Abstract
The identification of essential genetic elements in pathways governing the maintenance of fully established tumors is critical to the development of effective antioncologic agents. Previous studies revealed an essential role for H-RASV12G in melanoma maintenance in an inducible transgenic model. Here, we sought to define the molecular basis for RAS-dependent tumor maintenance through determination of the H-RASV12G-directed transcriptional program and subsequent functional validation of potential signaling surrogates. The extinction of H-RASV12G expression in established tumors was associated with alterations in the expression of proliferative, antiapoptotic, and angiogenic genes, a profile consistent with the observed phenotype of tumor cell proliferative arrest and death and endothelial cell apoptosis during tumor regression. In particular, these melanomas displayed a prominent RAS-dependent regulation of the epidermal growth factor (EGF) family, leading to establishment of an EGF receptor signaling loop. Genetic complementation and interference studies demonstrated that this signaling loop is essential to H-RASV12G-directed tumorigenesis. Thus, this inducible tumor model system permits the identification and validation of alternative points of therapeutic intervention without neutralization of the primary genetic lesion.
Malignant melanoma is a lethal malignancy due to its propensity to metastasize and near universal failure to respond to existing therapies (12, 33). Molecular genetic studies have identified multiple oncogenic alterations in sporadic melanomas, including frequent loss of the INK4a/ARF and PTEN tumor suppressor loci and activating mutations in B-RAF or in the H- and N-RAS GTPases (16, 26, 28, 62). In addition, germ line mutations in the INK4a locus are associated with familial malignant melanoma (29), and the tumors occurring in this syndrome appear to have a high incidence of N-RAS activation (19). We have shown that in mice, melanocyte-specific expression of oncogenic H-RASV12G cooperates with germ line deficiency of either component of the Ink4a/Arf locus or with p53 deficiency to promote melanoma (5, 13, 14, 32), as does the combined loss of Ink4a/Arf and Pten (63). In line with the epidemiological link between childhood UV exposure and melanoma in humans, Ink4a/Arf deficiency promotes UV-induced melanoma development in the mouse (50, 55). Mice deficient in Ink4a/Arf are also prone to the development of melanomas after carcinogen treatment (34, 54). These observations support the utility of the mouse as a genetic model system for validating the role of candidate melanoma genes and elucidating the interactions of molecular signaling pathways involved in the pathogenesis of human melanoma.
The transforming activity of RAS oncogenes involves the regulation of diverse cellular signal transduction pathways that direct processes, such as proliferation, migration, and resistance to apoptosis (57). These processes are mediated by multiple RAS effectors that include the Raf serine/threonine kinases, phosphoinositide 3-kinases (PI3-K), and Ral GDP-GTP exchange factors (RalGEFs). Both the signaling pathways engaged by activated RAS proteins and the resulting biological readouts are modulated in complex ways by differences in cell type and associated genetic alterations. The impact of cell type is evidenced, for example, by the capacity of RAS to activate PI3-K in fibroblasts but not in lymphocytes (24). With respect to genetic context, RAS can induce either senescence or transformation of primary human and mouse fibroblasts depending upon the functional status of INK4a/ARF and p53 loci (38, 53, 64); at the same time, however, activated RAS provokes a proliferative response in primary human or rat thyroid epithelial cells regardless of p53 status (8, 10, 36). The analysis of RAS effector pathways has revealed a corresponding level of complexity. For example, while transformation of immortal mouse fibroblasts is efficiently effected by activated Raf alone, transformation of rat intestinal epithelial cell lines requires both Raf and PI3-K activation (9, 35, 44, 59). It has also been suggested that there exist species-specific differences in response to RAS, since activation of RalGEF may be central to transformation of human cells but not mouse cells (27). However, genetic observation of the high frequency of activating BRAF mutations and the mutually exclusive relationship of BRAF and RAS mutations in numerous human cancers, including melanoma, underscores the primacy of the Raf-mitogen-activated protein (MAP) kinase pathway in RAS-dependent transformation in a number of human cell lineages and derivative tumors (16). In addition to these cell type-dependent differences in cultured cell systems, in vivo analysis has shown that RAS signaling pathways are modulated by reciprocal interactions with the extracellular matrix and with the host stroma and vasculature (49, 52). Together, these studies point to a need for continued investigation of RAS-directed transformation pathways in distinct cell types as well as in various physiological contexts.
To address the mechanisms by which activated RAS regulates the pathogenesis of melanoma in vivo, we have generated an Ink4a/Arf−/− mouse model of cutaneous melanoma in which an H-RASV12G transgene expression is controlled by the reverse tetracycline transactivator system in a doxycycline-dependent and melanocyte-specific manner (Tyr/Tet-RAS mice) (14). This inducible system permits RAS expression to be extinguished at any stage during tumorigenesis, enabling experimental interrogation of the requirement of sustained RAS activity in the maintenance of the transformed state. This model demonstrated the critical role of sustained H-RASV12G activity for tumor maintenance as evidenced by complete tumor regression upon removal of doxycycline and down-regulation of H-RASV12G transgene expression (14). Remarkably, in contrast to the prominent effects of H-RASV12G extinction observed in vivo, the growth and survival of early passage tumor-derived cell lines are grossly unaffected upon modulation of H-RASV12G expression under monolayer growth conditions. In this study, we determined expression changes that were associated with loss of H-RASV12G activity during tumor regression in vivo and assayed candidate H-RASV12G-dependent factors for their ability to modulate the H-RASV12G-directed tumor maintenance program in vivo.
MATERIALS AND METHODS
Transgenic mice and tumor cell lines.
Tyr/Tet-RAS transgenic mice have doxycycline-inducible expression of H-RASV12G in melanocytes (14). Mice were kept on an Ink4a/Arf−/− genetic background and were given doxycycline in the drinking water to induce melanomas. Primary tumors were adapted to culture as described previously (14). Tumor cell lines were used at early passage (before passage 10) for all studies to avoid divergence from the primary tumors.
SCID explant tumors.
Melanoma cells were implanted in CB-17-scid (C.B-Igh-1b/IcrTac-Prkdcscid; Taconic) mice at 106 cells/site. In most experiments, doxycycline was supplied in the drinking water to activate H-RASV12G expression. For regression experiments, tumors were allowed to develop over 2 to 3 weeks, and then doxycycline was withdrawn. Specimens were isolated at successive time periods after doxycycline withdrawal and snap-frozen or fixed in formalin.
Plasmids, retroviral transduction, and siRNA transfection.
For retrovirus production, cDNAs were inserted in pBABE-Puro. Mouse Ereg cDNA was isolated by reverse transcription-PCR (RT-PCR). Dominant-negative EGFR was a gift of E. Wagner. Retroviral vectors were transfected into 293T cells using the pCL-Eco helper plasmid (42). Retroviral supernatants isolated 36 to 54 h after transfection were diluted 1:1 in culture medium and used to infect melanoma cell lines in the presence of 4 μg/ml Polybrene. At 24 h postinfection, the cells were selected for 2 days in growth medium containing 2.25 μg/ml puromycin. Cells were passaged no more than two times before subcutaneous injection into SCID mice. At least eight injections for each cell line were performed for tumor regression experiments. For small interfering RNA (siRNA) experiments, R545 cells were seeded at 2.5 × 105 cells/well in six-well plates and transfected the next day with either Block-It (Invitrogen) or siRNA against epidermal growth factor receptor (EGFR) (SMARTpool; Dharmacon). Protein lysates were collected 72 h after transfection and assessed for EGFR expression.
RNA and protein analyses.
