Abstract
TFIID, comprising the TATA box binding protein (TBP) and 13 TBP-associated factors (TAFs), plays a role in nucleation in the assembly of the RNA polymerase II preinitiation complexes on protein-encoding genes. TAFs are shared among other transcription regulatory complexes (e.g., SAGA, TBP-free TAF-containing complex [TFTC], STAGA, and PCAF/GCN5). Human TAF10, a subunit of both TFIID and TFTC, has three histone fold-containing interaction partners: TAF3, TAF8, and SPT7Like (SPT7L). In human cells, exogenously expressed TAF10 remains rather cytoplasmic and leptomycin B does not affect this localization. By using fluorescent fusion proteins, we show that TAF10 does not have an intrinsic nuclear localization signal (NLS) and needs one of its three interaction partners to be transported into the nucleus. When the NLS sequences of either TAF8 or SPT7L are mutated, TAF10 remains cytoplasmic, but a heterologous NLS can drive TAF10 into the nucleus. Experiments using fluorescence recovery after photobleaching show that TAF10 does not associate with any cytoplasmic partner but that once transported into the nucleus it binds to nuclear structures. TAF10 binding to importin β in vitro is dependent on the coexpression of either TAF8 or TAF3, but not SPT7L. The cytoplasmic-nuclear transport of TAF10 is naturally observed during the differentiation of adult male germ cells. Thus, here we describe a novel role of the three mammalian interacting partners in the nuclear localization of TAF10, and our data suggest that a complex network of regulated cytoplasmic associations may exist among these factors and that this network is important for the composition of different TFIID and TFTC-type complexes in the nucleus.
Initiation of transcription of certain protein-encoding genes by RNA polymerase II requires the transcription factor TFIID that comprises the TATA binding protein (TBP) and a series of TBP-associated factors (TAFs) (1, 3, 34). Another set of human transcriptional regulatory multiprotein complexes containing TAFs includes TBP-free TAF-containing complex (TFTC), STAGA, and PCAF/GCN5 (29, 36, 47). These complexes are functional homologues of the yeast SAGA complex, and all contain human homologues of the yeast histone acetyltransferase Gcn5 as well as a subset of SPT and ADA proteins, the 400-kDa TRRAP, and a number of TAFs (shared TAFs) also found in TFIID (28). Recently, ataxin-7 (ATX7), the human orthologue of the yeast SAGA Sgf73 subunit, was also shown to be a bona fide subunit of the human TFTC-like complexes (19). In agreement with the functional similarity between human TFTC and yeast SAGA, the structures of these complexes are evolutionarily conserved (5, 48). The spatial distributions of the shared TAFs were found to be similar in SAGA and TFIID and suggested that the 4-nm-wide groove of TFIID, which could be involved in DNA binding, is similar to the cleft formed by several domains of SAGA (25, 48). Interestingly, TFTC, although devoid of TBP, is capable of functionally replacing TFIID at both TATA-containing and TATA-less promoters in vitro (47).
TAF10 is not only an integral component of TFIID but is also found in the SAGA and TFTC-type coactivator complexes (28). Analysis of a variety of conditional Saccharomyces cerevisiae TAF10 mutants indicated that TAF10 regulates different subsets of genes, some of which are important for cell cycle progression and cell morphology (22, 23, 43). Drosophila melanogaster has two paralogue genes encoding TAF10 homologues (TAF10 and TAF10b), which are differentially expressed during Drosophila embryogenesis (14). D. melanogaster TAF10 was found to be present in both TFIID and TFTC-like complexes, but TAF10b was identified only in TFIID (14, 35). Similarly, in mammalian cells, TAF10 is shared between TFIID and the three closely related GCN5-containing multiprotein complexes: TFTC, PCAF/GCN5, and the STAGA complexes (28). Lack of TAF10 leads to cell cycle arrest and cell death by apoptosis in mouse F9 embryonic carcinoma cells (31). Moreover, in mouse F9 cells lacking TAF10, the integrity of most TFIID is compromised (32). Interestingly, in human HeLa cells, different TFIID complexes containing or lacking TAF10 exist and exhibit functionally distinct properties (6, 21). By generating a functionally null mutation of the TAF10 gene, we showed that TAF10 is required for early mouse development and survival of the pluripotent inner cell mass but not for survival of mouse trophoblast cells. Together, these data suggest that TAF10 is required for transcription of a subset of genes essential for specific cell types and differentiation pathways.
