Abstract
Neuronal leucine-rich repeat proteins (NLRRs) are type I transmembrane proteins and expressed in neuronal tissues, but their function remains unknown. Here, we describe the identification and characterization of a new member of the NLRR family, NLRR4, and its potential role in long-lasting memory. We generated NLRR4-deficient (NLRR4−/−) mice and found that they showed impaired memory retention. In hippocampus-dependent learning tasks, NLRR4−/− mice were able to learn and maintain the memories for one day but unable to retain the memories for four days after learning. In contrast, in a hippocampus-independent task, NLRR4−/− mice were able to retain the memory normally for at least seven days. These results suggest that NLRR4 plays a key role in hippocampus-dependent long-lasting memory.
The hippocampus plays a role in declarative memory such as episodic and spatial memories, but unimodal memories such as cued fear memories occur independently of the hippocampus. Hippocampal lesions result in severe amnesia of contextual and spatial learning and memory but do not affect normal learning, as seen by the cued fear-conditioning test (3, 19, 24). Memory formation can be divided into three phases by differences in the length of the time from acquisition; short-term memory, long-term memory, and long-lasting memory (22). New protein synthesis is not required for short-term memory but is required for long-term memory (18, 21).
The hippocampus plays a role in the acquisition of short-term memory and also for the conversion of short-term memory to long-term memory within several hours after acquisition, a process known as cellular memory consolidation (22). Long-term memory is converted to long-lasting memory that is permanently stored in the cortex (7, 8). This process is known as system consolidation, and the memory acquired in the past is kept intact even if the hippocampus is damaged. Some proteins implicated in cellular memory consolidation have been reported, such as N-methyl-d-aspartate receptor, calcium-calmodulin-dependent protein kinase IV (CaMKIV), and cyclic AMP (cAMP)-responsive element-binding protein (CREB) in mice (5, 16, 18, 26). Cellular consolidation requires new protein synthesis directed by CREB-mediated transcription. α-Calcium-calmodulin-dependent protein kinase II (αCaMKII)is the only molecule known to be involved in system consolidation, and αCaMKII heterozygous mice are defective in long-lasting memory and cortical long-term potentiation (7). However, how αCaMKII is involved in this process is not fully understood.
In this study, we describe a novel type I transmembrane protein termed NLRR4 which exhibits similarity to neuronal leucine-rich repeat proteins NLRR1 to NLRR3. NLRR1 and NLRR2 are expressed in the developing nervous system (4, 14, 28, 29). NLRR3 is induced by brain injury and regulated by Ras-mitogen-activated protein kinase signaling (9, 10, 15). However, their functions remain to be studied. To uncover the function of NLRR4 in vivo, we generated NLRR4-deficient (NLRR4−/−) mice by replacing its exons with the β-galactosidase gene by homologous recombination. Homozygous NLRR4−/− mice were viable and fertile. We found that their long-term memory in hippocampus-dependent tasks was severely impaired, while hippocampal long-term potentiation was intact. Interestingly, hippocampus-independent memory assessed by the cued fear conditioning in NLRR4−/− mice was normal. These data indicate that NLRR4 is important for maintenance of hippocampus-dependent memories.
MATERIALS AND METHODS
Mice.
C57/BL6J and ICR mice were purchased from Nippon CLEA (Tokyo, Japan) and mutant mice were maintained and mated in our animal facility. All mice were housed in a specific-pathogen-free barrier animal facility. NLRR4−/− mice were backcrossed to the C57BL/6J strain at least 6 times before being used for behavioral and electrophysiological experiments. All experiments were performed according to the guidelines of the Animal Care and Experimentation Committee of University of Tokyo and Wakayama Medical University.
Expression cloning of NLRR4 cDNA.
Expression cloning of a cDNA encoding the B61 antigen was carried out using COS7 cells and a cDNA library prepared from LO cells as previously described (13). We used anti-rat immunoglobulin G-conjugated magnetic beads and a magnetic cell sorter to enrich COS7 cells expressing the B61 antigen. Plasmids recovered from enriched COS7 cells were transfected in COS7 cells. By repeating this selection 3 times, we obtained a cDNA encoding the B61 antigen NLRR4.
Northern blotting.
Total RNA was extracted using Trizol reagent (Invitrogen) and polyadenylated RNA was prepared from various tissues using an Oligo-MAG mRNA purification kit (Takara) according to the manufacturer's protocol. Northern blot analysis was carried out as previously described by using digoxigenin-labeled single-stranded DNA probe.
Generation of NLRR4-deficient mice.
To construct the NLRR4 targeting vector, overlapping NLRR4 genomic clones were isolated from a phage library made from 129SVJ strain mice (Stratagene). The targeting construct contained the 5.5-kb 5′ and 4.5-kb 3′ homology regions and the 2.5-kb fragment containing the exon with initiation codon was replaced with the β-galactosidase gene. The neomycin resistance gene was inserted between the homologous regions, and the herpes simplex virus thymidine kinase cassette was ligated to the 3′ end of the vector. TT2 embryonic stem (ES) cells were electroporated and selected with G418 and 2′-deoxy-2′-fluoro-β-d-arabinofuranosyl-5′-iodouracil. Drug-resistant clones were analyzed by Southern blotting. BamHI- or EcoRI-digested genomic DNA was hybridized with the 400-bp 3′ genomic fragment that corresponded to genomic sequences outside of the targeting vector or 500-bp 5′ genomic fragment. Chimeric mice were generated by the aggregation method as previously described (25). Genotyping of progenies was performed by Southern blot analysis using the 3′ probe or by PCR. The PCR-specific primers used were NLRR4S (5′-TACGGACTCTCTCTGTGTAGGACTCCC-3′), NLRR4AS (5′-AGGTTGTGACTGAGGTCGAGCTCAC-3′), NeoS (5′-ACGACGGGCGTTCCTTGCGCAGCTGTG-3′), and NeoAS (5′-TCAGAAGAACTCGTCAAGAAGGCGATA-3′)
Flow cytometry.
