Abstract
Tumor suppressor CYLD is a deubiquitinating enzyme (DUB) that inhibits the ubiquitination of key signaling molecules, including tumor necrosis factor (TNF) receptor-associated factor 2 (TRAF2). However, how the function of CYLD is regulated remains unknown. Here we provide evidence that inducible phosphorylation of CYLD is an important mechanism of its regulation. Under normal conditions, CYLD dominantly suppresses the ubiquitination of TRAF2. In response to cellular stimuli, CYLD undergoes rapid and transient phosphorylation, which is required for signal-induced TRAF2 ubiquitination and activation of downstream signaling events. Interestingly, the CYLD phosphorylation requires IκB kinase gamma (IKKγ) and can be induced by IKK catalytic subunits. These findings suggest that CYLD serves as a novel target of IKK and that the site-specific phosphorylation of CYLD regulates its signaling function.
Protein ubiquitination is an important mechanism that regulates diverse cellular processes, including protein degradation, signal transduction, immune response, cell cycle control, endocytosis, and vesicle trafficking (12, 16). This molecular mechanism is under the control of both ubiquitin enzymes and deubiquitinating enzymes (DUBs). The DUBs are a family of cysteine proteases that specifically digest polyubiquitin chains (21). In addition to their housekeeping function in the regeneration of free ubiquitin molecules, the DUBs play an important regulatory role in determining the fate and function of specific ubiquitin-conjugated proteins (12). A new member of the DUB family, CYLD (6, 22, 39), was identified as a tumor suppressor mutated in familial cylindromatosis (4), an autosomal dominant predisposition to multiple tumors of the skin appendages (5, 37). Recent studies suggest that CYLD mediates deubiquitination of certain signaling molecules, including members of the tumor necrosis factor receptor (TNFR)-associated factor (TRAF) family (22, 28, 39).
TRAFs function as signaling adaptors of the TNFR superfamily (2) as well as a number of other immune receptors, such as toll-like receptors, interleukin-1 receptor, and T-cell receptor (1, 7, 38). A common structural feature of TRAFs, with the exception of TRAF1, is their possession of a ring finger domain known to mediate protein ubiquitination (24). It has been shown that TRAF2 and TRAF6 function as ubiquitin ligases that catalyze both self-ubiquitination and the ubiquitination of specific target molecules involved in signal transduction (10, 13, 18, 34, 41). Strong evidence suggests that the ubiquitination of TRAF2 and TRAF6 functions as a molecular trigger for initiating downstream signaling events, including activation of c-Jun N-terminal kinase (JNK) (13, 34) and IκB kinase (IKK) (38, 41).
IKK is known as a specific activator of the transcription factor NF-κB (20, 36). The IKK complex is composed of two catalytic subunits, IKKα and IKKβ, and a regulatory subunit termed IKKγ (19). Upon activation by various cellular stimuli, IKK phosphorylates the NF-κB inhibitor IκBα, triggering the proteolysis of IκBα and nuclear translocation of active NF-κB (19). The canonical substrates of JNK include c-Jun, JunD, and ATF2, components of the transcription factor AP-1 (9). Like IKK, JNK regulates diverse biological processes, ranging from immune responses, cell growth, and apoptosis to tumor formation (9, 17, 23, 25). A role for CYLD in IKK regulation is suggested by some recent studies, which reveal that CYLD inhibits the activation of an NF-κB reporter gene (4, 6, 22, 39). Using RNA interference (RNAi)-mediated CYLD knockdown, we have shown that CYLD is also a key negative regulator of JNK downstream of specific immune receptors (29). In agreement with these findings, CYLD inhibits the ubiquitination of both TRAF2 and TRAF6 in transfected cells (6, 22, 39). However, despite these important observations, the molecular mechanism regulating the function of CYLD remains unknown. In this study, we demonstrate that CYLD plays a dominant role in suppressing the ubiquitination of endogenous TRAF2. CYLD knockdown results in constitutive ubiquitination of TRAF2. Interestingly, signal-induced TRAF2 ubiquitination is associated with site-specific phosphorylation of CYLD, a molecular event that appears to prevent CYLD from inhibiting the ubquitination of TRAF2. We further demonstrate that the CYLD phosphorylation is dependent on IKKγ.
MATERIALS AND METHODS
Plasmid constructs.
Human CYLD was cloned by reverse transcription-PCR (RT-PCR) and inserted into the pcDNA-HA (hemagglutinin) vector (15). CYLD truncation mutants were generated by PCR and designated by the specific amino acid residues retained in the mutant proteins. The CYLD mutants harboring amino acid substitutions were created by site-directed mutagenesis (Stratagene) using the wild-type CYLD expression vector as template (the primer sequences are available upon request). An RNAi-resistant form of CYLD (CYLDR) and its mutants were produced by introducing sense mutations (no alteration of amino acid codons) at the small interfering RNA (siRNA)-binding site. To generate the retroviral vectors expressing CYLD and its mutants, the corresponding cDNAs were transferred from pcDNA-HA to the pCLXSN retroviral vector (provided by Inder Verma; see reference 26). In some experiments, the pCLXSN vector was modified by replacing its neomycin-resistant gene with the green fluorescence protein (GFP) gene. Myc-tagged IKKγ was generated by inserting the human IKKγ cDNA into pcDNA vector downstream of a myc epitope tag. pCLXSN-hCD40 was cloned by inserting the human CD40 cDNA into the pCLXSN retroviral vector. Expression vectors encoding HA-tagged IKKα and IKKβ (40), HA-tagged ubiquitin (42, 43), glutathione S-transferase (GST)-IκBα(1-54), and GST-c-Jun(1-79) (29) were described previously. GST-CYLD(403-513) was cloned by inserting a DNA fragment encoding amino acids 403 to 513 of human CYLD into the pGEX4T-1 vector (Amersham/Pharmacia Biotech). The point mutants of GST-CYLD(403-513) were generated by site-directed mutagenesis. All the DNA constructs were sequenced at the Core Facility of Hershey Medical Center.
Antibodies, cell lines, and other reagents.
The anti-CYLD antibody was generated by injection of rabbits with a GST fusion protein containing an N-terminal region of human CYLD (amino acids 136 to 301). The phospho-specific CYLD antibody (anti-P-CYLD S418) specifically recognizes CYLD containing phosphorylated serine 418. This antibody was generated by injection of rabbits with a keyhole limpet hemocyanin-conjugated peptide covering amino acids 413 to 428 of human CYLD, in which serine 418 was phosphorylated. Polyclonal antibodies for tubulin (TU-02), JNK1 (C-17), JNK2 (N18), TRAF2 (C-20), and IKKγ (FL-419) were purchased from Santa Cruz. Horseradish peroxidase (HRP)-conjugated HA antibody (3F10) and human recombinant TNF-α were from Roche Applied Science. Phorbol myristate acetate (PMA), ionomycin, and lipopolysaccharide (LPS; derived from Escherichia coli 0127:B8) were from Sigma. Recombinant IKKα and IKKβ proteins were provided by Michael Karin.
Human embryonic kidney 293 cells, human cervical carcinoma HeLa cells, human B-cell line BJAB, and human leukemia T-cell line Jurkat were obtained from American Type Culture Collection. IKKγ-deficient Jurkat cell line, JM4.5.2, and its IKKγ-reconstituted derivative, JM4.5.2-IKKγ, were described previously (14, 30).
