Abstract
Sphingosine-1-phosphate (S1P), a bioactive sphingolipid metabolite, is the ligand for five specific G protein-coupled receptors, named S1P1 to S1P5. In this study, we found that cross-communication between platelet-derived growth factor receptor and S1P2 serves as a negative damper of PDGF functions. Deletion of the S1P2 receptor dramatically increased migration of mouse embryonic fibroblasts toward S1P, serum, and PDGF but not fibronectin. This enhanced migration was dependent on expression of S1P1 and sphingosine kinase 1 (SphK1), the enzyme that produces S1P, as revealed by downregulation of their expression with antisense RNA and small interfering RNA, respectively. Although S1P2 deletion had no significant effect on tyrosine phosphorylation of the PDGF receptors or activation of extracellular signal-regulated kinase 1/2 or Akt induced by PDGF, it reduced sustained PDGF-dependent p38 phosphorylation and markedly enhanced Rac activation. Surprisingly, S1P2-null cells not only exhibited enhanced proliferation but also markedly increased SphK1 expression and activity. Conversely, reintroduction of S1P2 reduced DNA synthesis and expression of SphK1. Thus, S1P2 serves as a negative regulator of PDGF-induced migration and proliferation as well as SphK1 expression. Our results suggest that a complex interplay between PDGFR and S1P receptors determines their functions.
Sphingosine-1-phosphate (S1P) is a bioactive sphingolipid metabolite formed by activation of SphK by many stimuli, including platelet-derived growth factor (PDGF) (43, 48). As a specific ligand for a family of five G protein-coupled receptors (GPCRs), S1P1 to S1P5 (2, 48), S1P regulates a wide variety of important cellular processes, including cytoskeletal rearrangements and cell movement (17, 25, 45, 49, 57), angiogenesis and vascular maturation (14, 16, 26, 32, 57), and immunity and lymphocyte trafficking (33, 34). Interestingly, all of the S1P receptors (S1PRs) have been shown to play critical roles in cytoskeletal reorganization and cell migration (13, 26, 57). Activation of S1P1 or S1P3 increases directional or chemotactic migration (14, 27, 57), and both mediate activation of Rac via Gi (26, 38). In contrast, ligation of S1P2 decreases chemotaxis and membrane ruffling (49), due to suppression of Rac activation, probably by stimulation of a GTPase-activating protein for Rac (38). Interestingly, the repellant receptor S1P2 and the attractant receptor S1P3 similarly stimulate RhoA activity, likely via G12/13 (21). Recent studies suggest that the balance of counteracting signals from the Gi- and the G12/13-Rho pathways directs either positive or negative regulation of Rac and cell migration (49). Similar to its functions in lower organisms, including yeasts and plants, which do not have S1PRs, S1P may also have intracellular actions important for calcium homeostasis (36), cell growth (40, 56), and stress responses (9, 11, 37, 42).
S1P, like various other GPCR agonists, can activate growth factor tyrosine kinase receptors in the absence of added growth factors (also known as transactivation). For example, ligation of S1P1 leads to transactivation of VEGFR2/Flk-1 (52) and PDGF receptor (PDGFR) (53) and also produces PDGF (55), which in turn can stimulate signaling cascades important for vascular remodeling and maturation. Because PDGF, which stimulates SphK1 (18, 39) and increases intracellular S1P (43), also activated the S1P1 receptor, as measured by its phosphorylation and by translocation of β-arrestin (18), a reciprocal mechanism of receptor cross-communication has been proposed (18). According to this paradigm, stimulation of the tyrosine kinase PDGFR activates and translocates SphK1 to the plasma membrane, leading to spatially restricted formation of S1P, which then activates S1P1, a critical event for PDGF-directed cell movement (18, 45). Two other mechanisms for S1P1 and receptor tyrosine kinase cross-communication have been suggested (25, 58). In the integrative signaling model, the PDGF receptor and S1P1 form a complex that is cointernalized together by PDGF as a functional signaling unit to regulate extracellular signal-regulated kinase 1/2 (ERK1/2) (58). Moreover, the insulin-like growth factor 1 receptor can transactivate S1P1 through its Akt-dependent phosphorylation, in a manner that does not require the SphK pathway (25). Both of these models suggest that activation of SphK1 and intracellular generation of S1P do not play any role and introduce the concept of ligand-independent activation of S1P receptors.
Although it has long been known that S1P can inhibit PDGF-induced migration of human arterial smooth muscle cells (6), little is yet known of cross talk between PDGFR and the chemorepellant S1P2 receptor. Utilizing embryonic fibroblasts from S1P2-null mice, we uncovered an important role for S1P2 as a negative regulator of both migratory and proliferative responses to PDGF. Moreover, our results suggest that complex interplay between PDGFR and S1PRs determines their functions.
MATERIALS AND METHODS
Reagents.
S1P, sphingosine, and N,N-dimethylsphingosine (DMS) were from Biomol (Plymouth Meeting, PA). [γ-32P]ATP (3,000 Ci/mmol) was purchased from Amersham Pharmacia Biotech (Piscataway, NJ). Serum and medium were obtained from Biofluids (Rockville, MD). PDGF-BB, rabbit polyclonal anti-PDGFRβ, and anti-PDGFRα immunoglobulin G (IgG) were obtained from Upstate Biotechnology (Lake Placid, NY). Anti-phosphorylated (pThr202/pTyr204) ERK1 and -2, anti-phosphorylated (pSer473) Akt, anti-ERK2, anti-phospho-p38, anti-p38, and anti-phospho-PAK1 (Thr423) antibodies were obtained from Cell Signaling Technology (Beverly, MA). Anti-PAK1 (C-19), anti-Myc, and antitubulin antibodies were from Santa Cruz Biotechnology Lab (Santa Cruz, CA). Polyclonal anti-S1P1 and anti-S1P2 antibodies were from Exalpha (Watertown, MA). Antiphosphotyrosine (PY20) was obtained from Sigma.
Cell culture.
Mouse embryonic fibroblasts (MEFs) were derived from day 14 embryos generated from wild-type or knockout double intercrosses on a mixed background of 129SvJ and C57BL mice as described previously (21). MEFs were immortalized by transfection with SV40 genomic DNA (59) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS).
Sphingosine kinase assay.
Cells were harvested and lysed by freeze-thawing in SphK buffer (20 mM Tris, pH 7.4; 20% glycerol; 1 mM β-mercaptoethanol; 1 mM EDTA; 5 mM sodium orthovanadate; 40 mM β-glycerophosphate; 15 mM sodium fluoride; 10 μg/ml leupeptin, aprotinin, and soybean trypsin inhibitor; 1 mM phenylmethylsulfonyl fluoride, and 0.5 mM 4-deoxypyridoxine). Lysates were centrifuged at 700 × g for 10 min to remove unbroken cells. SphK1 activity was determined in the presence of 50 μM sphingosine in 0.25% Triton X-100 and [γ-32P]ATP (10 μCi, 1 mM) containing MgCl2 (10 mM) as described previously (41). [32P]S1P was separated by thin-layer chromatography on silica gel G60 with 1-butanol/ethanol/acetic acid/water (80:20:10:20 [vol/vol]) as solvent, and the radioactive spots corresponding to S1P were quantified with an FX Molecular Imager (Bio-Rad, Hercules, CA). SphK specific activity is expressed as pmol S1P formed per min per mg protein.
RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed as follows. Total RNA was isolated from MEFs with TRIzol reagent (Life Technologies, Gaithersburg, MD). RNA was reverse transcribed with Superscript II (Life Technologies). Specific primers listed in Table 1 were used to amplify cDNA. For real-time PCR, a premixed mouse SphK1 primer-probe set was purchased from Applied Biosystems (Foster City, CA).
TABLE 1.
Sequences of primers used for RT-PCR
| Primer | Sequence | Predicted size (bp) |
|---|---|---|
| S1P1 | ||
| Forward | GCTGCTTGATCATCCTAGAG | 318 |
| Reverse | GAAAGGAGCGCGAGCTGTTG | |
| S1P2 | ||
| Forward | CCAAGGAGACGCTGGACATG | 511 |
| Reverse | TGCCGTAGAGCTTGACCTTG | |
| S1P3 | ||
| Forward | GCAACTTGGCTCTCTGCGAC | 342 |
| Reverse | GACGATGGTCACCAGAATGG | |
| SphK1 | ||
| Forward | CAAGGCTCTGCAGCTCTTCCAGAG | 375 |
| Reverse | CAGGTTCATGGGTGACAGGCGCCG | |
| SphK2 | ||
| Forward | CTCTACATCGATCTGTGTCTGACCTGC | 340 |
| Reverse | CCAGTCTTGGGGCAGTGGAGAGCC | |
| GAPDH | ||
| Forward | ACCACAGTCCATGCCATCAC | 452 |
| Reverse | TCCACCACCCTGTTGCTGTA |
Transfection.
