhibiting the activity of PIPKIIβ, in vitro. The sequences of various PIPK isoforms are quite conserved, and yet it is not clear how their activity and PI(4,5)P2 synthesis are regulated (25). Nevertheless, since PI(4)P is the major PIP present in cells, the physiological significance of inhibiting PIPKIIβ activity is likely to be of minimal importance. This would appear to be true, as in vivo Ajuba's effect upon PIPKIα clearly predominates, since the levels of PI(4,5)P2 are decreased in Ajuba−/− cells, while if its inhibitory effect upon PIPKIIβ predominated, they would be increased.
Supplementary Material
Acknowledgments
We thank P. W. Majerus for helpful discussions and suggestions and S. J. Pratt and H. Epple for isolation of Ajuba-null MEFs.
This work was supported by grant CA75315 from the NIH (G.D.L.) and the Washington University/Pfizer biomedical research program (G.D.L.).
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alpin, A. E., and R. L. Juliano. 2001. Regulation of nucleocytoplasmic trafficking by cell adhesion receptors and the cytoskeleton. J. Cell Biol. 155:187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, R. A., I. V. Boronenkov, S. D. Doughman, J. Kunz, and J. C. Loijens. 1999. Phosphatidylinositol phosphate kinases, a multifaceted family of signaling enzymes. J. Biol. Chem. 274:9907-9910. [DOI] [PubMed] [Google Scholar]
- 3.Auger, K. R., L. A. Serunian, S. P. Soltoff, P. Libby, and L. C. Cantley. 1989. PDGF-dependent tyrosine phosphorylation stimulates production of novel phosphoinositides in intact cells. Cell 57:167-175. [DOI] [PubMed] [Google Scholar]
- 4.Bach, I. 2000. The LIM domain: regulation by association. Mech. Dev. 91:5-17. [DOI] [PubMed] [Google Scholar]
- 5.Beckerle, M. C. 1997. Zyxin: zinc fingers at sites of cell adhesion. Bioessays 19:949-957. [DOI] [PubMed] [Google Scholar]
- 6.Botelho, R. J., M. Teruel, R. Dierckman, R. Anderson, A. Wells, J. D. York, T. Meyer, and S. Grinstein. 2000. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J. Cell Biol. 151:1353-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, J. D., S. J. Field, L. E. Rameh, C. L. Carpenter, and L. C. Cantley. 2004. Identification and characterization of a phosphoinositide phosphate kinase homolog. J. Biol. Chem. 279:11672-11679. [DOI] [PubMed] [Google Scholar]
- 8.Chatah, N., and C. S. Abrams. 2001. G-protein-coupled receptor activation induces the membrane translocation and activation of phosphotidylinositol-4-phosphate 5 kinase 1a by a Rac- and Rho-dependent pathway. J. Biol. Chem. 276:34059-34065. [DOI] [PubMed] [Google Scholar]
- 9.Cho, S. Y., and R. L. Klemke. 2002. Purification of pseudopodia from polarized cells reveals redistribution and activation of Rac through assembly of a CAS/Crk scaffold. J. Cell Biol. 156:725-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke, J. H., A. J. Letcher, S. D'Santos, C., J. R. Halstead, R. F. Irvine, and N. Divecha. 2001. Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukaemia cells. Biochem. J. 357:905-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crawford, A. W., J. W. Michelson, and M. C. Beckerle. 1992. An interaction between zyxin and alpha-actinin. J. Cell Biol. 116:1381-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cullen, P. J., G. E. Cozier, G. Banting, and H. Mellor. 2001. Modular phosphoinositide-binding domains—their role in signaling and membrane trafficking. Curr. Biol. 11:R882-R893. [DOI] [PubMed] [Google Scholar]
- 13.Di Paolo, G., L. Pellegrini, K. Letinic, G. Cestra, R. Zoncu, S. Voronov, S. Chang, J. Guo, M. R. Wenk, and P. De Camilli. 2002. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1 gamma by the FERM domain of talin. Nature 420:85-89. [DOI] [PubMed] [Google Scholar]