Electron microscopy, yeast genetics, X-ray crystallography, and biochemical experiments have shown that histone fold (HF) motifs mediate many of the subunit interactions within the TFIID and SAGA complexes (11). It was shown that S. cerevisiae TAF3 and TAF8 contain putative HF motifs and that both of these domains selectively heterodimerize with S. cerevisiae TAF10, which also contains a putative HF (13). The mapping of histone-like TAFs in yeast TFIID revealed that these TAF pairs are also formed in three distinct lobes within endogenous TFIID (25). Furthermore, yeast Spt7p, a component of the yeast SAGA complex (17, 49), has an HF motif similar to that of S. cerevisiae TAF3, which selectively heterodimerizes with the HF domain of S. cerevisiae TAF10, defining an additional histone-like pair in the SAGA complex (13). Thus, yeast TAF10 has three HF-containing interaction partners: S. cerevisiae TAF3 and TAF8 and Spt7. The mammalian and Drosophila homologues of TAF3 contain an N-terminal HF domain, which selectively heterodimerizes with members of the TAF10 family (12). The isolated HF of human TAF3 heterodimerizes with that of human TAF10, whereas the HF of D. melanogaster TAF3 is more selective since it interacts only with D. melanogaster TAF10 and not with D. melanogaster TAF10b (12). Drosophila Prodos, also called D. melanogaster TAF8 (45), contains an HF motif, which, in contrast to that of D. melanogaster TAF3, selectively heterodimerizes with D. melanogaster TAF10b but not with D. melanogaster TAF10 (20). Consequently, it has been proposed that Drosophila Prodos is a Drosophila TFIID component (20). Recently, the human homologue of TAF8 (TAFII43) was described as an integral component of TFIID (18). Human TAF8 is an orthologue of mouse Taube Nuss, which is essential for early embryonic mouse developmental events (46). Interestingly, Taube Nuss−/− and TAF10−/− mice show the same very early embryonic lethality, indicating that the lack of either TAF8 or TAF10 leads to selective inner cell mass death (32, 46). These knockout-mouse results further underline the possibility that these two mammalian proteins interact in vivo and have similar roles in the respective TAF-containing complexes. A human factor with significant similarity to yeast Spt7, called STAGA-associated factor 65γ or human SPT7Like (hereafter SPT7L), has also been described as a STAGA or TFTC subunit (30).
With the use of in vitro interaction assays, human TAF10 has been shown to associate with its three HF-containing interaction partners: TAF8, TAF3, and SPT7L. However, nothing was known about the associations of these factors in living cells. Here we investigate the roles of the mammalian TAF10-interacting partners in the nuclear localization of TAF10. We show that human TAF10 does not have a nuclear localization signal and its nuclear transport is dependent entirely on interaction(s) with TAF3, TAF8, or SPT7L. The potential implications of the regulation of the nuclear localization of TAF10 by its partners are discussed.
MATERIALS AND METHODS
Plasmid constructions.
The eukaryotic expression plasmids for TAF3, TAF6, TAF8, TAF9, and TAF10 have been previously described (2, 10, 12, 21, 26). Yellow fluorescent protein (YFP) and cyan fluorescent protein (CFP) fusions were generated by subcloning the different cDNA fragments from these vectors either by appropriate restriction enzyme digestion, purification, and insertion of the given fragment into the corresponding site of the pEYFP-C1 and pECFP-C1 (Clontech) vectors or by PCR amplification of the fragments with appropriate restriction sites, digestion, purification of the fragments, and insertion into the pEYFP-C1 and pECFP-C1 (Clontech) vectors. The pCFP-Nuc expression vector containing a nuclear localization signal (NLS) from simian virus 40 (SV40) was from Clontech. The SPT7L open reading frame was PCR amplified from the cDNA clone HK04750 (gene name, KIAA0764; kind gift from T. Nagase, Kazusa Research Institute, Japan) by using primers containing BamHI and EcoRI sites, and the digested PCR products were cloned into the appropriate baculovirus and CFP as well as YFP expression vectors. All plasmids have been verified by sequencing. Details on the cloning strategies are available upon request (see also Fig. 2).
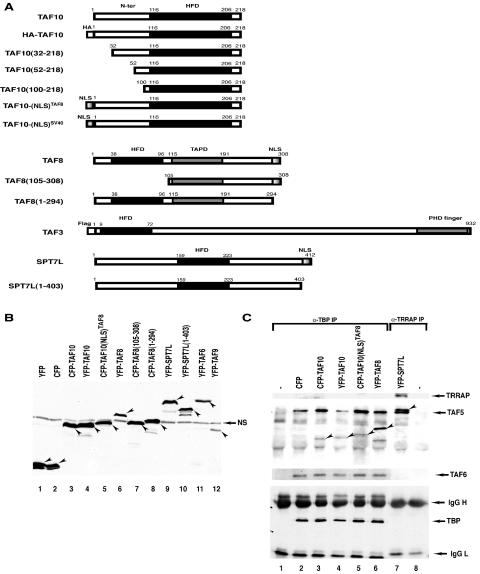
FIG. 2.
