Abstract
The 2μm circle plasmid confers no phenotype in wild-type Saccharomyces cerevisiae but in a nib1 mutant, an elevated plasmid copy number is associated with cell death. Complementation was used to identify nib1 as a mutant allele of the ULP1 gene that encodes a protease required for removal of a ubiquitin-like protein, Smt3/SUMO, from protein substrates. The nib1 mutation replaces conserved tryptophan 490 with leucine in the protease domain of Ulp1. Complete deletion of ULP1 is lethal, even in a strain that lacks the 2μm circle. Partial deletion of ULP1, like the nib1 mutation, results in clonal variations in plasmid copy number. In addition, a subset of these mutant cells produces lineages in which all cells have reduced proliferative capacity, and this phenotype is dependent upon the presence of the 2μm circle. Segregation of the 2μm circle requires two plasmid-encoded proteins, Rep1 and Rep2, which were found to colocalize with Ulp1 protein in the nucleus and interact with Smt3 in a two-hybrid assay. These associations and the observation of missegregation of a fluorescently tagged 2μm circle reporter plasmid in a subset of ulp1 mutant cells suggest that Smt3 modification plays a role in both plasmid copy number control and segregation.
The Saccharomyces cerevisiae 2μm circle is a rare example of a eukaryotic plasmid and has been exploited by molecular biologists as the basis for most high-copy-number yeast vectors (14). Replication of the 2μm circle is coordinately regulated with replication of the host chromosomal DNA, and utilizes the host cell enzymatic machinery, whereas segregation of the plasmid requires the concerted action of the 2μm-encoded Rep1 and Rep2 proteins and a cis-acting stability locus, STB, that bears no resemblance to well-characterized yeast chromosome centromeric sequences (6, 14). The 2μm circle also encodes a site-specific recombinase, Flp, that recognizes inverted repeat target sites (FRT) within the plasmid and can increase copy number if it falls below normal levels (approximately 60 copies per haploid cell) (49). Futcher (15) has proposed that Flp-mediated recombination during DNA replication inverts one of the replication forks, thereby allowing multimeric forms of the plasmid to be formed by a rolling circle mechanism from a single firing of the replication origin. Subsequent action of the Flp recombinase would allow replication to terminate and resolve the multimers into multiple monomeric copies of the plasmid. In support of this model, overexpression of the FLP gene leads to uncontrolled plasmid replication, which in turn causes longer cell cycles and cell death, while combined overexpression of the Rep1 and Rep2 proteins can repress the FLP gene and may limit amplification of the plasmid once it has reached its normal copy number (35, 39).
A mutation in the chromosomal NIB1 gene, nib1, leads to elevated plasmid copy numbers, which suggests that plasmid levels may also be regulated by the host (17). We report here that NIB1 is the ULP1 gene. The Ulp1 protein is a protease specific for removal of a ubiquitin-like protein, Smt3/SUMO, from other proteins to which it is covalently attached as a posttranslational modification (21, 23, 27, 28, 43). Ulp1 also converts the Smt3/SUMO primary translation product to the mature form that is conjugated to target proteins (23). A second yeast Smt3-specific deconjugating enzyme, Ulp2/Smt4, does not perform this essential processing function (24).
Smt3/SUMO modification apparently serves different functions depending on the substrate protein, either protecting the protein from ubiquitin-mediated proteolysis as in the case of IκBα (8) or targeting the protein to a particular subcellular location. For example, SUMO modification of Ran GTPase-activating protein (RanGAP1) targets the otherwise cytosolic protein to the nuclear pore complex during interphase and to the spindle apparatus during mitosis (28). Modification of PML (acute promyelocytic leukemia) protein is required for its localization to discrete matrix-associated subnuclear domains, termed PML nuclear bodies (5, 11, 34).
Numerous SUMO-conjugated proteins have recently been identified and are involved in diverse cellular processes that include transcription, chromatin remodeling, DNA replication and repair, metabolic processes, chromosome segregation, and cellular responses to DNA damage and stress (19, 36, 50, 53). Different Smt3/SUMO-modified conjugates are observed at different cell cycle stages, suggesting that cycling between conjugated and unmodified forms of target proteins is cell cycle regulated (20, 23). Disruption of the UBC9 gene encoding the Smt3/SUMO-specific conjugating enzyme, Ubc9, leads to a G2/M-phase cell cycle arrest (45), as does deletion of the ULP1 gene (23), indicating that modulating Smt3/SUMO modification of target proteins is essential for normal cell cycle progression. We report here that in yeast with a partial deletion of the ULP1 gene, as in nib1 mutants, 2μm plasmid levels are elevated, implicating Ulp1-mediated Smt3 deconjugation in plasmid copy number control. Furthermore, a subset of cells produces lineages in which all cells cease proliferation. The colocalization of Rep1 and Rep2 with Ulp1 in the nucleus, the interaction of both plasmid proteins with Smt3 in a two-hybrid genetic assay, and missegregation of a fluorescently tagged 2μm circle reporter plasmid in a subset of ulp1 mutant cells suggest that Smt3 modification is involved in plasmid segregation.
MATERIALS AND METHODS
Strains and media.
Yeast strains used in this study are given in Table 1. Strains lacking the 2μm circle, designated [cir0], were derived from strains containing the 2μm circle, [cir+], either by first driving out the native 2μm circle with a selectable 2μm-based plasmid and then screening for cells that had lost the hybrid plasmid (9) or by expression of a defective Flp recombinase from plasmid pBIS-GALkFLP-(TRP1) (47). Standard methods were used for growth and manipulation of yeast and bacteria (40, 42). Yeasts were grown in synthetic defined (SD) medium (0.67% Difco yeast nitrogen base without amino acids, 2% glucose, 0.003% adenine, 0.002% uracil, and all required amino acids), or in YPD medium (1% yeast extract, 2% Bacto Peptone, 2% glucose) (40).
TABLE 1.
Yeast strains used in this study
| Name | Relevant genotypea | Reference or parental strain |
|---|---|---|
| CH568 | MATα ade leu2 NIB1 [cir0] | 17 |
| CH569 | MATaade1 leu2 cyh2 nib1 [cir+] | 17 |
| W303a/α | MATa/α ade2-1/ade2-1 ura3-1/ura3-1 leu2-3,112/leu2-3,112 his3-11,15/his3-11,15 trp1-1/trp1-1 ULP1/ULP1 [cir+] | 41 |
| W303/1a | MATaade2-1 ura3-1 leu2-3,112 his3-11,15 trp1-1 ULP1 [cir+] | 41 |
| MD83a/α | MATa/α ulp1ΔES::URA3/ULP1 | W303a/α |
| MD85a/α | MATa/α ulp1Δ::URA3/ULP1 | W303a/α |
| MD83/1a | MATα ulp1ΔES::URA3 [cir0] | MD83 [cir0] |
| MD83/1d | MATaulp1ΔES::URA3 [cir0] | MD83 [cir0] |
| MD83/1c | MATα ULP1 [cir0] | MD83 [cir0] |
| MD83/1b | MATaULP1 [cir0] | MD83 [cir0] |
| MD83/7a | MATα ULP1 [cir+] | MD83 [cir+] |
| MD83/7b | MATaulp1ΔES::URA3 [cir+] | MD83 [cir+] |
| MD83/33 | MATα ULP1 GFP-LacI::HIS3 [cir+] | MD83/7a |
| MD83/34 | MATaulp1ΔES::URA3 GFP-LacI::HIS3 [cir+] | MD83/7b |
| ML83/4 | MATaulp1ΔES::LEU2 GFP-LacI::HIS3 [cir+] | ML83/10a |
| ML83a/α | MATa/α ulp1ΔES::LEU2/ULP1 | W303a/α |
| ML83/10a | MATaulp1ΔES::LEU2 [cir+] | ML83a/α [cir+] |
| AS2 | MATα his3Δ1 ura3-52 leu2-3,112 trp1-289 ade2ΔULP1 | 44 |
| MD82 | MATα his3Δ1 ura3-52 leu2-3,112 trp1-289 ade2Δ ulp1ΔES::URA3 | AS2 |
| CTY10/5d | MATagal4 gal80 his3-200 trp1-901 ade2 ura3-52 leu2-3,112 URA3::(lexAop)8-lacZ) [cir+] | 3 |
Both [cir0] and [cir+] versions of strains were created unless only one type is indicated.
