Summary
Artezomibs (ATZs), dual-pharmacophore molecules comprising of artemisinin and a parasite proteasome inhibitor, hijack parasite ubiquitin proteasome system to transform into new proteasome inhibitors following the activation of artemisinin by heme.1 Here, we present a protocol for using a fluorescent activity-based broad-spectrum proteasome inhibitor probe to study intracellular conversion of ATZ molecules into new proteasome inhibitors in malaria parasites. We describe steps for drug treatment and washout, parasite lysis, proteasome labeling, and visualization.
For complete details on the use and execution of this protocol, please refer to Zhan et al.1
Subject areas: Microbiology, Chemistry
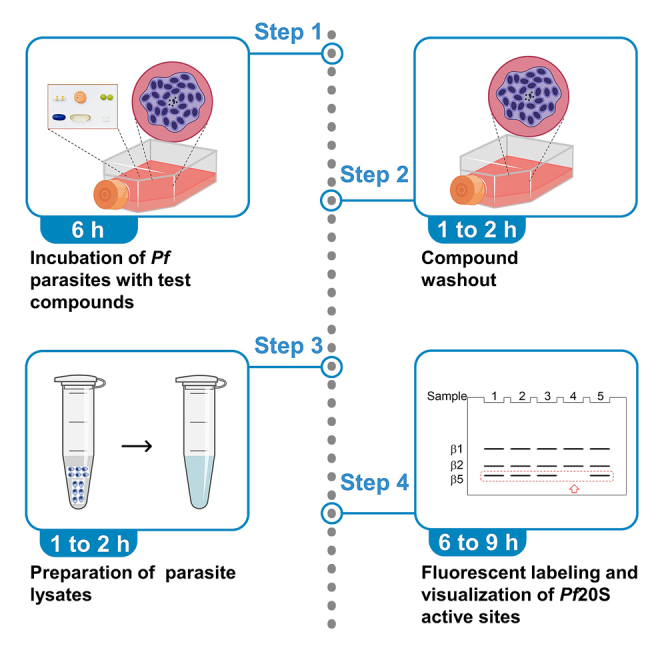
Graphical abstract
Highlights
-
•
An assay for assessing intracellular artezomib conversion to novel 20S inhibitors
-
•
Detailed steps of removing free compound from both media and cells
-
•
In-gel fluorescence scanning to visualize 20S active subunit labeling
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Artezomibs (ATZs), dual-pharmacophore molecules comprising of artemisinin and a parasite proteasome inhibitor, hijack parasite ubiquitin proteasome system to transform into new proteasome inhibitors following the activation of artemisinin by heme. Here, we present a protocol for using a fluorescent activity-based broad-spectrum proteasome inhibitor probe to study intracellular conversion of ATZ molecules into new proteasome inhibitors in malaria parasites. We describe steps for drug treatment and washout, parasite lysis, proteasome labeling, and visualization.
Before you begin
Plasmodium falciparum cell culture
Timing: 2 weeks
-
1.
Maintain P. falciparum laboratory lines at 5% hematocrit in RPMI 1640 medium with 0.5% Albumax II (Invitrogen), 0.25% sodium bicarbonate and 0.1 mg/mL gentamicin in an incubator under standard conditions: 37°C, 5% oxygen, 5% carbon dioxide and 90% nitrogen.
-
2.
Culture synchronized P. falciparum (Dd2, resistant Dd2 β5A49S 2 and Dd2 β6A117D 3). Perform sorbitol treatment once a week to keep the parasites synchronized. If tighter synchronization is desired, perform the sorbitol treatment after one parasite cycle (48 h).
-
3.Synchronize parasite culture.
-
a.Take parasites when they are mostly at the ring stage (at least 50% of infected RBCs must be in ring stage). They must not be later than 10–12 h post invasion when the sorbitol treatment is done.
-
b.Take 10 mL of a culture of >5% parasitemia.
-
c.Spin down the parasite culture at 2,521 × g at r.t. for 2 min.
-
d.Add 10 mL of 5% sorbitol (prewarmed to 37°C) and incubate for 10 min, shaking at 37°C and 55 rpm.
-
e.Spin down the parasite culture at 2,521 × g, RT for 2 min.