(A) Schematic representation of the TAF10-, TAF8-, TAF3-, and SPT7L-containing fusion proteins used in the HeLa cell transfection experiments. Most of the constructs were generated as CFP and/or YFP fusion proteins. The histone fold domain (HFD), the proline-rich domain of TAF8 (TAPD), the PHD finger, and the NLSs of the factors are indicated. The numbers refer to amino acid positions in each protein. N-ter, N terminus. (B) The YFP- and CFP-containing expression vectors express the correct fusion proteins in transfected HeLa cells. HeLa cells were transfected (as indicated), WCEs prepared, and 50 μg protein separated by SDS-PAGE and analyzed by Western blotting using an anti-GFP antibody. In each lane, the specifically expressed protein is labeled with an arrowhead. NS, nonspecific protein also present in nontransfected cell extracts. (C) The expressed YFP and CFP fusion transcription factors are functional since they integrate into their respective complexes. From the indicated extracts analyzed as described for panel B, either anti-TBP (lanes 1 to 6) or anti-TRRAP (lanes 7 to 8) IP was carried out, and bound proteins were analyzed as described in the legend to Fig. 1. −, antibody control without extract. The CFP or YFP fusion proteins incorporated into either a TBP- or a TRRAP-containing complex are labeled with arrowheads. Western blotting with anti-TRRAP, anti-TAF5, anti-TAF6, and anti-TBP was carried out to verify that the anti-TBP (α-TBP) and anti-TRRAP IPs worked as expected. IgG, immunoglobulin G (L, light chain; H, heavy chain).
HeLa cell transfections.
HeLa cells (1.5 × 105) were transfected in 6-well plates or 30-mm dishes with 250 ng of the expression vectors indicated in the figures, completed with pBSK to achieve a final volume of 3 μg of total DNA by using JetPEI (PolyPlus Transfection, France), and fixed, harvested, or visualized 24 to 30 h after transfection. If several proteins had to be expressed in the same experiment, then 250 ng of each expression plasmid was used.
Immunization and antibody production.
The QSPDDSDSSYGSHSTDSLMG(C) peptide corresponding to SPT7L amino acids 377 to 396 was synthesized, coupled to ovalbumin, and used for generation of mouse monoclonal antibodies as described in reference 6. All the other antibodies used were previously described (2, 12, 26, 32, 47).
Immunoprecipitation and Western blot analysis.
Sf9 cell lysates were prepared as described in reference 26. Proteins of Sf9 cell lysates (from a 75-cm2 Falcon flask) were subjected to immunoprecipitation (IP) with 100 μl protein G-Sepharose (Pharmacia) and approximately 5 μg of the different antibodies (as indicated in the figures). Antibody-protein G-Sepharose-bound protein complexes were washed three times with IP buffer (25 mM Tris-HCl [pH 7.9], 10% [vol/vol] glycerol, 0.1% NP-40, 0.5 mM dithiothreitol, 5 mM MgCl2) containing 0.5 M KCl and twice with IP buffer containing 100 mM KCl. After washing, protein G-antibody-bound proteins were either eluted by an excess of the corresponding epitope peptide or directly boiled in sodium dodecyl sulfate (SDS) sample buffer and separated by SDS-polyacrylamide gel electrophoresis (PAGE), transferred onto nitrocellulose membrane, and probed with the primary antibodies as indicated in the figures. Chemiluminescence detection was performed according to the manufacturer's instructions (Amersham).
Immunofluorescence.
Indirect immunofluorescence tests were performed on cells growing on coverslips. Cells were fixed in 2% paraformaldehyde for 10 min and permeabilized in 0.5% Triton X-100 for 10 min. After blocking in 1% bovine serum albumin-phosphate-buffered saline (PBS), they were incubated with anti-TAF10 monoclonal antibody (mAb; 2B11; diluted 1:1,000), antihemagglutinin (anti-HA) mAb (12CA5; diluted 1:1,000), or anti-Flag mAb (M5; diluted 1:1,000) followed by Cy3-labeled anti-mouse secondary antibody (diluted 1:1,000; Jackson Laboratories). When applicable, nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride; 50 ng/ml). Images were analyzed by using either a wide-field fluorescence Leica (DMIRBE) microscope with a Cool Snap Ropers camera or a Leica laser-based confocal microscope.