Cells harvested from cultures were resuspended in phosphate-buffered saline and filtered through 70-μm nylon mesh (cell strainer, Falcon) to remove cell debris. The cells were then incubated with monoclonal antibodies for 30 min on ice and analyzed by fluorescence-activated cell sorting (Becton Dickinson)
β-Galactosidase staining of whole brains.
For β-galactosidase staining, brains were fixed in phosphate-buffered saline containing 2% paraformaldehyde, 0.2% glutaraldehyde, and 0.02% NP-40 and sectioned at 300-μm thickness by vibratome. After washing, the sections were placed in staining buffer (44 mM HEPES [pH 7.4], 3 mM potassium ferricyanide, 15 mM NaCl, 1.3 mM MgCl2, 0.5 mg/ml 5-bromo-4-chloro-3-indolyl-β- d-galactopyranoside [X-Gal] in phosphate-buffered saline) at room temperature.
Contextual fear-conditioning tests.
For each experiment, a mouse was placed in the conditioning chamber and given a series of foot shocks alone (2-s duration, 0.3 mA, 1 min apart). To test its memory, the mouse was placed in the same conditioning chamber and freezing responses were analyzed using Image FZC (O'Hara & Co.), with modified software available in the public-domain National Institutes of Health Image program. In the experiment, a mouse was trained by three foot shocks and then its behavior was tested 1, 4, or 7 days after training.
Cued fear-conditioning tests.
For training, a mouse was placed in the conditioning chamber for 2 min before the onset of conditioned stimuli, a tone which lasted for 30 s at 10 kHz and 70 dB. The last 2 s of the conditioned stimulus was paired with am unconditioned stimulus, 0.3 mA of a continuous electric foot shock. After an additional 30 s in the chamber, the mouse was returned to its home cage. Four and seven days after training, the mouse was placed in a different chamber in which the same tone as the conditioned stimuli was applied for 120 s after a 1-min habituation period (preconditioned stimulus).
Morris water maze test.
The water pool used in these experiments was 1 m in diameter and made up of white polyvinyl chloride and the temperature of the water was 27 ± 1°C. For the hidden-platform task, an acrylic transparent platform (10 cm in diameter) was submerged 5 mm below the surface of water that was made opaque by adding nontoxic odorless white paint. The location of the platform was fixed during the trials for each mouse. If it located the platform within 60 s, it was kept on it for 30 s. Mice that failed to find the platform within 60 s were guided to the platform and allowed to stay there for 30 s. Each mouse was trained four trials per day for 5 consecutive days. A different starting point was used for each of the trials. The time to reach the platform was recorded (escape latency). Probe tests were performed at three time points (retention day) after the last training. In the probe test, the platform was removed and the mice were allowed to swim for 60 s. The swimming speed was calculated based on movement data. For all experiments, the movement of each mouse was monitored by a charge-coupled device camera and the data were processed with NIH Image WM 2.12r (O'Hara & Co.).
Electrophysiology.
All experiments were performed to compare NLRR4−/− mice with wild-type mice in a blind fashion using male littermates. Nine- to 11-week-old NLRR4−/− and wild-type mice were decapitated under deep halothane anesthesia, and both hippocampi were removed. Hippocampal slices (400 μm thick) were cut with a Vibratome tissue slicer and placed in a humidified holding chamber for at least 2 h. A single slice was then transferred to a recording chamber maintained at 25°C, and submerged beneath a continuously perfusing medium that had been saturated with 95% O2/5% CO2. The medium comprised 119 mM NaCl, 2.5 mM KCl, 1.3 mM MgSO4, 2.5 mM CaCl2, 1.0 mM NaH2PO4, 26.2 mM NaHCO3 and 11 mM glucose. All perfusing solutions contained picrotoxin (100 μM) to block gamma-aminobutyric acid A receptor-mediated inhibitory synaptic responses. Field potential recordings were made using a glass electrode filled with 3 M NaCl and placed in the stratum radiatum in the hippocampal CA1 region. An Axopatch-1D amplifier was used, and the signal was filtered at 1 kHz and digitized at 10 kHz.
To evoke synaptic responses, a bipolar tungsten electrode was placed in the stratum radiatum, and Schaffer collateral and commissural fibers were stimulated at 0.1 Hz (test pulses). A single high-frequency stimulus train (100 Hz, 1 s) was applied with the test pulse intensity to induce long-term potentiation. The input-output relationship of basal synaptic responses was examined in the presence of d-2-amino-5-phosphonovaleric acid (25 μM) to block N-methyl-d-aspartate receptor-mediated synaptic responses. A low concentration of 6-cyano-7-nitroquinoxaline-2,3-dione (1 μM) was also present to partially block alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA) receptors. This enables more accurate measurements of the input-output relationship, since the presence of low concentrations of 6-cyano-7-nitroquinoxaline-2,3-dione reduces the nonlinear summation of field excitatory postsynaptic potentials when strong stimulus strengths are used. For measurements of paired-pulse facilitation, afferent fibers were stimulated twice at intervals of 50, 100, and 200 ms in the presence of 25 μM d-2-amino-5-phosphonovaleric acid. All values were expressed as the mean ± standard errors of the mean. Student's t test (two-tailed, unpaired) was used to determine whether there was a significant difference in the means between two sets of data.
RESULTS
Molecular cloning of the NLRR4 gene.