MEFs were electroporated with a Gene Pulser (Bio-Rad) at 250 mV and 500 μF using 25 μg DNA and 200,000 cells/μl in Dulbecco's modified Eagle's medium supplemented with 10% FBS and 50 mM HEPES (pH 7.4). For transient expression, cells were allowed to recover 24 h.
Small interfering RNA (siRNA) for mouse SphK1 (CUGGCCUACCUUCCUGUAGdTT and CUACAGGAAGGUAGGCCAGdTT) targeting a region located 644 bp from the start codon (siSphK1a) and control siRNA were synthesized by Xeragon-QIAGEN (Valencia, CA). Cells (1 × 105) were transfected in six-well dishes for 3 h with the 21-nucleotide duplexes, using Oligofectamine (Invitrogen) as recommended by the manufacturer. To rule out off-target effects, where indicated, experiments were repeated with another siRNA targeted at a different SphK1 sequence (GGCAGAGAUAACCUUUAAAdTT and UUUAAAGGUUAUCUCUGCCdTT) 150 bp from the start codon (siSphK1b) obtained from Ambion (Austin, TX). A total of 65% ± 5% of the cells were transfected as determined with siGLO RISC-Free siRNA (Dharmacon).
Expression of S1P1 was downregulated by transfection with 18-mer phosphothioate oligonucleotides as previously described (18, 26). Briefly, cells (7 × 105) in six-well plates were transfected with antisense S1P1 (5′-GACGCTGGTGGGCCCCAT-3′) or scrambled S1P1 (5′-TGATCCTTGGCGGGGCCG-3′) (Integrated DNA Technologies, Coralville, IA) at a final concentration of 1 μM, using Oligofectamine.
Western blotting.
MEFs were scraped into lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 0.1% Triton X-100, 1.5 mM MgCl2, 1 mM EDTA, 2 mM sodium orthovanadate, 4 mM sodium pyrophosphate, 100 mM NaF, 1 mM phenylmethlysulfonyl flouride, 5 μg/ml leupeptin, 5 μg/ml aprotinin). Equal amounts of proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transblotted to nitrocellulose, blocked with 5% nonfat dry milk for 2 h at room temperature, and then incubated with primary antibodies overnight. Immunoreactive signals were visualized by enhanced chemiluminescence.
Immunoprecipitation.
Cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.25% deoxycholate, 1 mM NaF, 1 mM orthovanadate, and 1:200 diluted protease inhibitor cocktail). Cell lysates were clarified by centrifugation at 10,000 × g for 10 min at 4°C and then incubated with protein A/G beads and 0.5 μg of rabbit IgG for 1 h at 4°C. Precleared lysates were incubated with 10 μg of either PDGFRα or PDGFRβ antibodies overnight at 4°C and then with protein A/G beads for 2 h. Immune complexes were analyzed by Western blotting.
Rac activation.
Rac activation was assessed as previously described (18). Briefly, wild-type and S1P2−/− MEFs were serum starved overnight, treated as indicated in the figure legends, and then lysed at 4°C in buffer containing 25 mM HEPES (pH 7.5), 1% Triton X-100, 150 mM NaCl, 10 mM MgCl2, 1 mM Na3VO4, 10 μg/ml aprotinin, 10 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride. Lysates were cleared by centrifugation, and soluble fractions were incubated with 10 μg of glutathione S-transferase (GST)-fused CRIB-containing the N-terminus binding domain of p21-activated kinase (PAK) precoupled to glutathione-Sepharose beads, followed by three washes with lysis buffer. Proteins were extracted from the beads by boiling in SDS sample buffer, separated by 15% SDS-PAGE, transferred to nitrocellulose, and blotted with anti-Rac antibody (1:1,000; Upstate Biotechnology, Lake Placid, NY). An aliquot of total cell lysate was immunoblotted to determine total Rac levels.
PAK activation.
Cells cultured on poly-d-lysine-coated plates were serum starved for 18 h. After stimulation, cells were scraped in lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 5 mM MgCl2, 1 mM dithiothreitol, 10 mM NaF, 1 mM Na3VO4, 10% glycerol, 1% Nonidet P-40, and Complete protease inhibitor cocktail). Lysate proteins were analyzed by immunoblotting with anti-phospho-PAK1 (Thr423) antibody followed by anti-PAK1 (C-19) antibody.
Incorporation of BrdU.
Transfected MEFs were starved for 8 h before stimulation with serum or growth factors. Bromodeoxyuridine (BrdU) incorporation was determined as described previously (31). In brief, cells were incubated for 3 h with BrdU (10 μM) and fixed in 4% paraformaldehyde containing 5% sucrose (pH 7.0) for 20 min at room temperature, and nuclei incorporating BrdU were counted using a Zeiss fluorescence microscope. At least 500 cells were scored per point, which included at least five different randomly chosen fields.
Cell growth assays.
MEFs were seeded in 24-well plates at a density of 10,000 cells/well and grown in 1% FBS. At the indicated times, cell numbers were determined by the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) dye reduction assay (40). In some experiments, MEFs were cultured in the presence of 5% FBS and after treatment with mitogens for the indicated times, WST-1 reagent (Roche, Rockford, Ill.) was added and cells were incubated at 37°C for 3 h. Absorbance was measured at 450 nm with background subtraction at 650 nm. Values are means ± standard error of five to six determinations.
Chemotaxis.
Chemotaxis was measured in a modified Boyden chamber, using polycarbonate filters (25 by 80 mm, 12 μM pore size) (45). Chemoattractants were added to the lower chamber, and cells were added to the upper chamber at 5 × 104 cells/well. After 4 h, unless indicated otherwise, nonmigratory cells on the upper membrane surface were mechanically removed and the cells that traversed and spread on the lower surface of the filter were fixed and stained with Diff-Quik (Fisher Scientific, Pittsburgh, PA). The migrated cells were counted with a microscope and a 10× objective (57). Each data point is the average number of cells in four random fields, each counted twice, and is the average ± standard deviation (SD) of three individual wells.
Immunofluorescence microscopy.
MEFs grown on glass coverslips were fixed in 4% paraformaldehyde-5% sucrose and then permeabilized in 0.5% Triton X-100 in phosphate-buffered saline for 5 min. Cells were then incubated for 20 min with Cy2-phalloidin (1:150 dilution; Molecular Probes, Eugene, OR) to visualize the actin cytoskeleton and/or with antipaxillin antibody (1:100) to stain for focal complexes, followed by secondary antimouse antibody conjugated with Texas red or fluorescein isothiocyanate, respectively. Coverslips were mounted on glass slides using an Anti-Fade kit (Molecular Probes) and examined by confocal microscopy (LSM 510 Carl Zeiss Micro Imaging). Image analysis was performed using LSM image processing software. At least 50 cells were examined in each experiment.
Statistical analysis.
Experiments were repeated at least three times with consistent results. For each experiment, the data from triplicate samples were calculated and expressed as means ± SD. Differences between groups were determined with Student's t test or a one-way analysis of variance with a Tukey post hoc test, and P < 0.05 was considered significant.
RESULTS
Cell migration toward PDGF is markedly enhanced in S1P2 receptor-null fibroblasts.
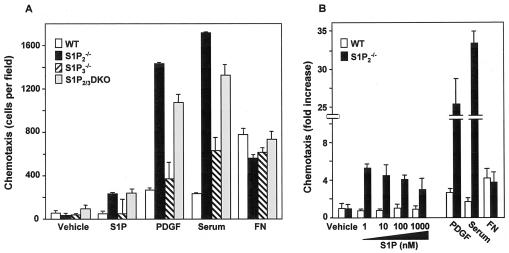
Previous studies demonstrated that the S1P2 receptor inhibits membrane ruffling and cell migration toward S1P (38). In agreement, migration of MEFs, which express transcripts for S1P1 to S1P3, but not S1P4 and S1P5 (21), toward S1P was increased when the S1P2 receptor was deleted (Fig. 1A). Surprisingly, we found that deletion of S1P2 also dramatically increased migration of MEFs toward PDGF and serum (Fig. 1A). In contrast, motility of S1P3-null MEFs (20) toward S1P or PDGF was not significantly different from that of wild-type cells. Additionally, migration of MEFs lacking both S1P2 and S1P3 toward S1P and PDGF was enhanced almost to the same extent as the S1P2-null MEFs, suggesting that loss of the S1P2 receptor was responsible for the increase in migration. To further examine the role of S1P2 in cell migration, we established immortalized fibroblast lines from wild-type and S1P2−/− MEFs. The immortalized S1P2-null cell line retained the enhanced migratory effects, not only toward S1P, but also toward PDGF and serum, which contains S1P and stimulates SphK (48), whereas haptotactic migration toward fibronectin was unaltered (Fig. 1B).
FIG. 1.