RNAi.
siRNAs specific for human CYLD and luciferase were synthesized by Dharmacon Research, Inc. (Lafayette, CO). The sense strand sequences of the oligonucleotides are as follows: CYLD siRNA, AAGUACCGAAGGGAAGUAUAG; and luciferase siRNA, AACTTACGCTGAGTACTTCGA.
For siRNA delivery, 293 and HeLa cells were transfected in six-well plates with 140 pmol of siRNA, using Oligofectamine (Invitrogen). At 16 to 24 h following the first transfection, the cells were transfected again with the same amount of siRNA together with 300 ng carrier DNA, using Lipofectamine 2000. At about 30 h after the second transfection, the cells were used for different experiments.
For stable gene knockdown using the small hairpin RNA (shRNA) technique, a double-stranded oligonucloetide corresponding to the CYLD siRNA was cloned into the pSUPER-retro-puromycin vector (Oligoengine) downstream of the U6 promoter. The generated retroviral construct, named pSUPER-shCYLD, was used to produce recombinant viruses and infect the indicated cells. The infected cells were enriched by selection using puromycin. The bulk-infected cells were used in the experiments to avoid clonal variations.
IB, IP, and in vitro kinase assays.
Cell lysates were prepared by lysing the cells in a kinase lysis buffer supplemented with phosphatase inhibitors and immediately subjected to immunoblotting (IB) and in vitro kinase assays as described previously (40). To detect phosphorylated CYLD based on its band shift, the CYLD proteins were first concentrated by immunoprecipitation (IP) (using anti-CYLD) followed by fractionation using 6% sodium dodecyl sulfate (SDS) gels and IB. coimmunoprecipitation (coIP) was performed to determine the interaction of CYLD with TRAF2 and IKKγ.
In vivo ubiquitin conjugation assays.
The indicated cells were transfected in six-well plates with pcDNA-HA-ubiquitin (0.5 μg). At 30 to 40 h following transfection, the cells were stimulated as indicated and lysed in radioimmunoprecipitation assay buffer (42) supplemented with inhibitors of ubiquitin hydrolases (20 mM N-ethylmaleimide and 5 μM ubiquitin aldehyde). The cell lysates were immediately subjected to IP using anti-TRAF2, and the ubiquitin-conjugated TRAF2 was analyzed by IB using HRP-conjugated anti-HA antibody.
Retroviral infection.
Retroviral particles were produced using the pCLXSN vector system as previously described (8, 30). The infected cells were enriched by drug selection using either puromycin (for CYLD knockdown with pSUPER-retro-puro vector) or neomycin (for infection with pCLXSN vector). For reconstitution of the Jurkat-shCYLD cells with RNAi-resistant CYLD (CYLDR) or mutant M4 (M4R), the pCLXSN(GFP) vector was used and the infected cells were enriched by fluorescence cell sorting (Core Facility of Hershey Medical Center). The bulk-infected cells were used in the experiments to avoid clonal variations.
RPA.
Total cellular RNA was isolated from the indicated cells using the TRI reagent (Molecular Research Center, Inc., Cincinnati, OH). The RNase protection assay (RPA) was performed using the BD RiboQuant reagents and a custom template set according to the manufacturer's instructions (BD Biosciences).
Luciferase reporter gene assay.
Luciferase assays were performed using Jurkat T cells expressing wild-type CYLD (Jurkat-shCYLD-CYLD WTR) or a phosphorylation-defective CYLD mutant (Jurkat-shCYLD-CYLD M4R). To create these cells, Jurkat T cells were first infected with pSUPER-shCYLD to stably knock down endogenous CYLD. The generated Jurkat-shCYLD cells were then reconstituted, by retroviral infection, with an RNAi-resistant form of either the wild type (WTR) or phosphorylation-deficient mutant (M4R) of CYLD. For luciferase assays, the cells (5 × 105) were seeded in 12-well plates and transfected using Fugene 6 transfection reagent (Roche Applied Science) with κB-TATA-luc reporter (250 ng) together with 250 ng of either an empty pCLXSN vector or the same vector containing human CD40 cDNA (pCLXSN-CD40). After 40 h, the cells were lysed in a cell lysis buffer (passive buffer) and subjected to dual-luciferase assays (Promega; dual-luciferase reporter assay system).
RESULTS
CYLD knockdown results in constitutive ubiquitination of TRAF2.
Recent studies suggest that TRAF2 undergoes rapid ubiquitination in response to TNF-α stimulation, which is involved in JNK activation (13, 34). Since CYLD functions as a negative regulator of JNK in the TNF-α signaling pathway (29), we analyzed the role of CYLD in regulating the uibquitination of TRAF2 under endogenous conditions. For these studies, endogenous CYLD was knocked down in 293 cells by RNAi followed by examining the ubiquitination of endogenous TRAF2 under untreated or TNF-α-stimulated conditions. As expected, the expression level of CYLD was greatly reduced in cells transfected with a CYLD-specific siRNA (siCYLD; Fig. 1A, panel 2, lanes 4 to 6) but was not affected in cells transfected with a control siRNA for luciferase (siLuc; lanes 1 to 3). In the control cells, only a trace amount of ubiquitinated TRAF2 was detected under untreated conditions (Fig. 1A, panel 1, lane 1) but the TRAF2 ubiquitination was markedly induced by TNF-α (lanes 2 and 3). Remarkably, when CYLD was knocked down by its specific siRNA, TRAF2 became costitutively ubiquitinated (lane 4). Stimulation of these cells with TNF-α failed to further enhance TRAF2 ubiquitination (lanes 5 and 6). Parallel studies using HeLa cells obtained similar results (Fig. 1B). These findings suggest that CYLD plays a critical role in suppressing the in vivo ubiquitination of TRAF2 and raise the possibility that signal-mediated induction of TRAF2 ubiquitination may involve functional modification of CYLD.
FIG. 1.
CYLD knockdown results in constitutive ubiquitination of TRAF2. 293 (A) or HeLa (B) cells were transfected using Lipofectamine 2000 with control or CYLD-specific siRNA together with pcDNA-HA-ubiquitin. The cells were stimulated with TNF-α (50 ng/ml) and then lysed in a buffer containing inhibitors of ubiquitin hydrolases. Endogenous TRAF2 was isolated by IP using anti-TRAF2 antibody, and the ubiquitinated TRAF2 was detected by IB using an HRP-conjugated anti-HA antibody (top panel). The intracellular level of CYLD and TRAF2 was analyzed by IB using anti-CYLD and anti-TRAF2 antibodies (middle and bottom panels). M, molecular mass. NT, not treated.
Endogenous CYLD undergoes phosphorylation in response to immune stimuli.