DNA manipulations.
A 5,130-bp BamHI restriction fragment containing the YPL020C/ULP1 gene and YPL019C (an uncharacterized open reading frame [ORF] flanking ULP1) was amplified by PCR from CH568 genomic DNA with the primers LPB1 (5′-CGGGATCCATATCCAATGATCTG-3′) and LPB2 (5′-CGCGGATCCAACTTTCGAGTGGG-3′) (details available on request) and cloned into the BamHI site in the CEN/ARS/LEU2 yeast/Escherichia coli shuttle vector pRS315 (46). Subsequently, the 2,872-bp BamHI/PstI fragment was subcloned into BamHI/PstI doubly digested pRS315 to give pRS-ULP1. Complementation was determined by transforming nib1 [cir+] strain CH569 to leucine prototrophy at the permissive temperature of 37°C and then scoring colony morphology of transformants after growth at 30°C. Deletions of the chromosomal ULP1 gene were carried out by transforming yeast with an HpaI/BamHI restriction fragment from pRS-ULP1 in which either the EcoRV/SalI fragment encoding the first 479 bp (ulp1ΔES) or an EcoRV/AvaII encoding the first 1,605 bp (ulp1Δ) of the 1,866-bp ULP1 ORF were replaced by standard DNA manipulation techniques with a SmaI/SalI fragment encoding the yeast URA3 gene. The URA3 gene was replaced by the LEU2 gene with a linearized marker swap plasmid as described by Cross (7). Gene deletions were verified by PCR using the above primers and by using a PCR amplicon of the ULP1 ORF generated with primers LPB1 and LPB3 (LPB3, 5′-CGGAATTCATAATAATGTCAGTTG-3′) as a radiolabeled probe in a Southern blot analysis of genomic DNA isolated from parent and transformed strains (42).
For two-hybrid analyses, the YPL020C ORF was PCR amplified using primers LPB1 and LPB3, digested with EcoRI and BamHI, and cloned in EcoRI/BamHI-digested pGAD424 (Clontech) to give pGAD-ULP1. The SMT3 ORF was amplified using primers 5′-CAGGATCCCGATGTCGGACTCA-3′ and 5′-AACTGCAGCTAACCACCAATCTGTTCTC-3′, digested with BamHI and PstI, and cloned in BamHI/PstI-digested pGAD424 to give pGAD-SMT3. In yeast, this plasmid directs expression of an in-frame fusion of the Gal4 transcription activation domain (Gal4AD) with the mature form of Smt3 lacking the three carboxy-terminal amino acids of the precursor Smt3 primary translation product (23). Two-hybrid plasmids directing expression of LexA-Rep fusion proteins have been previously described (44).
Plasmid copy number.
For Southern blot determination of plasmid copy number, total DNA was extracted from yeast growing logarithmically in SD medium as previously described (10), digested with EcoRI, resolved by electrophoresis on 1% agarose Tris-borate-EDTA gels, and transferred to nylon membranes. A genomic 1.45-kb EcoRI fragment encoding the TRP1 gene and a DNA fragment encoding the REP1 ORF were 32P labeled by random priming (41) and hybridized with the membranes. Signals were quantified using a Bio-Rad phosphorimager, and the copy number of the 2μm plasmid was determined by normalizing the 2μm signal to the TRP1 signal obtained from the same DNA sample. For determination of plasmid copy number relative to chromosomal content in colonies, a 2μm plasmid sequence and a chromosomal locus were amplified by PCR using primers 5′-CCTTAACGGACCTACAG-3′ and 5′-CGGGATCCTCGCATCCCCGGTT-3′ and primers 5′-CTCTGGATCCGAATGAGTAAGCGGGGTAG-3′ and 5′-CACCGGATCCGTAAAAGAACTTCTCCTC-3′, respectively, from serial dilutions of total DNA isolated by glass bead breakage from suspensions of cells from colonies of different sizes and morphologies (40). Amplicon yields were determined by densitometry of ethidium bromide-stained agarose gels on which the products had been resolved with Molecular Analyst software from Bio-Rad.
Nuclear staining.
Yeast cells growing logarithmically in YPD medium were harvested, washed in sterile phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), and fixed in 70% ethanol for 30 min at room temperature. Cells were then washed twice in PBS, stained with 0.001% DAPI (4′,6′-diamidino-2-phenylindole) (Sigma) in PBS for 15 min, washed again in PBS, and lightly sonicated to resolve clumping before being mounted on glass slides and photographed with a Leitz Laborlux visible UV-fluorescence microscope. All cells in randomly selected fields were scored for morphology and nuclear staining as described by Holm (17). For fluorescence-activated cell sorting, yeasts were fixed and stained with propidium iodide as described by Rose et al. (40). Cell cycle distribution was analyzed using Modfit LT for Mac V3.0 software. Images were processed with Photoshop 5.5.
Immunofluorescence.
The Ulp1 protein was Myc13 tagged at the carboxy terminus by homologous integration at the chromosomal ULP1 locus of a Myc13-TRP1 cassette (26). Yeast grown to mid-log phase were fixed, spheroplasted, and prepared for immunofluorescence staining essentially as previously described (38). Blocking was done using 1 mg ml−1 bovine serum albumin for 15 min. All primary and secondary antibodies were diluted in PBS containing 1 mg ml−1 bovine serum albumin and 0.02% sodium azide. Incubations with primary and secondary antibodies were done at room temperature for 60 and 30 min, respectively, and visualized with a Leica confocal system, TCS4D. Rep1 and Rep2 polyclonal antisera have been described previously (44).
Rep protein and plasmid localization.
Green fluorescent protein (GFP)-Rep fusion proteins were expressed under the control of the GAL10 promoter from the CEN/ARS plasmids pTS408-Rep1 and pTS408-REP2, respectively, as previously described (1). The TRP1-marked 2μm reporter plasmid, pSV5, containing the 2μm circle origin of replication, STB locus, and 256 repeats of the lac operator sequence (30) was visualized by expressing GFP-LacI repressor fusion protein in yeast as previously described (48). Cells were observed with a Zeiss Axiovert 200 fluorescence microscope (see Fig. 7A to C) and a Nikon inverted microscope with recommended excitation and emission filters for DAPI and GFP (see Fig. 7D and E). Images were captured with an Axiocam HRc camera and Axiovision 3.1 software or with a Photometrics Quantix camera and Universal imaging Corp. Metamorph software and analyzed using Adobe Photoshop 5.5.