Deletion of S1P2 increases chemotaxis toward S1P and growth factors. (A) Primary cultures of wild-type (WT), S1P2, S1P3, and single- and S1P2/3 double-knockout (DKO) MEFs were serum starved overnight and then allowed to migrate toward vehicle, S1P (10 nM), PDGF (20 ng/ml), serum (20%), or fibronectin (FN; 20 μg/ml) for 4 h. (B) Cultures of immortalized wild-type and S1P2 knockout MEFs were serum starved overnight and then allowed to migrate for 4 h toward vehicle, S1P, PDGF (20 ng/ml), serum (20%), or fibronectin (20 μg/ml). The data are expressed as fold increase and are means ± SD of triplicate determinations. Migrations of control wild-type and S1P2 knockout MEFs were 26 ± 4 and 29 ± 10 cells per field, respectively. Similar results were obtained in three independent experiments.
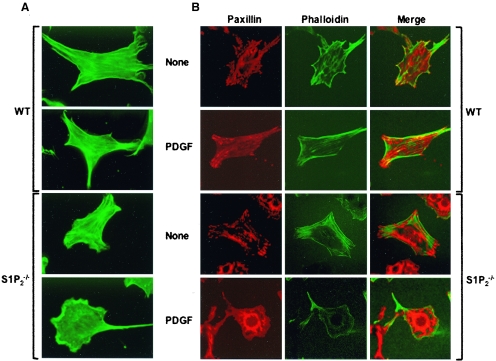
S1P2 receptor deletion increases PDGF-induced membrane ruffling.
When cells move, they use actin polymerization to push the plasma membrane outward, forming localized protrusions known as lamellipodia (44). To better understand the enhanced migration of S1P2-null fibroblasts toward PDGF, we next examined the architecture of the actin cytoskeleton. No obvious differences between quiescent wild-type and S1P2-null MEFs were revealed by phalloidin staining of actin filaments. PDGF, as expected, caused extension of lamellipodia at the cell periphery of wild-type fibroblasts, which was markedly enhanced in the S1P2-null MEFs (Fig. 2A), correlating with their significantly increased migration (Fig. 1B).
FIG. 2.
Changes in cytoskeletal architecture and focal complexes of S1P2-deleted MEFs induced by PDGF. Wild-type (WT) and S1P2−/− MEFs were grown on coverslips for 24 h in serum-free medium and then stimulated without (None) or with 20 ng/ml PDGF-BB for 30 min. Cells were then fixed, permeabilized, and stained with (A and B) Cy2-labeled phalloidin to detect actin fibers (green), and (B) adhesion complexes were visualized by confocal microscopy with antipaxillin antibody and Texas red-conjugated secondary antibody. Representative fields of more than 50 cells examined are shown.
Cell movement is orchestrated by the complex interplay of leading edge formation and continuous formation and disassembly of focal adhesions. Focal adhesions and complexes were visualized by staining for the scaffolding protein paxillin. Paxillin is a multidomain adaptor protein found at the interface between the plasma membrane and the actin cytoskeleton, and it has been implicated in focal adhesion turnover (54). In unstimulated cells, very prominent focal adhesions were seen in both wild-type and S1P2-null MEFs (Fig. 2B). Upon treatment with PDGF, there was rapid turnover of these focal adhesions, especially in the S1P2-null MEFs, and these cells showed polarity and smaller focal complexes, which are characteristics of more motile cells (Fig. 2B).
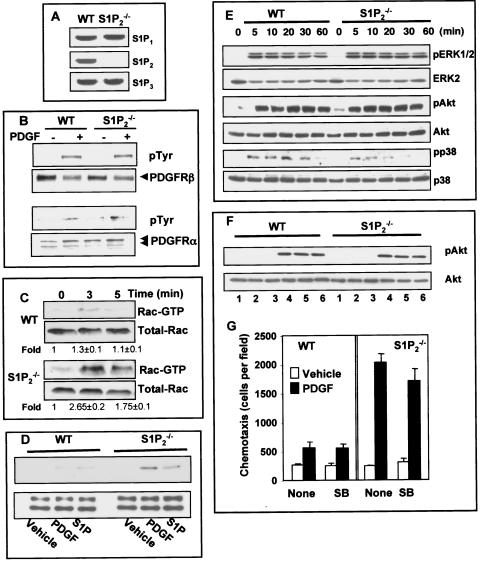
S1P2 is important for PDGF-induced activation of Rac.
Although deletion of S1P2 markedly enhanced PDGF-directed motility, there were no significant increases in expression of S1P1 or S1P3, the S1P receptors that positively regulate motility (Fig. 3A). Nor were there any differences in PDGFRα or PDGFRβ expression or enhancement of PDGF-stimulated tyrosine phosphorylation of its receptors in S1P2-null fibroblasts compared to wild-type cells (Fig. 3B).
FIG. 3.
Effect of deletion of S1P2 on PDGF-induced signaling pathways. (A) Expression of S1P1, S1P2, and S1P3 was determined by RT-PCR in wild-type (WT) and S1P2-null MEFs. No PCR products were observed in the absence of RT. (B) Wild-type and S1P2-null MEFs were serum starved overnight, treated with PDGF (20 ng/ml) for 5 min, and lysed, and equal amounts of proteins were immunoprecipitated with anti-PDGRβ (upper panel) or anti-PDGFRα (lower panel) and analyzed by Western blotting with antiphosphotyrosine antibody. Blots were stripped and reprobed with anti-PDGRβ or anti-PDGFRα to demonstrate equal loading. Arrows indicate a PDGFRβ 190-kDa band (upper panel) or a PDGFRα doublet of 150/170 kDa (lower panel). (C) Serum-starved wild-type and S1P2-null MEFs were treated with PDGF for the indicated times, and active Rac was specifically pulled down from cell lysates containing equal amounts of proteins with immobilized recombinant PAK-CRIB and analyzed by Western blotting using anti-Rac antibody. Relative activated levels were normalized to total Rac, and the fold stimulation ± SD from three independent experiments is indicated. Representative results are shown. (D) Serum-starved wild-type and S1P2-null MEFs were treated with vehicle, PDGF, or S1P for 5 min, and activation of PAK1 was determined by Western blot analysis with phospho-PAK1 (Thr423) antibodies (top panel). Equal loading was confirmed by reprobing with antibodies against PAK1, which also cross-react with PAK2 (bottom panel). (E) Wild-type and S1P2-null MEFs were treated with PDGF for the indicated times, and activation of ERK1/2, Akt, and p38 was determined by Western blotting analysis with phospho-specific antibodies. Equal loading was confirmed by reprobing with antibodies against ERK2, Akt, and p38, respectively. Similar results were obtained in three additional experiments. (F) Serum-starved wild-type and S1P2-null MEFs were treated without (lanes 1 and 4) or with SB202190 (10 μM, lanes 2 and 5) or SB202190 (25 μM, lanes 3 and 6) for 15 min and then stimulated with vehicle (lanes 1 to 3) or 20 ng/ml PDGF (lanes 4 to 6) for 10 min. Equal amounts of cell lysate proteins were separated by SDS-PAGE and probed with phospho-specific Akt S473 antibody. Blots were stripped and reprobed with anti-Akt antibody to show equal loading. (G) Serum-starved wild-type and S1P2-null MEFs were treated without or with 10 μM SB202190 (SB) and then allowed to migrate toward vehicle (open bars) or 20 ng/ml PDGF (filled bars) for 4 h.
Because binding of S1P to S1P2 in vascular smooth muscle cells was previously shown to inhibit PDGF-induced activation of Rac (46), a member of the Rho family of small GTPases that plays a critical role in cell motility by regulating formation of new lamellipodial protrusions at the leading edge (7), it was of interest to examine PDGF-induced Rac activation in the S1P2-null cells. While PDGF only slightly stimulated Rac activation in wild-type MEFs (Fig. 3C), it caused a considerable increase in activated Rac in the S1P2-null MEFs (Fig. 3C). The Rac effector PAK1 serves as an important regulator of cytoskeletal dynamics and cell motility (5). PAK1 activation can be determined using an antibody that recognizes phosphorylated threonine 423 in the activation loop of its catalytic domain, which is important for its activation (5). In agreement with the Rac activation, in wild type MEFs, PDGF barely activated PAK1 at 5 min while PDGF markedly stimulated PAK1 in S1P2-null cells at this time point (Fig. 3D). S1P also rapidly activated PAK1 in the S1P2-null cells but not in wild-type cells (Fig. 3D).
Proteins of the mitogen-activated protein kinase family (ERK, SAPK/JNK, p38) and Akt also play important roles in PDGF-mediated signaling (8). In agreement with previous studies (35), in wild-type fibroblasts, PDGF induced robust activation of ERK1/2 (Fig. 3E) and Akt and less vigorous activation of p38 (Fig. 3E), while SAPK1/JNK was not stimulated at all (data not shown). Although S1P2 deletion had no significant effect on activation of ERK1/2 or Akt induced by PDGF (Fig. 3E), it reduced p38 activation, particularly at later time points (Fig. 3E). Previous studies in HEY ovarian cancer cells found that S1P and PDGF stimulate phosphorylation of S473 on Akt, which is essential for its full activation, in a p38-dependent manner (3). However, SB202190, which inhibits p38, had no effect on Akt S473 phosphorylation induced by PDGF (Fig. 3F). Moreover, in agreement with a recent report that p38 inhibition has no major effect on the degree of lamellipodia formation and cellular protrusions in response to PDGF-BB (23), SB202190 did not affect PDGF-induced migration of wild-type or S1P2-null fibroblasts (Fig. 3G).