-
f.Wash once with 10 mL culture medium, pellet red cells using same conditions as above.
-
g.Resuspend cells back in 10 mL culture media.
-
a.
Preparation drug stock solutions
Timing: 10 min
-
4.
This protocol describes the activation of an ATZ, ATZ4, producing new proteasome inhibitors capable of inhibiting proteasome β5 subunit either in P. falciparum Dd2 wild type or in its two PI resistant strains. To demonstrate this unique MOA of ATZ, we set up experiments of ATZ4 together with a series of control compounds, including direct control deoxy-ATZ4 lacking the endoperoxide moiety in ATZ4, proteasome inhibitor PI01, DHA and its analog ART1. Approximately 50 μL of a 10 mM stock of each compound in DMSO is sufficient, and each molecule should be over 95% pure.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| PI01 | https://doi.org/10.1016/j.chembiol.2023.04.006 | N/A |
| ART1 | https://doi.org/10.1016/j.chembiol.2023.04.006 | N/A |
| ATZ4 | https://doi.org/10.1016/j.chembiol.2023.04.006 | N/A |
| Deoxy-ATZ4 | https://doi.org/10.1016/j.chembiol.2023.04.006 | N/A |
| Dihydroartemisinin (DHA) | Selleck Chemicals LLC | Catalog # S2290 |
| Activity-based probe MV151 | https://doi.org/10.1016/J.Chembiol.2006.09.013 | N/A |
| Bovine serum albumin | Sigma-Aldrich | Catalog # 3117057001 |
| Dimethyl sulfoxide (DMSO) 99.8+% for molecular biology, DNase, RNase, and protease-free | Thermo Fisher Scientific | Catalog # 10397841 |
| 100 mM Tris-HCl, pH 7.4, molecular biology grade | Sigma-Aldrich (Millipore) | Catalog # 648315 |
| 1 M Tris-HCI, pH 6.5 | Sigma-Aldrich | Catalog # 632417 |
| Sodium dodecyl sulfate | Sigma-Aldrich | Catalog # L3771 |
| Bromophenol blue | Sigma-Aldrich | Catalog # B0126 |
| Dithiothreitol | Sigma-Aldrich | Catalog # 10197777001 |
| Glycerol, for molecular biology, ≥99.0% | Sigma-Aldrich | Catalog # G5516 |
| Saponin | Sigma-Aldrich | Catalog # S7900 |
| D-Sorbitol | Sigma-Aldrich | Catalog # S1876 |
| Magnesium chloride solution, for molecular biology, 1.00 M ± 0.01 M | Sigma-Aldrich | Catalog # 63069 |
| 12% Novex Bis-Tris protein gel | Thermo Fisher Scientific | Catalog # NP0341BOX |
| NuPage MOPS SDS running buffer (20X) | Thermo Fisher Scientific | Catalog # NP0001 |
| Precision Plus Protein WesternC Blotting Standards | Bio-Rad | Catalog # 1610376 |
| Critical commercial assays | ||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Catalog # 23225 |
| Experimental models: Organisms/strains | ||
| Plasmodium falciparum Dd2 | BEI Resources | MRA-150 |
| Plasmodium falciparum Dd2 β6A117D | https://doi.org/10.1073/pnas.1806109115 | N/A |
| Plasmodium falciparum Dd2 β5A49S | https://doi.org/10.1021/acs.jmedchem.9b00363 | N/A |
| Software and algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| GraphPad Prism 9 | GraphPad Software Inc. | https://www.graphpad.com/ |
| Microsoft Office Suite | Microsoft, Inc. | N/A |
| Other | ||
| Mini gel tank | Thermo Fisher Scientific | Catalog # A25977 |
| Belly Dancer/Hybridization water bath | Sigma-Aldrich | Catalog #Z367621 |
| Protein LoBind tubes, 1.5 mL | Eppendorf | Catalog # 0030108442 |
| 80-place tube rack, natural | USA Scientific, Inc. | Catalog # NC9204348 |
| Pierce 96–well plate | Thermo Scientific | Catalog # 15041 |
| Typhoon 9410 Variable Mode Imager | GE Healthcare | |
Materials and equipment
-
•
Malaria culture media
| Reagent | Final concentration | Amount |
|---|---|---|
| Albumax II | 0.5% (w/v) | 5 g |
| RPMI 1640 | N/A | 10.5 g |
| Sodium bicarbonate | 12 mM | 8.06 g |
| Gentamicin | 20 mg/L | 4 mL |
| Hypoxanthine solution | 50 mg/L | .05 g |
| ddH2O | N/A | To 1 L |
| Total | N/A | 1 L |
Note: Ensure Albumax II has dissolved fully before adding other reagents. Adjust pH to 7.3 with sodium hydroxide. Filter sterilize and store at 4°C for up to 2 months. Warm to 37°C before use.
