Abstract
Ctk1 is a Saccharomyces cerevisiae cyclin-dependent protein kinase (CDK) that assembles with Ctk2 and Ctk3 to form an active protein kinase complex, CTDK-I. CTDK-I phosphorylates Ser2 within the RNA polymerase II C-terminal domain, an activity that is required for efficient transcriptional elongation and 3′ RNA processing. Ctk1 contains a conserved T loop, which undergoes activating phosphorylation in other CDKs. We show that Ctk1 is phosphorylated on Thr-338 within the T loop. Mutation of this residue abolished Ctk1 kinase activity in vitro and resulted in a cold-sensitive phenotype. As with other yeast CDKs undergoing T-loop phosphorylation, Ctk1 phosphorylation on Thr-338 was dependent on the Cak1 protein kinase. Ctk1 isolated from cak1Δ cells was unphosphorylated and exhibited low protein kinase activity. Moreover, Cak1 directly phosphorylated Ctk1 in vitro. Unlike wild-type cells, cells expressing Ctk1T338A delayed growth at early stationary phase, did not show the increase in Ser2 phosphorylation that normally accompanies the transition from rapid growth to stationary phase, and had compromised transcriptional activation of two stationary-phase genes, CTT1 and SPI1. Therefore, Ctk1 phosphorylation on Thr-338 is carried out by Cak1 and is required for normal gene transcription during the transition into stationary phase.
Cyclin-dependent protein kinases (CDKs) are best known for their roles in cell cycle progression. CDK activity is regulated by phosphorylation, cyclin availability, and binding to inhibitory proteins. Full activation of CDKs requires phosphorylation on a conserved threonine residue within the so-called T loop (or activation segment) that is mediated by Cdk-activating kinases (CAKs) (23). The CAK in the budding yeast Saccharomyces cerevisiae is Cak1 (11, 22, 41). Cak1 phosphorylates the main cell cycle CDK in yeast, Cdc28, on Thr-169. This phosphorylation stabilizes cyclin binding and is essential for protein kinase activity (37). Cak1 functions as a monomer and does not require posttranslational modification (21). In addition to Cdc28, Cak1 phosphorylates a number of other CDKs (see below), including those that regulate transcription by RNA polymerase II (pol II).
The largest subunit of RNA pol II contains a conserved C-terminal domain (CTD) composed of tandem repeats of the consensus heptapeptide sequence YSPTSPS (7, 9). Phosphorylation of Ser2 and Ser5 within the heptapeptide functions during transcriptional initiation, elongation, and RNA processing (5, 32, 34, 40). Phosphorylations of the two serine residues have essential nonredundant functions (45). Phosphorylation of Ser5 regulates the transition from transcriptional initiation to elongation and is limited to polymerases within the promoter region. In contrast, phosphorylation of Ser2 occurs after transcriptional initiation and remains high until transcriptional termination (4, 28). Phosphorylation of Ser2 increases the affinity of the polymerase for polyadenylation factors, enabling efficient processing of the pre-mRNA 3′ end (1, 38). Phosphorylated Ser5 and Ser2 also provide binding sites for the histone methyltransferases Set1 and Set2, respectively, facilitating passage of the polymerase through chromatin (25, 33, 43).
Four different cyclin-dependent kinases (CDKs), Kin28, Srb10, Bur1, and Ctk1, phosphorylate the CTD in S. cerevisiae. Kin28 phosphorylates Ser5, and this phosphorylation is required for transcription of most RNA pol II genes (6, 13, 18). Srb10 phosphorylates Ser5 and Ser2, but the effect of this phosphorylation is inhibitory (3, 17). Ctk1 is the major kinase acting on Ser2; recent findings indicate that it also phosphorylates Ser5 (20, 30, 36). Dephosphorylation of Ser5 and Ser2 is accomplished by the Ssu72 and Fcp1 phosphatases at the end of the transcription cycle (27, 29).
Phosphorylation of Ser2 by Ctk1 has been implicated in the control of transcript elongation (4, 19, 31). Recent studies revealed that it is also required for cotranscriptional recruitment of polyadenylation factors (1). In the absence of Ctk1, transcription termination and processing of RNA 3′ ends are compromised. Surprisingly, CTK1 is not an essential gene but its mutation confers a conditional cold-sensitive phenotype (30). Ctk1 assembles with Ctk2 and Ctk3 into an active protein kinase complex that phosphorylates RNA pol II on Ser2 (39). Ctk2 and Ctk3 are unstable proteins degraded through the ubiquitin-proteasome pathway (15, 16). Ctk1 activity and Ser2 phosphorylation of RNA pol II increase under a variety of stress conditions (35, 36).
Like the cell cycle CDKs, most CDKs functioning during transcription undergo activating phosphorylation within their T loops. Thus, previous studies demonstrated that Cak1 phosphorylates Kin28 and Bur1 within their T loops on Thr162 and Thr240, respectively (12, 26, 44). These phosphorylations are not essential under normal conditions but provide growth advantages when CDK-cyclin interactions are weakened by additional mutations (24, 26). Substitutions within the T loops of Kin28 and Bur1 compromise CTD phosphorylation in vitro (24, 26, 44).
We noticed that Ctk1 contains a typical T loop and examined whether it is phosphorylated. We found that Ctk1 was indeed phosphorylated on a conserved Thr-338 in vivo. Mutation of Thr-338 conferred a cold-sensitive phenotype and compromised the protein kinase activity of Ctk1. Several lines of evidence indicated that this phosphorylation was carried out by Cak1. We also found that Ctk1 phosphorylation and the increased phosphorylation of RNA pol II on Ser2 are essential to activate transcription of stationary-phase genes during the diauxic shift.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Yeast strains were derivatives of W303a (ade2-1 trp1-1 leu2-3,112 his3-11,15 ura3-1). ctk1Δ (W303a [ctk1Δ::HIS3]), ctk2Δ (W303a [ctk2Δ::URA3]), and ctk3Δ (W303a [ctk3Δ::URA3]) were previously described (35). ctk1T338A and ctk1D324N were generated by QuikChange mutagenesis (Stratagene) with a YCplac22-CTK1-HA plasmid as template. The parental and mutant plasmids were transformed into ctk1Δ cells, and TRP+ clones corresponding to wild type and the ctk1T338A and ctk1D324N mutants were collected. To improve the resolution of Ctk1 on two-dimensional gels, a stretch of negatively charged amino acids, DEDEDEDE, was introduced at its C terminus by PCR amplification, resulting in CTK1-8DE-HA. For analysis of the cold-sensitive phenotype and of RNA pol II phosphorylation, CTK1 and ctk1T338A were used without tags. All PCR products were sequenced to verify that no additional mutations were introduced. ctk1Δ deletion strain 1834-2A (cdc28::HIS3 cak1::LEU2/pCDC28-169-43244B) was described previously (8).