During the course of CYLD IB assays, we noticed that when the cell lysis buffer was supplemented with phosphatase inhibitors, a more slowly migrating CYLD band could be detected in Jurkat cells stimulated with mitogens (PMA plus ionomycin) or TNF-α (Fig. 2A). Interestingly, the slower-migrating form of CYLD could be converted to its basal form by in vitro incubation with a nonspecific phosphatase (Fig. 2A, lane 13), thus revealing phosphorylation as the nature of CYLD modification. The CYLD phosphorylation was also readily detected in TNF-α-stimulated 293 (Fig. 2C) and HeLa cells (Fig. 2D) as well as in LPS-stimulated BJAB B cells (Fig. 2E). Notably, the kinetics of CYLD phosphorylation was correlated with the phosphorylation of IκBα, both appearing as early as 5 min in cells stimulated with mitogens and TNF-α (Fig. 2A, C, and D, lane 2; P-CYLD and P-IκBα). Consistently, the activation of IKK was also tightly associated with CYLD phosphorylation (compare Fig. 1A and B). The slightly delayed kinetics of IκBα degradation, compared to that of IKK activation and phosphorylation of CYLD, is consistent with the involvement of postphosphorylation steps (e.g., ubiquitination and proteasome targeting) in this proteolytic event. Together with the previous finding that CYLD physically interacts with the IKK regulatory subunit, IKKγ (22, 39), these results raise the intriguing possibility that IKK may be involved in the inducible phosphorylation of CYLD.
FIG. 2.
CYLD is phosphorylated in response to diverse cellular stimuli. (A) Jurkat T cells were stimulated with either T-cell mitogens (50 ng/ml of PMA plus 1 μM of ionomycin) or TNF-α (20 ng/ml) for the indicated times. To prevent loss of the phosphorylated CYLD, the cells were lysed in a kinase lysis buffer supplemented with phosphatase inhibitors. CYLD proteins were concentrated by IP (using anti-CYLD) and then fractionated in low-percentage (6%) SDS gels in order to separated the basal and phosphorylated CYLD bands. In lanes 11 to 13, the CYLD immune precipitates isolated from TNF-α-stimulated cells (15 min) were either left on ice (NT) or incubated at 37°C for 30 min in calf intestinal alkaline phosphatase (CIP; lane 13) or buffer control (lane 12). The phosphorylated (P-CYLD) and basal (CYLD) forms of CYLD were detected by IB using anti-CYLD (upper panel). The cell lysates were also subjected to IB to detect IκBα degradation (lower panel). (B) Immune complex kinase assays to detect the activation of IKK. Jurkat cells were stimulated with PMA plus inomycin (Iono) as described for panel A. IKK complex was isolated by IP using anti-IKKγ antibody and subjected to kinase assays using GST-IκBα(1-54) as a substrate. The phosphorylated substrate (P-GST-IκBα) is indicated. (C to E) CYLD phosphorylation (upper panel) and IκBα degradation (lower panel) were analyzed in TNF-α-stimulated 293 and HeLa cells and LPS-stimulated BJAB B cells as described for panel A.
CYLD phosphorylation requires IKKγ and can be induced by IKK catalytic subunits.
To determine the importance of IKK in CYLD phosphorylation, we analyzed this signaling event using an IKKγ-deficient Jurkat T-cell line, JM4.5.2, which was created in our laboratory (14) and has since been used in numerous studies by others. Due to the lack of IKKγ, the IKK complex cannot be activated by various known IKK stimuli, such as mitogens and cytokines, and this defect can be rescued by expression of the exogenous IKKγ (30, 31). As expected, mitogens failed to induce the degradation of IκBα (target of IKK) in JM4.5.2 cells (Fig. 3A, lower panel, lane 4), but this signaling defect was rescued when the mutant cells were reconstituted with exogenous IKKγ (lane 6). Remarkably, the inducible phosphorylation of CYLD was also dependent on IKKγ, which was blocked in the IKKγ-deficient cells and rescued upon IKKγ reconstitution (Fig. 3A, upper panel). In concert with this finding, CYLD was phosphorylated in 293 cells by transfected IKK catalytic subunits, IKKβ (Fig. 3B, lane 2) or IKKα (lane 4). Notably, optimal phosphorylation of CYLD by both IKKα and IKKβ required their cotransfection with IKKγ (lane 6) (data not shown). Since IKKγ physically interacts with CYLD (22, 39), it is likely that IKKγ is required for recruiting CYLD to the IKK catalytic subunits.
FIG. 3.
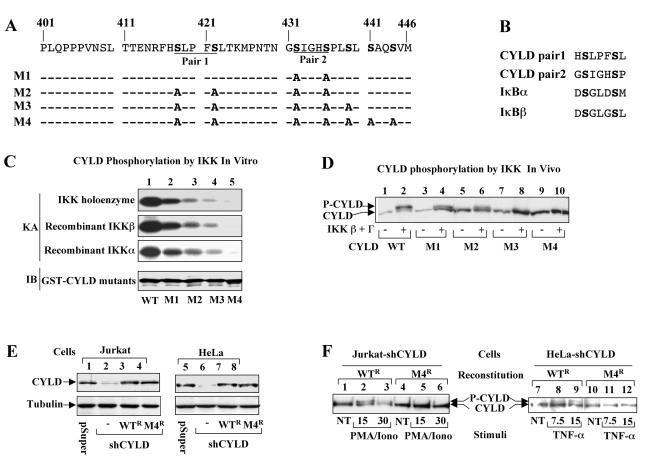
CYLD phosphorylation is mediated by IKK. (A) Inducible phosphorylation of CYLD requires IKKγ. Parental Jurkat cells, IKKγ-deficient Jurkat mutant (JM4.5.2), and IKKγ-reconstituted JM4.5.2 cells were either not treated (−) or were stimulated with PMA plus ionomycin for 15 min. Cell lysates were subjected to CYLD phosphorylation and IκBα degradation analyses as described for Fig. 2A. (B) CYLD phosphorylation by transfected IKK. 293 cells were transfected with HA-tagged CYLD together with either empty vector or expression vectors encoding IKKβ (0.5 μg), IKKα (0.5 μg), or IKKβ (0.5 μg) plus IKKγ (25 ng). CYLD phosphorylation was analyzed by IB using anti-HA antibody. (C) Phosphorylation of CYLD truncation mutants. CYLD truncation mutants covering different lengths of its C terminus (indicated by the amino acid numbers) were expressed in 293 cells in either the absence (−) or presence (+) of HA-IKKβ plus HA-IKKγ. Phosphorylation of CYLD (upper panel) and expression of IKKβ (middle panel) and IKKγ (bottom panel) were analyzed by IB using anti-HA. Phosphorylated CYLD bands are indicated by an arrowhead. (D) CYLD/IKKγ physical interaction. Full-length (FL) or truncated forms of CYLD (tagged with HA) were coexpressed with myc-tagged IKKγ. The IKKγ complex was isolated by IP using anti-myc followed by detecting the associated CYLD proteins by IB using anti-HA-HRP (upper panel). The expression level of IKKγ was analyzed by IB using anti-myc (lower panel). Lane 1 is a negative control that was transfected with CYLD only. (E) In vitro kinase assays to demonstrate CYLD phosphorylation by IKK homoenzyme. IKK holoenzyme was isolated by IP (using anti-IKKγ) from untreated (NT), PMA-ionomycin-stimulated (7.5 min), or TNF-α-stimulated (7.5 min) Jurkat cells and subjected to in vitro kinase assays using GST-IκBα(1-54) (lower panel) or GST-CYLD(403-513) (upper panel) as a substrate. (F) In vitro kinase assays were performed using IKK complex isolated from PMA-ionomycin-stimulated Jurkat cells and GST-CYLD(403-513) or GST substrate. GST-CYLD(403-513), but not GST, was phosphorylated by IKK. (G) CYLD phosphorylation by recombinant IKKβ. In vitro kinase assays were performed using the indicated amounts of purified IKKβ recombinant protein and GST-IκBα(1-54) (lanes 1 to 5) or GST-CYLD(403-513) (lanes 6 to 10) substrate. Autophosphorylated IKKβ (P-IKKβ) and phosphorylated substrates are indicated.