Total RNA was isolated using Trizol reagent (Invitrogen). Differential display/RT-PCR (DD/RT-PCR) was performed using RNAimage kits 1 to 6 (GenHunter) according to the manufacturer's instructions. RNA was prepared at intervals after doxycycline withdrawal from tumor specimens from two independent series of SCID injections. DD/RT-PCR products showing differential expression in both injection series were cloned and sequenced. Northern blot analysis was performed as described previously (14). For real-time quantitative RT-PCR, relative expression levels were determined by real-time PCR using SYBR green I detection chemistry and the ABI Prism 7700 sequence detection system (Applied Biosystems). Amplification reaction mixtures contained 1× QuantiTect SYBR green PCR buffer (QIAGEN), 1/40 volume of RT reaction mixture from 1 μg total RNA, and 300 nM of each primer in a final volume of 25 μl. Thermal cycling parameters were as follows: 15 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 56°C, and 30 s at 72°C. The comparative cycle threshold method was used to quantify target mRNA copy number in the tumor RNA sample relative to that of an endogenous control gene R15 (assay Z) and a time zero sample as a reference. RNA was prepared from two SCID explant tumors per time point for each cell line and analyzed by RT-PCR in triplicate. Expression was normalized relative to the levels at the zero time point. Ribosomal protein R15 expression was utilized as an internal control. For RNA in situ hybridization, digoxigenin-labeled antisense riboprobes were hybridized to cryostat sections of tissues as described previously (40). For immunoblots, lysates were resolved on 4 to 12% Nu-Page minigels (Invitrogen) or 7% sodium dodecyl sulfate-polyacrylamide gels (for EGFR). Western blots were probed with antibodies against AKT and phospho-AKT (Ser473) (Cell Signaling Technology), Bcl-xl (Pharmingen), phospho-EGFR (Biosource) or EGFR (sc-03), cyclin D1 (sc-450), and PTEN (sc-7974) (Santa Cruz Biotechnology).
Histological analysis and immunohistochemistry.
Tissue samples were formalin fixed and paraffin embedded. Apoptosis was measured by counting nuclear bodies in hematoxylin and eosin (H&E)-stained sections and by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay (ApopTag kit; Intergen). Proliferative indices were measured by counts of mitotic nuclei in H&E-stained sections and by KI-67 immunohistochemistry (Novocastra). Phospho-Erk1/2 (Thr202/Tyr204) was detected by immunohistochemistry (Cell Signaling Technology).
Soft-agar assay and siRNA transfection.
R545 cells were transfected with siRNA for 20 h, trypsinized, and suspended in 0.35% agarose-containing medium, and seeded at 10,000 cells/well in six-well plates in triplicate on top of 0.5% agarose-containing medium for soft-agar assay. Colonies were stained with p-iodonitrotetrazolium violet (Sigma) and counted on day 7.
Gene expression analysis.
Expression profiling was performed using the Affymetrix Mgu74av2 chip.
(i) Data processing.
The CEL files were obtained using Affymetrix Microarray Suite software. The DNA-Chip Analyzer (dChip) (www.dchip.org, version 1.3) was used to normalize all CEL files to the control array (see Fig. 2D, 72hrs b), and the model-based expression (PM-only model) was used to compute the expression values (26, 27).
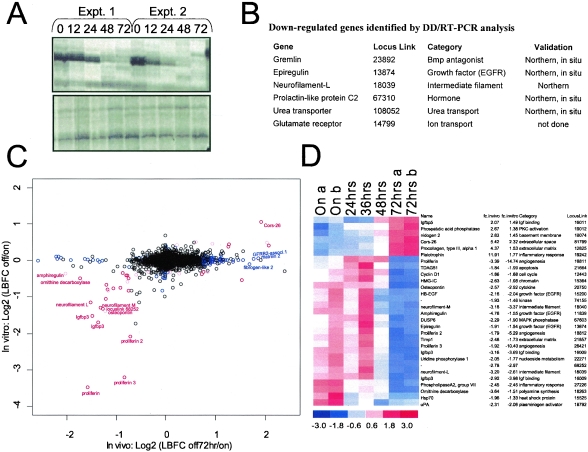
FIG. 2.
Profiling the H-RASV12G-dependent expression program. (A) Representative DD/RT-PCR analyses of melanomas at successive intervals following doxycycline withdrawal. Upper panel, identification of a down-regulated RT-PCR product. Lower panel, RT-PCR products unchanged during tumor regression. The DD/RT-PCR method uses arbitrary primers to profile expression of numerous mRNAs simultaneously (37). Expt., experiment. (B) Down-regulated genes identified by DD/RT-PCR. (C) Graph comparing genes significantly regulated by H-RASV12G in vivo (x axis) and in vitro (y axis) as determined by microarray analysis (see Materials and Methods). Genes in red are significantly modulated both in vivo and in vitro. Genes in blue are regulated only in vivo. The black circles represent genes without significant expression changes. (D) Genes with alterations in expression in both the in vivo and in vitro data sets. Blue color in 72 h specimens represents decreased expression following doxycycline withdrawal, while red color represents increased expression. The standardized expression values (mean of 0 and standard deviation of 1) for each gene is displayed in a range of from −3 to 3. Expression profiling analysis of two tumors is shown for the On and 72-h time points.
(ii) Selection of genes with strong up- and down-regulation both in vivo and in vitro.
Ten samples were used from three in vitro pairs (on/off) and two in vivo pairs (on/off 72 h). Only the 6,396 genes that have at least one sample with >200 expression index and “Present” in at least three samples were considered in the analysis (see Fig. 2B). Red-colored genes in Fig. 2D are differentially expressed both in vivo (lower bound fold change [LBFC] of >1.62) and in vitro (LBFC of >1.24). LBFC is a conservative measure of fold change that takes into account the variation at the expression level as well as the probe level of a gene in each of the groups compared (27). These thresholds are 97 percentile of in vivo absolute LBFC and 98.5 percentile in vitro absolute LBFC over 6,396 genes. LBFCs (or upper bound fold changes [UBFCs]) are averages of all pairwise (i.e., two pairs in vivo and three pairs in vitro) LBFCs (or UBFCs). By these analyses, 23 genes are highly down-regulated and 6 are highly up-regulated both in vivo and in vitro.
(iii) Selection of genes with graded expression changes.
The gene selection procedure with respect to the fold changes may overlook genes with moderate but consistent changes over time. We enlarged our gene list by selecting genes with consistent monotonic temporal changes in vivo. This analysis identified 70 down-regulated genes and 472 up-regulated genes. The selected genes (i) are called present by dChip in at least two of seven samples, (ii) have at least one sample with >200 expression index by dChip, and (iii) have the maximum absolute correlation with the four templates, which is designed to detect monotonically increasing (or decreasing) genes over time, >0.94 (estimated median false discovery rate [FDR] is 0.05). The median FDR is estimated by permuting the sample labels 100 times and calculating correlation with the four templates. The distribution of the maximum (minimum) correlation and the null distribution (see Fig. S2 in the supplemental material) obtained by permuting the sample labels reveal the asymmetric nature of the expression profile and explains the difference in the number of identified up- and down-regulated genes. Among these “gradual” in vivo genes, genes that were differentially expressed in vitro (LBFC of >1.24) are colored pink in the scatter plot. Blue-colored genes are only differentially expressed in vivo but unchanged in vitro (either “strong” or “gradual” regulation in vivo but UBFC of <1.2 in vitro) or only differentially expressed in vitro but unchanged in vivo (“strong” regulation in vitro but UBFC of <1.2 in vivo). Northern in situ hybridization or real-time RT-PCR analysis validated the differential expression of all candidates tested (n = 15).
RESULTS
Upon doxycycline withdrawal, primary de novo melanomas in the inducible Tyr/Tet-RAS model undergo regression within days, culminating in gross and microscopic tumor elimination by 3 weeks (14). Since the de novo tumors are not amenable to serial biopsies and analyses, the explant model—where isogenic tumor cell lines can be used to generate multiple samples at sequential time points—represents an attractive alternative for detailed molecular characterization of the melanoma maintenance program of activated RAS. To this end, we first assessed the in vitro and in vivo properties of several melanoma cell lines independently derived from the inducible Tyr/Tet-RAS transgenic model (n = 7) upon RAS inactivation via doxycycline withdrawal. In parallel, melanoma cells derived from the constitutive Tyr-RAS model were used as a control for the pharmacological effect of doxycycline.