-
•
Saponin lysis solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Saponin | 0.1% (w/v) | 1 mg |
| PBS | N/A | To 10 mL |
| Total | N/A | 10 mL |
Note: Use chilled at 4°C.
-
•
Parasite pellet lysis buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 100 mM Tris-HCl, pH 7.4 | 20 mM | 20 mL |
| 1 M magnesium chloride | 5 mM | 0.5 mL |
| 1 M dithiothreitol | 1 mM | 0.1 mL |
| ddH2O | N/A | To 100 mL |
| Total | N/A | 100 mL |
Note: Prepare fresh on day of lysate preparation (dithiothreitol must be added fresh). Use chilled at 4°C.
-
•
Labeling reaction buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 100 mM Tris-HCl, pH 7.4 | 50 mM | 50 mL |
| 1 M magnesium chloride | 5 mM | 0.5 mL |
| 1 M dithiothreitol | 1 mM | 0.1 mL |
| ddH2O | N/A | To 100 mL |
| Total | N/A | 100 mL |
Note: Prepare fresh on day of labeling reactions (dithiothreitol must be added fresh).
-
•
Dithiothreitol (DTT)
| Reagent | Final concentration | Amount |
|---|---|---|
| DTT | 1 M | 770 mg |
| ddH2O | - | 5 mL |
| Total | N/A | 5 mL |
Note: Make 0.5 mL aliquots and stored at ‒80°C for up to 6 months.
-
•
4X SDS loading buffer.
| Reagent | Final concentration | Amount |
|---|---|---|
| 1 M Tris-HCl, pH 6.5 | 200 mM | 1 mL |
| SDS (sodium dodecyl sulfate) | 277 mM (w/v, 8%) | 0.4 g |
| Bromophenol blue | 6 mM | 20 mg |
| 1 M dithiothreitol | 400 mM | 2 mL |
| Glycerol | 4.3 M | 1.6 mL |
| ddH2O | N/A | To 5 mL |
| Total | N/A | 5 mL |
Note: Vortex gently to aid dissolution. Store in 1 mL aliquots at −80°C, for up to 6 months. Warm up to room temperature (r.t.) before use.
-
•
MOPS SDS running buffer (1X)
| Reagent | Final concentration | Amount |
|---|---|---|
| MOPS SDS running buffer (20X) | - | 100 mL |
| ddH2O | N/A | To 2 L |
| Total | N/A | 2 L |
Note: Store at 4°C, use within 3 weeks.
Step-by-step method details
Drug treatment of malarial parasites
Timing: 6 h
-
1.Add indicated volumes of drug at desired concentration to 5 mL of parasite-infected red blood cells for each treatment condition.
-
a.Dilute the 10 mM compound stock to 1000 μM stock with DMSO, such as.
-
i.[5 μL] of 10 mM PI01 + [45 μL] of DMSO = [50 μL] of 1000 μM PI01 stock.
-
ii.Make 10 μL aliquots and store at ‒20°C for repeat experiments within 6 months.
-
i.
-
b.Prepare 50 μL of 100X drug solution by diluting the 1000 μM drug stock with media; volumes of stock used at each group are outlined in the following table:
Group Final con. (nM) 100 X (μM) Stock (μM) Vol. Of stock Vol. Of media Total Vol. DMSO / / / 4.0 μL 46 μL 50 μL PI01 800 80 1000 4.0 μL 46 μL 50 μL ART1 700 70 1000 3.5 μL 46.5 μL 50 μL ATZ4 700 70 1000 3.5 μL 46.5 μL 50 μL PI01/ART1 800/800 80/80 1000/1000 4.0 μL/4.0 μL 42 μL 50 μL DHA 700 70 1000 3.5 μL 46.5 μL 50 μL Deoxy-ATZ4 700 70 1000 3.5 μL 46.5 μL 50 μL -
c.Add above 50 μL of 100X drug solution to 5 mL cell culture in a 75 cm2 flask, swirl the flask several times to mix well, then keep in incubator for 6 h.