To localize the region within CYLD that is phosphorylated by IKK, CYLD mutants harboring sequential truncations were subjected to phosphorylation analysis (Fig. 3C). Deletion of 419 amino acids from the N terminus of CYLD has no significant effect on its phosphorylation (Fig. 3C, upper panel, lane 4). Interestingly, further deletion of 27 amino acids generated a CYLD mutant (amino acids 447 to 956) that was no longer phosphorylated (lane 6). The defect of this CYLD mutant in phosphorylation was not due to loss of its IKKγ-binding activity, since all the truncation mutants were competent in IKKγ binding (Fig. 3D). Thus, the phosphorylation site of CYLD is likely located between amino acids 420 and 446.
To determine whether IKK directly phosphorylates CYLD, we generated a GST-CYLD fusion protein containing a region of CYLD (amino acids 403 to 513) that covers its phosphorylation site. We then performed in vitro kinase assays using the GST-CYLD substrate and IKK holoenzyme isolated from Jurkat cells or recombinant IKKs. The GST-CYLD was not significantly phosphorylated by inactive IKK isolated from untreated cells but was potently phosphorylated by the active IKK isolated from mitogen- and TNF-α-stimulated cells (Fig. 3E, upper panel). The relative level of GST-CYLD phosphorylation was similar to that of the phosphorylation of GST-IκBα (lower panel). The CYLD phosphorylation was specific, since the IKK complex did not phosphorylate the GST protein (Fig. 3F). Further, the GST-CYLD was also phosphorylated by recombinant IKKβ (Fig. 3G, lanes 6 to 10) and IKKα (data not shown) (see Fig. 4C). Parallel dose-dependent kinase assays using GST-CYLD (Fig. 3G, lanes 6 to 10) and GST-IκBα (lanes 1 to 5) revealed that IKK phosphorylates these two substrates with comparable efficiencies.
FIG. 4.
Site-specific phosphorylation of CYLD by IKK. (A) Amino acid sequence of CYLD phosphorylation region. The putative phosphorylation sites (serines) are boldface, and two serine pairs are indicated. (B) Sequence homology between the CYLD serine pairs and the IKK phosphorylation sites within IκBα and IκBβ. (C) Phosphorylation of CYLD mutants by IKK holoenzyme (top panel) and recombinant IKKs (middle panels). GST-CYLD(403-513) with a wild-type phosphorylation site (WT) or the various mutations (indicated in panel A) were subjected to in vitro kinase assays (KA) using IKK holoenzyme isolated from mitogen-stimulated Jurkat cells (top panel), recombinant IKKβ (second panel), or recombinant IKKα (third panel). The substrate amounts were monitored by IB using anti-GST (bottom panel). (D) In vivo phosphorylation of CYLD mutants by IKK. HA-tagged full-length CYLD, either wild type or the indicated mutants, were expressed in 293 cells in the absence (−) or presence (+) of IKKβ plus IKKγ. The phosphorylation of CYLD was analyzed by IB using anti-HA. (E) CYLD knockdown by shRNA and reconstitution. Jurkat and HeLa cells were infected with either the empty pSUPER retroviral vector (lanes 1 and 5) or the same vector encoding CYLD-specific shRNA (shCYLD; lanes 2 and 6). The CYLD-knockdown cells were reconstituted by infection with retroviruses encoding RNAi-resistant wild-type CYLD (WTR) or its phosphorylation-deficient mutant M4 (M4R). Expression of CYLD and the housekeeping protein tubulin was detected by IB using anti-CYLD and antitubulin, respectively. (F) Phosphorylation of CYLD by cellular stimuli. The CYLD-knockdown Jurkat (Jurkat-shCYLD) and HeLa (HeLa-shCYLD) cells reconstituted with CYLD WTR or M4R were stimulated with mitogens or TNF-α as indicated, and the phosphorylation of CYLD was analyzed as described for Fig. 2A. Wild-type CYLD, but not CYLD M4, was phosphorylated.
A serine cluster of CYLD is involved in its phosphorylation.
The region of CYLD phosphorylation contains a cluster of serines (Fig. 4A, boldface), including two serine pairs that exhibit homology with the IKK phosphorylation site in IκBα and IκBβ (Fig. 4A, underlined, and Fig. 4B). Since IKK is known to be a serine kinase (11), one or more of these serines are likely involved in CYLD phosphorylation. To test this hypothesis, serine-to-alanine (S/A) substitutions were introduced into the GST-CYLD construct to generate mutants harboring various S/A substitutions (Fig. 4A, M1 to M4). In vitro kinase assays revealed that mutation of one pair of the serines (see Fig. 4A, M1) partially affected the CYLD phosphorylation by IKK (Fig. 4C, lane 2). Combined mutation of the two pairs of serines (M2) significantly diminished the CYLD phosphorylation (Fig. 4C, lane 3), and the phosphorylation of CYLD was completely abolished by the combined mutations of the entire serine cluster (M4, lane 5). Similar results were obtained with the IKK holoenzyme (Fig. 4C, panel 1), recombinant IKKβ (panel 2), and recombinant IKKα (panel 3).
To determine whether the result of the in vitro kinase assays is consistent with the in vivo phosphorylation of CYLD, the various mutations illustrated in Fig. 4A were introduced directly into full-length CYLD and the resulting CYLD mutants were examined for their in vivo phosphorylation by transfected IKKβ and IKKγ. Similar to the in vitro phosphorylation results, the in vivo phopshorylation of CYLD was progressively attenuated along with mutation of more serines and was completely abolished by the M4 mutation (Fig. 4D). The phosphorylation defect of M4 was not due to its loss of IKKγ-binding activity, as demonstrated by coIP assays (data not shown).
We next determined whether the serine cluster was responsible for CYLD phosphorylation induced by cellular stimuli. For these studies, we stably knocked down the endogenous CYLD in Jurkat and HeLa cells using a pSUPER retroviral vector encoding CYLD-specific shRNA (shCYLD). The generated CYLD-deficient cells (named Jurkat-shCYLD and HeLa-shCYLD) were then reconstituted with the RNAi-resistant form of wild-type CYLD (WTR) or its phosphorylation-deficient M4 mutant (M4R). As expected, CYLD expression was extremely low in cells infected with pSUPER-shCYLD (Fig. 4E, lanes 2 and 6) but could be readily detected in the control cells infected with the empty pSUPER vector (lanes 1 and 5). Further, the CYLD deficiency in the Jurkat-shCYLD and HeLa-shCYLD cells was efficiently rescued by retroviral infection with the RNAi-resistant wild-type CYLD (WTR; lanes 3 and 7) and M4 mutant (M4R; lanes 4 and 8). In vivo phosphorylation analysis revealed that like endogenous CYLD, the exogenous wild-type CYLD was phosphorylated upon cellular stimulation by mitogens (Fig. 4F, lanes 1 to 3) and TNF-α (lanes 7 to 9). In contrast, the CYLD M4 mutant was not phosphorylated in response to either inducer (lanes 4 to 6 and 10 to 12). Thus, the IKK phosphorylation sites within CYLD are responsible for its in vivo phosphorylation stimulated by immune stimuli.