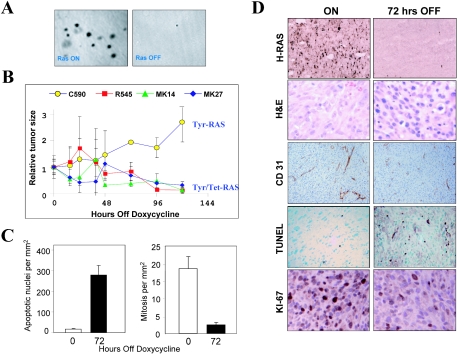
In line with previous observations, none of the cell lines examined required H-RASV12G activity for proliferation under monolayer conditions (14) (data not shown). In contrast, anchorage-independent growth in soft agar was critically dependent on continued H-RASV12G expression (Fig. 1A). Moreover, all seven cell lines demonstrated strict H-RASV12G dependency for tumorigenicity in immunodeficient mice (Materials and Methods; also data not shown). Similar to the de novo melanomas (14), these xenograft tumors also required H-RAS activity for maintenance; gross tumor shrinkage was observed within days after doxycycline withdrawal (Fig. 1B, cell lines R545, MK14, and MK27). In comparison, melanoma cell lines that constitutively express H-RASV12G (13) continued to grow in SCID mice following doxycycline withdrawal (Fig. 1B, cell line C590). Activated RAS expression was nearly extinguished by 72 h after doxycycline withdrawal (as measured by RNA in situ hybridization [Fig. 1D]) and by Northern blotting (see Fig. 3A) at which point the melanoma xenografts still displayed intact tumor parenchyma but had an increasingly benign tumor cell cytoarchitecture accompanied by dramatic loss of vascular integrity as measured by CD31 staining (Fig. 1D). This collapse in tumor vasculature was accompanied by reduction in vascular flow as measured in real-time in vivo magnetic resonance imaging (M. Kim and L. Chin, unpublished data). In addition, loss of RAS activity was associated with induction of apoptosis and reduction in proliferative capacity as measured by TUNEL assay and Ki67 staining, respectively. These immunohistochemical observations were confirmed by quantitative counts of apoptotic and mitotic bodies in H&E-stained tumor sections (Fig. 1C). Since these growth and regression phenotypes were indistinguishable from those observed in the de novo melanoma tumors in the Tyr/Tet-RAS model (see Fig. S1 in the supplemental material) (14), explanted melanomas derived from congenic cell lines and harvested at different time points after doxycycline withdrawal were used in the following studies aimed at characterizing the molecular profile of H-RASV12G-directed tumor maintenance.
FIG. 1.
Regression of transplanted melanomas following extinction of H-RASV12G expression. (A) Soft-agar assay of early passage R545 melanoma cells expressing H-RASV12G (on) or not expressing H-RASV12G (off), showing that colony formation is dependent on H-RASV12G activation. (B) Impact of doxycycline withdrawal on tumor size. Early passage melanoma cell lines were injected subcutaneously into SCID mice given doxycycline in the drinking water. After 14 days, doxycycline was removed from the water, and tumors were isolated at the indicated time points following doxycycline withdrawal. Melanomas with an inducible H-RASV12G transgene regress, while the C590 line that constitutively expresses H-RASV12G continues to grow. The data shown are from a representative experiment consisting of four tumors per time point. Comparable results were obtained in multiple repeat experiments. (C) Apoptotic and proliferative indices of tumors from R545 cells at 0 and 72 h following doxycycline withdrawal. (D) Impact of doxycyline withdrawal on tumor histopathology. Tumors from mice on doxycycline (left panels) or 72 h following doxycycline withdrawal (right panels) were analyzed by RNA in situ hybridization for H-RASV12G expression, H&E staining, CD-31 immunostaining to detect vasculature, TUNEL staining to detect apoptosis, and Ki-67 immunostaining to measure proliferation.
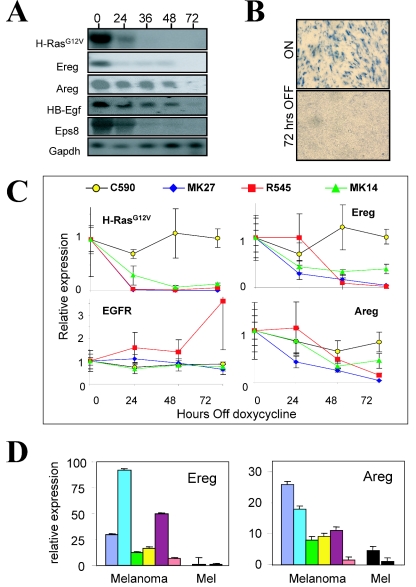
FIG. 3.
H-RASV12G-dependent regulation of EGFR signaling. (A) Northern blot analyses showing loss of expression of H-RASV12G and EGFR pathway components in vivo after doxycycline withdrawal. The time (in hours) after doxycycline withdrawal is shown above the lanes. HB-Egf, heparin-binding EGF-like growth factor; Gapdh, glyceraldehyde-3-phosphate dehydrogenase. (B) RNA in situ hybridization showing Ereg expression during tumor regression. (C) Real-time RT-PCR analysis of H-RASV12G, Ereg, Areg, and EGFR expression in melanomas at intervals following doxycycline withdrawal in vivo. The data are from a representative experiment. A second independent experiment showed comparable results. The y axis shows expression relative to time zero expression in the first experiment for each cell line. C590 expresses H-RASV12G constitutively. (D) In vitro measurement of Ereg and Areg expression in a series of independently derived Tyr/Tet-RAS melanoma cell lines grown in the presence of doxycycline (Melanoma) and in cultured melanocytes (Mel) (in the presence [left bar] or absence [right bar] of doxycycline). The y axis shows expression relative to that for cultured melanocytes in the absence of doxycycline (rightmost bar).
RAS-dependent transcriptome in the regression of established melanomas.
As an initial assessment of the RAS-directed tumor maintenance program, we sought to determine the gene expression changes following extinction of RAS transgene expression in established tumors. Total RNA was isolated at 0 (i.e., RAS on), 24, 36, 48, and 72 h following withdrawal of doxycycline in tumors generated by the R545 melanoma cell line (all experiments employed cells cultivated <10 passages in vitro [see Materials and Methods]). RNA specimens were analyzed by two complementary methods, DD/RT-PCR (37) and oligonucleotide-based microarray approaches; the former was anticipated to identify a set of highly regulated and potentially novel genes, while the latter was expected to provide a broad view of the maintenance program. DD/RT-PCR analysis using 144 different arbitrary primers/anchor primer pairs (Materials and Methods) revealed six mRNAs showing strong differences between the 0- and 72-h time points and graded changes at the intermediate stages (Fig. 2A and B). Notably, this gene set shows pronounced regulation of secreted factors, such as the EGFR ligand, epiregulin, and the bone morphogenetic protein (BMP) antagonist, Gremlin.
In parallel, the mRNA expression profiling analysis of approximately 12,488 oligonucleotide probe sets (approximately 9,800 genes) using Affymetrix MG_U74Av2 chips revealed 542 differentially regulated genes (472 increased and 70 decreased; median FDR of 0.05) (see Fig. S3A and B in the supplemental material) that showed monotonic temporal changes in vivo coinciding with detectable decline in H-RASV12G (see Fig. S4 in the supplemental material for gene list and Materials and Methods for gene selection criteria). Generally, the differentially expressed genes could be grouped into functional categories, consisting of secreted growth factors as well as regulators of signal transduction, angiogenesis, extracellular matrix, metabolism, and inflammation, matching well with the tumor regression phenotype. For example, consistent with previous evidence of H-RASV12G promoting a robust angiogenic state in these tumors, expression profiles showed a significant and acute decline of prolactin/proliferin family genes, several of which have been implicated in the migration and proliferation of vascular endothelial cells (31). Similarly, in line with a role of H-RASV12G in metabolic control, we observed strongly diminished expression of uridine phosphorylase (47) and ornithine decarboxylase (41), regulators of nucleoside and polyamine biosynthesis, respectively. The prominent decreases in neurofilament-L and -M suggest that H-RASV12G imposes a more primitive, neural crest-like phenotype on the tumor cells, a difference that is evident in the morphology observed on H&E-stained sections (Fig. 1D). Overall, the rapid expression changes in these diverse classes of genes are consistent with the concept that H-RASV12G regulates diverse biological processes in fully established tumors and these processes involve both cell autonomous and paracrine growth/survival networks.