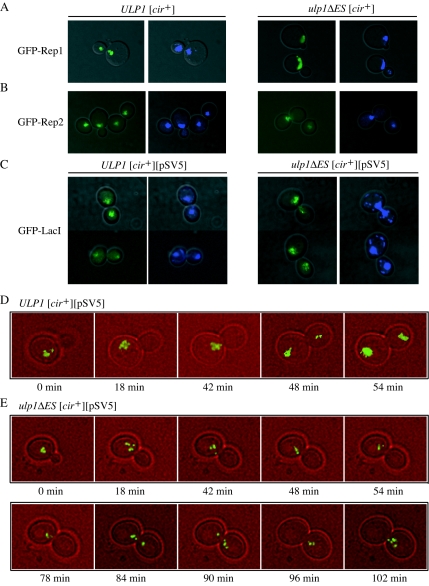
FIG. 7.
Localization of Rep proteins and fluorescently tagged 2μm reporter plasmid in [cir+] ULP1 and ulp1ΔES yeast strains. GFP-Rep1 (A), GFP-Rep2 (B), or GFP-LacI (C) fusion proteins were expressed in [cir+] ULP1 and ulp1ΔES yeast strains. The yeasts shown in panel C were also transformed with the 2μm-derived reporter plasmid pSV5, containing lac operator repeats. Merged bright-field-GFP (left) and merged bright-field-DAPI (right) fluorescence images of representative cells are shown. [cir+] ULP1 (D) and [cir+] ulp1ΔES (E) yeast cells expressing GFP-LacI and transformed with pSV5 were analyzed by time-lapse fluorescence microscopy. Bright-field and GFP fluorescence images of a single cell for each strain were taken every 6 min with the small-budded stage taken as the starting time point. Merged bright-field-GFP images are shown for all time points during the period when plasmids moved from mother to daughter cells and from representative earlier and subsequent times.
Two-hybrid assays.
Transformants and cotransformants in the two-hybrid yeast host CTY10/5d (3) were assayed for β-galactosidase expression by a permeabilized cell assay (40). Specificity of interactions was confirmed by coexpressing LexA-Rep fusion proteins with Gal4AD-Snf4 fusion proteins (12) and Gal4AD-Smt3 fusion proteins with unfused LexA.
RESULTS
NIB1 is the ULP1 gene.
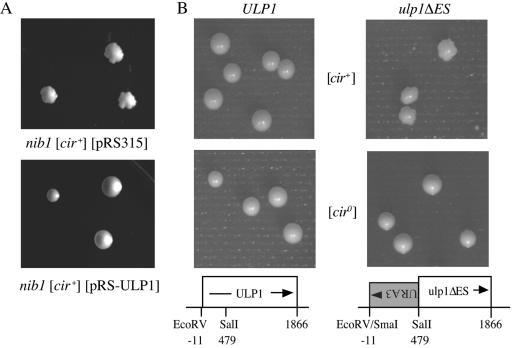
Studies of yeast carrying the recessive nib1 mutation suggest that host-encoded proteins may play a critical role in regulating the 2μm circle copy number. The nib1 mutation confers a “nibbled” colony morphology only in [cir+] yeast strains (Fig. 1A), while nib1 [cir0] strains and wild type NIB1 [cir+] yeast form smooth colonies (17). The nibbled morphology arises due to the production of lineages within the colony in which the 2μm circle copy number is more than double normal levels (17). All further cells in that lineage eventually become mortal, arresting with a single large bud, a phenotype associated with yeast cell cycle mutants blocked in some aspect of DNA replication or mitosis (16). The dead lineages are observed as “lethal sectors” in the growing colony. This nibbled phenotype was used to map the NIB1 gene to the left arm of chromosome XVI, tightly linked to the RAD1 gene (18). To isolate the NIB1 gene, stretches of DNA that contained ORFs in the vicinity of RAD1 were identified from the complete yeast genome sequence (Saccharomyces Genome Database; http://www.yeastgenome.org/). Selected ORFs with their flanking sequences were amplified by PCR using primers based on the published sequence and cloned in a single-copy yeast LEU2/ARS/CEN vector, pRS315 (46). The resulting clones, containing the YPL026C/STS1, YPL022W/RAD1, YPL018W/CTF19, YPL019C and YPL020C/ULP1 genes, respectively, were tested for their ability to complement the lethal sectoring phenotype when transformed into a nib1 [cir+] yeast strain, CH569 (18). Only plasmids containing the ULP1 gene were able to restore normal smooth-colony morphology to the nib1 strain (Fig. 1A).
FIG. 1.
Colony morphology of wild-type and nib1 mutant yeasts. (A) Complementation of the nib1 “nibbled” colony morphology by the ULP1 gene. The nib1 [cir+] yeast strain CH569 was transformed with either the vector pRS315 or the plasmid pRS-ULP1, and the transformants were grown on solid SD medium lacking leucine at 30°C for 5 days. (B) Congenic haploid [cir+] and [cir0] yeast strains with either the wild-type ULP1 or mutant ulp1ΔES allele were grown on solid YPD medium at 30°C for 5 days.
Mutations in ULP1 confer a 2μm-dependent nibbled colony phenotype.
Deletion of the entire ULP1 ORF in either a [cir+] or [cir0] haploid yeast strain results in nonviability (23 and data not shown). Viable transformants were obtained in both [cir+] and [cir0] haploid yeast strains when only the first 479 bp of the 1,866-bp genomic copy of the ULP1 ORF, including the initiating methionine codon, were replaced with the URA3 gene running in an antisense orientation (Fig. 1B). Although the [cir+] transformants with this partial deletion of the ULP1 gene (hereafter referred to as the ulp1ΔES allele) were viable, the colonies had a nibbled morphology, similar to that of the CH569 [cir+] nib mutants (Fig. 1A) (18), whereas the [cir0] ulp1ΔES yeast had normal colony morphology (Fig. 1B). This dependence of the lethal sectoring phenotype on the presence of the 2μm plasmid and the ability of the wild-type YPL020C ORF to complement the nib1 defect support the identification of ULP1 as the NIB1 gene. Sequencing of the ULP1 ORF from the original CH569 [cir+] nib1 mutant yeast strain revealed a single base change from the wild-type sequence, a G-to-T transversion at position 1469. This mutation would result in a nonconservative substitution of a leucine for a tryptophan residue at position 490 in the Ulp1 protein, a mutation previously reported as conferring a temperature-sensitive phenotype in yeast (33).
Both [cir0] and [cir+] ulp1ΔES yeast mutants lag at G2/M phase of the cell cycle.