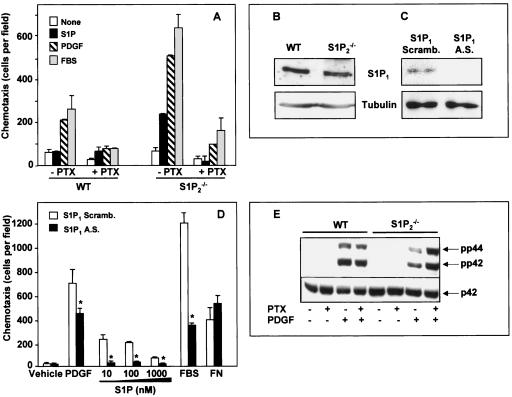
Chemotaxis of S1P2-null fibroblasts is dependent on S1P1.
Previously, we suggested that S1P1, which couples exclusively to Gi, was crucial for PDGF-induced migration (18, 45). In agreement, migration of wild-type MEFs toward PDGF and serum was pertussis toxin (PTX) sensitive (Fig. 4A). PTX treatment also markedly reduced migration of S1P2-null MEFs toward PDGF and serum as well as toward S1P (Fig. 4A). Of note, deletion of S1P2 does not significantly alter expression of the S1P1 receptor (Fig. 4B). In addition, transfection with antisense, but not scrambled, S1P1 oligonucleotides downregulated S1P1 expression (Fig. 4C) and also significantly inhibited migration toward PDGF, S1P, and serum without affecting migration toward fibronectin (Fig. 4D).
FIG. 4.
Involvement of S1P1 in PDGF-induced chemotaxis. (A) Wild-type (WT) and S1P2−/− MEFs were cultured in serum-free medium for 12 h in the absence or presence of PTX (200 ng/ml). Cells were allowed to migrate for 4 h toward vehicle, S1P (10 nM), PDGF (20 ng/ml), or FBS (20%), and chemotaxis was measured. (B) Expression of S1P1 receptor in wild-type and S1P2−/− MEFs was determined by Western blotting with an antibody against S1P1. The membranes were stripped and reprobed with an antibody against tubulin to show equal loading. (C and D) S1P2−/− cells were transfected with either scrambled (Scramb.; open bars) or antisense (A.S., filled bars) oligonucleotides against S1P1 receptor and (C) analyzed for the expression of S1P1 receptor by Western blotting. Tubulin expression was used as a loading control. (D) Duplicate cultures were serum starved for 12 h and then allowed to migrate toward PDGF (20 ng/ml), serum (20%), or the indicated concentrations of S1P or fibronectin (20 μg/ml). Similar results were obtained in two additional experiments. *, P < 0.05. (E) Wild-type and S1P2−/− MEFs were serum starved overnight in the absence or presence of PTX (100 ng/ml) and then stimulated without or with PDGF (20 ng/ml) for 5 min as indicated. Equal amounts of cell lysate proteins were analyzed by Western blotting with anti-phospho p42/44 antibodies. Blots were stripped and reprobed with anti-ERK2 as a loading control.
Recently, it was suggested that S1P1 is required for PDGF-induced ERK activation in airway smooth muscle cells (58). However, PTX at a concentration that markedly inhibited migration of MEFs toward PDGF (Fig. 4A) did not significantly affect its ability to stimulate ERK1/2 (Fig. 4E). This is consistent with previous studies showing that S1P1 deletion does not significantly affect activation of ERK induced by PDGF (45).
Enforced expression of S1P2 inhibits chemotaxis toward PDGF.
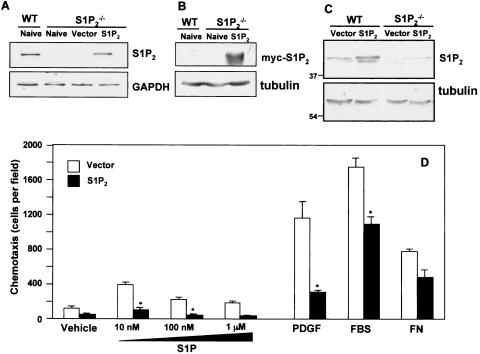
To confirm that the migratory differences observed between wild-type and S1P2-null fibroblasts were due to the loss of the S1P2 receptor rather than a general defect in migratory responses, we examined the effect of restoring S1P2 expression in these cells. Transient transfection of S1P2 restored mRNA expression to a similar level as in wild-type MEFs (Fig. 5A). Moreover, myc-tagged S1P2 protein was readily detectable 48 h after transfection (Fig. 5B). Western blotting with anti-S1P2 antibody also revealed a single band with the predicted molecular size of approximately 42 kDa in wild-type MEFs that was absent in the S1P2−/− MEFs (Fig. 5C). Moreover, transfection with S1P2 increased its levels in the null cells to comparable levels in wild-type MEFs (Fig. 5C).
FIG. 5.
Ectopic expression of S1P2 reverses the migratory phenotype of S1P2-deleted cells. Wild-type (WT) and S1P2-null MEFs were transiently transfected with vector or myc-S1P2, as indicated. Cells were lysed, and mRNA (A) and proteins (B and C) were analyzed. (A) S1P2 and GAPDH mRNA were determined by RT-PCR. (B and C) Protein expression was examined by Western blotting with anti-myc antibody (B) or anti-S1P2 antibody (C). Blots were stripped and reprobed with antitubulin to ensure equal loading. (D) One day after transfection, vector (open bars) and myc-S1P2 (filled bars) cells were serum starved for 6 h and allowed to migrate toward vehicle, PDGF (20 ng/ml), FBS (20%), fibronectin (FN; 20 μg/ml), or the indicated concentrations of S1P for 4 h. The data are expressed as means ± SD of triplicate determinations. Similar results were obtained in three independent experiments. *, P < 0.05.
Reconstitution of S1P2 expression markedly attenuated both S1P- and PDGF-induced migration of S1P2 knockout MEFs (Fig. 5D). However, haptotaxis toward fibronectin was not significantly affected by overexpression of S1P2 (Fig. 5D). These results further substantiate the negative regulatory role of S1P2 receptor on PDGF-induced migration.
Sphingosine kinase type 1 is required for migration toward PDGF.
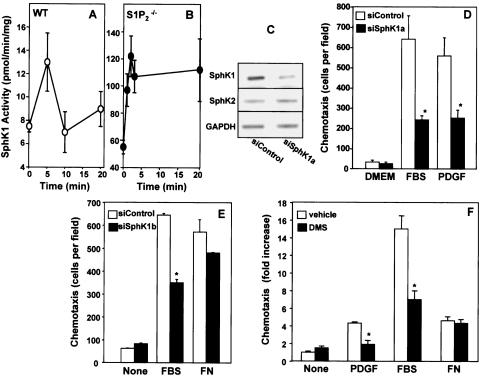
Previously, we suggested that spatially and temporally localized generation of S1P via PDGF-induced SphK activation results in transactivation of S1P1, which in turn stimulates downstream signaling important for cell locomotion (18, 45). Thus, we next examined the role of SphK in migration toward PDGF. MEFs express both SphK1 and SphK2 (Fig. 6C), which can be distinguished on the basis of differential activity measured when the substrate sphingosine is added either as a bovine serum albumin complex in the presence of high salt or in a micellar form with Triton X-100 (24, 30). Whereas, Triton X-100 stimulates SphK1 and strongly inhibits SphK2, high salt is optimal for SphK2 and drastically inhibits SphK1 (30). Consistent with previous results (18, 39), PDGF rapidly stimulated SphK activity in wild-type and S1P2-null MEFs measured in the presence of Triton X-100 (Fig. 6A, B), which was not evident when the activity was measured in the presence of high salt without Triton X-100 (data not shown), suggesting that only SphK1 is activated by PDGF. It is important to note that basal SphK activity in S1P2-null MEFs (Fig. 6B) was much greater than in the wild-type MEFs (Fig. 6A) and its activation by PDGF was more robust and prolonged. Although PDGF stimulates SphK1, similar to previous results with NIH 3T3 fibroblasts (40, 42), MEFs from S1P1 knockouts (45), and HEK 293 cells (18), there was no detectable secretion of S1P from wild-type or S1P2−/− MEFs. However, recent studies suggest that, even in the absence of detectable S1P secretion, localized formation of S1P at membrane ruffles due to translocation and activation of SphK1 by PDGF was sufficient to activate S1P receptors leading to downstream signaling important for cytoskeletal changes and migratory responses (18, 42, 45).