-
a.
Thorough washing of red blood cells
Timing: 1–2 h
These steps detail how to remove the test compound from the parasite culture flask prior to erythrocyte lysis. Note that this step is critical to analyzing the presentation of ATZ4-derived PIs inside the parasites.
-
2.Centrifuge the cell culture, remove the supernatant, and resuspend the pelleted red cells in fresh complete media, then shake in the incubator.
-
a.Transfer the cell culture to 15 mL centrifuge tube, cap the tube.
-
b.Centrifuge (2,521 × g, 3 min, r.t.) and then carefully remove the supernatant.
-
c.Refill the centrifuge tube with fresh complete media (12 mL), and cap the tube, then resuspend the cells well by inverting several times.
-
d.Transfer the resuspended cell culture to a new 25 cm2 flask carefully, invert and shake the flask gently, and place back in the incubator on a rotating shaker.
-
e.Set shaking speed at 55 rpm and shake for 10 min.
-
f.Repeat steps 2a∼2e additional times.
-
a.
CRITICAL: Carefully remove the supernatant to avoid loss of pelleted erythrocytes, it is important to repeat the washing steps at least 3 times to minimize interference by residual compound in the following labeling step.
Preparation of parasite pellets
Timing: 1–2 h
This section describes how to lyse the red cell and obtain the drug-treated parasite pellets in each group. Before you begin, pre-chill the PBS and saponin solution on ice.
-
3.Following the final wash, place red blood cell pellets on ice, washed with PBS and then lyse with 10% saponin to obtain parasite pellets.
-
a.Following step 2a and 2b, resuspend pelleted cells in cold PBS (10 mL), and pellet cells again by centrifugation (2521 × g, 5 min, r.t.).
-
b.Aspirate off the supernatant.
-
c.Resuspend pelleted erythrocytes in 5 pellet volumes of cold saponin solution and incubate on ice for 10 min, with vigorous mixing every 3 min to lyse the erythrocytes.
-
d.Set centrifuge to 4°C, and centrifuge the resuspended cells (4,863 × g, 10 min).
-
e.Carefully aspirate off the supernatant and wash the parasites 4 times with cold PBS until supernatant is free of red color.
-
i.Aspirate off supernatant and resuspend parasite pellet in 1.2 mL PBS.
-
ii.Centrifuge the resuspended pellet (4,863 × g, 4°C, 10 min).
-
iii.Repeat 3 times more. Supernatant should be pale-yellow solution after the final wash. If the supernatant is still pink, additional wash step is needed.
-
i.
-
f.Aspirate off the supernatant and proceed with the preparation of lysate of the resulting pellet in the next step.
-
a.
CRITICAL: After saponin treatment and 4 times PBS washing, the parasite pellet should be almost black and there should not be any remaining intact red blood cells. Saponin lysis can be repeated if needed.
Pause point: The parasite pellet can be stored at −80°C for later use.
Lysis of the parasite pellet
Timing: 2–4 h
These steps describe how to perform lysis of parasite pellet and prepare lysate sample for electrophoresis. Before you begin, pre-chill the parasite pellet lysis buffer on ice and set centrifuge to chill to 4°C.
-
4.Resuspend the parasite pellet of each group in 2x pellet volume of lysis buffer, vortexed every 10 min for 1 h on ice, centrifuged at high speed to give supernatant lysate.
-
a.Add 2x pellet volume of parasite pellet lysis buffer into 1.5 mL Eppendorf tube containing parasite pellet in each group on ice.
-
b.Tap the sides of tube firmly by finger and mix well ensuring no clumps remain.
-
c.Vortex the tube vigorously every 10 min for 1 h.
-
d.Spin down at 21,136 × g for 20 min at 4°C.