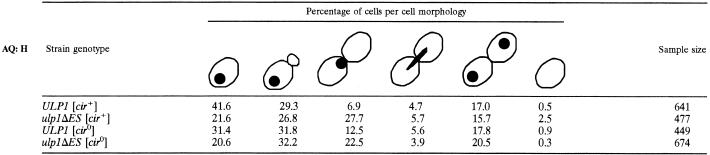
To further characterize the defect conferred by the ulp1ΔES allele, the nuclear and cellular morphology of [cir+] and [cir0] yeast carrying either the mutant or wild-type ULP1 gene was examined (Table 2). In a logarithmically growing culture, 28% of [cir+] ulp1ΔES mutant cells had a large bud and a single nucleus at the bud neck, compared to 7% for the congenic [cir+] ULP1 strain. These results are similar to those previously reported for [cir+] nib1 mutant yeast (17) and support the involvement of the encoded protein in cell cycle progression (23). Surprisingly, the [cir0] ulp1ΔES yeast mutant also showed a distribution of cellular morphologies similar to that obtained with the [cir+] ulp1ΔES mutant, despite the apparent lack of effect of this mutation on colony morphology (Fig. 1B). The results suggest that both [cir+] and [cir0] ulp1ΔES cells are delayed either in DNA replication or in the G2/M stage in the cell cycle. The presence of the 2μm circle seems to exacerbate the defect, with a higher percentage of anucleate cells being observed with the [cir+] ulp1ΔES culture than with the others. These anucleate cells suggest cytokinesis may have occurred despite mitosis not having been properly executed. The [cir+] ulp1ΔES cells were often enlarged, had elongated buds, and displayed an apical chain-like budding pattern, phenotypes similar to those produced by overexpression of FLP in a [cir+] yeast strain (35).
TABLE 2.
Cell morphology and nuclear staining of ULP1 and ulp1ΔES yeasta
Yeast were harvested during logarithmic growth and fixed, and the DNA was stained with DAPI before being observed microscopically under UV illumination. The percentage of cells of a given morphology are shown (from left to right): mononucleate, small budded, large budded, mitotic, binucleate budded, or anucleate. The sample size is given.
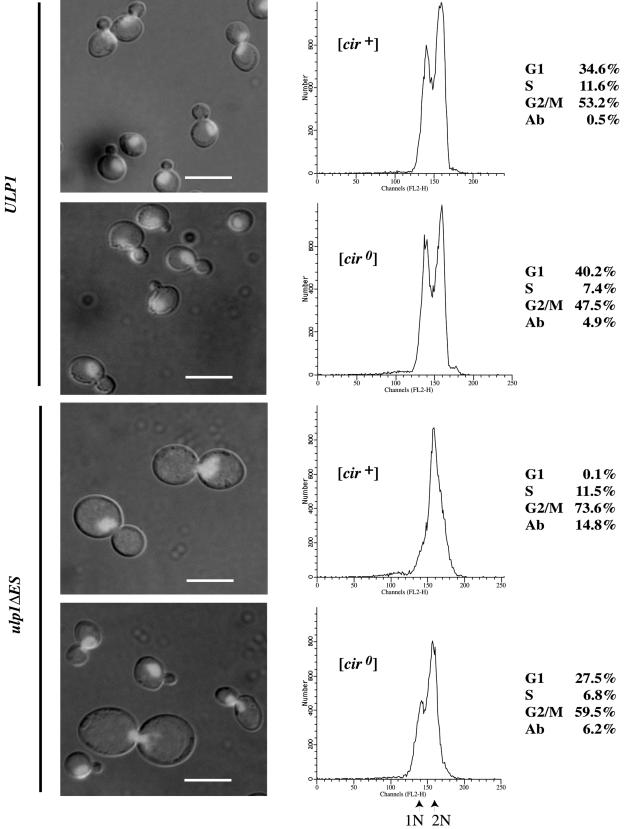
To establish whether the cell cycle lag observed with the ulp1ΔES yeast was due to a delay in DNA replication or was at the G2/M boundary, the cells were analyzed by flow cytometry (Fig. 2). The [cir+] and [cir0] ULP1 cells both displayed the distribution expected for a population of unsynchronized wild-type cells, with approximately half the cells having the 2N DNA content of G2, 40% having the 1N DNA content of G1, and the remainder undergoing DNA replication (S). In contrast, a higher percentage of the population for both [cir0] and [cir+] ulp1ΔES yeast mutants had a 2N DNA content, consistent with the cell cycle delay at the G2/M boundary. This was most pronounced for the [cir+] ulp1ΔES yeast strain where almost no cells with a 1N DNA content were observed. In addition, a significant proportion of the [cir+] ulp1ΔES yeast cells had a DNA content higher than expected for a 2N chromosome complement.
FIG. 2.
G2/M lag is exacerbated in [cir+] ulp1ΔES yeast. Unsynchronized cultures of congenic ULP1 and ulp1ΔES yeast, [cir+] and [cir0], were grown to early log phase in YPD medium, and the cells were fixed, stained with propidium iodide, visualized by fluorescence microscopy (left), and analyzed by flow cytometry (center), with the percentage of cells at each phase of the cell cycle and the percentage of aberrant cells (Ab), those with DNA content greater than 2N or less than 1N, shown at right. Scale bar, 5 μm.
Yeasts with partial deletion of ULP1 have elevated 2μm plasmid levels.
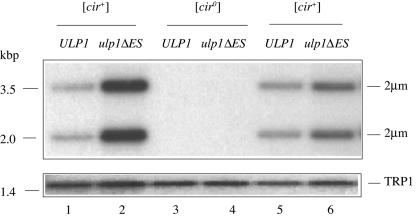
Consistent with the greater than diploid DNA content observed by flow cytometry, [cir+] yeast cells carrying the ulp1ΔES allele were found by Southern blotting to have a two- to fivefold amplification of the 2μm plasmid copy number relative to their parental ULP1 strain (Fig. 3 and data not shown). This amplification is similar to that previously observed with [cir+] nib1 mutants (17). Copy numbers for independent cultures of a given transformant varied. In addition, smooth colonies occasionally arose spontaneously in the [cir+] ulp1ΔES strain and were found by Southern blotting to have become [cir0] (data not shown). This clonal variability in plasmid copy number suggests a stochastic process in which some cells acquire either higher- or lower-than-average numbers of plasmid molecules at cellular division and that the frequency of these events may be increased in ulp1ΔES yeast.
FIG. 3.
Amplification of 2μm circle copy number in ulp1ΔES yeast. Southern blot analysis of EcoRI-restricted genomic DNA isolated from the indicated yeast strains. Phosphorimages of the blots after sequential hybridization with radiolabeled probes recognizing either the genomic TRP1 locus or the 2μm circle (the 2μm circle exists in equimolar amounts of two forms, giving the bands detected with this probe) are shown. TRP1-probed images are longer exposures to allow the weaker signal from this single-copy genomic probe to be visualized.
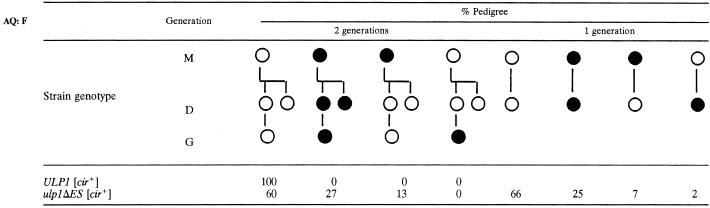
Commitment to cease proliferation is inherited.