Expression profiling of melanoma cell lines grown in vitro—in the presence or absence of doxycycline—revealed a set of RAS-regulated genes that only partially overlaps with that observed in the in vivo experiments (Fig. 2C and D; see Fig. S3 and S4 in the supplemental material). These differences in H-RASV12G-regulated gene expression likely relate to the expression changes in the stromal component of the tumor and/or to the contextual differences of cell culture versus in vivo environments. Notably, the in vivo profiles showed elevated expression of factors relating to fibrosis and inflammation characteristic of a host response, and a number of these factors showed sudden increases at the 72-h time point (see Fig. S3A in the supplemental material). Several of these genes are known to be expressed by stromal cells, including factors associated with the vascular injury (e.g., fibrinogen-like protein and CD59a) (25, 39), lymphocyte activation and gamma interferon stimulation (e.g., T-cell-specific GTPase, guanylate nucleotide-binding protein 2) (7, 11), and extracellular matrix deposition (e.g., matrillin 2) (46). In addition, a series of regulators of apoptosis were induced only in the in vivo setting (e.g., caspase 12 and catalase), consistent with the observed apoptosis in the tumor cell and vascular compartments in vivo. A small number of expression changes were seen only in vitro, including the gap junction protein connexin43, integrin alpha 6, and chemokine CCL9/MIP-1gamma, all of which showed diminished expression following extinction of H-RASV12G expression. These in vitro-only changes presumably reflect the modulating effects of the tissue culture growth environment (e.g., exposure to serum, oxidative stress, growth on plastic substrate) on cellular signaling pathways. Overall, there was a significant concordance of the in vivo and in vitro data sets, suggesting that most of the expression changes observed in vivo related to factors expressed by the tumor cells rather than the stroma.
RAS-directed autocrine EGFR signaling is required for melanoma maintenance.
To begin to identify critical effectors of the H-RASV12G-directed tumor maintenance program, initial studies focused primarily on genes that are regulated in a cell-autonomous fashion; such factors are readily identified by searching for genes showing significant and acute changes in both the in vitro and in vivo data sets (Materials and Methods and Fig. 2C and D). Among 29 candidates (23 increased and 6 decreased) in this category, there was a striking overrepresentation of the EGF family ligands. Specifically, many of the EGF ligands are regulated by H-RASV12G, including epiregulin (Ereg), amphiregulin (Areg), and heparin-binding EGF-like growth factor (HB-EGF) (Fig. 2D) (43). Profiles of the regressing tumors in vivo also showed loss of EGF substrate 8 (Eps8), a factor that links EGFR signaling to the activation of Rac GTPase (see Fig. S3B in the supplemental material). These expression changes were confirmed by Northern blotting (Fig. 3A) and by RNA in situ hybridization; the latter also confirmed that the EGF family ligand expression was tumor cell derived (Fig. 3B and data not shown). Real-time quantitative RT-PCR analysis of regressing tumors originating from three other independently derived Tyr/Tet-RAS melanoma cell lines also showed prominent RAS-dependent regulation of Areg and Ereg, while EGFR itself was largely unchanged (Fig. 3C). These expression changes were specific to declines in H-RASV12G activity, since they were not observed in the C590 constitutive H-RASV12G-expressing melanoma line following doxycycline withdrawal. In addition, analysis of the steady-state expression of Areg and Ereg in the melanoma cells in vitro compared to cultured primary nontransformed melanocytes from Tet/Tyr-RAS mice demonstrated that the melanomas had elevated expression of these factors (Fig. 3D).
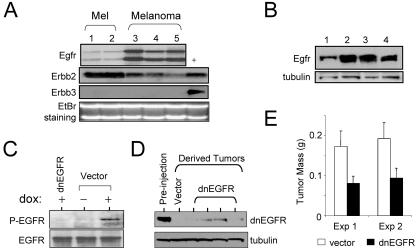
To determine whether autocrine activation of EGFR is operative in the Tyr/Tet-RAS melanomas, we evaluated Egfr expression in primary melanocyte cultures and melanomas derived from this model. As shown in Fig. 4A, all cultured melanocytes (two independent cultures) and melanomas (five independent lines) examined showed Egfr and Erbb2 (Her2/neu) expression, but not the other EGFR family receptors (Erbb3 and Erbb4) (data not shown). Immunoblot analysis confirmed the expression of EGFR protein (Fig. 4B). The expression of both EGFR family receptors and their ligands suggest that H-RASV12G-induced autocrine activation of EGFR signaling may contribute to melanoma genesis in this model. Thus, we directly assessed the presence of autocrine EGFR signaling by immunoblot analysis of phospho-EGFR levels in lysates from cultured melanoma cells. Phospho-EGFR levels were low in melanoma cells maintained in low serum without doxycycline but were strongly induced upon administration of doxycycline (Fig. 4C, compare the middle and rightmost lanes), a finding consistent with induction of EGFR autophosphorylation via an H-RASV12G-dependent autocrine loop.
FIG. 4.
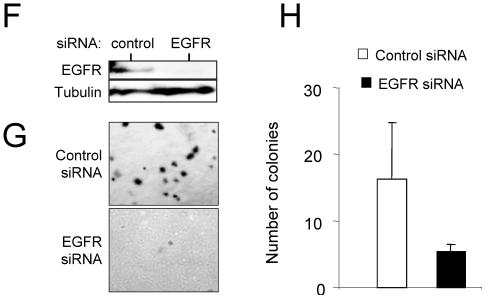
EGFR signaling is required for melanoma tumor maintenance. (A) Northern blot analysis of expression of EGFR family receptors in independent lines of cultured melanocytes (lanes 1 and 2) and independent lines of cultured melanomas from Tyr/Tet-RAS mice (lanes 3 to 5). Lane + contains positive controls for Erbb2 and Erbb3. The ethidium bromide (EtBr)-stained gel is shown as a control for equal loads in the lanes. (B) Immunoblot analysis of EGFR expression in Tyr/Tet-RAS melanomas. (C) Immunoblot analysis of endogenous levels of phospho-EGFR and total EGFR in R545 melanoma cells in vitro. Cells were transduced with dnEGFR or empty vector and grown in the presence (+) or absence (−) of doxycycline (dox). (D) Levels of ectopically expressed dnEGFR in the parental cell line (Pre-injection lane) or in tumors that emerged following injection of these cells in SCID mice (dnEGFR lanes). Lysates for the vector control are shown in the Vector lane. (E) Tumor size 14 days after subcutaneous injection with R545 melanoma cells expressing dnEGFR or empty vector. Eight tumors per time point were analyzed. Exp, experiment. (F) Western blot analysis of R545 cells transfected with control or EGFR siRNA (cell lysates were isolated 72 h posttransfection). (G) Photomicrographs showing soft-agar colony formation by siRNA-transfected R545 cells expressing H-RASV12G. (H) Tabulation of total number of colonies in the soft-agar assay.
It has been shown that EGFR autocrine loops are required for RAS transformation in cultured fibroblasts and epithelial cells (23, 57, 58). To determine whether EGFR signaling contributes to the maintenance of Tyr/Tet-RAS melanomas in vivo, we engineered R545 cells to express wild-type EGFR (wtEGFR) or a dominant-negative EGFR (dnEGFR) mutant that comprises only the ligand binding domain (51). To verify the inhibitory activity of dnEGFR, we performed immunoblot analysis of lysates from dnEGFR-expressing cultures exposed to doxycycline in vitro and found that EGFR autophosphorylation was completely abrogated, consistent with the attenuation of EGFR signaling in these cells (Fig. 4C). Next we generated subcutaneous tumor explants from R545 cells transduced with empty vector, dnEGFR, or wild-type EGFR (wtEGFR). While wtEGFR had no effect on tumorigenesis (i.e., comparable size after 14 days [data not shown]), dnEGFR strongly inhibited tumor formation (P = 0.03) (Fig. 4E). Consistent with the inhibitory impact of dnEGFR, tumors that eventually emerged from the dnEGFR-transduced cells showed greatly diminished levels of dnEGFR protein compared with the levels present in the preinjection cultures—a pattern consistent with strong selective pressure against expression of this dominant-negative mutant protein in developing tumors (Fig. 4D). This inhibitory effect of dnEGFR on the tumorigenicity of Tyr/Tet-RAS melanoma cell line was also observed in another independently derived line, AT21 (data not shown).
To more fully address the tumorigenic role of autocrine EGFR signaling, we assessed whether this autocrine loop contributes to the growth of melanoma cells in soft agar. The ability of R545 cells to form anchorage-independent colonies is strictly dependent on H-RASV12G expression (Fig. 1A). To determine the requirement of EGFR signaling in this process, we employed siRNA against EGFR and against a control protein (green fluorescent protein). Transfection of EGFR siRNA resulted in a specific and potent inhibition of EGFR expression in H-RASV12G-expressing R545 cells (Fig. 4F). Cells transfected with EGFR siRNA showed a diminished ability to form robust foci in soft agar compared to controls (Fig. 4G and H), consistent with a cell-autonomous role of EGFR signaling in efficient transformation of the Tyr/Tet-RAS melanomas.