Yeast cultures were grown in YPD medium and in complete minimal medium as previously described (2). For analysis of cold-sensitive phenotypes, yeast cultures were serially diluted fivefold and spotted on YPD plates that were then incubated at 30°C and 17°C for 4 days.
Protein analyses and immunoprecipitation.
Cells from 25-ml cultures (optical density at 600 nm [OD600], ∼0.6) were collected, washed with ice-cold TBS (10 mM Tris-Cl [pH 8.0], 150 mM NaCl), and suspended in 0.4 ml lysis buffer (50 mM Tris-Cl [pH 8.0], 10 mM MgCl2, 150 mM NaCl, 10 mM EDTA, 10% glycerol, 10 μg/ml each of leupeptin, chymostatin, and pepstatin [Chemicon], 1 mM dithiothreitol [DTT]). Cell lysis was achieved by shaking yeast suspensions with 0.4 g of glass beads (0.5-mm diameter; Sigma) in a bead beater (Biospec Products) for five 30-s pulses with 30 s on ice between pulses. Glass beads and cell debris were removed by centrifugation at 14,000 rpm in a Microfuge at 4°C for 10 min. The supernatant was clarified by centrifugation at 65,000 rpm in a TLA 100.2 rotor (Beckman) for 10 min at 4°C. Protein concentrations were determined using the Bradford assay (Bio-Rad) with bovine serum albumin as the standard. Yeast extracts were aliquoted, frozen in liquid nitrogen, and stored at −80°C.
For immunoprecipitation of Ctk1-HA, 1 μg of monoclonal antibody 12CA5 (Covance) was bound to 50 μl protein A-agarose (Invitrogen) at 4°C for 2 h. One hundred fifty microliters of yeast extract containing 10 mg/ml of protein was incubated with 50 μl of 12CA5 beads in 300 μl immunoprecipitation buffer (50 mM Tris-Cl [pH 8.0], 15 mM MgCl2, 10 mM EDTA, 1% NP-40, 1 mM DTT, 10 μg/ml each of leupeptin, chymostatin, and pepstatin [Chemicon], 0.5 mg/ml ovalbumin) at 4°C for 2 h. The beads were washed three times with 0.5 ml immunoprecipitation buffer for 5 min each at 4°C and stored at −20°C.
To detect Ctk1-HA by immunoblotting, 20 μg of yeast extract proteins was separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and transferred to an Immobilon-P membrane (Millipore). The membrane was incubated with rabbit anti-HA antibodies (100 ng/ml; Santa Cruz). Cak1 was detected using monoclonal anti-Cak1 antibody as described previously (21). Phosphorylated and unphosphorylated forms of RNA pol II were detected with H5 and 8WG16 antibodies (Covance) as described previously (35, 36). Proteins were visualized by chemiluminescence (SuperSignal; Pierce).
To fractionate Ctk1-containing complexes, yeast extracts were prepared from 400 ml of cells (OD600, ∼0.6), as described above. Yeast extracts (1 ml, 10 mg/ml protein) were fractionated on a Superdex-200 gel filtration column (Amersham Biosciences) in 25 mM HEPES (pH 7.5)-350 mM NaCl-1 mM DTT-10 μg/ml each of leupeptin, chymostatin, and pepstatin (Chemicon). A 10-μl aliquot of each 1-ml fraction was resolved by SDS-PAGE, and Ctk1-HA was detected by immunoblotting with rabbit anti-HA antibodies as described above.
Two-dimensional gel electrophoresis.
Ctk1-8DE-HA was resolved by nonequilibrium pH gel electrophoresis. One hundred microliters of yeast extract containing 1 mg of protein was treated with 2 μl DNase and 2 μl RNase (10 U/μl each; Roche) in 80 mM β-glycerophosphate-60 mM NaCl-10 mM MgSO4-10 mM EDTA-10% NP-40-10 μg/ml each of leupeptin, chymostatin, and pepstatin (Chemicon) for 30 min on ice. The proteins were precipitated with 9 volumes of cold acetone for 1 h at −20°C, centrifuged at 7,000 rpm for 10 min in a Microfuge at 4°C, and air dried. The protein pellets were dissolved in 200 μl 9.5 M urea-2% Triton X-100-1% ampholyte 3/10-1% ampholyte 7/9 (Bio-Rad)-100 mM DTT. Ten-microliter samples were loaded onto 8-mm tube gels (Bio-Rad) containing 9.8 M urea, 2% Triton, 4% acrylamide-bisacrylamide (19:1), 1% ampholyte 3/10, and 1% ampholyte 7/9 (Bio-Rad). Proteins were focused for 1,900 V-h with 10 mM phosphoric acid and 20 mM NaOH in the upper and lower buffer chambers, respectively. Following separation in the first dimension, gels were equilibrated in 62 mM Tris-Cl (pH 8.0)-2% SDS-10% glycerol, and proteins were separated by SDS-PAGE. Ctk1-8DE-HA was detected by immunoblotting using rabbit anti-HA antibodies as described above.
Protein kinase assays. (i) CTD kinase assay.
Immunoprecipitated Ctk1-HA complexes (8 μl; see above) were incubated with 4 μg CTD peptide (YSPTSPS)4 in the presence of 5 μCi [γ-32P]ATP and 10 μM ATP in 20 μl KB buffer (50 mM Tris-Cl [pH 8.0], 15 mM MgCl2, 150 mM NaCl, 0.1% Tween, 10 mM DTT, 0.1 mg/ml ovalbumin, 10 μg/ml each of leupeptin, chymostatin, and pepstatin [Chemicon]) for 60 min at 23°C. The reaction products were separated by SDS-PAGE and analyzed by PhosphorImaging (Molecular Dynamics) and by autoradiography.
(ii) Phosphorylation of Ctk1.
Ctk1-HA was expressed in the Coupled Transcription/Translation System (Promega) according to the manufacturer's protocol. Synthesized Ctk1-HA was immunoprecipitated (see above). GST-Cak1 was purified from insect cells as described previously (42). Ctk1-HA beads (8 μl) were incubated with 0.4 μg recombinant GST-Cak1 in the presence of 5 μCi [γ-32P]ATP and 10 μM ATP in 20 μl KB buffer for 90 min at 23°C. The reaction products were separated by SDS-PAGE and analyzed by PhosphorImaging (Molecular Dynamics) and autoradiography.