-
e.Carefully transfer the supernatant by a 200 μL pipette to a fresh labeled 1.5 mL Eppendorf tube and keep on ice.
-
a.
CRITICAL: Very carefully transfer the lysate to avoid the pipette touching/aspirating the settled parasite debris, err on the side of caution, it is better to lose excess lysate than to pick up the black pellet debris, that otherwise interference with following concentration determination of the lysate protein.
BCA protein assay of Pf lysate
Timing: 2–4 h
We closely followed the instructions of Pierce BCA Protein Assay Kit in determination of the protein concentrations in each sample (https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0011430_Pierce_BCA_Protein_Asy_UG.pdf). We thus are omitting the description of this step, and readers are encouraged to refer to the instruction. Protein concentrations of each sample were determined and listed in Table 1.
Alternatives: Other protein quantitation methods can be used, such as Bicinchoninic acid or Bradford or UV.
Table 1.
Calculations of protein concentrations and amounts of the proteins used in the proteasome labeling experiments
| PI 01 group | Net A (562 nM) | Net A (562 nM) _repeat1 |
Net A (562 nM) _Average |
Calculated con. (μg/mL) | Vol of lysate for 6 μg protein (μL) | Vol. Of reaction buffer (μL) |
|---|---|---|---|---|---|---|
| no wash | 0.3087 | 0.3106 | 0.3097 | 2656 | 2.3 | 9.7 |
| 1X | 0.3205 | 0.314 | 0.3173 | 2746 | 2.2 | 9.8 |
| 2X | 0.3138 | 0.3072 | 0.3105 | 2661 | 2.3 | 9.7 |
| 3X | 0.347 | 0.3468 | 0.3469 | 3116 | 1.9 | 10.1 |
| 4X | 0.2899 | 0.2934 | 0.2917 | 2425 | 2.5 | 9.5 |
Labeling of Pf lysate by MV151
Timing: 2–3 h
This step describes how to label Pf proteasome (Pf20S) in each Pf lysate sample with MV151. MV151 is an activity-based proteasome probe with a fluorescent vinyl sulfone warhead that irreversibly labels the three catalytic subunits of the parasite proteasome.4 We synthesized and purified MV151 in house, and kept it as 10 mM DMSO stock aliquots at ‒80°C. Before you begin, take out one MV151 aliquot to r.t and set Belly Dancer/Hybridization water bath to heat to 37°C.
-
5.Treat equal amounts of each lysate (Table 1) with MV151 at a final concentration of 2 μM for 1 h at 37°C in a 1.5 mL Eppendorf tube wrapped in aluminum foil.
-
a.Label 1.5 mL Eppendorf tubes and add 12 μL of labeling reaction buffer containing 10 μg of total lysate in each group.
-
b.Dilute the 10 mM MV151 stock to 10 μM (5X MV151) solution as follows:[2 μL] of 10 mM MV151 stock + [18 μL] of DMSO = [20 μL] of 1000 μM MV151 stock (100% DMSO).[2 μL] of 1000 μM MV151 stock + [198 μL] of labeling reaction buffer = [200 μL] of 10 μM MV151 solution (5X MV151, 1% DMSO).Note: Prepare fresh 5X MV151 in a brown/black tube, use immediately after preparation.
-
c.Add 3 μL of 5X MV151 to each tube and mix (Final con. of MV151 at 2 μM, and 0.2% DMSO in the reaction system).
-
d.Spin down at 94 × g for 30 s.
-
e.Wrap the tubes with aluminum foil and incubate at 37°C in a Belly Dancer/Hybridization water bath for 1 h.
-
f.After incubation, spin down at 94 × g for 30 s.
-
g.Place each tube in a tube rack at r.t. and proceed to the next step immediately.
-
a.
Protein electrophoresis
Timing: 3–4 h
This section details how to perform gel electrophoresis of the lysate samples labeled by MV151. Before you begin, thaw a 4X SDS loading buffer aliquot and prepare sufficient MOPS SDS running buffer (1X).
-
6.Heat the lysate with 4X SDS loading buffer at 95°C for 10 min, run samples on 12% Novex Bis-Tris Protein Gels with MOPS SDS running buffer.