Sustained EGFR signaling delays tumor regression but cannot replace H-RASV12G.
We next sought to determine whether sustained EGFR signaling could substitute for H-RASV12G activity in the establishment or maintenance of melanomas. Ereg binding to EGFR family receptors transmits more potent mitogenic signals compared to other EGF family ligands, by inducing prolonged receptor activation (56). Melanoma cells overexpressing Ereg or a constitutively active form of EGFR (48) were generated by retroviral transduction and subsequently injected into SCID mice. In the absence of doxycycline administration, these cells did not form tumors, indicating that EGFR signaling is not functionally equivalent to H-RASV12G in melanoma development (data not shown).
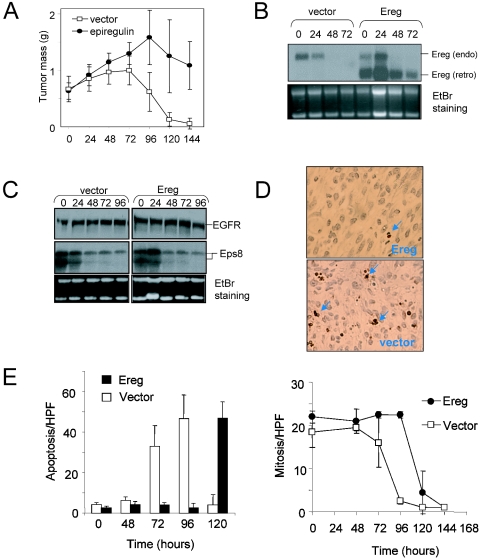
To evaluate the impact of enforced Ereg expression on tumor regression, Ereg-expressing melanoma cells were injected subcutaneously into mice maintained on doxycyline. These cells formed tumors with latencies and sizes similar to those of vector-transduced cells (Fig. 5A). Upon withdrawal of doxycycline, the Ereg-expressing tumors showed delayed regression relative to control tumors transduced with empty vector (Fig. 5A), although the Ereg-expressing tumors eventually regressed completely (data not shown). Consistent with delayed regression kinetics, measurement of apoptotic index by TUNEL staining at successive time points following doxycycline withdrawal demonstrated that enforced Ereg expression was associated with a marked delay in the onset of apoptosis (Fig. 5D and E, left panel) and a less pronounced decline in melanoma cell proliferation (Fig. 5E, right panel), indicating that the Ereg-mediated delay in tumor regression involves both enhanced proliferation and survival signaling.
FIG. 5.
Sustained EGFR signaling partially rescues melanoma survival following extinction of H-RASV12G. (A) R545 cells expressing Ereg or empty vector were injected subcutaneously into SCID mice provided with doxycycline in the drinking water. After 14 days, doxycycline was removed (time zero), and tumors were harvested and weighed at successive time points. Mean tumor mass is represented. Four tumors were analyzed per time point. (B and C) Northern blot analysis of regressing melanomas for expression of Ereg (the bands corresponding to endogenous [endo] and retroviral [retro] Ereg are indicated) (B) and EGFR and Eps8 (C). Ethidium bromide (EtBr)-stained gels were used as loading controls. (D) TUNEL staining (arrows) of apoptotic cells at 72 h following doxycycline withdrawal in control (vector) and Ereg-expressing tumors. (E) Measurement of the apoptotic and mitotic indices of during tumor regression. The apoptosis and mitosis rates per high-power field (HPF) are shown.
One possible mechanism to account for the eventual regression of the Ereg-expressing tumors is that endogenous EGFR expression is lost following extinction of RAS activity. To address this possibility, EGFR was measured at successive time points, which showed that it was maintained through late phases of regression (Fig. 5C, top panel), along with transduced Epg (Fig. 5B, top panel), reinforcing the conclusion that sustained EGFR activity is not equivalent to RAS activation in this system. It remains possible, however, that higher levels of EGFR signaling achieved by the combined overexpression of both autocrine ligands and EGFR may lead to oncogenic growth of melanomas in the absence of activated RAS. Alternatively, the expression of critical components of EGFR signaling—other than ligand-receptor complexes—may require H-RASV12G activity and not be effectively induced by Ereg. Along these lines, ectopic expression of Ereg did not stimulate expression of endogenous Ereg or of Eps8 (Fig. 5B and C).
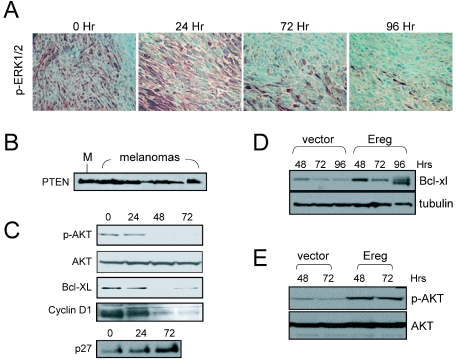
In order to better understand the molecular basis for tumor regression in this model and the specific role of EGFR signaling, we audited the regulation of key RAS effectors following doxycycline withdrawal. The loss of phospho-Erk1/2 staining following doxycycline withdrawal demonstrated that there was a rapid decline in the RAF-MAP kinase signaling during tumor regression (Fig. 6A). Although the expression of PTEN was retained in all melanomas analyzed (Fig. 6B), PI3-K signaling was similarly down-regulated following extinction of H-RASV12G as demonstrated by the loss of phospho-AKT (Fig. 6C). Consistent with these alterations, cyclin D1, a target of both the PI3-K and Erk pathways, showed a rapid loss of expression upon RAS inactivation (Fig. 6C). That AKT activation is lost following RAS down-regulation was further reenforced by the observations that there was a marked decrease in the expression of Bcl-xl and an increase in p27KIP2 levels (Fig. 6C), two proteins known to be induced or repressed, respectively, by activated AKT. Notably, melanomas constitutively expressing Ereg showed sustained phospho-AKT and Bcl-xl levels following withdrawal of doxycycline (Fig. 6D and E), providing a potential molecular basis for the delayed regression of Ereg-overexpressed tumors (Fig. 5A).
FIG. 6.
Survival and proliferative pathways in melanoma maintenance. (A) Immunohistochemistry of phospho-ERK1/2 (p-ERK1/2) levels in regressing melanomas. The time (in hours) after doxycycline withdrawal is shown above the panels. (B) Immunoblot showing expression of PTEN in cultured mouse melanocytes (M) and in melanoma cell lines in vitro. (C) Immunoblots of lysates from regressing melanomas for levels of phospho-AKT (p-AKT), total AKT, Bcl-xl, cyclin D1, and p27. (D and E) Constitutive expression of Ereg partially sustains phospho-Akt (D) and Bcl-xl (E) levels. The time (in hours) after doxycycline withdrawal is shown above the lanes in panels C to E.
RAS-driven transformation of melanocytes involves RAF-MAP kinase.
To further address the contribution of various RAS effector pathways to the melanoma maintenance phenotype and to induction of the EGF family ligands, we transduced early passage melanoma cell lines with retroviruses expressing known H-RAS effector loop mutants, H-RASV12S35,H-RASV12G37, and H-RASV12C40. These mutants have been shown historically to predominantly activate the Raf-MAPK, RalGEF, and PI3-K pathways, respectively. Real-time RT-PCR and Northern blot analysis of these transduced melanoma cells grown in vitro in the absence of doxycycline demonstrated that only RASV12S35 activated Ereg expression (data not shown). The tumorigenicity of these cell lines was assessed by injection into SCID mice without doxycycline administration. Expression of RASV12S35, but not the other RAS effector loop mutants, was able to induce tumors with latencies similar to those of parental cells injected in the presence of doxycycline (Table 1). In addition, expression of RASV12S35, but not the other RAS alleles, in cultured Ink4a/Arf−/− melanocytes produced tumorigenic growth following implantation in SCID mice (data not shown). It should be noted, however, that expression of H-RASV12G with an unmodified effector loop produces tumors with slightly shorter latencies than the S35 effector loop mutant does (∼7 days versus ∼10 days; data not shown), suggesting that the RASV12S35 mutant may be deficient in some melanoma-promoting activities.
TABLE 1.