Pull downs.
Six-His-tagged glutathione S-transferase (GST), six-His-tagged importin α, and six-His-tagged importin β were expressed, purified, and immobilized on nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagene) as described previously (7, 15). HeLa cells were transfected with 0.5 μg of the expression vectors described above, and whole-cell extracts were prepared in buffer B (20 mM HEPES [pH 7.9], 300 mM KCl, 10% glycerol, 15 mM imidazole, 0.1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml aprotinin). Nickel-based pull-down assays were performed by incubating the importin-containing beads with whole-cell extracts for 5 h at 4°C in buffer B. The beads were washed five times with the same buffer. The presence of the CFP, YFP, or Flag fusion proteins in the input or bound to the His-tagged importin α and His-tagged importin β was detected by Western blot analysis using the anti-GFP antibody (NEB) or anti-Flag antibody.
Imaging and fluorescence recovery after photobleaching (FRAP).
We used the Leica TCS SP2 (AOBS) confocal microscope operating with a 40-mW argon laser. The laser was tuned to lines of 458 nm and 514 nm. Cells were examined with a 63×/1.3-numerical-aperture oil immersion objective and 2.5× zoom. The gain of photomultiplier tubes and the emission intervals were adjusted to eliminate cross talk between the CFP (470 to 500 nm) and YFP (530 to 600 nm) channels.
Live-cell microscopy was performed on a Zeiss Meta confocal microscope using the 405-nm diode and 514-nm laser line of an Ar laser (nominal output, 40 mW; beam width at specimen, 0.2 μm). All experiments were done at 37°C, and imaging was done with a 63× objective and numerical aperture of 1.3. Scanning was bidirectional at the highest possible rate with a 4× zoom and a pinhole of 1 Airy unit. Laser power for bleaching was maximal. For imaging, the laser power was attenuated to 0.1% of the bleach intensity. FRAP experiments were performed as described in reference 38 with few modifications. Briefly, six single-prebleach images were acquired, followed by two iterative bleach pulses of 223 ms each. Single-section images were then collected at 250-ms intervals for 60 s. Recovery of signal in the bleached region and loss of signal in the unbleached region were measured as average intensity signals in a region comprising at least 50% of the bleached or unbleached area. The sizes of this measurement region were identical in all experiments. All recovery curves were generated from background-subtracted images. The fluorescence signal measured in a region of interest was individually normalized to the prebleach signal in the region of interest according to the following equation: R = (It − Ibg)/(Io − Ibg), where Io is the average intensity in the region of interest during prebleaching, It is the average intensity in the region of interest at time point t, and Ibg is the background signal determined in a region outside of the cell nucleus.
Bleaching of the whole nuclear fluorescent pools was performed using the same settings as before except that two single-prebleach images were acquired, followed by two iterative bleach pulses, and pictures were collected at 2-min intervals for 30 min.
Immunohistochemistry on squash preparations from microdissected seminiferous tubules.
Immunofluorescence tests were performed on squash preparations of microdissected tubules from adult mice (37). The slides were fixed with 4% paraformaldehyde in 1× PBS and permeabilized with 0.2% Triton-1× PBS. After blocking for 1 h in 5% bovine serum albumin in 1× PBS-0.05% Tween 20, the slides were incubated overnight at 4°C with the antibody indicated in the figures (anti-TAF10 [4G2] mAb, diluted 1:300; anti-TAF8 [1FR 1A2] mAb, diluted 1:300; or anti-TBP [3G3] mAb, diluted 1:1,000) (6, 21, 32). The slides were then washed with 1× PBS and incubated with a secondary antibody (Cy3-conjugated rabbit anti-mouse, diluted 1:500). Immunostained samples were counterstained with DAPI (Boehringer) before being subjected to microscopy.
RESULTS
Human TAF10 interacts with TAF3, TAF8, or SPT7L in vitro.