-
a.Add 5 μL of 4X SDS loading buffer to each tube, mix and spin down at 94 × g for 30 s.
-
b.Heat the samples at 95°C in a heat block for 10 min.
-
c.Remove the samples from the heat block, cool to r.t. and spin down at 94 × g for 30 s.
-
d.Load the samples onto a 12% Novex Bis-Tris Protein Gel, and add 5 μL of Precision Plus Protein Western Standards in one empty lane to monitor electrophoretic separation.
 CRITICAL: Based on our experience, 12% Novex Bis-Tris Protein Gel is best option for separation of 3 active Pf20S subunits.
CRITICAL: Based on our experience, 12% Novex Bis-Tris Protein Gel is best option for separation of 3 active Pf20S subunits. -
e.Electrophorese the gel at 90 V for 20 min, and then 130 V for 100 min in NuPAGE 1X MOPS SDS Running Buffer.
 CRITICAL: To ensure reproducibility, strictly keep the voltage consistent between experiments. Electrophoresis MOPS buffer CAN NOT be re-used.
CRITICAL: To ensure reproducibility, strictly keep the voltage consistent between experiments. Electrophoresis MOPS buffer CAN NOT be re-used. -
f.Carefully remove the gel from its cassette and then gently rinse the gel with ddH2O.
-
g.Keep the gel submerged in ddH2O before imaging in the next step.
-
a.
Fluorescence scanning of the gel
Timing: 1–2 h
This step describes how to scan the gel using a Typhoon Scanner. Protocol steps in this section have been partially modified from the Typhoon User’s Guide V3.0. Please read the Typhoon User’s Guide for detailed instructions before starting.5
-
7.Scan the gel at the TAMRA channel on a Typhoon Scanner by using the Cy3/TAMRA settings (λex 532 nM, λem 560 nM).
-
a.Decontaminate and clean the glass platen and sample lid of the Typhoon scanner.
-
b.In subdued light, place the gel on the glass platen. Make a note of the grid coordinates on the glass platen. Close the sample lid.
-
c.Select scan parameters using the Scanner control software.Note: In the Fluorescence Setup window, select the emission filter (580 BP 30Cy3, TAMRA, Alexa Fluor 546), PMT voltage (700), laser (Green (532)), and sensitivity (Normal) for each scan. Optionally, you can select scans that can be linked. Click OK. In the Scanner Control window, type comments you want saved with the image.
-
d.Click Scan. Type a file name and click Save.Note: During scanning, check the image in the ImageQuant Preview window for saturation. Saturated pixels appear in red. If the image appears saturated, you might need to repeat the scan. If the image appears usable, continue with the next scan. If you are scanning another sample, make sure you clean the glass platen and sample lid before you place the next sample in the instrument.
-
e.Analyze and process the scanned image, send data to a Flash Drive.
-
f.Remove the gel from the Typhoon instrument and clean the instrument.
-
a.
Expected outcomes
This protocol was developed to capture the intracellular transformation of artezomib following the activation of artemisinin moiety of the artezomib molecule. The protocol also takes advantage of the in-situ generated prolonged peptide-based proteasome inhibitors are not diffused out but retaining inside the cells. In contrast, deoxy-artezomib molecules and proteasome inhibitors are washed out in the step-wise washing schedule with an incubation step between the washes. This protocol allows a quantification of inhibition of parasite proteasomes by the novel proteasome inhibitors without the interference of the starting compounds.
Quantification and statistical analysis
We use ImageJ to quantify blot fluorescence signals. Data represent means ± standard error from two or three independent experiments. Statistical analysis was performed using unpaired two-tailed t-tests to compare the DMSO and treatment groups for p-values.
Limitations
The method described here is developed specifically to evaluate the transformation of artezomibs into new proteasome inhibitors inside malaria parasites. We do not know the degree of the transformation of the artezomibs during the 6 h treatment, we chose 6 h as it was used in the ring stage survival assay that was designed to evaluate the artemisinin resistance of parasites with Kelch13 mutations.6 Because of the noncovalent nature of the newly formed proteasome inhibitors, using this labeling method for covalent but reversible proteasome inhibitor based artezomibs would need to be optimized.
Troubleshooting
Problem 1
Residual inhibition of Pf20S labeling in control samples (related to step 2).