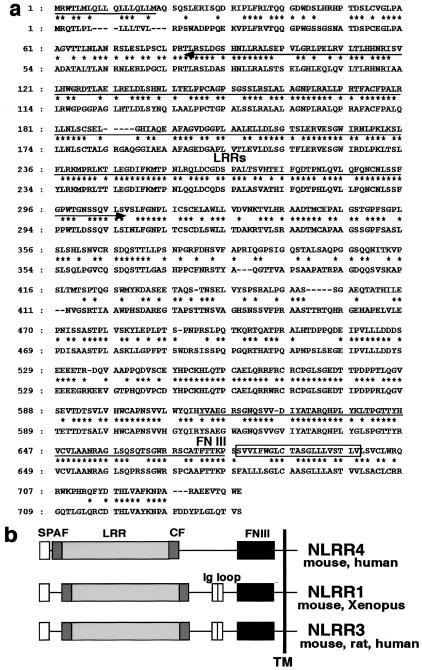
In our studies on the development of hematopoiesis, we generated a number of monoclonal antibodies against LO cells (12), a hemangioblast-like cell line. B61 is one such antibody and recognizes a cell surface molecule on LO cells. We employed an expression cloning strategy to identify the B61 antigen by using COS7 cells and an expression library of LO cells, and we obtained a 2.7-kb cDNA encoding the B61 antigen. While the B61 antigen was highly expressed in the hemangioblast-like cell line LO, B61 mRNA was not found in hematopoietic tissues (data not shown). Instead, we found its expression in neuronal tissues as described below. Sequence analysis revealed that the B61 cDNA has an open reading frame encoding 735 amino acid residues with two hydrophobic segments, one at the amino terminus (amino acids 1 to 21) and the second (amino acids 679 to 704) in the internal region. There are two distinct motifs in the extracellular region, leucine-rich repeats and fibronectin type III-like repeats and a small intracellular region consisting of only 30 amino acid residues without any known motifs. Due to the similarity to the previously identified neuronal leucine rich-repeat proteins NLRR1 to NLRR3 (4, 14, 28, 29), we named B61 NLRR4. These members are type I transmembrane proteins with multiple leucine-rich repeats and a fibronectin type III-like domain.
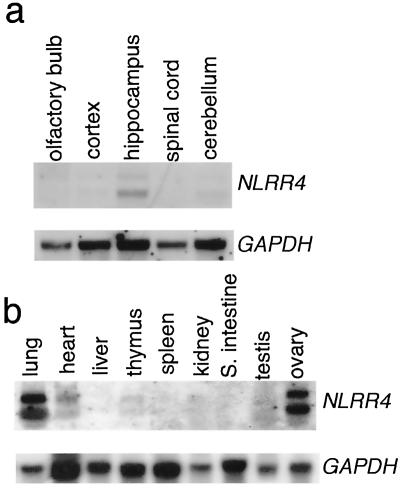
An NLRR4-homologous gene was found in the human genome but not in Caenorhabditis elegans or Drosophila melanogaster. A human cDNA homologous to NLRR4 encoded a type I transmembrane protein with 640 amino acids (Fig. 1a) with an overall homology of 64% at the amino acid level. NLRR4 showed 21% and 20% homology at the amino acid level to NLRR1 and NLRR3 (28, 29), respectively. Although there are two immunoglobulin C2 loops in NLRR1 and NLRR3, no such structure was found in NLRR4 (Fig. 1b). Northern blot analysis revealed that NLRR4 mRNA was expressed in lung, heart, and ovary (Fig. 2b). In the adult brain, NLRR4 was strongly expressed in the hippocampus and weakly in the cerebellum (Fig. 2a).
FIG. 1.
Structural properties of NLRR4. (a) The deduced amino acid sequences of mouse (upper) and human (lower) NLRR4. The putative signal peptide is underlined, and the transmembrane region is boxed. Identical amino acid residues are indicated by dots, and the leucine-rich repeats (LRRs) and fibronectin III (FN III) domains are indicated by the arrows underneath the sequences. (b) Structures of the NLRR family members. All members are type I transmembrane proteins with leucine-rich repeats and fibronectin III motifs in their extracellular region. SP, signal peptide; AF, amino-flanking region; CF, carboxy-flanking region; TM, transmembrane.
FIG. 2.
Expression of NLRR4. (a) Northern blot analysis of NLRR4 mRNA in the adult brain. (b) Northern blot analysis of NLRR4 in various adult mouse tissues. Polyadenylated RNA was prepared from various parts of the adult brain or various adult tissues. One μg of mRNA for each sample was separated on an agarose gel and then transferred to a nylon membrane. After UV cross-linking, the membrane was hybridized with a digoxigenin-labeled full-length NLRR4 probe. The lower panel shows equal loading of the sample hybridized with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe.
Targeted disruption of the NLRR4 gene.
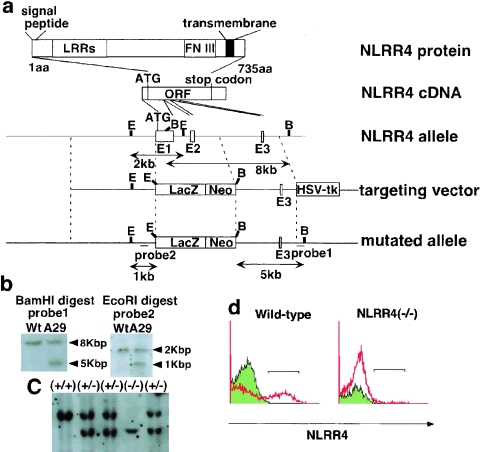
To uncover the function of NLRR4 in vivo, we generated an NLRR4-deficient mouse line. As shown in Fig. 3a, we replaced the exons of the NLRR4 gene with the β-galactosidase gene and the neomycin resistance gene (neo). The deleted exon encoded the initiation codon, the signal peptide, and the leucine-rich repeat domains, and the targeting vector was designed to express β-galactosidase from NLRR4 regulatory elements. We isolated six independent ES clones with homologous recombination (Fig. 3b), and chimeric male mice were generated from three of the clones. Heterozygous embryos from three independent chimeric mice exhibited the same β-galactosidase expression pattern. Genotyping by Southern blotting showed successful targeting of the NLRR4 gene (Fig. 3c).
FIG. 3.