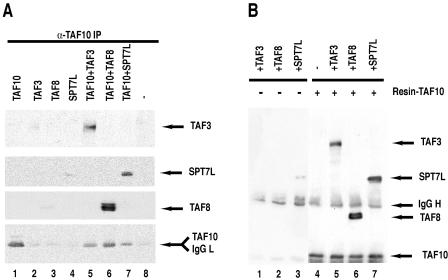
Since it has been shown by two-hybrid and genetic experiments with yeast that the HF motif of TAF10 (Taf25p) can interact with the HF domains of three different factors, TAF3 (Taf47p), TAF8 (Taf65p), and Spt7 (12, 13), and since all the mammalian homologues of these factors are by now known (see the introduction), we wanted to test whether human TAF10 would interact with the full-length mammalian homologues of these yeast factors. To this end, either full-length TAF10, TAF3, TAF8, and SPT7L were expressed individually or TAF10 was coexpressed with one of its putative partners by a baculovirus expression system in insect Sf9 cells (9, 26). From these cells, total protein extracts were obtained and proteins were immunoprecipitated. Coimmunoprecipitation experiments with an anti-TAF10 mouse mAb (23TA 1H8) showed that TAF10 could interact with either of its putative partners (Fig. 1A). To further study the specificity of these interactions, TAF10 was immunopurified by the anti-TAF10 mAb prior to the addition of extracts in which TAF3, TAF8, or SPT7L was individually expressed (Fig. 1B). In agreement with the coexpression results, TAF10 interacted with all its putative HF motif-containing partners even under these more-stringent conditions. In control experiments, TAF3, TAF8, or SPT7L did not bind, or bound only very weakly, to the control beads (Fig. 1A, lanes 2 to 4, and B, lanes 1 to 3). These data demonstrate that in vitro human TAF10 can form pairs with full-length TAF3, TAF8, or SPT7L.
FIG. 1.
Human TAF10 interacts with TAF3, TAF8, and SPT7L in vitro. (A) TAF3, TAF8, TAF10, and SPT7L were expressed individually or TAF10 was coexpressed with one of its potential partners (as indicated) in Sf9 cells by using the baculovirus expression system. Forty-eight hours after infection, WCEs were made, TAF10-associated proteins were subjected to IP with an anti-TAF10 mAb, the beads were extensively washed, boiled, and loaded onto an SDS-polyacrylamide gel, and TAF10-associated proteins were analyzed by Western blotting using the indicated antibodies. (B) TAF10 was first bound to protein G-Sepharose-anti-TAF10 antibody beads, the beads were extensively washed, and then protein extracts containing one of the three partners were incubated (as indicated) with the TAF10-containing beads (lanes 5 to 7) or with protein G-Sepharose antibody beads alone as controls (lanes 1 to 3) for 1 h at room temperature, and beads were extensively washed and bound proteins analyzed as described for panel A. IgG, immunoglobulin G (L, light chain; H, heavy chain).
Exogenously expressed TAF10 localizes mostly to the cytoplasm of the cells.
To verify whether TAF10 associates with the above-described TFIID and TFTC subunits in living cells, we generated a series of eukaryotic expression vectors, which express TAF10, TAF8, SPT7L, and their truncated derivatives (Fig. 2A and B), as fusions with either CFP or YFP. HeLa cells were first transfected with constructs expressing TAF10, TAF8, or SPT7L as YFP and CFP fusion proteins individually, whole-cell extracts (WCEs) were prepared, and the expression of the fusion proteins was tested by Western blot analysis using an anti-GFP antibody (Fig. 2B). The tested CFP and/or YFP fusion proteins were expressed in the corresponding WCEs and were able to associate with endogenous TBP-containing complexes (in the case of TAF8 and TAF10) or TRRAP-containing complexes (in the case of SPT7L) since they coimmunoprecipitated with TBP as well as TAF5 and TAF6 or with TRRAP, respectively (Fig. 2C). These results suggest that the expressed YFP and CFP fusion proteins are functional. The Flag-tagged full-length TAF3 used was previously characterized (12).
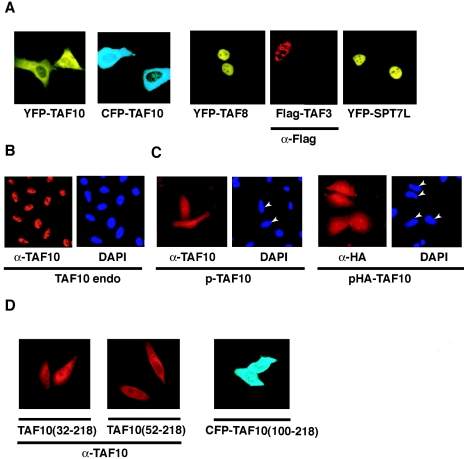
When the cellular localization of these proteins was analyzed by fluorescence microscopy, as expected the TAF3-, TAF8-, and SPT7L-containing fusion proteins all localized exclusively to the nucleus (Fig. 3A and data not shown). Surprisingly, and in contrast to endogenous TAF10 protein (Fig. 3B), neither the CFP-TAF10 nor the YFP-TAF10 fusion protein localized exclusively to the nucleus (Fig. 3A). The expression of these proteins was cytoplasmic rather than nuclear (Fig. 3A).