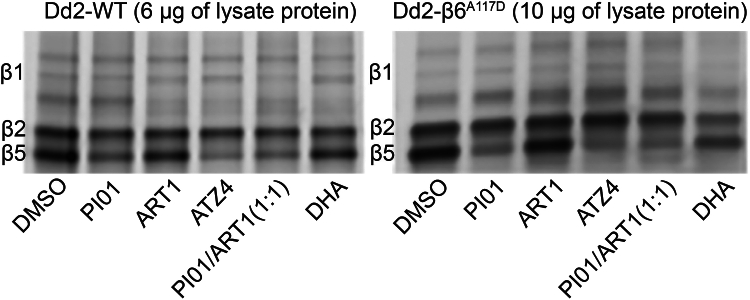
If the compounds in each treated samples were not completed removed, residual inhibition of the Pf20S or Pf20Sβ6A117D labeling will be observed. See examples in Figure 1.
Figure 1.
Fluorescent scanning images of representative SDS-PAGE gels of Pf20S labeling by MV151 of samples from Dd2 (left) and Dd2 β6A117D (right) pellets without wash
Potential solution
Optimization of washout step is strongly recommended.
Problem 2
Weak fluorescence intensity (related to step 4)
MV151 loses fluorescence in aqueous solution even when stored at ‒80°C.
Potential solution
Always makes the MV151 fresh from DMSO stock immediately prior to the labeling.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Gang Lin (gal2005@med.cornell.edu).
Technical contact
WZhan2016@outlook.com.
Materials availability
All new chemical compounds are available on request on condition of stock availability.
Data and code availability
This study did not generate/analyze datasets/code.
Acknowledgments
This work was supported in part by the National Institutes of Health grant R21AI153485, R01AI177635, and R01AI143714 to G.L.
Author contributions
W.Z. wrote the initial draft of the manuscript, created the figures, and participated in the revision. G.L. created graphical abstract and revised and finalized the manuscript. L.A.K. revised and finalized the manuscript.
Declaration of interests
Cornell University’s Center for Technology Licensing has filed a patent application on these artemisinin proteasome inhibitor hybrids. G.L., W.Z., and L.A.K. are listed as inventors.
Contributor Information
Laura A. Kirkman, Email: lak9014@med.cornell.edu.
Gang Lin, Email: gal2005@med.cornell.edu.
References
- 1.Zhan W., Li D., Subramanyaswamy S.B., Liu Y.J., Yang C., Zhang H., Harris J.C., Wang R., Zhu S., Rocha H., et al. Dual-pharmacophore artezomibs hijack the Plasmodium ubiquitin-proteasome system to kill malaria parasites while overcoming drug resistance. Cell Chem. Biol. 2023;30:457–469.e11. doi: 10.1016/j.chembiol.2023.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhan W., Visone J., Ouellette T., Harris J.C., Wang R., Zhang H., Singh P.K., Ginn J., Sukenick G., Wong T.T., et al. Improvement of Asparagine Ethylenediamines as Anti-malarial Plasmodium-Selective Proteasome Inhibitors. J. Med. Chem. 2019;62:6137–6145. doi: 10.1021/acs.jmedchem.9b00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirkman L.A., Zhan W., Visone J., Dziedziech A., Singh P.K., Fan H., Tong X., Bruzual I., Hara R., Kawasaki M., et al. Antimalarial proteasome inhibitor reveals collateral sensitivity from intersubunit interactions and fitness cost of resistance. Proc. Natl. Acad. Sci. USA. 2018;115:E6863–E6870. doi: 10.1073/pnas.1806109115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdoes M., Florea B.I., Menendez-Benito V., Maynard C.J., Witte M.D., Van der Linden W.A., Van den Nieuwendijk A.M.C.H., Hofmann T., Berkers C.R., van Leeuwen F.W.B., et al. A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem. Biol. 2006;13:1217–1226. doi: 10.1016/J.Chembiol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Typhoon User’s Guide v3.0.
- 6.Witkowski B., Amaratunga C., Khim N., Sreng S., Chim P., Kim S., Lim P., Mao S., Sopha C., Sam B., et al. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet. Infect. Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate/analyze datasets/code.