Targeted disruption of the NLRR4 gene. (a) Structures of the NLRR4 protein, cDNA, genomic locus, targeting vector, and mutated allele. Three boxes in the NLRR4 genome represent exons. The locations of the probes used for Southern blotting are shown (probes 1 and 2). E, EcoRI; B, BamHI; lacZ, Escherichia coli β-galactosidase cDNA; neo, neomycin resistance gene; HSV-tk, herpes simple virus thymidine kinase. (b) Identification of a homologous recombinant clone (A29) by southern blot analysis of BamHI- or EcoRI-digested genomic DNA from ES clones. (c) Southern blot analysis of BamHI-digested genomic DNA from mouse tails using probe 1. (d) Flow cytometric analysis using an NLRR4 antibody was performed on cells derived from wild-type and NLRR4−/− embryos. NLRR4 protein was not detected in NLRR4−/− cells.
NLRR4 protein expression was not detected in NLRR4−/− embryos by flow cytometric analysis of primary culture cells derived from wild-type and NLRR4−/− embryos with an anti-NLRR4 monoclonal antibody (Fig. 3d). We genotyped 372 pups born from heterozygous matings to assess whether the expected number of knockout and heterozygote pups was affected. The number of NLRR4+/+ to NLRR4+/− to NLRR4−/− was 83:196:93, as expected for unbiased Mendelian frequencies (NLRR4−/−), and the mice grew with normal body weights. Both males and females are fertile, and no increased mortality was observed up to 1 year of age.
Expression of NLRR4 in the adult brain and anatomy of the NLRR4−/− hippocampus.
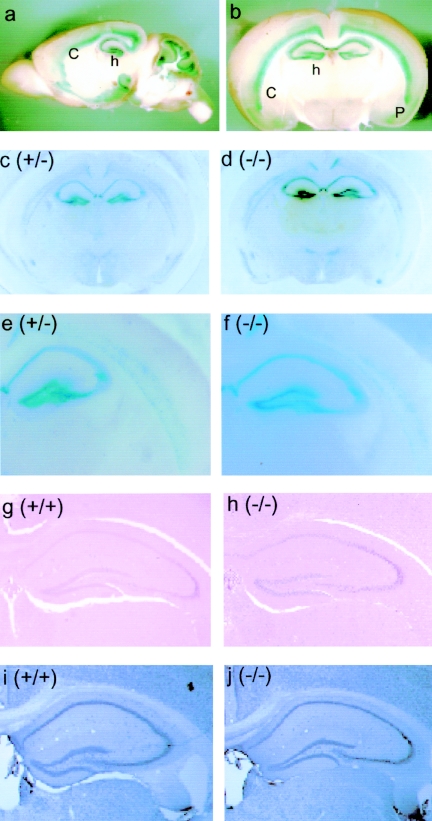
Since NLRR4 mRNA was found in the brain by Northern blotting analysis (Fig. 2), we precisely analyzed the expression of NLRR4 in the brain. As the β-galactosidase gene was inserted in the exon of NLRR4, we performed β-galactosidase staining of the adult brain of two-month-old heterozygous mice (NLRR4+/−) (Fig. 4a, b, c, and e). NLRR4 was expressed in the CA1 and CA3 regions and dentate gyrus of the hippocampus. In addition, NLRR4 was expressed in layers V and VI in the neocortex and piriform cortex. It was also expressed in the inner granular layer and Purkinje cells of the cerebellum. The NLRR4−/− brain was also stained for β-galactosidase activity, and there was no difference in the staining pattern between NLRR4+/− and NLRR4−/− mice (Fig. 4c, d, e, and f).
FIG. 4.
Expression of NLRR4 and hippocampal anatomy in the NLRR4−/− adult brain. NLRR4 expression was assessed by β-galactosidase staining of a sagittal section (a) and a coronal section (b) of the adult brain of 2-month-old heterozygous mice. β-Galactosidase activity was detected in the hippocampus, layers V and VI in the cortex, the piriform cortex, the inner granule layer, and Purkinje cells in the cerebellum. β-Galactosidase staining was performed in coronal sections of the NLRR4+/− and NLRR4−/− cerebrum. Similar staining patterns were observed in both NLRR4+/− and NLRR4−/− mice (c, d, e, and f). Gross hippocampal anatomy in NLRR4−/− mice was evaluated by histochemical and immunohistochemical staining. Hematoxylin-eosin staining (g and h) and immunostaining of a neuron-specific maker, NeuN (i and j), showed that there were no significant differences between the two genotypes. Abbreviations: h, hippocampus; c, cortex; p, piriform cortex.
We performed histochemical staining to examine hippocampal anatomy in wild-type and NLRR4−/− mice. Hematoxylin-eosin staining and immunohistochemical analysis of NeuN, a neuron-specific DNA binding protein, showed no difference between wild-type and NLRR4−/− mice (Fig. 4g, h, i, and j). These results indicated that the gross anatomy of hippocampus was not affected by the NLRR4 mutation.
Defective contextual fear memory in NLRR4−/− mice.
Since NLRR4 is expressed in the hippocampus, which plays a role in learning and memory (3, 24), we considered the possibility that NLRR4 plays a role in memory formation. To address this, we examined the associative emotional memory of the mutant mice using the contextual fear-conditioning test. If mice learn fear such as that resulting from an electric foot shock in a spatial context, they show some defensive responses, including freezing behavior in the conditioning chamber (3).
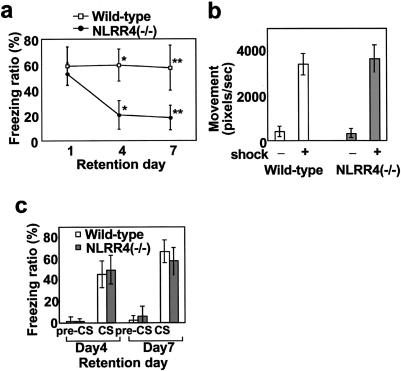
Without experiencing the electric shock, wild-type and NLRR4−/− mice showed little of this behavior. Mice with either genotype spent the same time freezing in the context at 24 h after the training, indicating that NLRR4−/− mice are able to learn fear by contextual fear-conditioning. However, NLRR4−/− mice froze significantly less than wild-type mice 4 and 7 days after the training (Fig. 5a). There were no significant differences between the two genotypes in terms of nociceptive sensitivity to an electrical foot shock (Fig. 5b). For the contextual fear-conditioning test, fear-eliciting experiences such as an electric foot shock are input into the amygdala and spatial information such as the conditioning chamber is sent to the hippocampus. Both sets of information are consolidated and stored in a manner dependent on the hippocampus and amygdala. As NLRR4−/− mice showed normal freezing behavior during the 1-day retention period, cellular consolidation in NLRR4−/− mice was normal. However, as the freezing time was significantly reduced in NLRR4−/− mice 4 days after the training, their memory was impaired after long retention delay.