What remains unexplained is why the presence of the 2μm circle in ulp1ΔES yeast results in clonal lethality of a subpopulation of the cells, observed as a nibbled colony morphology. Lethal sectors would not be produced if random cells in the colony arrested as a consequence of the mutation. Proliferation of neighboring cells would obscure their loss. Instead, cells that produce lethal sectors must continue to proliferate after they have become committed to death, pass their commitment on to their progeny, and then die during a later cell division. To test this hypothesis, individual [cir+] ulp1ΔES and ULP1 cells were assessed by a pedigree assay for their colony-forming ability (Table 3). As expected, all cells in the completed [cir+] ULP1 pedigrees produced normal large smooth colonies. Similarly, for over half of all pedigrees completed for the [cir+] ulp1ΔES mutant, all cells within the pedigree produced large but (in this case) variably nibbled colonies. In contrast, for about one-quarter of the completed [cir+] ulp1ΔES mutant pedigrees, all cells produced microcolonies of <100 cells. In the latter pedigrees, the results suggest that the initial mother cell had already become committed to cease proliferation and that daughter and the granddaughter cells inherited the commitment. Commitment might reflect an elevated plasmid copy number in these mother cells.
TABLE 3.
Pedigree analysis of colony-forming ability of ULP1 and ulp1ΔES [cir+] yeasta
Daughter buds were removed from small-budded mother cells (M) for the first generation (D) and from the first daughter cell for the second generation (G) and each moved to a new position on a YPD plate by micromanipulation. The ability of cells to form colonies was scored after 3 days of incubation at 30°C. Cells were scored as having proliferative capacity (open circles) if the colony formed was visible to the naked eye or lacking proliferative capacity (closed circles) if there were <100 cells in colony. The percentages of pedigrees of the indicated viabilities are shown. Total numbers of pedigrees were 6 and 15 of two generations for the wild type and mutant, respectively, and 59 of one generation for the mutant. Pedigrees in which any of the cells failed to form a bud after micromanipulation were excluded.
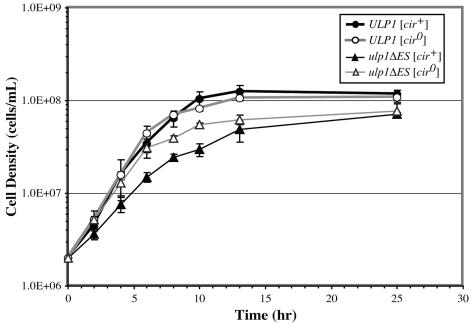
The 2μm circle exacerbates the effect of the ulp1ΔES mutation on growth.
To further investigate the basis of the ulp1ΔES defect, we compared the rate of growth of [cir0] and [cir+] ulp1ΔES strains to ULP1 yeast in liquid culture (Fig. 4). Both the [cir0] and [cir+] ulp1ΔES strains underwent a diauxic shift and entered stationary phase at a lower cell density than their respective ULP1 strains. Since the average size of a ulp1ΔES cell is greater than that of a ULP1 cell (Fig. 2), the lower cell density at diauxic shift may result from the larger mutant cells, exhausting the glucose at a lower cell density, irrespective of their plasmid content. In addition to an earlier entry into stationary phase, the [cir+] ulp1ΔES strain grew significantly slower during the logarithmic phase than either the [cir0] or [cir+] ULP1 strains or the [cir0] ulp1ΔES mutant strain. This growth difference is consistent with the [cir+] ulp1ΔES strain having a colony morphology that differs from that of the others. The reduced rate of logarithmic growth in the [cir+] ulp1ΔES yeast may represent the production of cells that constitute the lethal sectors in colonies formed by these strains.
FIG. 4.
The 2μm plasmid impairs growth of ulp1ΔES yeast. Yeast cells were grown to stationary phase in liquid SD medium, diluted into fresh medium, and incubated with shaking at 30°C. Initial and subsequent cell densities at the indicated times after dilution were measured by hemocytometer counts. Results are the averages with standard deviations for four cultures of each strain.
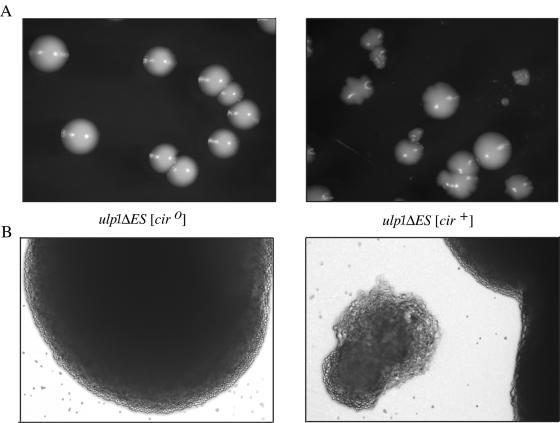
Plasmid copy number varies with colony size and morphology.
Replating of [cir+] ulp1ΔES yeast cells obtained from a nibbled colony results in a variety of different colony sizes ranging from large normal-sized colonies to microcolonies of <100 cells and colony morphologies ranging from smooth to extensively nibbled (Fig. 5A). Cell size and shape are also more variable for the [cir+] ulp1ΔES yeast, even for cells within a single colony (Fig. 5B). To determine whether the different-sized and shaped colonies observed with [cir+] ulp1ΔES yeast reflected differences in plasmid levels within the cells of the colony, a semiquantitative PCR approach was undertaken (Table 4). For the DNA isolated from nibbled ulp1ΔES mutant colonies, 2μm plasmid levels showed a trend of increasing with decreasing colony size relative to the level of 2μm plasmid in a ULP1 strain. The smooth ulp1ΔES mutant colonies had only a twofold increase in 2μm plasmid levels. This less-dramatic increase in plasmid copy number might explain the loss of the nibbled colony phenotype. The correlation between the increase in 2μm plasmid levels and the loss of proliferative capacity, as reflected in reduced colony size, supports the hypothesis that elevated plasmid levels lead to cell death in ulp1ΔES yeast. Consistent with this hypothesis is the observation that [cir0] ulp1ΔES yeast does not produce nibbled colonies or microcolonies.
FIG. 5.
Variable cell and colony size and morphology in [cir+] ulp1ΔES yeast. (A) [cir0] and [cir+] ulp1ΔES yeast mutants were grown on solid SD medium for 5 days. Colony size and morphology varied only for the [cir+] strain. Several large- and medium-sized nibbled colonies, one large smooth colony, and a small nibbled colony are shown. (B) Bright-field images (magnification, ×100) are of a [cir0] ulp1ΔES colony (left) and [cir+] ulp1ΔES yeast colonies (right), with one small nibbled colony adjacent to a large nibbled colony, from the respective plates above in panel A.
TABLE 4.
Variation in plasmid levels in colonies with different morphologies
| Strain genotype | Plasmid | Colony size, morphology | Fold increase in plasmid-to- chromosome ratio relative to 2μm in ULP1 straina |
|---|---|---|---|
| ULP1 | 2μm | Large, smooth | 1 |
| ulp1ΔES | 2μm | Large, smooth | 2 |
| Large, nibbled | 17 | ||
| Medium, nibbled | 19 | ||
| Small, nibbled | 27 |
Plasmid and chromosomal targets were amplified by PCR from serial dilutions of total DNA isolated from the indicated strains and yields determined by agarose gel analysis and densitometry. The ratio of plasmid to chromosome amplicon yields obtained from limiting amounts of target DNA was normalized relative to the ratio of 2μm plasmid to chromosome amplicons obtained from ULP1 [cir+] strain. Colonies representative of different sizes and/or morphologies from which DNA was obtained are shown in Fig. 5A.