Tumorigenicity of melanoma cell lines in mice injected with retroviruses expressing known H-RAS effector loop mutants
| H-RAS mutant | Doxa | No. of tumors/no. of mice |
|---|---|---|
| Vector | + | 4/4 |
| − | 0/4 | |
| H-RASV12S35 | − | 4/4 |
| H-RASV12G37 | − | 1/4 |
| H-RASV12C40 | − | 0/4 |
Melanoma cells were grown in vitro in the presence (+) or absence (−) of doxycycline (Dox).
Together, these results demonstrate that activation of the downstream effectors in response to the RASV12S35 mutant, such as the Raf-Erk1 pathway, are critical for RAS-dependent melanoma genesis and that the oncogenic and tumor maintenance program directed by this pathway involves activation of EGFR signaling.
DISCUSSION
In this study, we have examined the genetic program of RAS-directed melanoma tumor maintenance in vivo. These studies led to the identification of a functionally relevant EGFR autocrine loop. In addition to these molecular findings, the changing transcriptional profile following RAS extinction mirrored well the biological features observed in the regressing tumors, providing insights into the mechanisms underlying the regression process. For example, there is an H-RASV12G-directed angiogenic transcriptional program with prominent regulation of the family of prolactin-related/proliferin hormones. Proliferin genes are reactivated in a fibrosarcoma model in which they are essential for neovascularization (60). The potent induction of these hormones by H-RASV12G suggests that this oncogene appropriates a normal developmental placental program of gene expression to promote tumor angiogenesis.
Our demonstration of a role for EGFR signaling in RAS-dependent melanomas is consistent with observations made in other cell systems, including fibroblasts, keratinocytes, and intestinal epithelial cells, that autocrine EGFR signaling is required for transformation by activated RAS (18, 23, 58). The EGFR pathway provides important survival signals in these cell types involving PI3-K-dependent activation of AKT. In the absence of such signaling, RAS is unable to overcome the apoptotic stimulus associated with low growth factor concentrations and detachment from extracellular matrix. Our results suggest that these pathways are operative in melanocyte transformation. It is noteworthy that the contribution of EGFR signaling to melanoma is evolutionarily conserved, since activating mutations in the EGFR homologue, Xmrk, lead to melanoma susceptibility in Xiphophorus fish (17).
It is of interest to consider the genetics of the mouse melanoma model in light of the distinct mutational patterns of human melanomas. Specifically, like murine tumors, human melanomas show mutually exclusive mutations in the NRAS and PTEN genes (62). A similar relationship between PTEN and KRAS occurs in endometrial cancer and between Pten and H-RAS in a mouse model of squamous cell carcinoma (30). Moreover, NRAS and BRAF mutations occur reciprocally in human melanomas, while BRAF mutations are detected in approximately 80% of cultured melanomas harboring loss-of-function PTEN mutations (15, 61). These data appear to indicate RAS mutations are redundant to PTEN loss in tumorigenesis. It is likely that RAS-induced stimulation of survival pathways downstream of PTEN, for example via direct PI3-K activation and through induction of autocrine EGFR signaling, may obviate the requirement of PTEN mutations. PTEN loss may act synergistically more effectively with genetic lesions other than RAS in the pathogenesis of melanomas.
Recent studies have presented contradictory evidence on the relative importance of RAS effector pathways in tumorigenesis. On the basis of in vitro studies with RAS effector loop mutants in immortal human cell lines, one report has suggested that the RalGEF pathway is the major mediator of RAS transformation and that RAF-MAP kinase and PI3-K are dispensable (27). On the other hand, cancer genetics data, most prominently of melanomas, has revealed a very high incidence of BRAF mutations that occur mutually exclusive to RAS mutations in a given tumor (16). Such data lend support to the importance of RAF-MAP kinase signaling for RAS-driven melanoma pathogenesis. Consistent with this notion, the H-RASV12S35 mutant alone is capable of rescuing tumorigenesis in the absence of H-RASV12 in our system, demonstrating genetically that the effectors engaged by this mutant, such as the RAF-MAP kinase pathway, do indeed play important roles in mediating H-RASV12-dependent transformation. What role the RAS effector pathways play in melanoma initiation rather than maintenance is an important question requiring additional investigation.
There is increasing enthusiasm for target-specific therapies driven in part by recent successes in the use of agents directed to specific molecular targets. The effectiveness of this target-specific approach is reinforced further by observations of inducible mouse tumor models directed by single oncogenes, such as H-RAS in melanomas (14), K-RAS in lung cancers (22), Her2/neu in breast cancers and c-Myc in hematopoietic malignancies (20), and islet cell carcinomas (45). The regression of these tumors following deactivation of the specific oncogene demonstrates that advanced malignancies can remain critically dependent on the activity of their initiating lesions and thus constitute attractive candidates for drug discovery and development. On the other hand, many of these oncogenes are not readily targeted by current drug development strategies. Thus, many current therapeutic approaches are directed against molecular targets other than those with specific oncogenic mutations (1, 21). Along these lines, implementation of inducible tumor model systems in conjunction with expression profiling could expand the therapeutic options by targeting signaling pathways linked to an oncogene. Our study illustrates this scenario by uncovering a role for autocrine EGFR signaling in RAS-directed melanoma survival. This raises the possibility that melanomas harboring mutant RAS may show sensitivity to EGFR inhibitors in treatment regimens. Notably, human melanomas harboring RAS mutations appear to have elevated EGFR immunoreactivities than melanomas with wild-type RAS alleles (2). On the other hand, some studies have shown that EGFR inhibitors do not block growth of human melanoma cell lines in vitro (4), suggesting there may be differences in the molecular circuitry of human and mouse melanomas. Beyond the EGFR ligands, our expression profiles reveal other genes highly regulated by H-RASV12G that are alternative useful targets. For example, the chromatin component HMG-IC is necessary for RAS-induced transformation of thyroid cells (6) and hence may also be a requisite component of the transformation program of Tyr/Tet-RAS melanomas. Similarly, ornithine decarboxylase is required for RAS transformation of fibroblasts (3). Overall, these observations suggest that this and similar model systems may provide versatile platforms for the identification and validation of improved molecular therapies.
Supplementary Material
Acknowledgments
We thank E. Wagner and W. Cavenee for reagents and R. DePinho for critical review of the manuscript.