FIG. 5.
Contextual and cued fear memory in NLRR4−/− mice. (a) Freezing ratio for contextual fear at different retention delays for NLRR4−/− and wild-type mice (n = 13 for each genotype and each time point). No difference was found in the time spent on freezing behavior after 1 day of retention (wild-type, 58.7 ± 15.1%; NLRR4−/−, 59.5 ± 12.6%). Freezing behavior was significantly reduced in the NLRR4−/− mice after 4 days (wild-type, 59.5 ± 12.6%, NLRR4−/−, 20.1 ± 11.3%, Student's t test P < 0.001) and 7 days retention (wild-type, 57.2 ± 17.5%; NLRR4−/−, 17.8 ± 9.8%, Student's t test, P < 0.001). All data points represent the mean (± standard error of the mean). *, P < 0.001; **, P < 0.001. (b) Normal pain sensitivity of NLRR4−/− and wild-type mice. Pain sensitivities were calculated by the total pixels that mice moved before and after electric foot shock. All data points represent the mean (± standard error of the mean). (c) Percent of freezing time at preconditioned or conditioned stimuli after cued fear conditioning for NLRR4−/− and wild-type mice. Tests were performed 4 and 7 days after training. No significant difference between the genotypes was found for either time points (day 4 after training: wild-type, 45.1 ± 12.5%; NLRR4−/−, 49.2 ± 13.4%; day 7 after training: wild-type, 66.4 ± 20.7%; NLRR4−/−, 57.4 ± 27.7% [n = 10 for each genotype and each time point]). All data points represent the mean (± standard error of the mean).
Normal tone-dependent cued fear memory in NLRR4−/− mice.
As contextual fear-conditioning tasks are dependent on the hippocampus and amygdala, we examined hippocampus-independent fear memory to assess the function of amygdala in NLRR4−/− mice. The auditory cued fear-conditioning test is an amygdala-dependent but hippocampus-independent task (19). Wild-type and NLRR4−/− mice were exposed simultaneously to a tone (conditioned stimulus) and an electric foot shock (unconditioned stimulus). After this learning process, we measured the freezing time when the tone was replayed in a different chamber. Mice with either genotype did not show any freezing behavior before presenting conditioned stimuli (preconditioned stimuli). In the presence of a conditioned stimulus, mice of either genotype showed the same level of freezing time at both time points after the conditioning (Fig. 5c). No significant differences were observed in freezing time between the two genotypes 4 and 7 days after the training, indicating that NLRR4 is dispensable for tone information processing and fear memory is fully integrated into the amygdala without NLRR4 after long retention.
Defective spatial memory in NLRR4−/− mice.
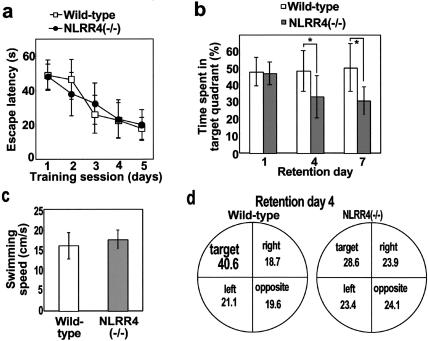
For the fear memory tests, NLRR4−/− mice showed defects in the hippocampus-dependent memory (contextual fear-conditioning test), but not the hippocampus-independent memory (tone fear-conditioning test). These results indicated that NLRR4−/− mice are defective in hippocampus-dependent fear memories. We then examined another type of hippocampus-dependent memory without the experience of fear in NLRR4−/− mice. We employed the hidden-platform Morris water maze test, which is a hippocampus-dependent learning task. Wild-type and NLRR4−/− mice improved their performance during 5 days of training at 4 trials/day (Fig. 6a). There was no significant difference in escape latencies between the two genotypes on any day during training. For the probe test on 1 day after training, no significant difference between the two genotypes was observed for the time spent in the target quadrant, indicating that cellular consolidation of long-term memory in NLRR4−/− mice was normal. However, the NLRR4−/− mice spent less time in the target quadrant than that of wild-type mice 4 days after the training (Fig. 6b). NLRR4−/− mice spent almost the same time in all quadrants 4 days after the training (Fig. 6d), showing that NLRR4−/− mice randomly swam in all quadrants. Swimming speed in the training session was not different between the two genotypes (Fig. 6c). These results indicate that NLRR4 is required for hippocampus-dependent long-lasting memory.
FIG. 6.
Spatial learning and memory in the hidden-platform Morris water maze task. (a) Average of the escape latency for each day for hidden-platform Morris water maze training sessions. Four trials were performed each day for 5 consecutive days (n = 20 for each genotype). No difference was observed between the two genotypes, indicating that the mutant mice learned equally well. All data points represent the mean (± standard error of the mean). (b) The probe tests were performed at three different retention time points (1, 4, and 7 days after the last day of acquisition for the hidden-platform task). Different groups of mice were used at each time point. One day after training, no difference was observed between the genotypes (n = 10 for each genotype), indicating that the learning and acquisition of initial memory are normal for NLRR4−/− mice. Four and seven days after the last training, the time that NLRR4−/− mice spent in the target quadrant was significantly less than that of wild-type mice. Statistical significance was calculated by the Scheffe's test following two-way analysis of variance. *, P < 0.05. (n = 10 for each genotype and each time point). (c) Swimming speeds were calculated from these movement data. No differences were observed between the genotypes during the training sessions. (d) Time spent in each quadrant of the pool 4 days after training. The time spent by NLRR4−/− mice in the target quadrant was shorter than that by the wild-type control mice.