- 14.Doughman, R. L., A. J. Firestone, and R. A. Anderson. 2003. Phosphatidylinositol phosphate kinases put PI4,5P(2) in its place. J. Membr. Biol. 194:77-89. [DOI] [PubMed] [Google Scholar]
- 15.Doughman, R. L., A. J. Firestone, M. L. Wojtasiak, M. W. Bunce, and R. A. Anderson. 2003. Membrane ruffling requires coordination between type Iα phosphatidylinositol phosphate kinase and Rac signaling. J. Biol. Chem. 278:23036-23045. [DOI] [PubMed] [Google Scholar]
- 16.Goyal, R. K., P. Lin, J. Kanungo, A. S. Payne, A. J. Muslin, and G. D. Longmore. 1999. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol. Cell. Biol. 19:4379-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 18.Hirota, T., N. Kunitoku, T. Sasayama, T. Marumoto, D. Zhang, M. Nitta, K. Hatakeyama, and H. Saya. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114:585-598. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman, L. M., D. A. Nix, B. Benson, R. Boot-Hanford, E. Gustafsson, C. Jamora, A. S. Menzies, K. L. Goh, C. C. Jensen, F. B. Gertler, E. Fuchs, R. Fassler, and M. C. Beckerle. 2003. Targeted disruption of the murine zyxin gene. Mol. Cell. Biol. 23:70-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda, A., M. Nogami, T. Yokozeki, M. Yamazaki, H. Nakamura, H. Watanabe, K. Kawamoto, K. Nakayama, A. J. Morris, M. A. Frohman, and Y. Kanaho. 1999. Phosphatidylinositol 4-phosphate 5-kinase a is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99:521-532. [DOI] [PubMed] [Google Scholar]
- 21.Ikonomov, O. C., D. Sbrissa, K. Mlak, M. Kanzaki, J. Pessin, and A. Shisheva. 2002. Functional dissection of lipid and protein kinase signals of PIKfyve reveals the role of PtdIns 3,5-P2 production for endomembrane integrity. J. Biol. Chem. 277:9206-9211. [DOI] [PubMed] [Google Scholar]
- 22.Irvine, R. 2000. Nuclear lipid signaling. Sci. STKE 2000:RE1. [DOI] [PubMed] [Google Scholar]
- 23.Kanungo, J., S. J. Pratt, H. Marie, and G. D. Longmore. 2000. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell 11:3299-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kisseleva, M. V., L. Cao, and P. W. Majerus. 2002. Phosphoinositide-specific inositol polyphosphate 5-phosphatase IV inhibits Akt/protein kinase B phosphorylation and leads to apoptotic cell death. J. Biol. Chem. 277:6266-6272. [DOI] [PubMed] [Google Scholar]
- 25.Kunz, J., M. P. Wilson, M. Kisseleva, J. H. Hurley, P. W. Majerus, and R. A. Anderson. 2000. The activation loop of phosphatidylinositol phosphate kinases determines signaling specificity. Mol. Cell 5:1-11. [DOI] [PubMed] [Google Scholar]
- 26.Ling, K., R. L. Doughman, A. J. Firestone, M. W. Bunce, and R. A. Anderson. 2002. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature 420:89-93. [DOI] [PubMed] [Google Scholar]
- 27.Marie, H., S. J. Pratt, M. Betson, E. H., J. Kittler, L. Meek, S. Moss, S. Troyanosky, D. Attwell, G. D. Longmore, and V. M. M. Braga. 2003. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with α-catenin. J. Biol. Chem. 278:1220-1228. [DOI] [PubMed] [Google Scholar]
- 28.McLaughlin, S., J. Wang, A. Gambhir, and D. Murray. 2002. PIP2 and proteins: interactions, organization, and information flow. Annu. Rev. Biophys. Biomol. Struct. 31:151-175. [DOI] [PubMed] [Google Scholar]
- 29.Nix, D. A., and M. C. Beckerle. 1997. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 138:1139-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petit, M. M., J. Fradelizi, R. M. Golsteyn, T. A. Y. Ayoubi, B. Menichi, D. Louvard, W. J. Van de Ven, and E. Friederich. 2000. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell 11:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility is driven by assembly and disassembly of actin filaments. Cell 112:453-465. [DOI] [PubMed] [Google Scholar]