N.B. was supported by a Charles A. King Trust postdoctoral award and an DF/HCC SPORE in Skin Cancer Development grant (P50 CA93683). This work was supported by grants from NIH to M.W.B. (K08 CA89124), W.H.W. (P20 CA96470), and L.C. (R01 CA93947; U01 CA84313). L.C. is a V Foundation Scholar.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adams, J. 2004. The development of proteasome inhibitors as anticancer drugs. Cancer Cell 5:417-421. [DOI] [PubMed] [Google Scholar]
- 2.Albino, A. P., M. J. Vidal, N. S. McNutt, C. R. Shea, V. G. Prieto, D. M. Nanus, J. M. Palmer, and N. K. Hayward. 1994. Mutation and expression of the p53 gene in human malignant melanoma. Melanoma Res. 4:35-45. [DOI] [PubMed] [Google Scholar]
- 3.Auvinen, M., A. Paasinen, L. C. Andersson, and E. Holtta. 1992. Ornithine decarboxylase activity is critical for cell transformation. Nature 360:355-358. [DOI] [PubMed] [Google Scholar]
- 4.Baguley, B. C., E. S. Marshall, K. M. Holdaway, G. W. Rewcastle, and W. A. Denny. 1998. Inhibition of growth of primary human tumour cell cultures by a 4-anilinoquinazoline inhibitor of the epidermal growth factor receptor family of tyrosine kinases. Eur. J. Cancer 34:1086-1090. [DOI] [PubMed] [Google Scholar]
- 5.Bardeesy, N., B. C. Bastian, A. Hezel, D. Pinkel, R. A. DePinho, and L. Chin. 2001. Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol. Cell. Biol. 21:2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berlingieri, M. T., G. Manfioletti, M. Santoro, A. Bandiera, R. Visconti, V. Giancotti, and A. Fusco. 1995. Inhibition of HMGI-C protein synthesis suppresses retrovirally induced neoplastic transformation of rat thyroid cells. Mol. Cell. Biol. 15:1545-1553. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Boehm, U., L. Guethlein, T. Klamp, K. Ozbek, A. Schaub, A. Futterer, K. Pfeffer, and J. C. Howard. 1998. Two families of GTPases dominate the complex cellular response to IFN-gamma. J. Immunol. 161:6715-6723. [PubMed] [Google Scholar]
- 8.Bond, J. A., F. S. Wyllie, J. Rowson, A. Radulescu, and D. Wynford-Thomas. 1994. In vitro reconstruction of tumour initiation in a human epithelium. Oncogene 9:281-290. [PubMed] [Google Scholar]
- 9.Bonner, T. I., S. B. Kerby, P. Sutrave, M. A. Gunnell, G. Mark, and U. R. Rapp. 1985. Structure and biological activity of human homologs of the raf/mil oncogene. Mol. Cell. Biol. 5:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns, J. S., J. P. Blaydes, P. A. Wright, L. Lemoine, J. A. Bond, E. D. Williams, and D. Wynford-Thomas. 1992. Stepwise transformation of primary thyroid epithelial cells by a mutant Ha-ras oncogene: an in vitro model of tumor progression. Mol. Carcinog. 6:129-139. [DOI] [PubMed] [Google Scholar]
- 11.Carlow, D. A., J. Marth, I. Clark-Lewis, and H. S. Teh. 1995. Isolation of a gene encoding a developmentally regulated T cell-specific protein with a guanine nucleotide triphosphate-binding motif. J. Immunol. 154:1724-1734. [PubMed] [Google Scholar]
- 12.Chin, L. 2003. The genetics of malignant melanoma: lessons from mouse and man. Nat. Rev. Cancer 3:559-570. [DOI] [PubMed] [Google Scholar]
- 13.Chin, L., J. Pomerantz, D. Polsky, M. Jacobson, C. Cohen, C. Cordon-Cardo, J. W. Horner II, and R. A. DePinho. 1997. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 11:2822-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin, L., A. Tam, J. Pomerantz, M. Wong, J. Holash, N. Bardeesy, Q. Shen, R. O'Hagan, J. Pantginis, H. Zhou, J. W. Horner II, C. Cordon-Cardo, G. D. Yancopoulos, and R. A. DePinho. 1999. Essential role for oncogenic Ras in tumour maintenance. Nature 400:468-472. [DOI] [PubMed] [Google Scholar]
- 15.Daniotti, M., M. Oggionni, T. Ranzani, V. Vallacchi, V. Campi, D. Di Stasi, G. D. Torre, F. Perrone, C. Luoni, S. Suardi, M. Frattini, S. Pilotti, A. Anichini, G. Tragni, G. Parmiani, M. A. Pierotti, and M. Rodolfo. 2004. BRAF alterations are associated with complex mutational profiles in malignant melanoma. Oncogene 23:5968-5977. [DOI] [PubMed] [Google Scholar]
- 16.Davies, H., G. R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M. J. Garnett, W. Bottomley, N. Davis, E. Dicks, R. Ewing, Y. Floyd, K. Gray, S. Hall, R. Hawes, J. Hughes, V. Kosmidou, A. Menzies, C. Mould, A. Parker, C. Stevens, S. Watt, S. Hooper, R. Wilson, H. Jayatilake, B. A. Gusterson, C. Cooper, J. Shipley, D. Hargrave, K. Pritchard-Jones, N. Maitland, G. Chenevix-Trench, G. J. Riggins, D. D. Bigner, G. Palmieri, A. Cossu, A. Flanagan, A. Nicholson, J. W. Ho, S. Y. Leung, S. T. Yuen, B. L. Weber, H. F. Seigler, T. L. Darrow, H. Paterson, R. Marais, C. J. Marshall, R. Wooster, M. R. Stratton, and P. A. Futreal. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949-954. [DOI] [PubMed] [Google Scholar]
- 17.Dimitrijevic, N., C. Winkler, C. Wellbrock, A. Gomez, J. Duschl, J. Altschmied, and M. Schartl. 1998. Activation of the Xmrk proto-oncogene of Xiphophorus by overexpression and mutational alterations. Oncogene 16:1681-1690. [DOI] [PubMed] [Google Scholar]
- 18.Dlugosz, A. A., L. Hansen, C. Cheng, N. Alexander, M. F. Denning, D. W. Threadgill, T. Magnuson, R. J. Coffey, Jr., and S. H. Yuspa. 1997. Targeted disruption of the epidermal growth factor receptor impairs growth of squamous papillomas expressing the v-ras(Ha) oncogene but does not block in vitro keratinocyte responses to oncogenic ras. Cancer Res. 57:3180-3188. [PubMed] [Google Scholar]
- 19.Eskandarpour, M., J. Hashemi, L. Kanter, U. Ringborg, A. Platz, and J. Hansson. 2003. Frequency of UV-inducible NRAS mutations in melanomas of patients with germline CDKN2A mutations. J. Natl. Cancer Inst. 95:790-798. [DOI] [PubMed] [Google Scholar]
- 20.Felsher, D. W., and J. M. Bishop. 1999. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol. Cell 4:199-207. [DOI] [PubMed] [Google Scholar]
- 21.Ferrara, N., K. J. Hillan, H. P. Gerber, and W. Novotny. 2004. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat. Rev. Drug Discov. 3:391-400. [DOI] [PubMed] [Google Scholar]
- 22.Fisher, G. H., S. L. Wellen, D. Klimstra, J. M. Lenczowski, J. W. Tichelaar, M. J. Lizak, J. A. Whitsett, A. Koretsky, and H. E. Varmus. 2001. Induction and apoptotic regression of lung adenocarcinomas by regulation of a K-Ras transgene in the presence and absence of tumor suppressor genes. Genes Dev. 15:3249-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gangarosa, L. M., N. Sizemore, R. Graves-Deal, S. M. Oldham, C. J. Der, and R. J. Coffey. 1997. A raf-independent epidermal growth factor receptor autocrine loop is necessary for Ras transformation of rat intestinal epithelial cells. J. Biol. Chem. 272:18926-18931. [DOI] [PubMed] [Google Scholar]
- 24.Genot, E., K. Reif, S. Beach, I. Kramer, and D. Cantrell. 1998. p21ras initiates Rac-1 but not phosphatidyl inositol 3 kinase/PKB, mediated signaling pathways in T lymphocytes. Oncogene 17:1731-1738. [DOI] [PubMed] [Google Scholar]
- 25.Ghanekar, A., M. Mendicino, H. Liu, W. He, M. Liu, R. Zhong, M. J. Phillips, G. A. Levy, and D. R. Grant. 2004. Endothelial induction of fgl2 contributes to thrombosis during acute vascular xenograft rejection. J. Immunol. 172:5693-5701. [DOI] [PubMed] [Google Scholar]
- 26.Guldberg, P., P. thor Straten, A. Birck, V. Ahrenkiel, A. F. Kirkin, and J. Zeuthen. 1997. Disruption of the MMAC1/PTEN gene by deletion or mutation is a frequent event in malignant melanoma. Cancer Res. 57:3660-3663. [PubMed] [Google Scholar]
- 27.Hamad, N. M., J. H. Elconin, A. E. Karnoub, W. Bai, J. N. Rich, R. T. Abraham, C. J. Der, and C. M. Counter. 2002. Distinct requirements for Ras oncogenesis in human versus mouse cells. Genes Dev. 16:2045-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herlyn, M., and K. Satyamoorthy. 1996. Activated ras. Yet another player in melanoma? Am. J. Pathol. 149:739-744. [PMC free article] [PubMed] [Google Scholar]