RNA isolation and analysis.
Total RNA was isolated from 100-ml cultures (OD600, ∼0.6) by an acid lysis protocol. Briefly, cells were pelleted, resuspended in 1.5 ml lysis buffer (10 mM Tris-Cl [pH 7.5], 10 mM EDTA, 0.5% SDS), and incubated with 1.5 ml acidic phenol, pH ∼4.5 (Gibco BRL) for 1 h at 65°C with brief vortexing every 10 min. Aqueous phases were extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and chloroform-isoamyl alcohol (24:1). The RNA was ethanol precipitated for 3 h at −20°C, centrifuged at 10,000 rpm for 15 min at 4°C, washed with 70% ethanol, and resuspended in 300 μl diethyl pyrocarbonate-treated H2O. RNA concentrations were determined spectrophotometrically. For hybridization analysis, 20 μg of RNA was electrophoresed in 1% formaldehyde-agarose gels and transferred to GeneScreen membranes (NEN Life Science Products). DNA probes were labeled with [α-32P]dCTP using a random priming kit (Roche). Hybridizations were carried out for 10 h at 42°C according to the manufacturer's protocol. Signals were detected by PhosphorImaging (Molecular Dynamics) and by autoradiography.
Microarray hybridization and analysis.
For microarray hybridization, mRNA was isolated using an Oligotex column (QIAGEN). cDNA probes were synthesized using Superscript II reverse transcriptase (Life Technologies) in the presence of Cy3- and Cy5-conjugated dCTP (Amersham). Cy3- and Cy5-labeled probes corresponding to wild-type and mutant samples were combined and hybridized with custom-made yeast whole-genome microarrays (Yale Microarray Facility). The microarrays were read on a laser scanner GenePix4000A (Axon Instruments Inc.). The fold change for every gene was calculated as an average of pixel-by-pixel ratios using the GenePix Pro software (Axon Instruments Inc.).
RESULTS
Ctk1p is phosphorylated on Thr-338 in vivo.
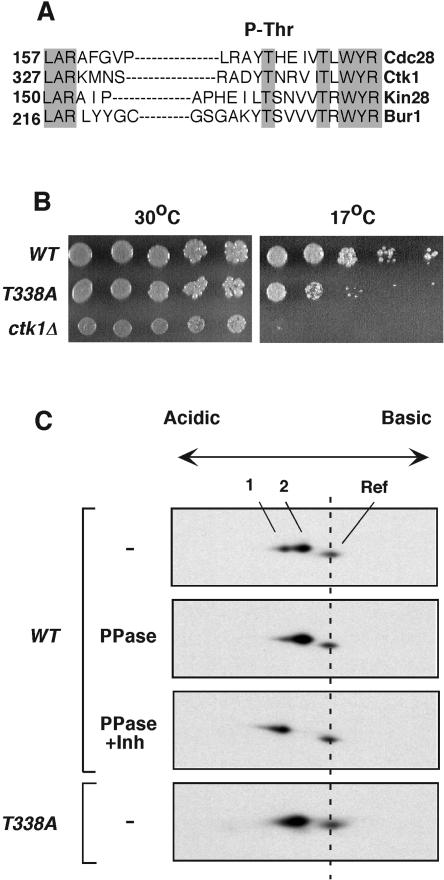
A comparison of the T-loop regions of yeast CDKs suggested that Ctk1 might be phosphorylated on Thr-338 (Fig. 1A). To assess this possibility, we mutated Thr-338 to the nonphosphorylatable alanine and examined the phenotype of the resulting ctk1T338A strain. CTK1 is not essential for viability, but its deletion causes cells to grow slowly at normal temperatures and to die when incubated at low temperatures. ctk1T338A cells grew as well as wild-type cells at 30°C. The growth of ctk1T338A cells decreased at 17°C, but they were still able to form colonies, unlike the ctk1Δ strain, which was inviable (Fig. 1B). We also noticed that ctk1T338A cells had a specific growth delay during the diauxic shift, when yeast cells prepare to enter stationary phase (see below). Taken together, the phenotypes of ctk1T338A cells suggested that Thr-338 was essential for Ctk1 activity under suboptimal conditions.
FIG. 1.
Ctk1 is phosphorylated on Thr-338. (A) Protein sequence alignment of T-loop regions in four S. cerevisiae CDKs. Grey shading denotes conserved residues. Numbers indicate positions of the T loops within the individual CDKs. The alignment was done using the Clustal algorithm. The site of activating phosphorylation is indicated at the top. (B) Serial dilutions of wild-type (WT) and ctk1T338A and ctk1Δ mutant cultures were spotted onto YPD plates. The plates were incubated at 30°C and at 17°C and photographed after 4 days. (C) Two-dimensional gel analysis of Ctk1. Yeast proteins were separated by nonequilibrium pH gel electrophoresis according to their charges and molecular weights. Ctk1-8DE-HA contained eight negatively charged amino acids to compensate for the large positive charge of the protein. Ctk1-8DE-HA was detected by immunoblotting with rabbit anti-HA antibodies. In wild-type yeast extracts, Ctk1-8DE-HA migrated as a doublet (spots 1 and 2, top). Yeast extracts were treated with λ phosphatase (PPase) in the absence (second panel) or presence (third panel) of phosphatase inhibitors (Inh). Ctk1T338A-8DE-HA is shown in the lower panel. A yeast extract containing Ctk1-HA with no added acidic residues was included in the loading mixtures to provide a reference (Ref) mark to monitor the positions of the Ctk1-8DE-HA spots. Ctk1-HA is more positively charged and smaller than Ctk1-8DE-HA. Ctk1-HA was not treated with phosphatase, and its position remained constant in the various experiments.