- 32.Pratt, S. J., H. Epple, M. Ward, Y. Feng, V. M. M. Braga, and G. D. Longmore. 2005. The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J. Cell Biol. 168:813-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2002. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren, X. D., G. M. Bokoch, A. Traynor-Kaplan, G. H. Jenkins, R. A. Anderson, and M. A. Schwartz. 1996. Physical association of the small GTPase Rho with a 68-kDa phosphatidylinositol 4-phosphate 5-kinase in Swiss 3T3 cells. Mol. Biol. Cell 7:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saito, K., K. F. Tolias, A. Saci, H. B. Koon, L. A. Humphries, A. Scharenberg, D. J. Rawlings, J.-P. Kinet, and C. L. Carpenter. 2003. BTK regulates PtdIns-4,5-P2 synthesis: importance for calcium signaling and PI3K activity. Immunity 19:669-678. [DOI] [PubMed] [Google Scholar]
- 36.Sbrissa, D., O. C. Ikonomov, and A. Shisheva. 2002. Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve endomembrane localization. J. Biol. Chem. 277:6073-6079. [DOI] [PubMed] [Google Scholar]
- 37.Sbrissa, D., O. C. Ikonomov, and A. Shisheva. 1999. PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin. J. Biol. Chem. 274:21589-21597. [DOI] [PubMed] [Google Scholar]
- 38.Tolias, K. F., L. C. Cantley, and C. L. Carpenter. 1995. Rho family GTPases bind to phosphoinositide kinases. J. Biol. Chem. 270:17656-17659. [DOI] [PubMed] [Google Scholar]
- 39.Tolias, K. F., A. D. Couvillon, L. C. Cantley, and C. L. Carpenter. 1998. Characterization of a Rac1- and RhoGDI-associated lipid kinase signaling complex. Mol. Cell. Biol. 18:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolias, K. F., J. H. Hartwig, H. Ishihara, Y. Shibasaki, L. C. Cantley, and C. L. Carpenter. 2000. Type Iα phosphatidylinositol-4-phosphate 5-kinase mediates Rac-dependent actin assembly. Curr. Biol. 10:153-156. [DOI] [PubMed] [Google Scholar]
- 41.Tolias, K. F., L. E. Rameh, H. Ishihara, Y. Shibasaki, J. Chen, G. D. Prestwich, L. C. Cantley, and C. L. Carpenter. 1998. Type I phosphatidylinositol-4-phosphate 5-kinases synthesize the novel lipids phosphatidylinositol 3,5-bisphosphate and phosphatidylinositol 5-phosphate. J. Biol. Chem. 273:18040-18046. [DOI] [PubMed] [Google Scholar]
- 42.Ward, M. E., J. Y. Wu, and Y. Rao. 2004. Visualization of spatially and temporally regulated N-WASP activity during cytoskeletal reorganization in living cells. Proc. Natl. Acad. Sci. USA 101:970-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weernink, P. A., K. Meletiadis, S. Hommeltenberg, M. Hinz, H. Ishihara, M. Schmidt, and K. H. Jakobs. 2004. Activation of type I phosphatidylinositol 4-phosphate 5-kinase isoforms by the Rho GTPases, RhoA, Rac1, and Cdc42. J. Biol. Chem. 279:7840-7849. [DOI] [PubMed] [Google Scholar]
- 44.Wei, Y. J., H. Q. Sun, M. Yamamoto, P. Wlodarski, K. Kunii, M. Martinez, B. Barylko, J. P. Albanesi, and H. L. Yin. 2002. Type II phosphatidylinositol 4-kinase beta is a cytosolic and peripheral membrane protein that is recruited to the plasma membrane and activated by Rac-GTP. J. Biol. Chem. 277:46586-46593. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, X., J. C. Loijens, I. V. Boronenkov, G. J. Parker, F. A. Norris, J. Chen, O. Thum, G. D. Prestwich, P. W. Majerus, and R. A. Anderson. 1997. Phosphatidylinositol-4-phosphate 5-kinase isozymes catalyze the synthesis of 3-phosphate-containing phosphatidylinositol signaling molecules. J. Biol. Chem. 272:17756-17761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.