To further confirm that the serine cluster of CYLD is phosphorylated in vivo, we generated an antibody that detects CYLD phosphorylation at one of the clustered serines (serine 418). As expected, this phospho-specific CYLD antibody (named anti-P-CYLD S418) did not react with the basal form of CYLD in untreated cells (Fig. 5A, lane 3). More importantly, the phosphorylated CYLD was efficiently detected by this antibody in stimulated cells (lane 4). Thus, serine 418 is phosphorylated in vivo. IB assays using a CYLD mutant harboring a serine-to-alanine mutation at serine 418 (S418A) showed that mutation of this phosphorylation site did not abolish the inducible phosphorylation of CYLD (Fig. 5B). This result further supports our idea that multiple serines within the serine cluster likely contribute to CYLD phosphorylation.
FIG. 5.
Serine 418 of CYLD is phosphorylated in vivo. (A) Jurkat cells were either not treated (NT) or stimulated with PMA plus ionomycin (Iono) for 10 min. Cell lysates were subjected to IB using either the regular anti-CYLD antibody or a phospho-specific anti-CYLD antibody (αP-CYLD) that recognizes CYLD with phosphorylated serine 418. As negative control, an IB was performed using the preserum of αP-CYLD. (B) 293 cells were transfected with wild-type CYLD (WT) or CYLD S418A together with IKKγ and IKKβ as described for Fig. 3D. Cell lysates were subjected to IB using either phospho-specific anti-CYLD (αP-CYLD) or regular anti-CYLD (αCYLD) antibodies.
CYLD phosphorylation serves as a critical mechanism for regulating signal-induced TRAF2 ubiquitination and JNK activation.
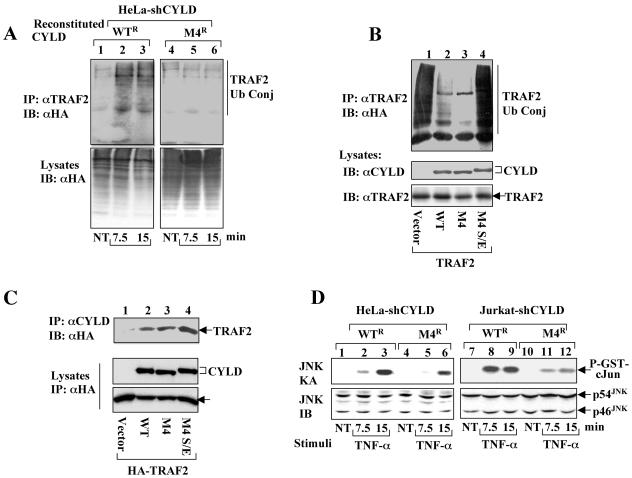
To understand the functional significance of CYLD phosphorylation, we first examined whether CYLD phosphorylation affected its ability to regulate TRAF2 ubiquitination. These studies were performed using the HeLa-shCYLD cells reconstituted with RNAi-resistant wild-type CYLD (WTR) or its phosphorylation-defective mutant (M4R). Upon stimulation with TNF-α, the endogenous TRAF2 was ubiquitinated in the CYLD WTR-reconstituted cells (Fig. 6A, upper panel, lanes 2 and 3). The inducible TRAF2 ubiquitination was not due to alteration in the level of overall protein ubiquitination in the cells (lower panel, lanes 1 to 3). Remarkably, the inducible ubiquitination of TRAF2 was largely defective in cells reconstituted with the phosphorylation-defective CYLD mutant (Fig. 6A, upper panel, lanes 5 and 6). Thus, the CYLD phosphorylation is required for TNF-α-stimulated TRAF2 ubiquitination. This finding, together with the results presented in Fig. 1, indicates that the CYLD phosphorylation may serve as a mechanism to inactivate its TRAF2 deubiquitination activity. To further examine this possibility, we generated a phospho-mimetic form of the CYLD M4 mutant by substituting the phosphorylation sites (serines) with glutamic acids (named M4 S/E). The constitutive TRAF2-deubiquitinating activity of CYLD and its mutants were analyzed by transient transfection in 293 cells. As previously reported, overexpressed TRAF2 underwent self-ubiquitination (Fig. 6B, lane 1), which was inhibited by coexpression with wild-type CYLD (lane 2). Consistent with its ability to block the inducible TRAF2 ubiquitination, the M4 mutant of CYLD retained the TRAF2-deubiquitinating activity (lane 3). Interestingly, however, the phospho-mimetic CYLD (M4 S/E) completely lost its ability to inhibit TRAF2 ubiquitination (Fig. 6B, lane 4). Parallel coIP assays revealed that both M4 and M4 S/E remained competent in association with TRAF2 (Fig. 6C). It is thus likely that the phosphorylation of CYLD does not prevent its binding to TRAF2 but may inactivate its DUB function by other mechanisms.
FIG. 6.
CYLD phosphorylation is required for signal-induced TRAF2 ubiquitination and optimal JNK activation. (A) Signal-induced TRAF2 ubiquitination requires CYLD phosphorylation. HeLa-shCYLD cells reconstituted with RNAi-resistant CYLD (WTR) or M4 (M4R) were transfected with HA-tagged ubiquitin. The cells were either not treated (NT) or stimulated with TNF-α. Ubiquitin-conjugated TRAF2 were isolated by IP using anti-TRAF2 followed by detection by IB using anti-HA-HRP (upper panel). The level of total ubiquitinated cellular proteins was analyzed by direct IB (lower panel). (B) Phospho-mimetic CYLD loses TRAF2-deubiquitinating activity. 293 cells were transfected with TRAF2 together with empty vector or expression vectors encoding wild-type CYLD (WT), M4, or a phospho-mimetic CYLD harboring serine/glutamic acid substitutions at the phosphorylation sites (M4 S/E). Ubiquitin-conjugated (Ub Conj) TRAF2 was isolated by IP using anti-TRAF2 and detected by IB using anti-HA-HRP (top panel). The expression of CYLD and TRAF2 proteins was monitored by IB using anti-CYLD (middle panel) and anti-TRAF2 (bottom panel). (C) coIP assays to detect the association of CYLD mutants with TRAF2. 293 cells were transfected with HA-tagged TRAF2 together with either an empty vector or expression vectors encoding HA-tagged wild-type (WT) CYLD, CYLD M4, or CYLD M4 S/E. The CYLD complexes were isolated by IP using anti-CYLD followed by detection of the associated HA-TRAF2 by IB using HRP-conjugated anti-HA (upper panel). The protein expression level was monitored by direct IB using HRP-conjugated anti-HA (lower panels). (D) Diminished activation of JNK in cells expressing the phosphorylation-defective CYLD mutant. CYLD-knockdown HeLa (HeLa-shCYLD) or Jurkat (Jurkat-shCYLD) cells were reconstituted with the RNAi-resistant form of wild-type CYLD (WTR) or the CYLD M4 mutant (M4R). Following TNF-α stimulation, JNK kinase activity and expression were determined by kinase assays (upper panel) and IB (lower panel), respectively.