We next determined whether Ctk1 is phosphorylated on Thr-338 in vivo. Protein phosphorylation is often detectable by mobility shift during migration of a protein through polyacrylamide gels. To this end, a tagged version of Ctk1 was expressed in ctk1Δ cells and analyzed on high-resolution gels by immunoblotting. Unfortunately, Ctk1-HA migrated through one-dimensional gels as a single band, which was consistent with previous reports. We then attempted to resolve Ctk1 on two-dimensional gels, separating proteins according to their charge in the first dimension and by their molecular weight in the second dimension. We found in preliminary experiments that the extremely basic isoelectric point of Ctk1-HA (pI = 9.4) significantly reduced resolution in the first dimension. To compensate for this effect, we appended eight negatively charged amino acids to the C terminus of Ctk1. The resulting protein, Ctk1-8DE-HA, was judged functional based on its ability to complement the cold-sensitive phenotype of ctk1Δ cells. Ctk1-8DE-HA could be resolved into two distinct spots, suggestive of posttranslational modification (Fig. 1C, top). A reference protein (Ctk1-HA without the introduced acidic residues) was added from a parallel extract to allow comparison of Ctk1-8DE-HA mobilities on different gels. Yeast extract containing Ctk1-8DE-HA was then treated with λ phosphatase in the absence or presence of phosphatase inhibitors. The acidic spot disappeared after phosphatase treatment, indicating that it corresponded to phosphorylated Ctk1-8DE-HA (Fig. 1C, spot 1, middle panel). The reference protein was mock treated, so that its mobility was unaltered. Conversely, Ctk1T338-8DE-HA migrated as a single spot with the same mobility as phosphatase-treated wild-type Ctk1 (Fig. 1C, spot 2, lower panel and second panel). More of the Ctk1-8DE-HA was present in the acidic spot after phosphatase inhibitor treatment (Fig. 1C, third panel) than in the untreated sample (Fig. 1C, first panel), suggesting that the inhibitors also prevented dephosphorylation by endogenous phosphatases (see also Fig. 3A). We concluded that spots 1 and 2 correspond to phosphorylated and unphosphorylated forms of Ctk1-8DE-HA, respectively. Taken together, these experiments indicated that Ctk1 is phosphorylated in vivo at a unique site corresponding to Thr-338. A recent proteomic study also identified Thr-338 as a site of Ctk1 phosphorylation (14).
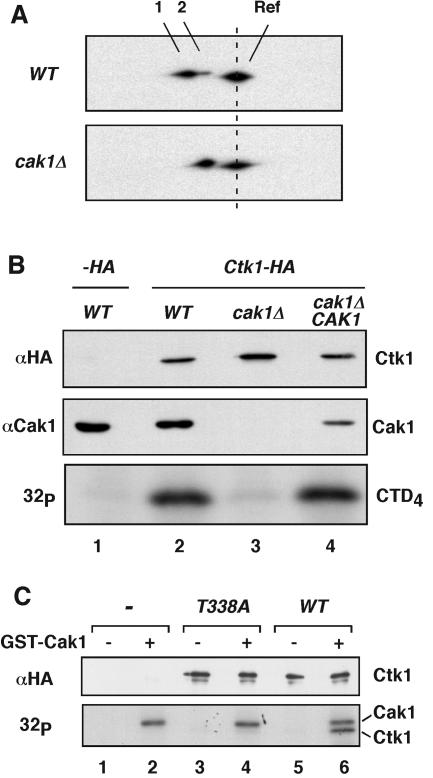
FIG. 3.
Cak1p phosphorylates Ctk1 on Thr-338. (A) Ctk1-8DE-HA was expressed in wild-type (WT) and cak1Δ cells and resolved on two-dimensional gels as described in the legend to Fig. 1C. Spots 1 and 2 correspond to phosphorylated and unphosphorylated forms of Ctk1-8DE-HA, respectively. Ctk1-HA was included as a reference (Ref) protein. (B) Yeast extracts were prepared from wild-type, cak1Δ, and cak1Δ/CAK1 cells also containing an empty plasmid (−HA, lane 1) or a plasmid expressing Ctk1-HA (lanes 2 to 4). The presence of Cak1p was determined by immunoblotting with an anti-Cak1p monoclonal antibody (middle). Ctk1-HA-containing complexes were immunoprecipitated. The level of precipitated Ctk1-HA was determined by immunoblotting using rabbit anti-HA antibodies (top). The immunoprecipitated Ctk1 was also used to phosphorylate CTD4 in the presence of [γ-32P]ATP (bottom). (C) Coupled transcription/translation reactions were programmed with an empty vector (−), ctk1T338A-HA, and CTK1-HA. The expressed proteins were immunoprecipitated with 12CA5 antibodies and incubated with [γ-32P]ATP in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of recombinant GST-Cak1p (bottom). The abundance of synthesized Ctk1-HA was verified by immunoblotting with rabbit anti-HA antibodies (top). Note that we used GST-Cak1p in this experiment, which has a molecular mass of 67 kDa and runs close to Ctk1-HA (63 kDa).
T-loop phosphorylation is required for Ctk1 activity.
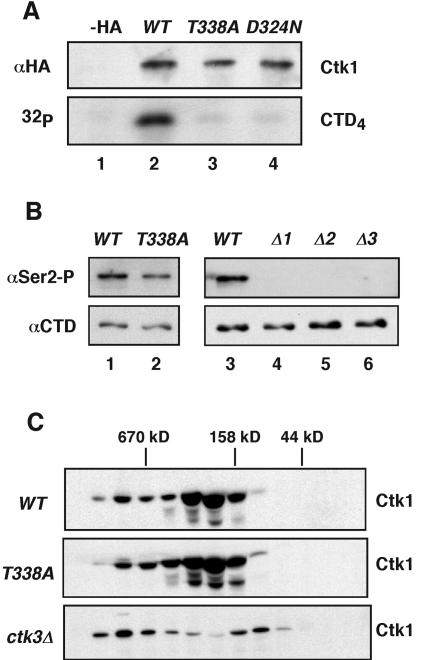
We next asked whether this phosphorylation is essential for Ctk1's protein kinase activity. To this end, wild-type and mutant forms of Ctk1 were immunoprecipitated from yeast extracts and their kinase activities were examined. We found that wild-type Ctk1 efficiently phosphorylated a CTD peptide in vitro, whereas Ctk1T338A displayed background activity (Fig. 2A, lanes 2 and 3) comparable to that of Ctk1D324N (Fig. 2A, lane 4). The D324N mutation alters a key catalytic residue and interferes with ATP binding, giving rise to cold-sensitive and slow-growth phenotypes as severe as those found in ctk1Δ cells. The three Ctk1 proteins were expressed at comparable levels (Fig. 2A, top; data not shown).
FIG. 2.
Thr-338 phosphorylation is required for Ctk1 activity. (A) Ctk1-HA was immunoprecipitated from yeast strains expressing vector (−HA), wild-type (WT) Ctk1, Ctk1T338A, and Ctk1D324A (lanes 1 to 4). The expression levels and efficiencies of immunoprecipitation were verified by immunoblotting with rabbit anti-HA antibodies (top). Immunoprecipitated Ctk1 complexes were assayed for their CTD kinase activity using a CTD4 peptide as the substrate in the presence of [γ-32P]ATP (bottom). The reaction products were resolved by SDS-PAGE and visualized by autoradiography. (B) Yeast strains corresponding to CTK1, ctk1T338A (lanes 1 and 2) and the wild type and ctk1Δ, ctk2Δ, and ctk3Δ mutants (lanes 3 to 6) were examined for their levels of RNA pol II phosphorylation. Proteins were resolved by SDS-PAGE and immunoblotted for the presence of phosphorylated serine 2 (Ser2-P) using H5 antibodies; unphosphorylated RNA pol II was detected using 8WG16 antibodies (bottom). (C) Yeast extracts from CTK1, ctk1T338A, and ctk3Δ strains were fractionated on a Sephadex-200 gel filtration column and resolved by SDS-PAGE. The distribution of Ctk1-HA among the fractions was determined by immunoblotting with rabbit anti-HA antibodies. The positions of molecular weight markers are indicated at the top.