FIG. 3.
Exogenously expressed TAF10 localizes mostly to the cytoplasm of the cells, and TAF3, TAF8, and SPT7L are homogenously distributed within the nucleus but excluded from the nucleolus. (A to D) HeLa cells were either nontransfected (B) or transfected with the indicated expression vectors, and the expressed proteins were visualized by fluorescence microscopy either directly or by using the indicated primary mouse antibodies and a Cy3-labeled secondary antibody. Where indicated, the nuclei of the cells were visualized with DAPI staining. In panel C, the nuclei of the transfected cells are indicated with arrowheads. If antibodies (α) were used to visualize the proteins, they are indicated under the corresponding photos. TAF10 endo, endogenous TAF10.
Both the YFP and the CFP tags were engineered onto the N-terminal end of the TAF10 protein because previous experiments showed that about 100 amino acids from the N-terminal end of TAF10 can be deleted without any functional consequences (23). To exclude the possibility that the observed cytoplasmic localization of the TAF10 fluorescent proteins was due to altered conformation of the fusion proteins, we examined the localization of overexpressed TAF10 by immunofluorescence after transfecting HeLa cells with eukaryotic expression vectors expressing either nontagged TAF10 (p-TAF10) or HA-tagged TAF10 (pHA-TAF10) (Fig. 3C). With the use of antibodies raised against either TAF10 itself or the HA tag, the exogenously expressed TAF10 derivatives showed both cytoplasmic and nuclear localization (Fig. 3C). These experiments indicated that even the nontagged version of the exogenously expressed full-length TAF10 localizes both to the cytoplasm and the nucleus.
Next, we examined whether the deletion of the nonessential N-terminal domain of TAF10 would allow its nuclear localization. When a series of truncated TAF10 mutants were used to transfect cells (Fig. 3D), they all remained mostly cytoplasmic, indicating that the N-terminal 100 amino acids of TAF10 do not influence its cellular localization (Fig. 3D). These results suggest that TAF10, in contrast with TAF3, TAF8, and SPT7L, is not able to efficiently enter the nucleus or is actively exported from there.
Leptomycin B does not change the localization of TAF10.
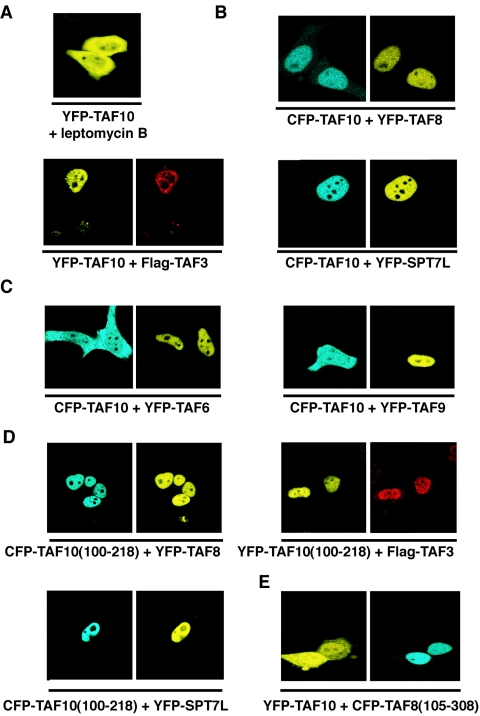
The above-described results suggested two possibilities concerning the lack of nuclear localization of exogenously expressed TAF10. It is possible that (i) TAF10 is able to enter by itself into the nucleus and that it is then retained there through interaction with one of its partners but that the excess of “free” TAF10 is exported from the nucleus to the cytoplasm through a nuclear export signal (NES) or that (ii) TAF10 is not able to enter efficiently by itself into the nucleus. The nuclear pore complex protein CRM1 is able to recognize specific leucine-rich NESs to mediate protein export from the nucleus (33). CRM1 is a target of the cytotoxin leptomycin B (LMB). To test the above-listed first possibility and to check whether blocking of CRM1-dependent nuclear export changes the cytoplasmic localization of TAF10, we incubated the pYFP-TAF10-transfected cells for 4 h with 20 ng/ml of LMB. There was no influence of LMB on the cellular localization of TAF10 (Fig. 4A), but a GFP-NES sequence fusion protein was retained in the nucleus under the same conditions (data not shown). These data suggest that if TAF10 shuttles between the nucleus and the cytoplasm, it is not exported into the cytoplasm through a CRM1-dependent pathway.