We have recently shown that CYLD is a negative regulator of JNK in the TNF-α signaling pathway (29). Based on the results described above, we reasoned that CYLD phosphorylation might also be required for optimal activation of JNK. This hypothesis was examined using the CYLD-knockdown cells reconstituted with wild-type or mutant forms of exogenous CYLD. In response to TNF-α stimulation, JNK was activated in HeLa-shCYLD cells reconstituted with wild-type CYLD (Fig. 6D, lanes 1 to 3) and the level of JNK activation was comparable to the parental HeLa cells (data not shown). Interestingly, JNK activation was significantly attenuated in the cells reconstituted with CYLD M4 (lanes 4 to 6). Similar results were obtained with the Jurkat-shCYLD cells, which revealed a significant defect in JNK activation in the M4R-reconstituted cells (Fig. 6D, lanes 10 to 12) as compared with the WTR-reconstituted cells (lanes 7 to 9). These data suggest that CYLD phosphorylation is required for disrupting its negative regulatory function in JNK activation.
Functional consequence of CYLD phosphorylation in regulating gene expression.
We next analyzed the functional consequence of CYLD phosphorylation in gene induction. Reporter gene assays were performed to assess the role of CYLD phosporylation in regulating NF-κB-specific gene expression. We used CD40 as an inducer, because CYLD plays an important role in regulating CD40-mediated NF-κB signaling (29). As expected, CD40 strongly induced the κB-dependent reporter gene expression in Jurkat cells expressing the wild-type CYLD (Fig. 7A, column 2). In contrast, the CD40-induced κB response was drastically reduced in cells expressing the phosphorylation-defective CYLD mutant M4 (column 4).
FIG. 7.
Functional role of CYLD phosphorylation in gene induction. (A) Luciferase reporter gene assays to determine NF-κB activation by CD40. CYLD-knockdown Jurkat cells reconstituted with the RNAi-resistant form of wild-type CYLD (WTR) or the M4 mutant (M4R) were transfected with a κB-luciferase reporter (κB-TATA-luc) and a control Renilla luciferase reporter driven by the constitutive thymidine kinase promoter (pRL-TK). The cells were also transfected with either an empty vector (−) or a cDNA expression vector encoding human CD40 (+). After 40 h of transfection, cell lysates were prepared and subjected to dual-luciferase assays. The κB-specific luciferase activity was normalized based on the control Renilla luciferase activity and is presented as fold induction relative to the basal level measured in cells transfected with empty vector. The data are representative of two independent experiments. (B) RNase protection assay to analyze cellular genes regulated by CYLD. CYLD-knockdown HeLa cells reconstituted with CYLD WTR or M4R were either not treated (−) or were stimulated with TNF-α for 30 min (+). Total RNA was isolated and subjected to RPA.
We also analyzed the expression of endogenous genes induced by TNF-α. Although CYLD has no significant role in TNF-α-mediated NF-κB activation, it negatively regulates the TNF-α-stimulated JNK signaling pathway. RPA was performed to examine the expression of several genes known to be induced by TNF-α (29). Whereas most of these genes were similarly induced in cells expressing the wild-type or M4 mutant CYLD, the induction of one gene (that coding for RANTES) was markedly attenuated in the M4-expressing cells (Fig. 7B, top panel, lane 4). This result was not due to the variation in RNA amounts, since it was observed only with RANTES. Further, expression of two housekeeping genes (coding for L32 and GAPDH) suggests that the level of M4 cell RNA was even slightly higher than that of the wild-type cell RNA. This result is consistent with the involvement of JNK in RANTES gene induction (32). Since RANTES is a critical chemokine that mediates septic shock (27), this finding suggests a possible role for CYLD in regulating inflammatory responses.
DISCUSSION
Protein ubiquitination is positively regulated by ubiquitin enzymes and counterregulated by DUBs. Although the ubiquin enzymes have been extensively studied, little is known about how the DUBs are functionally regulated. In the present study, we have examined the molecular mechanism regulating the function of CYLD, a DUB that functions as a key negative regulator of cell signaling. Our data suggest that CYLD plays a dominant role in suppressing the ubiquitination of TRAF2, since CYLD knockdown results in constitutive TRAF2 ubiquitination (Fig. 1). We provide evidence that signal-induced TRAF2 ubiquitination and JNK activation require phosphorylation of CYLD, which appears to serve as a mechanism that inactivates the TRAF2 deubiquitination function of CYLD. Moreover, we show that IKKγ, a key component of the canonical IKK, is required for the inducible phosphorylation of CYLD.
The constitutive TRAF2 ubiquitination in CYLD-knockdown cells explains the basal activation of JNK and IKK associated with CYLD knockdown (29). However, since the kinase activity (29), but not the TRAF2 ubiquitination (Fig. 1), is further induced by TNF-α, it suggests that TRAF2 ubiquitination is not sufficient for triggering optimal activation of JNK or IKK. The finding that CYLD knockdown results in constitutive TRAF2 ubiquitination not only suggests a dominant function of the DUB CYLD but also implies that the ubiquitin ligase TRAF2 may undergo spontaneous ubiquitination. This hypothesis is consistent with the finding that TRAF2 undergo self-ubiquitination in transfected cells, which is inhibited by contransfection with CYLD (22, 28, 39) (Fig. 6B). Of course, since these results were obtained with transformed cell lines, it remains to be determined whether CYLD-deficient primary cells also exhibit basal TRAF2 ubiquitination activity. Nevertheless, it is conceivable that under normal conditions, the ubiquitinated TRAF2 may be rapidly deubiquitinated by CYLD. Based on this hypothesis, one can predict that induction of TRAF2 ubiquitination by cellular stimuli is achieved by either enhancing the ubiquitin ligase activity of TRAF2 or inhibiting the DUB function of CYLD. Whereas the former possibility remains to be investigated, we have obtained strong evidence that supports the latter possibility. We show that signal-induced TRAF2 ubiquitination is associated with phosphorylation of CYLD. When the endogenous CYLD is replaced with a phosphorylation-defective CYLD mutant, the inducible ubiquitination of TRAF2 is severely attenuated (Fig. 6A). A logical explanation of this finding is that phosphorylation serves as a mechanism that temporarily inactivates the DUB activity of CYLD, thus allowing the accumulation of ubiquitin-conjugated TRAF2. In further support of this hypothesis, a phospho-mimetic form of CYLD completely lost its TRAF2-deubiquitinating function (Fig. 6B).