Since T-loop phosphorylation was important in vitro, we next examined whether it is also essential for Ctk1 activity in living cells. We used monoclonal antibodies that recognize different forms of RNA pol II to compare the extents of Ser2 phosphorylation in wild-type and ctk1T338A cells. In contrast to the absence of detectable activity in vitro, Ctk1T338A retained the ability to phosphorylate Ser2 in vivo, albeit at a reduced level (Fig. 2B, lanes 1 and 2). Deletion of CTK1 or of any of its cyclin subunits eliminated Ser2 phosphorylation (Fig. 2B, lanes 4 to 6). These results are consistent with the reduction in growth of ctk1T338A cells and the inability of ctk1Δ cells to grow at low temperatures.
Because phosphorylation of the T loop increases cyclin binding to some CDKs, we wondered whether Ctk2 and Ctk3 could associate stably with unphosphorylated Ctk1. To address this question, we fractionated yeast extracts from wild-type and ctk1T338A mutant cells on gel filtration columns and compared the sizes of Ctk1-containing complexes. We found that Ctk1 eluted as a complex of ∼160 kDa (Fig. 2C) that probably contained Ctk2 and Ctk3. Ctk1T338A showed the same elution pattern. In contrast, Ctk1 isolated from ctk3Δ cells appeared in fractions of lower molecular weight, indicating that Ctk3 was a component of the 160-kDa complex and that binding of Ctk3 was not affected by the absence of T-loop phosphorylation in cells expressing Ctk1T338A. Therefore, it is unlikely that loss of Ctk1T338A activity in vitro resulted from dissociation of its cyclin subunits. It is worth noting that the level of Ctk1 in ctk2Δ and ctk3Δ mutant cells was significantly lower than that in wild-type cells, suggesting that binding to Ctk2 and Ctk3 stabilized Ctk1 (Fig. 2C and data not shown).
Cak1 phosphorylates Ctk1 within the T loop.
The primary candidate for Ctk1 phosphorylation was Cak1, as it phosphorylates other yeast CDKs within their T loops. Therefore, we first examined whether Ctk1 phosphorylation is Cak1 dependent. Although CAK1 is normally an essential gene, it can be rendered nonessential in the presence of a form of Cdc28 (Cdc28-169-43244B) containing a number of mutations, including a T169E mutation of its site of activating phosphorylation, making its activation by cyclins independent of Cak1 (8). In this experiment, Ctk1-8DE-HA was expressed in wild-type and cak1Δ mutant cells and analyzed by two-dimensional gel electrophoresis. As demonstrated earlier, Ctk1-8DE-HA isolated from wild-type cells migrated as a doublet (Fig. 1C and 3A, spots 1 and 2). In contrast, Ctk1-8DE-HA expressed in cak1Δ cells migrated as a single spot corresponding to the unphosphorylated form of Ctk1-8DE-HA (Fig. 3A, bottom).
Using a similar approach, we then assessed whether Cak1 is required for Ctk1 activity. To this end, Ctk1 was immunoprecipitated from cak1Δ cells and assayed for its CTD kinase activity in vitro. We also immunoblotted cell extracts to determine their levels of Cak1 and Ctk1. As expected, Cak1 was absent from cak1Δ cells, whereas expression of Ctk1-HA was comparable in CAK1 and cak1Δ cells (Fig. 3B, lanes 2 and 3). Deletion of CAK1 eliminated detectable Ctk1 activity (Fig. 3B, bottom, compare lanes 2 and 3), and reintroduction of CAK1 restored Ctk1 activity (Fig. 3B, lane 4). Similar results were obtained using a conditional cak1-23 mutant at the nonpermissive temperature (data not shown). We conclude that Ctk1 activity depends strictly on the integrity of its T loop and the presence of Cak1.
Because Cak1 also phosphorylates Kin28 and Bur1, two CDKs that interact with RNA pol II transcription complexes and function in close proximity to Ctk1, it was formally possible that the Cak1-dependent phosphorylation of Ctk1 was mediated by Kin28 or Bur1. To rule out these possibilities, we asked whether Ctk1 activity requires the integrity of the Kin28 and Bur1 pathways. Ctk1 activity was examined after isolation from conditional kin28-ts and bur1-1 mutant strains, each of which lacks the corresponding protein kinase activity. Ctk1 retained its full activity in both strains (data not shown), suggesting that neither of these kinases is responsible for Ctk1 phosphorylation and that Cak1 phosphorylates Ctk1 directly.
We tested whether Cak1 could phosphorylate Ctk1 in vitro. In spite of numerous attempts, we were unable to detect Cak1-mediated phosphorylation or activation of Ctk1 isolated from cak1Δ cells (data not shown). Because Cak1 preferentially phosphorylates CDKs unbound to cyclin subunits (37), we suspected that Ctk1 isolated from yeast cells might be inaccessible for Cak1-mediated phosphorylation. To test whether a Ctk1 monomer might be a better substrate for Cak1, we expressed Ctk1-HA by translation in a reticulocyte lysate, immunoprecipitated it, and incubated it with recombinant Cak1 in the presence of radioactive ATP (Fig. 3C). In addition to Cak1 autophosphorylation (Fig. 3C, lane 2, bottom), we detected phosphorylation of wild-type Ctk1 (Fig. 3C, lane 6) but not of Ctk1T338A (Fig. 3C, lane 4). Thus, Cak1 can phosphorylate Ctk1 directly but its accessibility to the T loop appears to be restricted in the presence of Ctk2 and Ctk3.
Phosphorylation of Ctk1 promotes the transition into stationary phase.