FIG. 4.
Exogenously expressed TAF10 becomes exclusively nuclear when cells are cotransfected with one of TAF10's HF domain-containing interaction partners. The HF domain of TAF10 and the partner are required for nuclear localization. (A to E) HeLa cells were cotransfected with the indicated CFP- and YFP-containing expression vectors, and localization of the expressed proteins was visualized by fluorescence microscopy. Flag-TAF3 was visualized by immunofluorescence. In panel A, pYFP-TAF10-transfected cells were incubated for 4 h with 20 ng/ml leptomycin B. The images shown in each panel are representative of all the transfected cells.
The nuclear localization of TAF10 is dependent on the interaction of its HF with its partners.
To test the possibility that the nuclear localization of TAF10 in the cells could be regulated by its interaction partners, which localized to the nucleus when individually expressed (Fig. 3A), TAF10 fluorescent proteins were co-overexpressed with TAF3, TAF8, or SPT7L fusion proteins. Figure 4B shows that cotransfection of expression vectors for TAF10 with one of the three interaction partners resulted in the nuclear accumulation of either CFP- or YFP-TAF10. In contrast, coexpression of two other HF-containing TAFs, YFP-TAF6 and YFP-TAF9, which do not interact with TAF10 in vitro, did not result in nuclear translocation of TAF10 (Fig. 4C), indicating that the nuclear localization process of TAF10 is specifically due to its interacting partners. Thus, these data suggest that TAF10 associates also in vivo with its three partners and that for its exclusive nuclear localization TAF10 needs one of its three interaction partners.
To determine whether the highly conserved domain of TAF10, containing only the putative HF domain (14, 23), would be sufficient for the nuclear localization of the protein, we tested the subcellular localization of the TAF10 deletion mutant comprising amino acids 100 to 218 [TAF10(100-218)] in the presence of the three HF-containing interaction partners. TAF10(100-218) fused either to CFP or to YFP alone was found mainly in the cytoplasm (Fig. 3D), but in the presence of TAF3, TAF8, or SPT7L fusion proteins, the TAF10(100-218) deletion mutant localized to the nucleus (Fig. 4D). These results further suggest that TAF10 interacts in the cells with its three partners and that the domain of interaction with these three partners is within the evolutionarily conserved putative HF motif of TAF10.
Next, we tested whether the HF domains of the partners were necessary for the nuclear localization of TAF10. To this end, we cotransfected cells with full-length YFP-TAF10 and a TAF8 deletion mutant that lacks the putative HF of TAF8 [TAF8(105-308)] (Fig. 2 and Fig. 4E). When the HF was deleted from TAF8, the truncated protein still localized to the nucleus, but it could not participate in the nuclear translocation of YFP-TAF10. Similar results were obtained when the HF of SPT7L was deleted (data not shown). The above-described results suggest that the interactions of TAF10 with its partners through its HF domain are crucial for its nuclear translocation and/or retention process.
The nuclear localization of TAF10 is dependent on the NLS present in its interaction partners.
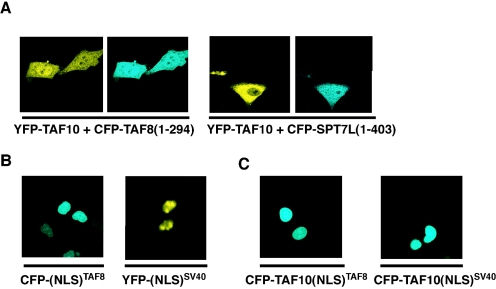
The hypothesis that the nuclear localization of TAF8, TAF3, and SPT7L is involved in the nuclear translocation of TAF10 prompted us to search for potential short peptides within the amino acid sequences of these proteins that are sufficient and necessary for their nuclear localization. Most frequently, an NLS comprises a short stretch of basic amino acids (16). Such putative NLS sequences in both TAF8 and SPT7L were predicted to be at the very C-terminal ends of the two proteins (297-KKPKIRRKKSLS-308 for TAF8 and 405-KKRMRKI-412 for SPT7L) by the PSORT II program. Thus, we constructed short C-terminal deletion mutants of TAF8 [TAF8(1-294)] and SPT7L [SPT7L(1-403)] fused with YFP or CFP (Fig. 2). First, we observed the localization of the mutants after transfection into HeLa cells. The deletion mutants CFP-TAF8(1-294) and SPT7L(1-403) were completely cytoplasmic (Fig. 5A, right panels; data not shown), suggesting that their NLS sequences are located within the deleted amino acid regions. In contrast, for TAF3 no single C-terminal NLS sequence has been identified; instead, several NLS sequences were predicted by the computer analysis. However, deleting most of them did not change the nuclear localization of TAF3 (data not shown).