FIG. 6.
Involvement of SphK1 isoform in PDGF-induced migration. Wild-type (WT) (A) and S1P2−/− (B) cells were treated with PDGF (20 ng/ml) for the indicated times, and SphK1 activity was measured in cell lysates with sphingosine presented in Triton X-100 micelles. Data are means ± SD of duplicate determinations. Similar results were obtained in two additional experiments. (C, D, and E) S1P2−/− cells were transfected with control siRNA or with siSphK1a (C and D) or siSphK1b (E) targeted to SphK1 for 48 h. (C) mRNAs of SphK1, SphK2, and GAPDH were determined in duplicate cultures by RT-PCR. (D and E) Cells were serum starved for 6 h and allowed to migrate for 4 h toward medium (None), PDGF (20 ng/ml), FBS (20%), or fibronectin (FN; 20 μg/ml) as indicated. (F) S1P2−/− cells were treated with vehicle (open bars) or 5 μM DMS (hatched bars) for 20 min and then allowed to migrate toward PDGF, FBS, or FN for 4 h. Data are expressed as fold increase ± SD of triplicate determinations relative to vehicle controls. *, P < 0.05.
To investigate the role of SphK activity in PDGF-induced migration, we utilized specific siRNAs targeted to SphK1 and SphK2 to downregulate their expression. Transfection of S1P2-null fibroblasts with siSphK1 reduced mRNA expression of SphK1 but not SphK2 as determined by RT-PCR analysis (Fig. 6C). Conversely, siSphK2 did not affect SphK1 expression (data not shown). No differences in migratory responses were seen in MEFs transfected with control siRNA or siSphK2 compared to untransfected cells (data not shown), whereas migration toward PDGF and serum was significantly reduced in siSphK1-treated S1P2-null MEFs (Fig. 6D). To ensure that the observed effects were specific to reduction of SphK1 expression, the effects of another siRNA targeted to a different SphK1 sequence (siSphK1b) were examined. Transfection with this siRNA reduced expression of SphK1 by 45%, as determined by real-time PCR, without significantly affecting expression of SphK2. siSphK1b also reduced migratory responses toward serum but not fibronectin (Fig. 6E). To confirm the siRNA results, we used a complementary pharmacological approach. The specific SphK inhibitor N,N-dimethylsphingosine, at a concentration of 5 μM, which has no effects on other signaling molecules (12), markedly inhibited chemotaxis toward serum and PDGF but not fibronectin-induced haptotaxis (Fig. 6F).
S1P2 is a negative regulator of SphK1.
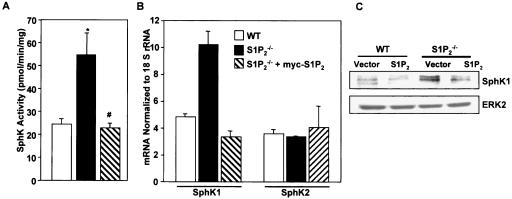
Surprisingly, SphK1 activity was significantly higher in S1P2−/− than in wild-type MEFs (Fig. 6A, B and 7A). This correlated with the higher SphK1 expression in the S1P2-null cells measured by real-time RT-PCR (Fig. 7B). Similarly, SphK1 protein levels were higher in S1P2 knockout cells compared to wild-type cells (Fig. 7C). Interestingly, expression of SphK2 was the same as in wild-type cells (Fig. 7B). To examine whether S1P2 negatively regulates SphK1 or whether this increase was due to nonspecific differences between these cell lines, the S1P2 receptor was reintroduced into the S1P2-null cells. Transient transfection of S1P2 markedly reduced SphK1 expression and activity to comparable levels determined in the wild-type cells without altering SphK2 expression (Fig. 7). These results suggest that S1P2 receptor expression specifically regulates the activity and expression of SphK1.
FIG. 7.
S1P2 negatively regulates expression and activity of SphK1. (A) Wild-type (WT) MEFs (open bar), S1P2−/− MEFs transfected with empty vector (filled bar), or S1P2−/− MEFs transfected with S1P2 (hatched bar) were serum starved for 16 h and lysed, and SphK1 activity was measured in the presence of 0.25% Triton X-100. Data are means ± SD of triplicate determinations. In duplicate cultures, expression of SphK1 and SphK2 mRNA (B) was determined by real-time quantitative PCR relative to 18S RNA and protein levels (C) were determined by Western blotting. Values are means ± SD of triplicate determinations.
S1P2 receptor deficiency causes increased proliferation of MEFs.
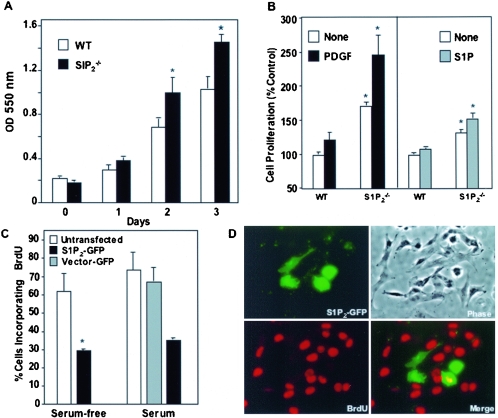
Because numerous studies link SphK1 to cell growth (40, 42, 60), it was of interest to examine the effect of S1P2 deletion on the proliferative potential of MEFs. Loss of this receptor led to a significant increase in cell growth compared to wild-type MEFs that was evident within 2 days of culture in the presence of a suboptimal serum concentration (Fig. 8A). Moreover, growth of the S1P2-null MEFs was stimulated to a significantly greater extent by both PDGF and S1P than wild-type cells, although PDGF was a more potent mitogen than S1P (Fig. 8B). To confirm that the increased proliferation of the knockout cells was the result of deletion of the S1P2 receptor, S1P2 expression was reconstituted in the S1P2-null MEFs by transient transfection with S1P2-GFP. Double immunofluorescence was used to visualize transfected cells and DNA synthesis, determined by measuring BrdU incorporation into nascent DNA. Expression of S1P2 markedly inhibited DNA synthesis of cells cultured in the absence or presence of serum, as the fraction of transfected cells positive for BrdU incorporation was significantly lower than in adjacent, nontransfected cells in the same field (Fig. 8C, D). In contrast, transfection with vector-GFP had no significant effect on DNA synthesis. These results imply that S1P2 expression negatively regulates the transition from G1 to S phase of the cell cycle.
FIG. 8.
S1P2 receptor deficiency leads to an increased proliferation of MEFs (A) Proliferation of wild-type (WT) and S1P2−/− MEFs grown in 2% FBS was measured by the MTT assay at the indicated times. (B) Wild-type and S1P2−/− MEFs were cultured for 2 days with vehicle (open bars), 20 ng/ml PDGF (black bars), or 100 nM S1P (gray bars), and proliferation was measured by WST-1 assay. Data are means ± SD of triplicate determinations. (C) S1P2−/− MEFs were transiently transfected with vector-GFP (open bars) or S1P2-GFP (filled bars), serum starved for 8 h, and incubated in either serum-free medium or medium containing 2% serum. After 16 h, BrdU was added for an additional 3 h and cells were fixed and stained with anti-BrdU antibody. Double immunofluorescence was used to visualize transfected cells and BrdU incorporation, and the proportion of cells incorporating BrdU among total transfected cells (expressing GFP) was determined. Data are means ± SD of duplicate cultures from a representative experiment. At least three different fields were scored with a minimum of 100 cells scored per field. Similar results were obtained in three independent experiments. *, P < 0.05. (D) Representative images of cells transfected with S1P2-GFP (green) and incorporating BrdU (red). Yellow in the merged images indicates a cell that is positive for both.
DISCUSSION
Cross-communication between growth factor tyrosine kinase receptors and GPCRs provides fine-tuning mechanisms for cells to respond to external clues. The present study revealed that cross-talk between PDGFR and S1P2 plays an important role in regulating migratory responses to PDGF. Loss of the S1P2 receptor led to enhanced migration of cells not only toward S1P and serum but also toward PDGF, which stimulates production of S1P, while fibronectin haptotactic migration was unaffected. There were no obvious differences in PDGFR expression, its tyrosine phosphorylation by PDGF, or activation of ERK1/2 or Akt in S1P2-null fibroblasts compared to wild-type cells. Although deletion of S1P2 reduced activation of p38 by PDGF, particularly at later time points, several lines of evidence suggest that these changes in p38 activation do not contribute to the enhanced migratory responses of S1P2-null fibroblasts toward PDGF. First, SB202190, a p38 inhibitor, had no effect on Akt S473 phosphorylation induced by PDGF or on chemotaxis toward PDGF of wild-type or S1P2-null fibroblasts. This is consistent with a recent report showing that p38 inhibition had no effect on the degree of lamellipodia formation and cellular protrusions in MEFs in response to PDGF-BB (23). Moreover, endothelial cell migration in response to S1P was blocked by pretreatment with PTX but was not affected by inhibition of ERK or p38 (27). Finally, a recent study suggests that although PDGFRβ plays a crucial role in PDGF-BB-induced fibroblast proliferation, survival, and migration, p38 is not involved in the biological effects of PDGF-BB (15).