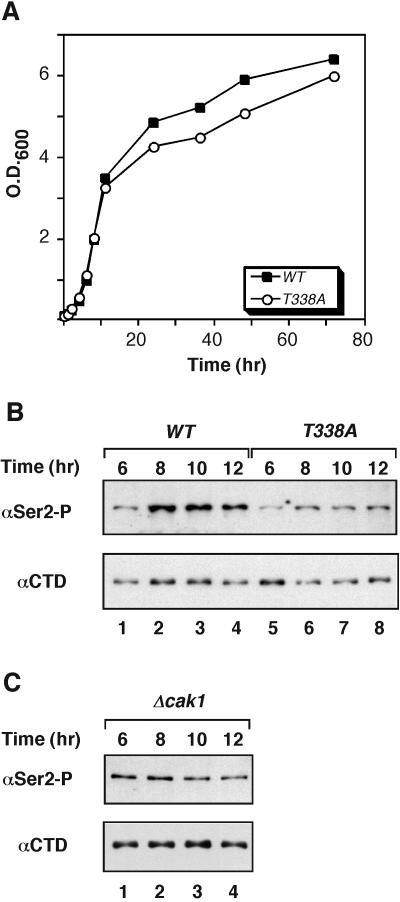
Phosphorylation of Ser2 of RNA pol II by Ctk1 increases during the diauxic shift, when nutrients become limiting and cells prepare to enter stationary phase (36). We wondered whether the role of T-loop phosphorylation might become more prominent under these conditions. To examine this possibility, we compared the growth of wild-type and ctk1T338A cells. The two strains grew at nearly identical rates during exponential phase, but their growth started to diverge as nutrients became limited (Fig. 4A). The ctk1T338A cells delayed growth and reached stationary phase later than wild-type cells. This small but reproducible difference in growth rates suggested that the activity of Ctk1T338A was limiting for cell growth during this period. We then examined the level of RNA pol II phosphorylation during the diauxic shift. In agreement with previous reports, we found that phosphorylation of Ser2 in wild-type cells increased and remained elevated during the transition from rapid growth to stationary phase (Fig. 4B, lanes 1 to 4). This effect was specific for Ser2 phosphorylation, as the amount of unphosphorylated RNA pol II changed little. The level of Ser2 phosphorylation and the total amount of RNA pol II declined at later times (data not shown). In contrast, ctk1T338A cells showed little change in Ser2 phosphorylation during the diauxic shift (Fig. 4B, lanes 5 to 8). The difference between wild-type and ctk1T338A cells was most apparent at the beginning of this interval (Fig. 4B, compare lanes 2 and 6), which just preceded the growth delay of ctk1T338A cells (Fig. 4A). Δcak1 cells also showed no change in RNA pol II phosphorylation at the diauxic shift (Fig. 4C). Thus, Cak1-mediated phosphorylation of Ctk1 on Thr-338 is essential for the elevated Ser2 phosphorylation and rapid cell growth during the transition into stationary phase.
FIG. 4.
Thr-338 phosphorylation is important for entry into stationary phase. (A) Growth rates of CTK1 and ctk1T338A cells. Yeast cultures were grown in YPD medium at 30°C, and cell density was measured during the transition from exponential to early stationary phase. The average of three independent experiments is presented. Similar results were obtained by direct counting of yeast cells. The time course started when cells reached an OD600 of 0.1. (B) Yeast extracts were prepared from CTK1 and ctk1T338A cells at different times during late exponential growth and early stationary phase. The indicated times are the same as in panel A. The proteins were resolved by SDS-PAGE and immunoblotted for the presence of Ser2-phosphorylated RNA pol II (Ser2-P) using H5 antibodies and for unphosphorylated RNA pol II (CTD) using 8WG16 antibodies. (C) Δcak1 cells were grown into early stationary phase and analyzed for the presence of RNA pol II isoforms as for panel B. WT, wild type.
Transcriptional activation of diauxic-shift genes requires Ctk1 phosphorylation.
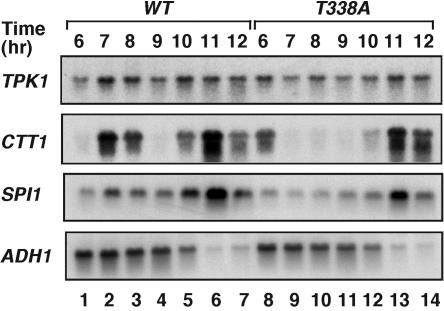
The diauxic shift marks a major transition in transcription by RNA pol II, where multiple genes involved in active growth are repressed and those required for survival in stationary phase are activated. We asked whether the deficiency in Ser2 phosphorylation in ctk1T338A cells compromises activation of stationary-phase-specific genes. To this end, mRNA was isolated from wild-type and ctk1T338A cells at the beginning of stationary phase, when the differences between the two strains became apparent (Fig. 4A). Using microarray hybridization, we compared the levels of individual transcripts in the two mRNA pools and found that transcription of 24 genes was altered at least threefold in ctk1T338A cells and that the expression of an additional 371 genes was altered by at least twofold. From these genes, we selected a few that were highly induced at the diauxic shift (10) and examined them in more detail by Northern blot hybridization. For this experiment, wild-type and ctk1T338A mutant cells were grown into early stationary phase and mRNA levels of TPK1, CTT1, and SPI1 were compared at different times. TPK1 is a catalytic subunit of protein kinase A, which is expressed during both exponential and stationary phases. CTT1 and SPI1, both of which are specific for stationary phase, encode catalase and a protein of unknown function, respectively. In contrast, ADH1 is a housekeeping gene; its transcription ceases when nutrients become limited. In wild-type cells, we found that activation of TPK1, CTT1, and SPI1 proceeded in two phases (Fig. 5, lanes 1 to 7). The initial phase of gene activation (peaking in lanes 2 to 3) was followed by a short repression period (lane 4) and a second stronger wave of induction (peaking in lane 6). This pattern of gene transcription was altered in ctk1T338A cells (Fig. 5, lanes 8 to 14), which lacked the initial phase of CTT1 and SPI1 induction (Fig. 5, compare lanes 2 and 9) and had a reduced level of expression in the second phase (Fig. 5, compare lanes 6 and 13). Note that the decline in ADH1 expression and the second wave of gene induction occurred at the same times in both strains. This transcription deficiency was not limited to these genes, as we examined only a few of the genes that were most prominent in microarray hybridization performed at a single time point. It is likely that the inefficient activation of these and other genes resulted in the growth delay of ctk1T338A cells. Therefore, phosphorylation of Ctk1 on Thr-338 is essential for the rapid and timely transcriptional activation of stationary-phase-specific genes.
FIG. 5.