Ulp1 colocalizes in the nucleus with the 2μm circle-encoded Rep proteins.
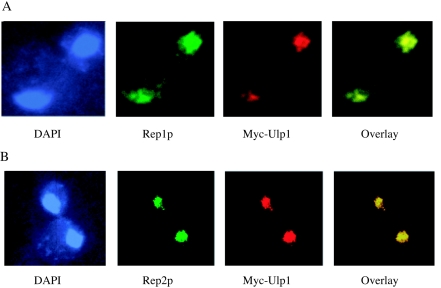
The possible role of the ULP1 gene in the interplay between the physiology of the nuclear resident plasmid and the cell cycle of its host led us to compare the intracellular distribution of the Ulp1 protein and the 2μm circle-encoded Rep1 and Rep2 proteins. Immunofluorescence staining of the Ulp1 protein tagged with the Myc epitope and expressed from the native ULP1 promoter showed that nearly all of the protein was internal to but not uniformly distributed within the nucleus (Fig. 6). This is in agreement with previous data showing that Ulp1 concentrated in the nuclear periphery-nuclear envelope region (24). Quite strikingly, there was near-perfect colocalization between the Ulp1 protein and the 2μm circle-encoded Rep proteins, with the Rep2 distribution in particular being indistinguishable from that of Ulp1.
FIG. 6.
In situ localization of the Ulp1 and Rep proteins. The Ulp1 protein carrying a carboxy-Myc13 tag was expressed from the native promoter at the ULP1 chromosomal locus. A Myc-specific monoclonal antibody conjugated to rhodamine was used to localize the protein while DNA was stained with DAPI. Native Rep1 (A) and native Rep2 (B) proteins were detected with Rep protein-specific primary polyclonal antibodies and visualized with secondary antibodies conjugated to FITC.
In vivo interaction of 2μm-encoded Rep proteins with Smt3.
The apparent colocalization of the 2μm circle-encoded Rep proteins with the Ulp1 protease suggested there might be a direct interaction between these proteins in the cell. A two-hybrid assay was used to test for this association (3). No interaction was detected between the Rep proteins and Ulp1 (data not shown), but both Rep1 and Rep2 gave specific interactions with Smt3, the ubiquitin-like protein removed from target proteins by the Ulp1 protease (Table 5) (23). The interaction suggests that the 2μm Rep proteins may either be modified by Smt3 or associate with the Smt3-modified protein(s).
TABLE 5.
Two-hybrid interaction of Smt3 with Rep1 and Rep2
| Gal4AD | LexA | U of β-galactosidase activity (10−4) |
|---|---|---|
| Smt3 | LexA | 0.2 ± 0.2 |
| Smt3 | Rep1 | 475 ± 123 |
| Smt3 | Rep2 | 194 ± 75 |
| Snf4 | Rep1 | 2.3 ± 0.7 |
| Snf4 | Rep2 | 1.1 ± 0.4 |
β-Galactosidase activities represent the average and standard deviation for three independent yeast transformants coexpressing the Gal4AD and LexA fusion proteins shown on the left.
Rep protein localization in ulp1ΔES yeast.
The Rep proteins have been shown to colocalize with the 2μm circle in the nucleus (48). To determine whether this localization was affected by the ulp1ΔES mutation, GFP-tagged versions of Rep1 (Fig. 7A) and Rep2 (Fig. 7B) were expressed in yeast. Both Rep fusion proteins colocalized with the DAPI-stained chromosomal DNA in the ulp1ΔES mutant cells. The nuclear distribution of the plasmid in the mutant did not significantly differ from that observed with ULP1 cells, indicating that the increase in plasmid copy number in the ulp1ΔES mutant was not due to altered localization of Rep1 and Rep2.
Plasmid segregation in ulp1ΔES yeast.
To determine whether 2μm plasmid distribution was affected by the ulp1ΔES mutation, a 2μm-derived reporter plasmid, pSV5, containing multiple lac operator sequences was visualized in cells expressing a GFP-LacI repressor fusion protein (48). The fluorescently tagged 2μm plasmids appeared as a small number of discrete foci colocalized with the DAPI-stained chromosomal DNA in both [cir+] ulp1ΔES and [cir+] ULP1 cells (Fig. 7C), indicating that plasmid localization within the nucleus did not require wild-type ULP1 function. Although the clustering of the plasmid in foci within the nucleus was unaffected, GFP-LacI fluorescence was not equally intense in some mother and daughter ulp1ΔES mutant cells, suggesting that the 2μm-derived reporter plasmid was not equally partitioned during mitosis (Fig. 7C). To assess this difference, cells with large buds having two distinct DAPI-stained masses, consistent with the mother cell having completed mitosis, were scored for their relative intensity of GFP-LacI repressor fluorescence (Table 6). Complete missegregation of the 2μm-derived reporter plasmid was observed with 6% of [cir+] ulp1ΔES mutant cells and not observed with [cir+] ULP1 cells, suggesting that the mutation in ULP1 impaired 2μm circle plasmid segregation.
TABLE 6.
Distribution of fluorescently tagged 2μm reporter plasmid in ULP1 and ulp1ΔES yeast
| Genotype | 2μm-derived pSV5 plasmid distributiona (%)
|
||
|---|---|---|---|
| Equal | Unequal | Complete missegregation | |
| ULP1 [cir+] | 90 | 9 | <1 |
| ulp1ΔES [cir+] | 83 | 11 | 6 |
Cells with large buds and two well-separated equal DAPI-stained masses were scored for GFP-LacI repressor fluorescence as a measure of distribution of the Lac operator-containing 2μm-derived pSV5 plasmid. A total of 400 cells were scored for each strain.
To further characterize the plasmid segregation defect, inheritance of the fluorescently tagged 2μm reporter plasmid was monitored in live cells by time-lapse photography (Fig. 7D and 7E). Plasmid delivery to the daughter cell took <6 min in the [cir+] ULP1 cells, with GFP-LacI repressor fluorescence equally partitioned between mother and daughter cells. In the [cir+] ulp1ΔES cell, the fluorescently tagged plasmids took >12 min to move from mother to daughter cell, and in some cells, all fluorescence was delivered to and retained in the daughter cell, consistent with complete missegregation of the plasmid in these mitotic divisions. The longer time taken for plasmid segregation in the [cir+] ulp1ΔES mutant is consistent with the G2/M lag observed for these mutant cells by flow cytometry (Fig. 2). The direction of plasmid missegregation contrasts with the maternal bias reported for plasmids that lack a functional partitioning system (6). However, more plasmid missegregation events need to be captured by time-lapse microscopy to establish whether missegregation is random or biased toward the daughter cell in the ulp1ΔES mutant.