Normal hippocampal long-term potentiation in NLRR4−/− mutants.
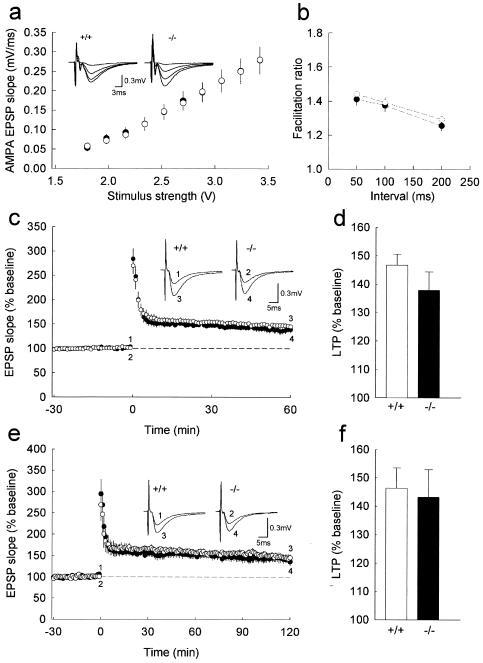
The finding that NLRR4−/− mice showed normal short-term memory but impaired long-lasting memory prompted us to examine whether the physiological properties of the hippocampus were altered by the lack of NLRR4. We first examined the input-output relationship of synaptic transmission to assess possible changes in basal synaptic properties, using hippocampal slices. AMPA receptor-mediated excitatory postsynaptic potentials in the CA1 region in response to various stimulus intensities were indistinguishable between NLRR4−/− and wild-type mice (Fig. 7a), indicating that the basal synaptic efficacy was not altered in NLRR4−/− mice.
FIG. 7.
Normal basal synaptic transmission, paired-pulse facilitation and long-term potentiation in NLRR4−/− mice. (a) The input-output relationships of AMPA receptor-mediated excitatory postsynaptic potentials of wild-type (open circles, n = 12) and mutant (closed circles, n = 9) mice. Shown are the mean synaptic responses to stimuli of various strengths for the NLRR4−/− and wild-type (+/+) slices. There was no significant difference between the two genotypes. Insets display representative traces evoked with four different stimulus intensities of 1.8 to 3.42 V. (b) Paired-pulse facilitation induced by stimulating afferent fibers twice at 50, 100, and 200 ms in wild-type (open circles) and NLRR4−/− (solid circles) mice. The ratio of the slope of the second excitatory postsynaptic potentials to that of the first excitatory postsynaptic potentials was calculated (at 50-ms interstimulus interval (ISI): NLRR4−/−, 1.41 ± 0.04%, n = 7; wild-type, 1.45 ± 0.05%, n = 10, P > 0.6; at 100-ms ISI: NLRR4−/−, 1.38 ± 0.04%, n = 7; wild-type, 1.39 ± 0.03%, n = 10, P > 0.8; at 200-ms ISI: NLRR4−/−, 1.26 ± 0.03%, n = 7; wild-type, 1.29 ± 0.04%, n = 10, P > 0.6). (c) The averaged time course of long-term potentiation for 60 min after tetanic stimulation for wild-type (open circle, n = 20) and NLRR4−/− (solid circle, n = 18) mice. At time zero, tetanic stimulation (100 Hz, 1 s) was delivered to the Schaffer collateral/commissural pathway. Initial excitatory postsynaptic potentials slopes were normalized for each experiment to the averaged slope value during the baseline period (−30 to 0 min). Representative traces (average of 10 consecutive responses) in the inset were excitatory postsynaptic potentials obtained at the times indicated by the numbers on the graph. (d) Summary of long-term potentiation calculated as the percent increase in the mean excitatory postsynaptic potential slope from 50 to 60 min after tetanic stimulation compared with the mean excitatory postsynaptic potential slope during the baseline period (NLRR4−/−, 137.8 ± 6.5% of baseline, n = 18; wild type, 146.7 ± 3.9% of baseline, n = 20, P > 0.2). (e) The averaged time course of long-term potentiation for 120 min after tetanic stimulation for wild-type (open circle, n = 5) and NLRR4−/− (solid circle, n = 6) mice. This graph consists of a part of the data shown in Fig. 8C, which was obtained from the slices with 1 h of additional recording. (f) Summary of long-term potentiation calculated as the percent increase in the mean excitatory postsynaptic potential slope from 110 to 120 min after tetanic stimulation compared with the mean excitatory postsynaptic potential slope during the baseline period (NLRR4−/−, 143.2 ± 9.8% of baseline, n = 6; wild-type, 146.3 ± 7.2% of baseline, n = 5, P > 0.8).
We then examined paired-pulse facilitation, which is a presynaptic form of short-term plasticity and is sensitive to presynaptic release probability (20). The paired-pulse facilitation observed in slices from NLRR4−/− mice did not significantly differ from that of wild-type mice at any interstimulus interval examined (Fig. 7b), indicating that presynaptic properties including transmitter release probability were intact in NLRR4−/− mice. Furthermore, we analyzed long-term synaptic plasticity and found that a single high-frequency stimulus train (100 Hz/s) produced long-term potentiation in mutant slices that was indistinguishable from that of wild-type slices (Fig. 7c and d), which is consistent with the results that NLRR4−/− mice performed normally hippocampus-dependent memory tasks 24 h after the training. Since there was a tendency of decreased long-term potentiation 60 min after the induction in NLRR4−/− mice, we also followed the time course of long-term potentiation for another hour, but found that long-term potentiation 120 min after the induction was also normal (Fig. 7e and f). Taken together, these results indicate that NLRR4 is not required for basal synaptic transmission or short-term and the early phase of long-term plasticity at the Schaffer collateral-CA1 synapse in the hippocampus.