Phosphorylation of Ctk1 is required for normal transcription of diauxic-shift genes. Total RNA was isolated from CTK1 (lanes 1 to 7) and ctk1T338A cells (lanes 8 to 14) during late exponential phase and early stationary phase (same time points as in Fig. 4A and B). Equal amounts of RNA were separated on a formaldehyde-agarose gel and probed for the presence of specific transcripts by Northern blot hybridization. TPK1, CTT1, and SPI1 transcripts were selected for analysis due to their differential expression in CTK1 and ctk1T338A cells in preliminary microarray hybridization experiments. ADH1 is a housekeeping gene and was used as a loading control. Its level is known to decline during stationary-phase growth. WT, wild type.
DISCUSSION
Although it has been known for some time that Ctk1 phosphorylates Ser2 of RNA pol II and that this phosphorylation is required for efficient transcriptional elongation and RNA processing, relatively little is known about the regulation of Ctk1 activity. In this work, we found that Ctk1 is phosphorylated on a conserved Thr-338 within its T loop and that this phosphorylation greatly stimulates its protein kinase activity. We detected the phosphorylated form of Ctk1 using two-dimensional gel electrophoresis in which we shifted the isoelectric point of the very basic Ctk1 toward neutrality by appending a stretch of acidic residues to its C terminus. This shift greatly improved the resolution of phosphorylated and unphosphorylated forms of Ctk1 from each other. This approach might be useful for the analysis of other highly basic proteins. The phosphorylated form of Ctk1 was eliminated upon phosphatase treatment and upon mutation of Thr-338 to Ala, indicating that Thr-338 is the only site of Ctk1 phosphorylation. Ctk1 phosphorylation on Thr-338 was also recently identified in a proteomic study of yeast phosphoproteins (14).
Is Thr-338 phosphorylation essential for Ctk1 activity? We demonstrated that mutation of Thr-338 compromised Ctk1 activity in vitro, which is similar to the effect of the equivalent mutations in Kin28 and Bur1, two other CDKs regulating CTD phosphorylation (26, 44). Surprisingly, the lack of T-loop phosphorylation in Ctk1 or Kin28 had little effect on cell growth. However, T-loop phosphorylation became essential when additional mutations were introduced into Kin28 or its cyclin subunit, indicating that activating phosphorylation was required for stability of the kinase complex (24, 26). In contrast, examination of Ctk1T338A-containing complexes by gel filtration chromatography indicated that mutation did not affect binding of Ctk2 and Ctk3. Ctk1 function was more sensitive to mutation of Thr-338 in vitro than in vivo. It is possible that the increased requirement for Ctk1 phosphorylation in vitro is due to substrate differences, as the CTD peptide used in vitro contained fewer repeats than the natural RNA pol II CTD and may not adopt its normal conformation. Alternatively, unstressed cells may require very little Ctk1 activity to phosphorylate the CTD to sufficient levels for normal growth. Regarding the need for Thr-338 phosphorylation for activity in vitro, Hautbergue and Goguel reported the reconstitution of a functional Ctk1-Ctk2-Ctk3 complex from recombinant proteins, suggesting that T-loop phosphorylation might not be required for activity (15). However, a comparison of their results and ours is difficult without knowing the specific activities of the respective complexes. It seems likely that a high concentration of unphosphorylated Ctk1 could display detectable activity in vitro but that its kinase activity could still be increased significantly upon T-loop phosphorylation.
Several lines of evidence indicate that Cak1 is responsible for Ctk1 phosphorylation. First, Ctk1 isolated from cak1Δ cells is unphosphorylated. Second, like Ctk1T338A from wild-type cells, Ctk1 isolated from cak1Δ cells was catalytically inactive in vitro. Third, Cak1 directly phosphorylated monomeric Ctk1 on Thr-338. Because Cak1 has a strong preference for cyclin-free CDKs, and because gel filtration experiments did not reveal the presence of any Ctk1 monomer, we suspect that our inability to phosphorylate Ctk1 isolated from yeast cells was due to its incorporation into Ctk1-Ctk2-Ctk3 complexes.
Ctk1 joins a growing list of Cak1 substrates. Phosphorylation by Cak1 is clearly essential for Cdc28 activity and cell cycle progression (11, 22). However, phosphorylation by Cak1 has more modest effects on the functioning of three other CDKs, Kin28, Bur1, and Ctk1. An intriguing possibility is that activating phosphorylation of these kinases becomes important under specific growth conditions. For example, Yao and Prelich demonstrated that cak1Δ cells are sensitive to caffeine, sharing this phenotype with a conditional bur1 mutant (44). Presumably, Cak1-mediated phosphorylation of Bur1 is important for the transcriptional activation of genes required for cell growth in the presence of caffeine. We found that ctk1T338A cells delayed growth during the diauxic shift. At a molecular level, ctk1T338A was unable to stimulate RNA pol II phosphorylation on Ser2 and displayed aberrant activation of diauxic-shift genes. Thus, a lack of activating phosphorylation resulted in diverse conditional phenotypes. Cak1-mediated phosphorylation of Bur1, Ctk1, and Kin28 may have been preserved to support transcriptional adaptation to variable growth conditions.
Acknowledgments
We thank Janet Burton and Aiyang Cheng for support, discussions, and critical reading of the manuscript. We thank Vasiliki Tsakraklides for providing recombinant GST-Cak1 protein and Jingsheng Si for technical assistance. We thank Ken Nelson for assistance with microarray printing.
The microarray facility was supported by grant CA77808 from the National Institutes of Health to Michael Snyder. This work was supported by grant GM47830 (to M.J.S.) from the National Institutes of Health.