DISCUSSION
The discovery that nib1, a mutation that results in elevated plasmid copy numbers, is a single amino acid change in a protease required for cell cycle progression provides another host function that the 2μm circle requires for its stable maintenance within the yeast nucleus. Partial deletion of the ULP1 gene encoding the protease was found to mimic the nib1 mutation (17). The G2/M lag in both [cir+] and [cir0] yeast strains with partial deletion of the ULP1 gene is consistent with the G2/M arrest observed for yeasts carrying a complete deletion of the ULP1 ORF (23). In our study, the ulp1ΔES allele retained the 3′ end of the ORF encoding the conserved protease domain (23) (33) and part of the noncatalytic regulatory domain required for retention of Ulp1 at nuclear pores (37). The remarkable similarity between the phenotypes conferred by this deletion allele and by nib1, a mutation that alters a single amino acid within the conserved carboxy-terminal protease domain, suggests that the ulp1ΔES allele expresses a truncated protein with protease activity sufficient for viability, but insufficient for normal plasmid maintenance. Both alleles may decrease the availability of free mature Smt3, either by impairing the proteolytic activity or by reducing expression of Ulp1. The loss of Ulp1 processing activity would reduce the level of some Smt3 conjugates, while other targets that are normally deconjugated by Ulp1 could accumulate (23, 25). The altered levels of these modified target proteins may be responsible for the phenotype of elevated 2μm circle plasmid levels common to both mutant ulp1 alleles.
Although not specifically indicated, yeast strains used in previous experiments where either the ULP1 or UBC9 genes were disrupted or deleted (23, 45) would, like most strains of Saccharomyces cerevisae, probably be [cir+]. Altered levels of the 2μm circle may occur in these mutant yeasts. The distinct domains of nuclear staining with propidium iodide, observed when yeasts carrying temperature-sensitive alleles of the ULP1 gene were shifted to and incubated at the nonpermissive temperature (23), may correspond to elevated plasmid levels.
Deletion of the SMT4/ULP2 gene encoding the second yeast Smt3-deconjugating protease, Ulp2 (24), or simultaneous deletion of MLP1 and MLP2, two genes encoding myosin-like proteins required for nuclear pore localization and stabilization of Ulp1 (52), also results in abnormal cell morphology, temperature-sensitive growth, and formation of nibbled colonies in [cir+] yeast, phenotypes remarkably similar to the 2μm circle-associated phenotypes observed with the ulp1ΔES mutant yeast. Accumulation of the 2μm circle in these mutant cells may also occur in response to loss or mislocalization of Smt3-deconjugating activity.
The lethality observed with [cir+] ulp1ΔES mutant yeast correlates with elevated plasmid copy numbers and may reflect titration of a host protein critical for progression through mitosis. Increased plasmid levels in the ulp1 mutant could arise from enhanced Flp recombinase-mediated amplification or from plasmid missegregation. Although plasmid missegregation seems more likely in light of the observed missegregation of the fluorescently tagged 2μm reporter plasmid, it is possible that altered Flp activity is the primary defect and that plasmid missegregation is a secondary response to elevated plasmid copy number. Altered Smt3 modification could stabilize the Flp protein or reduce Rep protein-mediated repression of FLP gene expression. In the alternative model, altered levels of a Smt3-modified protein(s) common to the pathways of both chromosome and plasmid segregation could be responsible for the G2/M cell cycle delay in the [cir0] ulp1ΔES mutant yeast and the missegregation of the multicopy plasmid observed with some [cir+] ulp1ΔES mutant cells. The higher-than-normal number of plasmids received by some cells could further impair plasmid segregation in subsequent divisions due to increased competition for limiting Smt3-conjugates. In cells that received less plasmid, the Flp recombinase would restore the normal plasmid copy number, the result being a continuous increase in plasmid levels in the culture.
Smt3 modification has been implicated in regulating chromosome segregation. The SMT3 gene was originally cloned on the basis of its ability, when overexpressed, to suppress the effects of mutation in Mif2, the yeast homolog of mammalian CENP-C, a kinetochore protein required for chromosome segregation and mitotic spindle integrity (31, 32). CENP-C may itself be SUMO modified (13). Smt3 modification may also participate in chromosome segregation by contributing to cohesion between chromosomes. Mcd1 and Smc1, two components of cohesin, a complex enriched at chromosomal centromeres that holds sister chromatids together until the onset of anaphase, are Smt3 modified (4, 22, 50). In a ulp2 mutant, centromeric cohesion is impaired in response to altered Smt3 modification of topoisomerase II (2), and a similar defect might be produced by loss of Ulp1 function. Since SUMO modification targets RanGAP1 to the spindle apparatus during mitosis (28), it is tempting to speculate that centromeric cohesin components and the 2μm circle use a similar approach, with Smt3/SUMO modification mediating attachment to the host spindle and conferring efficient segregation. In support of this hypothesis, the mitotic spindle has recently been shown to promote recruitment of the cohesin complex to the 2μm circle STB-partitioning locus (29).
The colocalization of the Ulp1 protease with the 2μm circle-encoded Rep proteins and the two-hybrid interaction of both Rep1 and Rep2 with Smt3 suggested the plasmid proteins might be targets of Smt3 conjugation or interact with Smt3-modified proteins. Evidence for Rep2, Flp, and possibly Rep1 being Smt3 modified has recently been obtained (E. Johnson, personal communication). The Rep proteins are essential for normal plasmid segregation at cytokinesis, are tightly associated with the 2μm circle DNA in a small number of discrete foci in live yeasts (48), and probably form a DNA-protein aggregate at the plasmid STB locus (6, 44). Efficient plasmid segregation involves recruitment of cohesin complexes to STB by the Rep proteins (30, 51). Cycling between Smt3-modified and unmodified forms of the plasmid proteins could allow their retention or release from a nuclear substructure in response to cell cycle signals, a process that may mediate both efficient plasmid segregation and copy number control. In the ulp1ΔES mutant, altered levels of Smt3-conjugated Rep proteins or cohesin components may affect plasmid association with the host spindle or impair separation of the 2μm plasmid-containing foci, structures that may colocalize with a yeast equivalent of the mammalian matrix-associated PML nuclear bodies (5, 11, 34). Identifying the targets of Ulp1 protease at the G2/M stage in the cell cycle may help reveal the mechanism by which a simple extrachromosomal DNA element parasitizes components of the sophisticated host cell cycle apparatus for its own stable, high-copy-number propagation.
Acknowledgments
We thank C. Holm for the gift of yeast strains CH568 and CH569, Marc Gartenberg for the plasmid pBIS-GALkFLP-(TRP1), Barbara Goettgens (Core Facility, Institute for Cell and Molecular Biology, University of Texas, Austin) for assistance with confocal microscopy, and J. Benjamin and Y. Zhang for technical assistance. We are grateful to Erica Johnson for communicating results prior to publication and for helpful comments on the manuscript.
This work was supported by an NSERC grant to M.J.D. and grants from the National Institutes of Health and the Robert Welch Foundation to M.J.
REFERENCES
- 1.Ahn, Y.-T., X.-L. Wu, S. Biswal, S. Velmurugan, F. C. Volkert, and M. Jayaram. 1997. The 2μm-encoded Rep1 and Rep2 proteins interact with each other and colocalize to the Saccharomyces cerevisiae nucleus. J. Bacteriol. 179:7497-7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachant, J., A. Alcasabas, Y. Blat, N. Kleckner, and S. J. Elledge. 2002. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of topisomerase II. Mol. Cell 9:1169-1182. [DOI] [PubMed] [Google Scholar]
- 3.Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Using the two-hybrid system to detect protein-protein interactions., p. 153-179. In D. A. Hartley (ed.), Cellular interactions in development: a practical approach. IRL Press, Oxford, United Kingdom.