The functional consequence of CYLD phosphorylation remains an interesting topic of investigation, since the in vivo physiological function of CYLD has not been well defined. Nevertheless, we have shown that interruption of CYLD phosphorylation markedly inhibits CD40-mediated induction of a κB-specific reporter gene (Fig. 7A). Further, in the TNF-α signaling pathway, the CYLD phosphorylation appears to be important for the expression of a chemokine gene, coding for RANTES (Fig. 7B). Future studies will employ the gene array technique to identify more genes under the regulation of CYLD. A prior study suggests that CYLD participates in TNF-α-induced apoptosis (6). We thus examined whether the phosphorylation of CYLD diminishes its proapoptotic function. To our surprise, we did not observe any significant effect of CYLD knockdown or CYLD phosphorylation on TNF-α-induced apoptosis in our system (see Fig. S1 in the supplemental material). Although the precise reason for this discrepancy is not clear, we have noticed that the previous study used a different protocol for apoptosis induction, which involves prestimulation of cells with PMA (6). Since CYLD knockdown has no obvious effect on TNF-α-stimulated NF-κB activation (29), we believe that CYLD may regulate cell survival downstream of specific receptors. The IKK-mediated CYLD phosphorylation likely provides a cross talk between the canonical NF-κB inducing signals and the CYLD-specific receptors. This idea is consistent with the fact that patients with CYLD genetic deficiency only develop a specific type of benign tumor, cylindromatosis (4), instead of having global abnormalities in cell growth/survival. Clearly, a better understanding of the functional consequence of CYLD phosphorylation requires more knowledge regarding the physiological function of CYLD, which in turn will rely on studies using an in vivo model system (e.g., CYLD knockout mice).
IKK is known as a kinase that specifically phosphorylates IκBα and related inhibitors, thereby mediating the nuclear translocation of NF-κB. One intriguing question is whether IKK has other substrates or mediates additional signaling functions. Novel functions have indeed been identified for the noncanonical IKK component IKKα. One such function of IKKα is regulation of epidermal differentiation and skeletal morphology, a process that does not require the catalytic activity of IKKα (35). Another interesting function of IKKα, which requires its kinase activity, occurs in the nucleus and involves phosphorylation of the histone H3 (3, 44, 45). This novel function of IKKα is required for transcriptional activation of NF-κB target genes. Our present study suggests that IKK also regulates the function of DUB CYLD. We have found that CYLD undergoes rapid and transient phosphorylation in cells stimulated with various known IKK inducers (Fig. 2). The CYLD phoshorylation is dependent on IKKγ, since it is blocked in IKKγ-deficient Jurkat T cells (Fig. 3A). Transfection and in vitro kinase assays reveal that both IKKα and IKKβ are able to phosphorylate CYLD (Fig. 3 and 4), although it remains unclear whether one or both of these IKK catalytic subunits are essential for CYLD phosphorylation. We attempted to address this question using mouse embryonic fibroblasts deficient in different IKK subunits. Unfortunately, the phosphorylation of CYLD could not be detected even in wild-type mouse embryonic fibroblasts (data not shown). Currently, we do not know whether this result is due to the variations in species (the anti-CYLD antibody is for human protein) or cell types. Nevertheless, at least in vitro, CYLD does not show preference to IKKα or IKKβ, a property that is different from IκBα and the IκB-like molecule p100, which are preferentially phosphorylated by IKKβ and IKKα, respectively (33). Another interesting feature of CYLD phosphorylation is the critical requirement of IKKγ. Although IKKα and IKKβ efficiently phosphorylate CYLD in vitro (Fig. 3G and 4C), induction of CYLD phosphorylation in vivo by these IKK catalytic subunits requires the assistance by IKKγ (Fig. 3B). One likely interpretation of this result is that IKKγ functions as both the regulatory subunit of IKK and an adaptor for recruiting CYLD to the IKK catalytic subunits. However, the possibility of the involvement of novel mechanisms in IKKγ function cannot be excluded. For example, IKKγ may regulate another kinase that cooperates with the classical IKK in CYLD phosphorylation. Notwithstanding, our findings clearly establish IKKγ as an essential factor in signal-induced CYLD phosphorylation. Phosphorylation of CYLD represents a novel aspect of IKK function, since it regulates the deubiquitination function of CYLD rather than triggering the degradation of this novel substrate.
Supplementary Material
Acknowledgments
We thank M. Karin and I. M. Verma for reagents and the Sun lab members for fruitful discussion. We also thank the Core Facility of Hershey Medical Center for oligonucleotide synthesis and DNA sequencing analyses.
This work was supported by research grants from the National Institutes of Health to S.-C.S (CA094922 and AI45045) and M.Z (AI45045) and the Four Diamond Fund at Penn State Children's Hospital to M.Z.W.R. was supported by a predoctoral/postdoctoral training grant (5 T32CA60395-09) from the National Institutes of Health.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal, B. B. 2003. Signalling pathways of the TNF superfamily: a double-edged sword. Nat. Rev. Immunol. 3:745-756. [DOI] [PubMed] [Google Scholar]
- 3.Anest, V., J. L. Hanson, P. C. Cogswell, K. A. Steinbrecher, B. D. Strahl, and A. S. Baldwin. 2003. Nucleosomal function for IkappaB kinase-alpha in NF-kappaB-dependent gene expression. Nature 423:659-663. [DOI] [PubMed] [Google Scholar]
- 4.Bignell, G. R., W. Warren, S. Seal, M. Takahashi, E. Rapley, R. Barfoot, H. Green, C. Brown, P. J. Biggs, S. R. Lakhani, C. Jones, J. Hansen, E. Blair, B. Hofmann, R. Siebert, G. Turner, D. G. Evans, C. Schrander-Stumpel, F. A. Beemer, A. van Den Ouweland, D. Halley, B. Delpech, M. G. Cleveland, I. Leigh, J. Leisti, and S. Rasmussen. 2000. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 25:160-165. [DOI] [PubMed] [Google Scholar]
- 5.Brooke, H. G. 1892. Epithelioma adenoides cysticum. Br. J. Dermatol. 4:269-287. [Google Scholar]
- 6.Brummelkamp, T. R., S. M. Nijman, A. M. Dirac, and R. Bernards. 2003. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424:797-801. [DOI] [PubMed] [Google Scholar]
- 7.Cao, Z., J. Xiong, M. Takeuchi, T. Kurama, and D. V. Goeddel. 1996. TRAF6 is a signal transducer for interleukin-1. Nature 383:443-446. [DOI] [PubMed] [Google Scholar]
- 8.Cvijic, M. E., G. Xiao, and S. C. Sun. 2003. Study of T-cell signaling by somatic cell mutagenesis and complementation cloning. J. Immunol. Methods 278:293-304. [DOI] [PubMed] [Google Scholar]
- 9.Davis, R. J. 2000. Signal transduction by the JNK group of MAP kinases. Cell 103:239-252. [DOI] [PubMed] [Google Scholar]