DISCUSSION
We isolated a cDNA encoding a new member of the NLRR family, a type I transmembrane protein with leucine-rich repeats and a fibronectin type III repeat. Leucine-rich repeats consist of multiple copies of the conserved sequence. Proteins with leucine-rich repeats mediate homophilic or heterophilic protein-protein interactions. Slits function as a ligand for the Robo receptors through its leucine-rich repeat (2, 11), and NgR and Trks are receptors for Nogo and neurotrophic factors (6, 17), respectively. CAPS and connectin function as cell recognition molecules in neuromuscular development via their homophilic interaction (23, 27). We therefore tested a homophilic interaction of NLRR4 protein. However, cell aggregation assays using NLRR4-expressing cells failed to show any interaction (data not shown), suggesting that NLRR4 may be involved in cell-cell interactions as a component or it may be a receptor or a ligand for an unknown molecule.
NLRR4-deficient mice generated by inserting the β-galactosidase cDNA into the NLRR4 gene locus developed normally and exhibited no anatomical abnormalities. The NLRR4 expression revealed by β-galactosidase activity indicates that NLRR4 is expressed in the hippocampus, layers V and VI in the cortex and piriform cortex, the internal granule layer, and Purkinje cell in the cerebellum. The hippocampus plays a role in the initial storage and the cellular consolidation of spatial information (22, 26). Since NLRR4 is highly expressed in the hippocampus, it is possible that NLRR4 is involved in learning and/or memory. In fact, we found that the memory of NLRR4−/− mice was severely impaired for two learning tests that are dependent on the hippocampus; the contextual fear-conditioning and hidden-platform Morris water maze tests.
In the contextual fear-conditioning test, NLRR4−/− mice were able to learn normally as shown by the normal freezing behavior 1 day after training. However, NLRR4−/− mice failed to retain the memory longer as their freezing behavior was significantly reduced 4 and 7 days after training. We obtained similar results in the hidden-platform Morris water maze test. Thus, in the hippocampus-dependent behavioral tests NLRR4−/− mice are able to learn and retain memories normally memory 1 day after training. Consistently, NLRR4−/− mice showed normal long-term potentiation in the hippocampus, indicating that initial memory and cellular consolidation are normal in NLRR4−/− mice. Contextual and spatial memories are known to be retained in the hippocampus for several days and long-lasting memory is permanently stored in the cortex (7, 8). As memory retention 4 days after training was severely impaired in NLRR4−/− mutant mice, it is likely that NLRR4 is required for long-lasting memory.
In contrast to hippocampus-dependent memories, NLRR4−/−mice showed normal cued fear memory in which a tone was paired with an electric foot shock. This task depends on the amygdala but is independent of the hippocampus. It is known that hippocampus lesion results in impaired spatial memory but normal memory in the cued fear-conditioning tests (19). Likewise, the hippocampal CA1 region-specific knockout of the NR1 gene encoding an essential subunit of the N-methyl-d-aspartate receptor results in amnesia of contextual fear memory without affecting cued fear memory (26). Axon of CA1 pyramidal cells mainly extend to the layer V in entorhinal cortex and NLRR4 is expressed in CA1, CA3, and dentate gyrus in the hippocampus and in layers V and VI in the cortex. These results collectively suggest that the impaired retention of memories in NLRR4−/− mice depends on the hippocampal function but is independent of the amygdala.
Amnesia in NLRR4−/− mice is similar to that in αCaMKII heterozygous mice, which exhibit severely impaired hippocampus-dependent long-lasting memory, while the molecular mechanism how αCaMKII is involved in long-lasting memory still remains to be elucidated, there may be a link between NLRR and αCaMKII. CREB is required for the cellular consolidation of long-term memory, indicating that a protein(s) regulated by CREB plays a role in long-term memory (1, 5, 18). Therefore, the impaired memory in NLRR4−/− mice might be due to an alteration in this pathway. As NLRR4 is a transmembrane protein with protein-protein interaction motifs, it is tempting to speculate that NLRR4 may generate an intracellular signal that affects the CREB pathway in the hippocampus. It will be of interest to find a molecule that interacts with NLRR4.
Acknowledgments
We thank S. Kida for helpful discussion and critical reading of the manuscript and T. Fukuda, T. Watanabe, H. Kawano, and T. Sato for technical advice. We are also grateful to T. Akiyama and his staff for their help for behavioral tests and to Y. Iwakura for providing us with a mouse genomic DNA library and the LacZ plasmid.
This work was supported in part by Grants-in-Aid for Scientific Research, the 21st Century COE program, the Center for Biosignaling and Integrated Brain Medical Science, and Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports and Technology, by RISTEX and CREST of the Japan Science and Technology Agency, and by the Inoue Foundation for Science.