On the other hand, PDGF-induced activation of the small G protein Rac, known to play an important role in formation of membrane ruffles, was strongly enhanced in S1P2-null cells. Similarly, activation of the Rac effector PAK1, which is recruited to the leading edge of motile cells, was also enhanced. Indeed, PDGF treatment led to increased membrane ruffling in these cells with cortical actin at the leading edge and rapid turnover of focal complexes. Importantly, the increased migration of the S1P2-null cells was eliminated by reintroduction of S1P2, further supporting a role for this receptor as a negative regulator of migration toward PDGF. This is consistent with the previous observation that binding of S1P to S1P2 stimulates Rho and inhibits Rac activation, leading to decreased membrane ruffling and chemotaxis (49).
In addition, we found that the enhanced PDGF-induced migratory responses in cells lacking the S1P2 receptor appeared to be intimately linked to SphK1. It has long been known that PDGF stimulates SphK1 and S1P formation (43). In agreement, PDGF rapidly and transiently increased SphK1 activity and silencing of SphK1 expression with siRNA directed against SphK1, but not the SphK2 isoform, diminished PDGF-induced migration of S1P2-null fibroblasts. However, migration toward fibronectin was not affected by siSphK1, suggesting that downregulation of SphK1 does not affect the cellular motility machinery. These results implicate SphK1 as a mediator of the cross talk between the PDGFR and the S1PRs.
In agreement with our previous proposal that transactivation of S1P1 was required for cell movement toward PDGF (18, 45), treatment of S1P2-null cells with PTX to inactivate Gi, the only G protein that S1P1 signals through, or downregulation of S1P1 expression markedly reduced migration. These results suggested that, in wild-type cells expressing S1P1 and S1P2, both receptors are transactivated by PDGF. Activation of S1P1 is necessary for motility, and S1P2 acts to dampen or regulate motility responses, and thus the net responses are dependent on the relative expression levels of these two receptors and their activation by PDGF. Hence a delicate balance between transactivation of S1P1 and S1P2 by PDGF is a critical factor that determines net movement toward PDGF (Fig. 9).
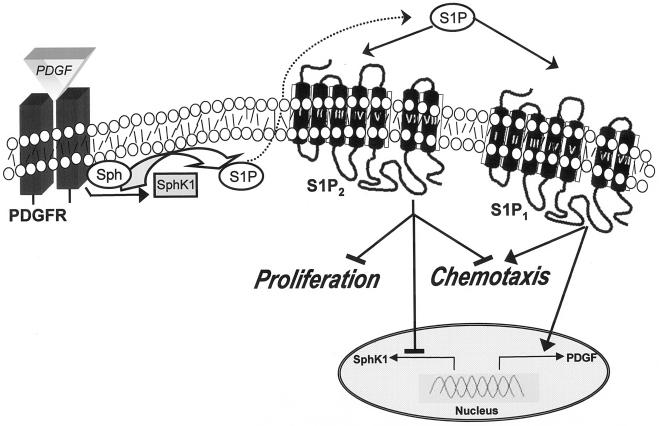
FIG. 9.
Schematic depicting the role of S1P2 in PDGF-induced proliferation and migration. Activation of PDGFRβ by PDGF stimulates and translocates SphK1 to membrane ruffles to increase phosphorylation of sphingosine to S1P. S1P, in turn, can bind to and activate its receptors. Activation of the S1P1 receptor stimulates downstream signals important for cell locomotion. Activation of the S1P2 receptor transduces signaling pathways that inhibit motility and also decreases cell proliferation and expression of SphK1. It is also shown that S1P1 can stimulate PDGF expression (55). Transactivation of PDGFR by S1P1 (53) is not depicted, nor are the numerous signaling pathways downstream of PDGF and S1P receptors which do not involve receptor cross-communication described in this study.
We found that S1P2 not only regulates migration induced by PDGF but also is a negative regulator of cell proliferation. Numerous studies have shown that S1P is a potent mitogen for diverse cell types (reviewed in reference 48). However, a few reports suggest that S1P can also be a growth inhibitor (10, 19, 22). Binding of S1P to S1P2 inhibits proliferation of rat hepatocytes by activating Rho, suggesting that S1P might be a negative regulator of liver regeneration (19). Loss of S1P2 receptor resulted in increased proliferation of cells in response to serum, S1P, and PDGF. Conversely, reintroduction of S1P2 into MEFs lacking this receptor decreased proliferation. In agreement, S1P2 but not S1P1 mRNA expression was enhanced in hepatocytes 24 to 72 h after partial hepatectomy, which coincides with decreasing hepatocyte proliferation, suggesting that activation of S1P2 by S1P negatively regulates liver regeneration (19).
An important observation that might explain how S1P2 acts as a negative regulator of proliferation induced by PDGF emerged from our unexpected finding that S1P2 inhibits expression and activity of SphK1 (Fig. 9). Many previous studies have shown that expression of SphK1 enhances cell growth and survival and inhibitors of SphK1 decrease proliferation and induce apoptosis (40, 42, 50, 51, 60). Consistent with this, S1P2-null cells have higher SphK1 activity than wild-type cells and their rate of proliferation is significantly greater. Moreover, reconstitution of S1P2 not only decreases proliferation, it also decreases expression and activity of SphK1 back to wild-type levels.
Our study adds another level of complexity to the intricate interplay between PDGF and S1P signaling. Similar to other GPCR ligands, S1P acting through S1P1 or S1P3 can transactivate PDGFR (4, 53). S1P also stimulates the synthesis of PDGF A and B polypeptides through S1P1-dependent signaling process (55). Then, according to our paradigm, binding of PDGF to its receptor causes activation and translocation of SphK1, increasing S1P production and subsequent transactivation of S1PRs. The balance between S1P1 and S1P2 receptor signaling is a critical regulator of PDGF-induced motility and proliferation (Fig. 9).
The complex interplay between PDGFR and S1PRs may have important implications for vascular maturation. Disruption of either PDGF-B (28, 29), PDGFR (47), or the S1P1 receptor (32) genes in mice results in lethal hemorrhage and edema in the perinatal stage due to incomplete coverage of blood vessels by vascular smooth muscle cells and pericytes. Interestingly, disruption of the S1P1 gene specifically in endothelial cells also produced the same phenotype (1). Stimulation of S1P1 on endothelial cells may regulate the recruitment of vascular smooth muscle cells by stimulating the secretion of recruitment factors, such as PDGF (55). Furthermore, our work suggests that in the absence of S1P1, the S1P2 receptor would dominate and inhibit migration of vascular smooth muscle cells toward PDGF, causing deficient coverage of vessels, a process that occurs during the last stages of angiogenesis and is important for stability of the nascent vascular network.
Acknowledgments
This work was supported by National Institutes of Health grant CA61774 (to S.S.) and, in part, by MH01723 (to J.C.).