- 10.Deng, L., C. Wang, E. Spencer, L. Yang, A. Braun, J. You, C. Slaughter, C. Pickart, and Z. J. Chen. 2000. Activation of the IkB kinase complex by TRAF6 requires a dimeric ubiqutitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351-361. [DOI] [PubMed] [Google Scholar]
- 11.DiDonato, J. A., M. Hayakawa, D. M. Rothwarf, E. Zandi, and M. Karin. 1997. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature 388:548-554. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, J. A. 2003. Deubiquitinating enzymes: their roles in development, differentiation, and disease. Int. Rev. Cytol. 229:43-72. [DOI] [PubMed] [Google Scholar]
- 13.Habelhah, H., S. Takahashi, S. G. Cho, T. Kadoya, T. Watanabe, and Z. Ronai. 2003. Ubiquitination and translocation of TRAF2 is required for activation of JNK but not of p38 or NF-kappaB. EMBO J. 23:322-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harhaj, E. W., L. Good, G.-T. Xiao, M. Uhlik, M. E. Cvijic, I. Rivera, and S.-C. Sun. 2000. Somatic mutagenesis studies of NF-kappa B signaling in human T cells: evidence for an essential role of IKK gamma in NF-kappa B activation by T-cell costimulatory signals and HTLV-I Tax protein. Oncogene 19:1386-1391. [DOI] [PubMed] [Google Scholar]
- 15.Harhaj, E. W., and S.-C. Sun. 1999. Regulation of RelA subcellular localization by a putative nuclear export signal and p50. Mol. Cell. Biol. 19:7088-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 17.Ip, Y. T., and R. J. Davis. 1998. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr. Opin. Cell Biol. 10:205-219. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, L. E., and A. S. Whitehead. 2003. Ubiquitin activated tumor necrosis factor receptor associated factor-6 (TRAF6) is recycled via deubiquitination. FEBS Lett. 553:190-194. [DOI] [PubMed] [Google Scholar]
- 19.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 20.Karin, M., and M. Delhase. 2000. The I kappa B kinase (IKK) and NF-kappa B: key elements of proinflammatory signalling. Semin. Immunol. 12:85-98. [DOI] [PubMed] [Google Scholar]
- 21.Kim, J. H., K. C. Park, S. S. Chung, O. Bang, and C. H. Chung. 2003. Deubiquitinating enzymes as cellular regulators. J. Biochem. 134:9-18. [DOI] [PubMed] [Google Scholar]
- 22.Kovalenko, A., C. Chable-Bessia, G. Cantarella, A. Israel, D. Wallach, and G. Courtois. 2003. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424:801-805. [DOI] [PubMed] [Google Scholar]
- 23.Lin, A., and B. Dibling. 2002. The true face of JNK activation in apoptosis. Aging Cell 1:112-116. [DOI] [PubMed] [Google Scholar]
- 24.Lorick, K. L., J. P. Jensen, S. Fang, A. M. Ong, S. Hatakeyama, and A. M. Weissman. 1999. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA 96:11364-11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manning, A. M., and R. J. Davis. 2003. Targeting JNK for therapeutic benefit: from junk to gold? Nat. Rev. Drug Discov. 2:554-565. [DOI] [PubMed] [Google Scholar]
- 26.Naviaux, R. K., E. Costanzi, M. Haas, and I. M. Verma. 1996. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J. Virol. 70:5701-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ness, T. L., K. J. Carpenter, J. L. Ewing, C. J. Gerard, C. M. Hogaboam, and S. L. Kunkel. 2004. CCR1 and CC chemokine ligand 5 interactions exacerbate innate immune responses during sepsis. J. Immunol. 173:6938-6948. [DOI] [PubMed] [Google Scholar]
- 28.Regamey, A., D. Hohl, J. W. Liu, T. Roger, P. Kogerman, R. Toftgard, and M. Huber. 2003. The tumor suppressor CYLD interacts with TRIP and regulates negatively nuclear factor kappaB activation by tumor necrosis factor. J. Exp. Med. 198:1959-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiley, W., M. Zhang, and S.-C. Sun. 2004. Tumor suppressor negatively regulates JNK signaling pathway downstream of TNFR members. J. Biol. Chem. 279:55161-55167. [DOI] [PubMed] [Google Scholar]
- 30.Rivera-Walsh, I., M. E. Cvijic, G. Xiao, and S. C. Sun. 2000. The NF-kappa B signaling pathway is not required for Fas ligand gene induction but mediates protection from activation-induced cell death. J. Biol. Chem. 275:25222-25230. [DOI] [PubMed] [Google Scholar]
- 31.Rivera-Walsh, I., M. Waterfield, G. Xiao, A. Fong, and S.-C. Sun. 2001. NF-κB signaling pathway governs TRAIL gene expression and HTLV-I Tax-induced T-cell death. J. Biol. Chem. 276:40385-40388. (First published 11 September 2001; doi:10.1074/jbc.C100501200.) [DOI] [PubMed] [Google Scholar]
- 32.Schwabe, R. F., R. Bataller, and D. A. Brenner. 2003. Human hepatic stellate cells express CCR5 and RANTES to induce proliferation and migration. Am. J. Physiol. Gastrointest. Liver Physiol. 285:G948-G958. [DOI] [PubMed] [Google Scholar]
- 33.Senftleben, U., Y. Cao, G. Xiao, G. Kraehn, F. Greten, Y. Chen, Y. Hu, A. Fong, S.-C. Sun, and M. Karin. 2001. Activation of IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science 293:1495-1499. [DOI] [PubMed] [Google Scholar]
- 34.Shi, C. S., and J. H. Kehrl. 2003. TNF-induced GCKR and SAPK activation depends upon the E2/E3 complex Ubc13-Uev1A/TRAF2. J. Biol. Chem. 278:15429-15434. [DOI] [PubMed] [Google Scholar]
- 35.Sil, A. K., S. Maeda, Y. Sano, D. R. Roop, and M. Karin. 2004. IkappaB kinase-alpha acts in the epidermis to control skeletal and craniofacial morphogenesis. Nature 428:660-664. [DOI] [PubMed] [Google Scholar]
- 36.Silverman, N., and T. Maniatis. 2001. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15:2321-2342. [DOI] [PubMed] [Google Scholar]
- 37.Spiegler, E. 1899. Ueber endotheliome der haut. Arch. Dermatol. Syphilol. 50:163-176. [Google Scholar]
- 38.Sun, L., L. Deng, C.-K. Ea, Z.-P. Xia, and Z. J. Chen. 2004. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Mol. Cell 14:289-301. [DOI] [PubMed] [Google Scholar]
- 39.Trompouki, E., E. Hatzivassiliou, T. Tsichritzis, H. Farmer, A. Ashworth, and G. Mosialos. 2003. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424:793-796. [DOI] [PubMed] [Google Scholar]
- 40.Uhlik, M., L. Good, G. Xiao, E. W. Harhaj, E. Zandi, M. Karin, and S.-C. Sun. 1998. NF-kappaB-inducing kinase and IkappaB kinase participate in human T-cell leukemia virus I Tax-mediated NF-kappaB activation. J. Biol. Chem. 273:21132-21136. [DOI] [PubMed] [Google Scholar]
- 41.Wang, C., L. Deng, M. Hong, G. R. Akkaraju, J.-I. Inoue, and Z. J. Chen. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412:346-351. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, G., M. E. Cvijic, A. Fong, E. W. Harhaj, M. T. Uhlik, M. Waterfield, and S. C. Sun. 2001. Retroviral oncoprotein Tax induces processing of NF-kappaB2/p100 in T cells: evidence for the involvement of IKKalpha. EMBO J. 20:6805-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao, G., E. W. Harhaj, and S. C. Sun. 2001. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell 7:401-409. [DOI] [PubMed] [Google Scholar]
- 44.Yamamoto, Y., and R. B. Gaynor. 2004. IkappaB kinases: key regulators of the NF-kappaB pathway. Trends Biochem. Sci. 29:72-79. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, Y., U. N. Verma, S. Prajapati, Y. T. Kwak, and R. B. Gaynor. 2003. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature 423:655-659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.