REFERENCES
- 1.Abel, T., and E. Kandel. 1998. Positive and negative regulatory mechanisms that mediate long-term memory storage. Brain Res. Brain Res. Rev. 26:360-378. [DOI] [PubMed] [Google Scholar]
- 2.Battye, R., A. Stevens, R. L. Perry, and J. R. Jacobs. 2001. Repellent signaling by Slit requires the leucine-rich repeats. J. Neurosci. 21:4290-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best, P. J., A. M. White, and A. Minai. 2001. Spatial processing in the brain: the activity of hippocampal place cells. Annu. Rev. Neurosci. 24:459-486. [DOI] [PubMed] [Google Scholar]
- 4.Bormann, P., L. W. Roth, D. Andel, M. Ackermann, and E. Reinhard. 1999. zfNLRR, a novel leucine-rich repeat protein is preferentially expressed during regeneration in zebrafish. Mol. Cell. Neurosci. 13:167-179. [DOI] [PubMed] [Google Scholar]
- 5.Bourtchuladze, R., B. Frenguelli, J. Blendy, D. Cioffi, G. Schutz, and A. J. Silva. 1994. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 79:59-68. [DOI] [PubMed] [Google Scholar]
- 6.Fournier, A. E., T. GrandPre, S. M. Strittmatter. 2001. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409:341-346. [DOI] [PubMed] [Google Scholar]
- 7.Frankland, P. W., C. O'Brien, M. Ohno, A. Kirkwood, and A. J. Silva. 2001. α-CaMKII-dependent plasticity in the cortex is required for permanent memory. Nature. 411:309-313. [DOI] [PubMed] [Google Scholar]
- 8.Frankland, P. W., B. Bontempi, L. E. Talton, L. Kaczmarek, and A. J. Silva. 2004. The involvement of the anterior cingulate cortex in remote contextual fear memory. Science 304:881-883. [DOI] [PubMed] [Google Scholar]
- 9.Fukamachi, K., Y. Matsuoka, C. Kitanaka, Y. Kuchino, and H. Tsuda. 2001. Rat neuronal leucine-rich repeat protein-3: cloning and regulation of the gene expression. Biochem. Biophys. Res. Commun. 287:257-263. [DOI] [PubMed] [Google Scholar]
- 10.Fukamachi, K., Y. Matsuoka, H. Ohno, T. Hamaguch, and H. Tsuda. 2002. Neuronal leucine-rich repeat protein-3 amplifies MAPK activation by epidermal growth factor through a carboxyl-terminal region containing endocytosis motifs. J. Biol. Chem. 277:43549-43552. [DOI] [PubMed] [Google Scholar]
- 11.Guthrie, S. 1999. Axon guidance: starting and stopping with slit. Curr. Biol. 9:R432-435. [DOI] [PubMed] [Google Scholar]
- 12.Hara, T., Y. Nakano, M. Tanaka, K. Tamura, T. Sekiguchi, K. Minehata, N. G. Copeland, N. A. Jenkins, M. Okabe, H. Kogo, Y. Mukouyama, and A. Miyajima. 1999. Identification of podocalyxin-like protein 1 as a novel cell surface marker for hemangioblasts in the murine aorta-gonad-mesonephros region. Immunity 11:567-578. [DOI] [PubMed] [Google Scholar]
- 13.Harada, N., B. E. Castle, D. M. Gorman, N. Itoh, J. Schreurs, R. L. Barrett, M. Howard, and A. Miyajima. 1990. Expression cloning of a cDNA encoding the murine interleukin 4 receptor based on ligand binding. Proc. Natl. Acad. Sci. USA. 87:857-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayata, T., T. Uochi, and M. Asashima. 1998. Molecular cloning of XNLRR-1, a Xenopus homolog of mouse neuronal leucine-rich repeat protein expressed in the developing Xenopus nervous system. Gene 221:159-166. [DOI] [PubMed] [Google Scholar]
- 15.Ishii, N., A. Wanaka, and M. Tohyama. 1996. Increased expression of NLRR-3 mRNA after cortical brain injury in mouse. Brain Res. Mol. Brain Res. 40:148-152. [DOI] [PubMed] [Google Scholar]
- 16.Kang, H., L. D. Sun, C. M. Atkins, T. R. Soderling, M. A. Wilson, and S. Tonegawa. 2001. An important role of neural activity-dependent CaMKIV signaling in the consolidation of long-term memory. Cell 106:771-783. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, D. R., and R. M. Stephens. 1994. Neurotrophin signal transduction by the Trk receptor. J. Neurobiol. 25:1404-1417. [DOI] [PubMed] [Google Scholar]
- 18.Kida, S., S. A. Josselyn, S. P. de Ortiz, J. H. Kogan, I. Chevere, S. Masushige, and A. J. Silva. 2002. CREB required for the stability of new and reactivated fear memories. Nat. Neurosci. 5:348-355. [DOI] [PubMed] [Google Scholar]
- 19.Kim, J. J., and M. S. Fanselow. 1992. Modality-specific retrograde amnesia of fear. Science 256:675-677. [DOI] [PubMed] [Google Scholar]
- 20.Manabe, T., D. J. Wyllie, D. J. Perkel, and R. A. Nicoll. 1993. Modulation of synaptic transmission and long-term potentiation: effects on paired pulse facilitation and EPSC variance in the CA1 region of the hippocampus. J. Neurophysiol. 70:1451-1549. [DOI] [PubMed] [Google Scholar]
- 21.Matthies, H. 1989. In search of cellular mechanisms of memory. Prog. Neurobiol. 32:277-349. [DOI] [PubMed] [Google Scholar]
- 22.McGaugh, J. L. 2000. Memory-a century of consolidation. Science 287:248-251. [DOI] [PubMed] [Google Scholar]
- 23.Nose, A., V. B. Mahajan, and C. S. Goodman. 1992. Connectin: a homophilic cell adhesion molecule expressed on a subset of muscles and the motoneurons that innervate them in Drosophila. Cell. 70:553-567. [DOI] [PubMed] [Google Scholar]
- 24.Scoville, W. B., and B. Milner. 1957. Loss of recent memory after bilateral hippocampal lesions. J. Neurochem. 20:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekine, K., H. Ohuchi, M. Fujiwara, M. Yamasaki, T. Yoshizawa, T. Sato, N. Yagishita, D. Matsui, Y. Koga, N. Itoh, and S. Kato. 1999. Fgf10 is essential for limb and lung formation. Nat. Genet. 21:138-141. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu, E., Y. P. Tang, C. Rampon, and J. Z. Tsien. 2000. NMDA receptor-dependent synaptic reinforcement as a crucial process for memory consolidation. Science 290:1170-1174. [DOI] [PubMed] [Google Scholar]
- 27.Shishido, E., M. Takeichi, and A. Nose. 1998. Drosophila synapse formation: regulation by transmembrane protein with Leu-rich repeats, CAPRICIOUS. Science 280:2118-2121. [DOI] [PubMed] [Google Scholar]
- 28.Taguchi, A., A. Wanaka, T. Mori, K. Matsumoto, Y. Imai, T. Tagaki, and M. Tohyama. 1996. Molecular cloning of novel leucine-rich repeat proteins and their expression in the developing mouse nervous system. Brain Res. Mol. Brain Res. 35:31-40. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi, H., M. Tohyama, and T. Takagi. 1996. Cloning and expression of a novel gene for a protein with leucine-rich repeats in the developing mouse nervous system. Brain Res. Mol. Brain Res. 36:45-52. [DOI] [PubMed] [Google Scholar]