REFERENCES
- 1.Allende, M. L., T. Yamashita, and R. L. Proia. 2003. G-protein coupled receptor S1P1 acts within endothelial cells to regulate vascular maturation. Blood 102:3665-3667. [DOI] [PubMed] [Google Scholar]
- 2.Anliker, B., and J. Chun. 2004. Lysophospholipid G protein-coupled receptors. J. Biol. Chem. 279:20555-20558. [DOI] [PubMed] [Google Scholar]
- 3.Baudhuin, L. M., K. L. Cristina, J. Lu, and Y. Xu. 2002. Akt activation induced by lysophosphatidic acid and sphingosine-1-phosphate requires both mitogen-activated protein kinase kinase and p38 mitogen-activated protein kinase and is cell-line specific. Mol. Pharmacol. 62:660-671. [DOI] [PubMed] [Google Scholar]
- 4.Baudhuin, L. M., Y. Jiang, A. Zaslavsky, I. Ishii, J. Chun, and Y. Xu. 2004. S1P3-mediated Akt activation and cross-talk with platelet-derived growth factor receptor (PDGFR). FASEB J. 18:341-343. [DOI] [PubMed] [Google Scholar]
- 5.Bokoch, G. M. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72:743-781. [DOI] [PubMed] [Google Scholar]
- 6.Bornfeldt, K. E., L. M. Graves, E. W. Raines, Y. Igarashi, G. Wayman, S. Yamamura, Y. Yatomi, J. S. Sidhu, E. G. Krebs, S. Hakomori, et al. 1995. Sphingosine-1-phosphate inhibits PDGF-induced chemotaxis of human arterial smooth muscle cells: spatial and temporal modulation of PDGF chemotactic signal transduction. J. Cell Biol. 130:193-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burridge, K., and K. Wennerberg. 2004. Rho and Rac take center stage. Cell 116:167-179. [DOI] [PubMed] [Google Scholar]
- 8.Chang, L., and M. Karin. 2001. Mammalian MAP kinase signalling cascades. Nature 410:37-40. [DOI] [PubMed] [Google Scholar]
- 9.Cuvillier, O., G. Pirianov, B. Kleuser, P. G. Vanek, O. A. Coso, S. Gutkind, and S. Spiegel. 1996. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 381:800-803. [DOI] [PubMed] [Google Scholar]
- 10.Davaille, J., C. Gallois, A. Habib, L. Li, A. Mallat, J. Tao, T. Levade, and S. Lotersztajn. 2000. Antiproliferative properties of sphingosine 1-phosphate in human hepatic myofibroblasts. A cyclooxygenase-2 mediated pathway. J. Biol. Chem. 275:34628-34633. [DOI] [PubMed] [Google Scholar]
- 11.Edsall, L. C., O. Cuvillier, S. Twitty, S. Spiegel, and S. Milstien. 2001. Sphingosine kinase expression regulates apoptosis and caspase activation in PC12 cells. J. Neurochem. 76:1573-1584. [DOI] [PubMed] [Google Scholar]
- 12.Edsall, L. C., J. R. Van Brocklyn, O. Cuvillier, B. Kleuser, and S. Spiegel. 1998. N,N-Dimethylsphingosine is a potent competitive inhibitor of sphingosine kinase but not of protein kinase C: modulation of cellular levels of sphingosine 1-phosphate and ceramide. Biochemistry 37:12892-12898. [DOI] [PubMed] [Google Scholar]
- 13.English, D., A. T. Kovala, Z. Welch, K. A. Harvey, R. A. Siddiqui, D. N. Brindley, and J. G. Garcia. 1999. Induction of endothelial cell chemotaxis by sphingosine 1-phosphate and stabilization of endothelial monolayer barrier function by lysophosphatidic acid, potential mediators of hematopoietic angiogenesis. J. Hematother. Stem Cell Res. 8:627-634. [DOI] [PubMed] [Google Scholar]
- 14.English, D., Z. Welch, A. T. Kovala, K. Harvey, O. V. Volpert, D. N. Brindley, and J. G. Garcia. 2000. Sphingosine 1-phosphate released from platelets during clotting accounts for the potent endothelial cell chemotactic activity of blood serum and provides a novel link between hemostasis and angiogenesis. FASEB J. 14:2255-2265. [DOI] [PubMed] [Google Scholar]
- 15.Gao, Z., T. Sasaoka, T. Fujimori, T. Oya, Y. Ishii, H. Sabit, M. Kawaguchi, Y. Kurotaki, M. Naito, T. Wada, S. Ishizawa, M. Kobayashi, Y. I. Nabeshima, and M. Sasahara. 2004. Deletion of the PDGFR-β gene affects key fibroblast functions important for wound healing. J. Biol. Chem. 280:9375-9389. [DOI] [PubMed] [Google Scholar]
- 16.Garcia, J. G., F. Liu, A. D. Verin, A. Birukova, M. A. Dechert, W. T. Gerthoffer, J. R. Bamberg, and D. English. 2001. Sphingosine 1-phosphate promotes endothelial cell barrier integrity by Edg-dependent cytoskeletal rearrangement. J. Clin. Investig. 108:689-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graeler, M., G. Shankar, and E. J. Goetzl. 2002. Cutting edge: suppression of T cell chemotaxis by sphingosine 1-phosphate. J. Immunol. 169:4084-4087. [DOI] [PubMed] [Google Scholar]
- 18.Hobson, J. P., H. M. Rosenfeldt, L. S. Barak, A. Olivera, S. Poulton, M. G. Caron, S. Milstien, and S. Spiegel. 2001. Role of the sphingosine-1-phosphate receptor EDG-1 in PDGF-induced cell motility. Science 291:1800-1803. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda, H., H. Satoh, M. Yanase, Y. Inoue, T. Tomiya, M. Arai, K. Tejima, K. Nagashima, H. Maekawa, N. Yahagi, Y. Yatomi, S. Sakurada, Y. Takuwa, I. Ogata, S. Kimura, and K. Fujiwara. 2003. Antiproliferative property of sphingosine 1-phosphate in rat hepatocytes involves activation of Rho via Edg-5. Gastroenterology 124:459-469. [DOI] [PubMed] [Google Scholar]
- 20.Ishii, I., B. Friedman, X. Ye, S. Kawamura, C. McGiffert, J. J. Contos, M. A. Kingsbury, G. Zhang, J. H. Brown, and J. Chun. 2001. Selective loss of sphingosine 1-phosphate signaling with no obvious phenotypic abnormality in mice lacking its G protein-coupled receptor, LPB3/EDG-3. J. Biol. Chem. 276:33697-33704. [DOI] [PubMed] [Google Scholar]
- 21.Ishii, I., X. Ye, B. Friedman, S. Kawamura, J. J. Contos, M. A. Kingsbury, A. H. Yang, G. Zhang, J. H. Brown, and J. Chun. 2002. Marked perinatal lethality and cellular signaling deficits in mice null for the two sphingosine 1-phosphate receptors, S1P2/LPB2/EDG-5 and S1P3/LPB3/EDG-3. J. Biol. Chem. 277:25152-25159. [DOI] [PubMed] [Google Scholar]
- 22.Jin, Y., E. Knudsen, L. Wang, Y. Bryceson, B. Damaj, S. Gessani, and A. A. Maghazachi. 2003. Sphingosine 1-phosphate is a novel inhibitor of T-cell proliferation. Blood 101:4909-4915. [DOI] [PubMed] [Google Scholar]
- 23.Kallin, A., J. B. Demoulin, K. Nishida, T. Hirano, L. Ronnstrand, and C. H. Heldin. 2004. Gab1 contributes to cytoskeletal reorganization and chemotaxis in response to platelet-derived growth factor. J. Biol. Chem. 279:17897-17904. [DOI] [PubMed] [Google Scholar]
- 24.Kohama, T., A. Olivera, L. Edsall, M. M. Nagiec, R. Dickson, and S. Spiegel. 1998. Molecular cloning and functional characterization of murine sphingosine kinase. J. Biol. Chem. 273:23722-23728. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M., S. Thangada, J. Paik, G. P. Sapkota, N. Ancellin, S. Chae, M. Wu, M. Morales-Ruiz, W. C. Sessa, D. R. Alessi, and T. Hla. 2001. Akt-mediated phosphorylation of the G protein-coupled receptor edg-1 is required for endothelial cell chemotaxis. Mol. Cell 8:693-704. [DOI] [PubMed] [Google Scholar]
- 26.Lee, M. J., S. Thangada, K. P. Claffey, N. Ancellin, C. H. Liu, M. Kluk, M. Volpi, R. I. Sha'afi, and T. Hla. 1999. Vascular endothelial cell adherens junction assembly and morphogenesis induced by sphingosine-1-phosphate. Cell 99:301-312. [DOI] [PubMed] [Google Scholar]
- 27.Lee, O. H., Y. M. Kim, Y. M. Lee, E. J. Moon, D. J. Lee, J. H. Kim, K. W. Kim, and Y. G. Kwon. 1999. Sphingosine 1-phosphate induces angiogenesis: its angiogenic action and signaling mechanism in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 264:743-750. [DOI] [PubMed] [Google Scholar]
- 28.Leveen, P., M. Pekny, S. Gebre-Medhin, B. Swolin, E. Larsson, and C. Betsholtz. 1994. Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 8:1875-1887. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl, P., B. R. Johansson, P. Leveen, and C. Betsholtz. 1997. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277:242-245. [DOI] [PubMed] [Google Scholar]
- 30.Liu, H., M. Sugiura, V. E. Nava, L. C. Edsall, K. Kono, S. Poulton, S. Milstien, T. Kohama, and S. Spiegel. 2000. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J. Biol. Chem. 275:19513-19520. [DOI] [PubMed] [Google Scholar]