- 4.Blat, Y., and N. Kleckner. 1999. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell 98:249-259. [DOI] [PubMed] [Google Scholar]
- 5.Boddy, M. N., K. Howe, L. D. Etkin, E. Solomon, and P. S. Freemont. 1996. Pic-1, a novel ubiquitin-like protein which interacts with the PML component of a multiprotein complex that is disrupted in acute promyelocytic leukaemia. Oncogene 13:971-982. [PubMed] [Google Scholar]
- 6.Broach, J. R., and F. C. Volkert. 1991. Circular DNA plasmids of yeasts: genome dynamics, protein synthesis and energetics., p. 297-331. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor Laboratory press, Cold Spring Harbor, N.Y.
- 7.Cross, F. R. 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast 13:647-653. [DOI] [PubMed] [Google Scholar]
- 8.Desterro, J. M. P., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkBα inhibits NF-kB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 9.Dobson, M. J., A. B. Futcher, and B. S. Cox. 1980. Loss of 2μm DNA from Saccharomyces cerevisiae transformed with the chimaeric plasmid pJDB219. Curr. Genet. 2:201-205. [DOI] [PubMed] [Google Scholar]
- 10.Dobson, M. J., F. E. Yull, M. Molina, S. M. Kingsman, and A. J. Kingsman. 1988. Reconstruction of the yeast 2μm plasmid partitioning mechanism. Nucleic Acids Res. 16:7103-7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 12.Durfee, T., K. Becherer, P. Chen, S. Yeh, Y. Yang, A. E. Kilburn, W. Lee, and S. J. Elledge. 1993. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 7:555-569. [DOI] [PubMed] [Google Scholar]
- 13.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Futcher, A. B. 1988. The 2μm circle plasmid of Saccharomyces cerevisiae. Yeast 4:27-40. [DOI] [PubMed] [Google Scholar]
- 15.Futcher, A. B. 1986. Copy number amplification of the 2μm circle plasmid of Saccharomyces cerevisiae. J. Theor. Biol. 119:197-204. [DOI] [PubMed] [Google Scholar]
- 16.Hartwell, L. H., J. Culotti, and B. Reid. 1970. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 66:352-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holm, C. 1982. Clonal lethality caused by the yeast plasmid 2μm DNA. Cell 29:585-594. [DOI] [PubMed] [Google Scholar]
- 18.Holm, C. 1982. Sensitivity to the yeast plasmid 2μm DNA is conferred by the nuclear allele nib1. Mol. Cell. Biol. 2:985-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, E. S., and G. Blobel. 1999. Cell cycle-regulated attachment of ubiquitin-related SUMO to the yeast septins. J. Cell Biol. 147:981-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, E. S., I. Schwienhorst, R. J. Dohmen, and G. Blobel. 1997. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16:5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laloraya, S., V. Guacci, and D. Koshland. 2000. Chromosomal addresses of the cohesin component Mcd1p. J. Cell Biol. 151:1047-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, S., and M. Hochstrasser. 1999. A new protease required for cell-cyle progression in yeast. Nature 398:246-251. [DOI] [PubMed] [Google Scholar]
- 24.Li, S., and M. Hochstrasser. 2000. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 20:2367-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, S. J., and M. Hochstrasser. 2003. The Ulp1 SUMO isopeptidase: distinct domains required for viability, nuclear envelope localization, and substrate specificity. J. Cell Biol. 160:1069-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtine, M. S., A. McKenzie, N. G. Shah, A. Wach, A. Brachant, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 27.Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88:97-107. [DOI] [PubMed] [Google Scholar]
- 28.Matunis, M. J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta, S., X. Yang, M. Jayaram, and S. Velmurugan. 2005. A novel role for the mitotic spindle during DNA segregation in yeast: promoting 2μm plasmid-cohesin association. Mol. Cell. Biol. 25:4283-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mehta, S., X. M. Yang, C. S. Chan, M. J. Dobson, M. Jayaram, and S. Velmurugan. 2002. The 2 micron plasmid purloins the yeast cohesin complex: a mechanism for coupling plasmid partitioning and chromosome segregation. J. Cell Biol. 158:625-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meluh, P. B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meluh, P. B., and D. Koshland. 1995. Suppressors of MIF2, a putative centromere protein gene in Saccharomyces cerevisiae. Mol. Biol. Cell 6S:360a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mossessova, E., and C. D. Lima. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865-876. [DOI] [PubMed] [Google Scholar]
- 34.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray, J. A. H., M. Scarpa, N. Rossi, and G. Cesareni. 1987. Antagonistic controls regulate copy number of the yeast 2μm plasmid. EMBO J. 6:4205-4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panse, V. G., U. Hardeland, T. Werner, B. Kuster, and E. Hurt. 2004. A proteome-wide approach identifies sumolyated substrate proteins in yeast. J. Biol. Chem. 279:41346-41351. [DOI] [PubMed] [Google Scholar]
- 37.Panse, V. G., B. Kuster, T. Gerstberger, and E. Hurt. 2003. Unconventional tehering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat. Cell Biol. 5:21-27. [DOI] [PubMed] [Google Scholar]
- 38.Pringle, J. R., A. E. M. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast, p. 565-602. In C. Guthrie, and G. R. Fink (ed.), Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds, A. E., A. W. Murray, and J. W. Szostak. 1987. Roles of the 2μm gene products in stable maintenance of the 2μm plasmid of Saccharomyces cerevisiae. Mol. Cell. Biol. 7:3566-3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Rothstein, R. 1991. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 194:281-301. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 43.Schwarz, S. E., K. Matuschewski, D. Liakopoulos, M. Scheffner, and S. Jentsch. 1998. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA 95:560-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sengupta, A., K. Blomqvist, A. Pickett, Y. Zhang, J. S. K. Chew, and M. J. Dobson. 2001. Functional domains of yeast plasmid-encoded Rep proteins. J. Bacteriol. 183:2306-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seufert, W., A. B. Futcher, and S. Jentsch. 1995. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature 373:78-81. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsalik, E. L., and M. R. Gartenberg. 1998. Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast 14:847-852. [DOI] [PubMed] [Google Scholar]
- 48.Velmurugan, S., X. Yang, C. Chan, M. J. Dobson, and M. Jayaram. 2000. Partitioning of the 2μm circle plasmid of Saccharomyces cerevisiae: functional coordination with chromosome segregation and plasmid-encoded Rep protein distribution. J. Cell Biol. 149:553-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volkert, F. C., and J. R. Broach. 1986. Site-specific recombination promotes plasmid amplification in yeast. Cell 46:541-550. [DOI] [PubMed] [Google Scholar]
- 50.Wohlschlegel, J. A., E. S. Johnson, S. I. Reed, and J. R. Yates 3rd. 2004. Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 279:45662-45668. [DOI] [PubMed] [Google Scholar]
- 51.Yang, X. M., S. Mehta, D. Uzri, M. Jayaram, and S. Velmurugan. 2004. Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation. Mol. Cell. Biol. 24:5290-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, X., C. Wu, and G. Blobel. 2004. Mlp-dependent anchorage and stabilization of a desumoylating enzyme is required to prevent clonal lethality. J. Cell Biol. 167:605-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou, W., J. J. Ryan, and H. Zhou. 2004. Global analyses of sumolylated proteins in Saccharomyces cerevisiae. J. Biol. Chem. 279:32262-32268. [DOI] [PMC free article] [PubMed] [Google Scholar]