- 29.Hussussian, C. J., J. P. Struewing, A. M. Goldstein, P. A. Higgins, D. S. Ally, M. D. Sheahan, W. H. Clark, Jr., M. A. Tucker, and N. C. Dracopoli. 1994. Germline p16 mutations in familial melanoma. Nat. Genet. 8:15-21. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda, T., K. Yoshinaga, A. Suzuki, A. Sakurada, H. Ohmori, and A. Horii. 2000. Anticorresponding mutations of the KRAS and PTEN genes in human endometrial cancer. Oncol. Rep. 7:567-570. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, D., O. V. Volpert, N. Bouck, and D. I. Linzer. 1994. Stimulation and inhibition of angiogenesis by placental proliferin and proliferin-related protein. Science 266:1581-1584. [DOI] [PubMed] [Google Scholar]
- 32.Kannan, K., N. E. Sharpless, J. Xu, R. C. O'Hagan, M. Bosenberg, and L. Chin. 2003. Components of the Rb pathway are critical targets of UV mutagenesis in a murine melanoma model. Proc. Natl. Acad. Sci. USA 100:1221-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koh, H. K. 1991. Cutaneous melanoma. N. Engl. J. Med. 325:171-182. [DOI] [PubMed] [Google Scholar]
- 34.Krimpenfort, P., K. C. Quon, W. J. Mooi, A. Loonstra, and A. Berns. 2001. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature 413:83-86. [DOI] [PubMed] [Google Scholar]
- 35.Leevers, S. J., H. F. Paterson, and C. J. Marshall. 1994. Requirement for Ras in Raf activation is overcome by targeting Raf to the plasma membrane. Nature 369:411-414. [DOI] [PubMed] [Google Scholar]
- 36.Lemoine, N. R., S. Staddon, J. Bond, F. S. Wyllie, J. J. Shaw, and D. Wynford-Thomas. 1990. Partial transformation of human thyroid epithelial cells by mutant Ha-ras oncogene. Oncogene 5:1833-1837. [PubMed] [Google Scholar]
- 37.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 38.Lin, A. W., M. Barradas, J. C. Stone, L. van Aelst, M. Serrano, and S. W. Lowe. 1998. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 12:3008-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, F., D. J. Salant, H. Meyerson, S. Emancipator, B. P. Morgan, and M. E. Medof. 2004. Respective roles of decay-accelerating factor and CD59 in circumventing glomerular injury in acute nephrotoxic serum nephritis. J. Immunol. 172:2636-2642. [DOI] [PubMed] [Google Scholar]
- 40.Ma, Q., Z. Chen, I. del Barco Barrantes, J. L. de la Pompa, and D. J. Anderson. 1998. Neurogenin1 is essential for the determination of neuronal precursors for proximal cranial sensory ganglia. Neuron 20:469-482. [DOI] [PubMed] [Google Scholar]
- 41.Marton, L. J., and A. E. Pegg. 1995. Polyamines as targets for therapeutic intervention. Annu. Rev. Pharmacol. Toxicol. 35:55-91. [DOI] [PubMed] [Google Scholar]
- 42.Naviaux, R. K., E. Costanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olayioye, M. A., R. M. Neve, H. A. Lane, and N. E. Hynes. 2000. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 19:3159-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oldham, S. M., G. J. Clark, L. M. Gangarosa, R. J. Coffey, Jr., and C. J. Der. 1996. Activation of the Raf-1/MAP kinase cascade is not sufficient for Ras transformation of RIE-1 epithelial cells. Proc. Natl. Acad. Sci. USA 93:6924-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelengaris, S., M. Khan, and G. I. Evan. 2002. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell 109:321-334. [DOI] [PubMed] [Google Scholar]
- 46.Piecha, D., S. Muratoglu, M. Morgelin, N. Hauser, D. Studer, I. Kiss, M. Paulsson, and F. Deak. 1999. Matrilin-2, a large, oligomeric matrix protein, is expressed by a great variety of cells and forms fibrillar networks. J. Biol. Chem. 274:13353-13361. [DOI] [PubMed] [Google Scholar]
- 47.Pizzorno, G., D. Cao, J. J. Leffert, R. L. Russell, D. Zhang, and R. E. Handschumacher. 2002. Homeostatic control of uridine and the role of uridine phosphorylase: a biological and clinical update. Biochim. Biophys. Acta 1587:133-144. [DOI] [PubMed] [Google Scholar]
- 48.Prigent, S. A., M. Nagane, H. Lin, I. Huvar, G. R. Boss, J. R. Feramisco, W. K. Cavenee, and H. S. Huang. 1996. Enhanced tumorigenic behavior of glioblastoma cells expressing a truncated epidermal growth factor receptor is mediated through the Ras-Shc-Grb2 pathway. J. Biol. Chem. 271:25639-25645. [DOI] [PubMed] [Google Scholar]
- 49.Rak, J., Y. Mitsuhashi, C. Sheehan, A. Tamir, A. Viloria-Petit, J. Filmus, S. J. Mansour, N. G. Ahn, and R. S. Kerbel. 2000. Oncogenes and tumor angiogenesis: differential modes of vascular endothelial growth factor up-regulation in ras-transformed epithelial cells and fibroblasts. Cancer Res. 60:490-498. [PubMed] [Google Scholar]
- 50.Recio, J. A., F. P. Noonan, H. Takayama, M. R. Anver, P. Duray, W. L. Rush, G. Lindner, E. C. De Fabo, R. A. DePinho, and G. Merlino. 2002. Ink4a/arf deficiency promotes ultraviolet radiation-induced melanomagenesis. Cancer Res 62:6724-6730. [PubMed] [Google Scholar]
- 51.Redemann, N., B. Holzmann, T. von Ruden, E. F. Wagner, J. Schlessinger, and A. Ullrich. 1992. Anti-oncogenic activity of signalling-defective epidermal growth factor receptor mutants. Mol. Cell. Biol. 12:491-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schulze, A., K. Lehmann, H. B. Jefferies, M. McMahon, and J. Downward. 2001. Analysis of the transcriptional program induced by Raf in epithelial cells. Genes Dev. 15:981-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 54.Sharpless, N. E., N. Bardeesy, K. H. Lee, D. Carrasco, D. H. Castrillon, A. J. Aguirre, E. A. Wu, J. W. Horner, and R. A. DePinho. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86-91. [DOI] [PubMed] [Google Scholar]
- 55.Sharpless, N. E., K. Kannan, J. Xu, M. W. Bosenberg, and L. Chin. 2003. Both products of the mouse Ink4a/Arf locus suppress melanoma formation in vivo. Oncogene 22:5055-5059. [DOI] [PubMed] [Google Scholar]
- 56.Shelly, M., R. Pinkas-Kramarski, B. C. Guarino, H. Waterman, L. M. Wang, L. Lyass, M. Alimandi, A. Kuo, S. S. Bacus, J. H. Pierce, G. C. Andrews, and Y. Yarden. 1998. Epiregulin is a potent pan-ErbB ligand that preferentially activates heterodimeric receptor complexes. J. Biol. Chem. 273:10496-10505. [DOI] [PubMed] [Google Scholar]
- 57.Shields, J. M., K. Pruitt, A. McFall, A. Shaub, and C. J. Der. 2000. Understanding Ras: ‘it ain't over ′til it's over.’ Trends Cell Biol. 10:147-154. [DOI] [PubMed] [Google Scholar]
- 58.Sibilia, M., A. Fleischmann, A. Behrens, L. Stingl, J. Carroll, F. M. Watt, J. Schlessinger, and E. F. Wagner. 2000. The EGF receptor provides an essential survival signal for SOS-dependent skin tumor development. Cell 102:211-220. [DOI] [PubMed] [Google Scholar]
- 59.Stokoe, D., S. G. Macdonald, K. Cadwallader, M. Symons, and J. F. Hancock. 1994. Activation of Raf as a result of recruitment to the plasma membrane. Science 264:1463-1467. [DOI] [PubMed] [Google Scholar]
- 60.Toft, D. J., S. B. Rosenberg, G. Bergers, O. Volpert, and D. I. Linzer. 2001. Reactivation of proliferin gene expression is associated with increased angiogenesis in a cell culture model of fibrosarcoma tumor progression. Proc. Natl. Acad. Sci. USA 98:13055-13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsao, H., V. Goel, H. Wu, G. Yang, and F. G. Haluska. 2004. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J. Investig. Dermatol. 122:337-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsao, H., X. Zhang, K. Fowlkes, and F. G. Haluska. 2000. Relative reciprocity of NRAS and PTEN/MMAC1 alterations in cutaneous melanoma cell lines. Cancer Res. 60:1800-1804. [PubMed] [Google Scholar]
- 63.You, M. J., D. H. Castrillon, B. C. Bastian, R. C. O'Hagan, M. W. Bosenberg, R. Parsons, L. Chin, and R. A. DePinho. 2002. Genetic analysis of Pten and Ink4a/Arf interactions in the suppression of tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 99:1455-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu, J., D. Woods, M. McMahon, and J. M. Bishop. 1998. Senescence of human fibroblasts induced by oncogenic Raf. Genes Dev. 12:2997-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.