- 31.Liu, H., R. E. Toman, S. Goparaju, M. Maceyka, V. E. Nava, H. Sankala, S. G. Payne, M. Bektas, I. Ishii, J. Chun, S. Milstien, and S. Spiegel. 2003. Sphingosine kinase type 2 is a putative BH3-only protein that induces apoptosis. J. Biol. Chem. 278:40330-40336. [DOI] [PubMed] [Google Scholar]
- 32.Liu, Y., R. Wada, T. Yamashita, Y. Mi, C. X. Deng, J. P. Hobson, H. M. Rosenfeldt, V. E. Nava, S. S. Chae, M. J. Lee, C. H. Liu, T. Hla, S. Spiegel, and R. L. Proia. 2000. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Investig. 106:951-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mandala, S., R. Hajdu, J. Bergstrom, E. Quackenbush, J. Xie, J. Milligan, R. Thornton, G. J. Shei, D. Card, C. Keohane, M. Rosenbach, J. Hale, C. L. Lynch, K. Rupprecht, W. Parsons, and H. Rosen. 2002. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science 296:346-349. [DOI] [PubMed] [Google Scholar]
- 34.Matloubian, M., C. G. Lo, G. Cinamon, M. J. Lesneski, Y. Xu, V. Brinkmann, M. L. Allende, R. L. Proia, and J. G. Cyster. 2004. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427:355-360. [DOI] [PubMed] [Google Scholar]
- 35.Matsumoto, T., K. Yokote, K. Tamura, M. Takemoto, H. Ueno, Y. Saito, and S. Mori. 1999. Platelet-derived growth factor activates p38 mitogen-activated protein kinase through a Ras-dependent pathway that is important for actin reorganization and cell migration. J. Biol. Chem. 274:13954-13960. [DOI] [PubMed] [Google Scholar]
- 36.Meyer zu Heringdorf, D., H. Lass, R. Alemany, K. T. Laser, E. Neumann, C. Zhang, M. Schmidt, U. Rauen, K. H. Jakobs, and C. J. van Koppen. 1998. Sphingosine kinase-mediated Ca2+ signalling by G-protein-coupled receptors. EMBO J. 17:2830-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita, Y., G. I. Perez, F. Paris, S. R. Miranda, D. Ehleiter, A. Haimovitz-Friedman, Z. Fuks, Z. Xie, J. C. Reed, E. H. Schuchman, R. N. Kolesnick, and J. L. Tilly. 2000. Oocyte apoptosis is suppressed by disruption of the acid sphingomyelinase gene or by sphingosine-1-phosphate therapy. Nat. Med. 6:1109-1114. [DOI] [PubMed] [Google Scholar]
- 38.Okamoto, H., N. Takuwa, T. Yokomizo, N. Sugimoto, S. Sakurada, H. Shigematsu, and Y. Takuwa. 2000. Inhibitory regulation of Rac activation, membrane ruffling, and cell migration by the G protein-coupled sphingosine-1-phosphate receptor EDG5 but not EDG1 or EDG3. Mol. Cell. Biol. 20:9247-9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olivera, A., L. Edsall, S. Poulton, A. Kazlauskas, and S. Spiegel. 1999. Platelet-derived growth factor-induced activation of sphingosine kinase requires phosphorylation of the PDGF receptor tyrosine residue responsible for binding of PLCgamma. FASEB J. 13:1593-1600. [DOI] [PubMed] [Google Scholar]
- 40.Olivera, A., T. Kohama, L. C. Edsall, V. Nava, O. Cuvillier, S. Poulton, and S. Spiegel. 1999. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J. Cell Biol. 147:545-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olivera, A., T. Kohama, Z. Tu, S. Milstien, and S. Spiegel. 1998. Purification and characterization of rat kidney sphingosine kinase. J. Biol. Chem. 273:12576-12583. [DOI] [PubMed] [Google Scholar]
- 42.Olivera, A., H. M. Rosenfeldt, M. Bektas, F. Wang, I. Ishii, J. Chun, S. Milstien, and S. Spiegel. 2003. Sphingosine kinase type 1 induces G12/13-mediated stress fiber formation yet promotes growth and survival independent of G protein coupled receptors. J. Biol. Chem. 278:46452-46460. [DOI] [PubMed] [Google Scholar]
- 43.Olivera, A., and S. Spiegel. 1993. Sphingosine-1-phosphate as a second messenger in cell proliferation induced by PDGF and FCS mitogens. Nature 365:557-560. [DOI] [PubMed] [Google Scholar]
- 44.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell 112:453-465. [DOI] [PubMed] [Google Scholar]
- 45.Rosenfeldt, H. M., J. P. Hobson, M. Maceyka, A. Olivera, V. E. Nava, S. Milstien, and S. Spiegel. 2001. EDG-1 links the PDGF receptor to Src and focal adhesion kinase activation leading to lamellipodia formation and cell migration. FASEB J. 15:2649-2659. [DOI] [PubMed] [Google Scholar]
- 46.Ryu, Y., N. Takuwa, N. Sugimoto, S. Sakurada, S. Usui, H. Okamoto, O. Matsui, and Y. Takuwa. 2002. Sphingosine-1-phosphate, a platelet-derived lysophospholipid mediator, negatively regulates cellular Rac activity and cell migration in vascular smooth muscle cells. Circ. Res. 90:325-332. [DOI] [PubMed] [Google Scholar]
- 47.Soriano, P. 1994. Abnormal kidney development and hematological disorders in PDGF β-receptor mutant mice. Genes Dev. 8:1888-1896. [DOI] [PubMed] [Google Scholar]
- 48.Spiegel, S., and S. Milstien. 2003. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4:397-407. [DOI] [PubMed] [Google Scholar]
- 49.Sugimoto, N., N. Takuwa, H. Okamoto, S. Sakurada, and Y. Takuwa. 2003. Inhibitory and stimulatory regulation of Rac and cell motility by the G12/13-Rho and Gi pathwaysintegrated downstream of a single G protein-coupled sphingosine-1-phosphate receptor isoform. Mol. Cell. Biol. 23:1534-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sukocheva, O. A., L. Wang, N. Albanese, S. M. Pitson, M. A. Vadas, and P. Xia. 2003. Sphingosine kinase transmits estrogen signaling in human breast cancer cells. Mol. Endocrinol. 17:2002-2012. [DOI] [PubMed] [Google Scholar]
- 51.Taha, T. A., W. Osta, L. Kozhaya, J. Bielawski, K. R. Johnson, W. E. Gillanders, G. S. Dbaibo, Y. A. Hannun, and L. M. Obeid. 2004. Down-regulation of sphingosine kinase-1 by DNA damage: dependence on proteases and p53. J. Biol. Chem. 279:20546-20554. [DOI] [PubMed] [Google Scholar]
- 52.Tanimoto, T., Z. G. Jin, and B. C. Berk. 2002. Transactivation of vascular endothelial growth factor (VEGF) receptor Flk-1/KDR is involved in sphingosine 1-phosphate-stimulated phosphorylation of Akt and endothelial nitric-oxide synthase (eNOS). J. Biol. Chem. 277:42997-43001. [DOI] [PubMed] [Google Scholar]
- 53.Tanimoto, T., A. O. Lungu, and B. C. Berk. 2004. Sphingosine 1-phosphate transactivates the platelet-derived growth factor β receptor and epidermal growth factor receptor in vascular smooth muscle cells. Circ. Res. 94:1050-1058. [DOI] [PubMed] [Google Scholar]
- 54.Turner, C. E. 2000. Paxillin and focal adhesion signalling. Nat. Cell Biol. 2:E231-E236. [DOI] [PubMed] [Google Scholar]
- 55.Usui, S., N. Sugimoto, N. Takuwa, S. Sakagami, S. Takata, S. Kaneko, and Y. Takuwa. 2004. Blood lipid mediator sphingosine 1-phosphate potently stimulates platelet-derived growth factor-A and -B chain expression through S1P1-Gi-Ras-MAPK-dependent induction of Kruppel-like factor 5. J. Biol. Chem. 279:12300-12311. [DOI] [PubMed] [Google Scholar]
- 56.Van Brocklyn, J. R., M. J. Lee, R. Menzeleev, A. Olivera, L. Edsall, O. Cuvillier, D. M. Thomas, P. J. P. Coopman, S. Thangada, T. Hla, and S. Spiegel. 1998. Dual actions of sphingosine-1-phosphate: extracellular through the Gi-coupled orphan receptor edg-1 and intracellular to regulate proliferation and survival. J. Cell Biol. 142:229-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, F., J. R. Van Brocklyn, J. P. Hobson, S. Movafagh, Z. Zukowska-Grojec, S. Milstien, and S. Spiegel. 1999. Sphingosine 1-phosphate stimulates cell migration through a Gi-coupled cell surface receptor. Potential involvement in angiogenesis. J. Biol. Chem. 274:35343-35350. [DOI] [PubMed] [Google Scholar]
- 58.Waters, C., B. S. Sambi, K. C. Kong, D. Thompson, S. M. Pitson, S. Pyne, and N. J. Pyne. 2003. Sphingosine 1-phosphate and platelet-derived growth factor (PDGF) act via PDGFβ receptor-sphingosine 1-phosphate receptor complexes in airway smooth muscle cells. J. Biol. Chem. 278:6282-6290. [DOI] [PubMed] [Google Scholar]
- 59.Wei, M. C., W. X. Zong, E. H. Cheng, T. Lindsten, V. Panoutsakopoulou, A. J. Ross, K. A. Roth, G. R. MacGregor, C. B. Thompson, and S. J. Korsmeyer. 2001. Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292:727-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia, P., J. R. Gamble, L. Wang, S. M. Pitson, P. A. Moretti, B. W. Wattenberg, R. J. D'Andrea, and M. A. Vadas. 2000. An oncogenic role of sphingosine kinase. Curr. Biol. 10:1527-1530. [DOI] [PubMed] [Google Scholar]