REFERENCES
- 1.Ahn, S. H., M. Kim, and S. Buratowski. 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 13:67-76. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Borggrefe, T., R. Davis, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 2002. A complex of the srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277:44202-44207. [DOI] [PubMed] [Google Scholar]
- 4.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cismowski, M. J., G. M. Laff, M. J. Solomon, and S. I. Reed. 1995. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corden, J. L. 1990. Tails of RNA polymerase II. Trends Biochem. Sci. 15:383-387. [DOI] [PubMed] [Google Scholar]
- 8.Cross, F. R., and K. Levine. 1998. Molecular evolution allows bypass of the requirement for activation loop phosphorylation of the Cdc28 cyclin-dependent kinase. Mol. Cell. Biol. 18:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahmus, M. E. 1996. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J. Biol. Chem. 271:19009-19012. [DOI] [PubMed] [Google Scholar]
- 10.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 11.Espinoza, F. H., A. Farrell, H. Erdjument-Bromage, P. Tempst, and D. O. Morgan. 1996. A cyclin-dependent kinase-activating kinase (CAK) in budding yeast unrelated to vertebrate CAK. Science 273:1714-1717. [DOI] [PubMed] [Google Scholar]
- 12.Espinoza, F. H., A. Farrell, J. L. Nourse, H. M. Chamberlin, O. Gileadi, and D. O. Morgan. 1998. Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18:6365-6373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feaver, W. J., J. Q. Svejstrup, N. L. Henry, and R. D. Kornberg. 1994. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell 79:1103-1109. [DOI] [PubMed] [Google Scholar]
- 14.Ficarro, S. B., M. L. McCleland, P. T. Stukenberg, D. J. Burke, M. M. Ross, J. Shabanowitz, D. F. Hunt, and F. M. White. 2002. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20:301-305. [DOI] [PubMed] [Google Scholar]
- 15.Hautbergue, G., and V. Goguel. 2001. Activation of the cyclin-dependent kinase CTDK-I requires the heterodimerization of two unstable subunits. J. Biol. Chem. 276:8005-8013. [DOI] [PubMed] [Google Scholar]
- 16.Hautbergue, G., and V. Goguel. 1999. The yeast C-type cyclin Ctk2 is phosphorylated and rapidly degraded by the ubiquitin-proteasome pathway. Mol. Cell. Biol. 19:2527-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengartner, C. J., V. E. Myer, S. M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 18.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 19.Jona, G., B. O. Wittschieben, J. Q. Svejstrup, and O. Gileadi. 2001. Involvement of yeast carboxy-terminal domain kinase I (CTDK-I) in transcription elongation in vivo. Gene 267:31-36. [DOI] [PubMed] [Google Scholar]
- 20.Jones, J. C., H. P. Phatnani, T. A. Haystead, J. A. MacDonald, S. M. Alam, and A. L. Greenleaf. 2004. C-terminal repeat domain kinase I phosphorylates Ser2 and Ser5 of RNA polymerase II C-terminal domain repeats. J. Biol. Chem. 279:24957-24964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaldis, P., Z. W. Pitluk, I. A. Bany, D. A. Enke, M. Wagner, E. Winter, and M. J. Solomon. 1998. Localization and regulation of the cdk-activating kinase (Cak1p) from budding yeast. J. Cell Sci. 111:3585-3596. [DOI] [PubMed] [Google Scholar]
- 22.Kaldis, P., A. Sutton, and M. J. Solomon. 1996. The Cdk-activating kinase (CAK) from budding yeast. Cell 86:553-564. [DOI] [PubMed] [Google Scholar]
- 23.Kaldis, P., V. Tsakraklides, K. E. Ross, E. Winter, and A. Cheng. 2001. Activating phosphorylation of cyclin-dependent kinases in budding yeast, p. 13-30. In P. Kaldis (ed.), The CDK-activating kinase (CAK). Landes Bioscience, Georgetown, Tex.
- 24.Keogh, M. C., E. J. Cho, V. Podolny, and S. Buratowski. 2002. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 22:1288-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keogh, M. C., V. Podolny, and S. Buratowski. 2003. Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23:7005-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kimmelman, J., P. Kaldis, C. J. Hengartner, G. M. Laff, S. S. Koh, R. A. Young, and M. J. Solomon. 1999. Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol. 19:4774-4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobor, M. S., J. Archambault, W. Lester, F. C. Holstege, O. Gileadi, D. B. Jansma, E. G. Jennings, F. Kouyoumdjian, A. R. Davidson, R. A. Young, and J. Greenblatt. 1999. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 4:55-62. [DOI] [PubMed] [Google Scholar]
- 28.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krishnamurthy, S., X. He, M. Reyes-Reyes, C. Moore, and M. Hampsey. 2004. Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 14:387-394. [DOI] [PubMed] [Google Scholar]
- 30.Lee, J. M., and A. L. Greenleaf. 1991. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. Gene Expr. 1:149-167. [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, J. M., and A. L. Greenleaf. 1997. Modulation of RNA polymerase II elongation efficiency by C-terminal heptapeptide repeat domain kinase I. J. Biol. Chem. 272:10990-10993. [DOI] [PubMed] [Google Scholar]
- 32.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bentley. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 33.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien, T., S. Hardin, A. Greenleaf, and J. T. Lis. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature 370:75-77. [DOI] [PubMed] [Google Scholar]
- 35.Ostapenko, D., and M. J. Solomon. 2003. Budding yeast CTDK-I is required for DNA damage-induced transcription. Eukaryot. Cell 2:274-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patturajan, M., N. K. Conrad, D. B. Bregman, and J. L. Corden. 1999. Yeast carboxyl-terminal domain kinase I positively and negatively regulates RNA polymerase II carboxyl-terminal domain phosphorylation. J. Biol. Chem. 274:27823-27828. [DOI] [PubMed] [Google Scholar]
- 37.Ross, K. E., P. Kaldis, and M. J. Solomon. 2000. Activating phosphorylation of the Saccharomyces cerevisiae cyclin-dependent kinase, Cdc28p, precedes cyclin binding. Mol. Biol. Cell 11:1597-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skaar, D. A., and A. L. Greenleaf. 2002. The RNA polymerase II CTD kinase CTDK-I affects pre-mRNA 3′ cleavage/polyadenylation through the processing component Pti1p. Mol. Cell 10:1429-1439. [DOI] [PubMed] [Google Scholar]
- 39.Sterner, D. E., J. M. Lee, S. E. Hardin, and A. L. Greenleaf. 1995. The yeast carboxyl-terminal repeat domain kinase CTDK-I is a divergent cyclin-cyclin-dependent kinase complex. Mol. Cell. Biol. 15:5716-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Svejstrup, J. Q., Y. Li, J. Fellows, A. Gnatt, S. Bjorklund, and R. D. Kornberg. 1997. Evidence for a mediator cycle at the initiation of transcription. Proc. Natl. Acad. Sci. USA 94:6075-6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thuret, J. Y., J. G. Valay, G. Faye, and C. Mann. 1996. Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell 86:565-576. [DOI] [PubMed] [Google Scholar]
- 42.Tsakraklides, V., and M. J. Solomon. 2002. Comparison of Cak1p-like cyclin-dependent kinase-activating kinases. J. Biol. Chem. 277:33482-33489. [DOI] [PubMed] [Google Scholar]
- 43.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao, S., and G. Prelich. 2002. Activation of the Bur1-Bur2 cyclin-dependent kinase complex by Cak1. Mol. Cell. Biol. 22:6750-6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuryev, A., and J. L. Corden. 1996. Suppression analysis reveals a functional difference between the serines in positions two and five in the consensus sequence of the C-terminal domain of yeast RNA polymerase II. Genetics 143:661-671. [DOI] [PMC free article] [PubMed] [Google Scholar]