FIG. 5.
The nuclear localization of exogenously expressed TAF10 depends on the presence of an NLS either located in the interaction partner itself or fused directly to TAF10. (A to C) HeLa cells were cotransfected with the indicated CFP- and YFP-containing expression vectors, and localization of the expressed proteins was visualized by fluorescence microscopy. The images shown in each panel are representative of all the transfected cells.
Next, we investigated whether the previously tested NLS-deficient mutants affect the partner-dependent nuclear accumulation of TAF10. To this end, we coexpressed these mutants with TAF10 in HeLa cells and tested the localization of TAF10. When TAF8 and SPT7L are cytoplasmic, TAF10 cannot be transferred to the nucleus (Fig. 5A), indicating that the nuclear localization of TAF10 is dependent on the NLS of either TAF8 or SPT7L.
A heterologous NLS sequence fused to TAF10 is sufficient to translocate TAF10 to the nucleus.
To test whether TAF10 alone can be efficiently imported into and/or retained in the nucleus in the absence of a coexpressed interaction partner, we fused either the NLS sequence derived from the SV40 T antigen (NLSSV40) or the above-characterized NLS sequence of TAF8 (NLSTAF8) to the N-terminal end of the full-length TAF10 (Fig. 2). As controls we used vectors expressing either CFP-NLSSV40, which contains the NLS from the SV40 T antigen alone, or CFP-NLSTAF8, which contains only the above-characterized NLS of TAF8 (Fig. 5B). When the CFP-TAF10-NLSSV40 and the CFP-TAF10-NLSTAF8 fusion proteins containing heterologous NLSs were expressed in HeLa cells, they were exclusively nuclear (Fig. 5C). The above-described in vivo results together indicate that TAF10 itself does not have an NLS but that once the protein is imported into the nucleus, either by a heterologous NLS or by one of its three partners, it remains nuclear. Thus, the in vivo nuclear localization of TAF10 seems to be regulated by its complex formation with one its HF motif-containing partners, TAF3, TAF8, or SPT7L.
FRAP measurements indicate that cytoplasmic TAF10 does not bind to any partner or structure until it is transported into the nucleus by TAF8 or an NLS.
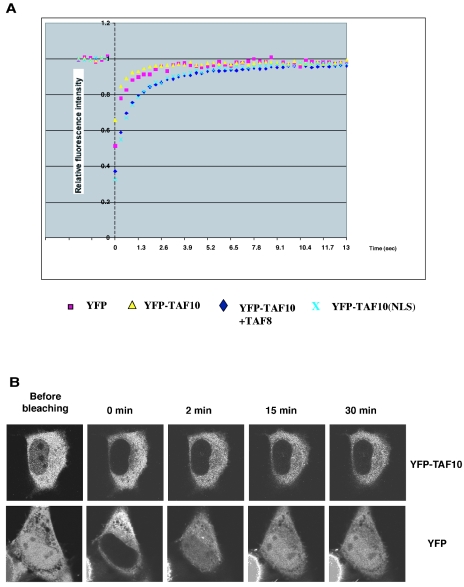
FRAP is a technique that can be used to quantitatively measure the kinetics of binding of proteins to unperturbed structures in living cells. The rationale for this approach is based on the property that FRAP kinetics reflect the overall mobility of a protein. For proteins that do not interact with any cellular structures, FRAP kinetics are a direct reflection of their translational motion properties. In contrast, for proteins that bind to relatively immobile structures such as chromatin, binding events slow down the protein's overall mobility (reference 39 and references therein). We used FRAP measurements to compare the binding properties of TAF10 in the cytoplasmic and nuclear compartments. To this end, after irreversible bleaching of a circular region of interest (ROI) by using a 250-ms pulse with a 514-nm laser, we measured the recovery of fluorescence intensity in the bleached spot. When YFP-TAF10 was overexpressed alone in HeLa cells, the ROI was selected in the cytoplasm and the time to reach half of the maximal recovery (t50) was short (t50 was less than 250 ms) and comparable to the time for recovery of the YFP protein (Fig. 6A), suggesting that TAF10 diffuses rather freely and does not bind to any cytoplasmic partner or compartment. When TAF8 was coexpressed along with YFP-TAF10, an ROI within the nucleoplasm was bleached and the fluorescence recovery of YFP-TAF10 was assessed. In this case, 50% recovery of YFP-TAF10 was obtained in 3 s and complete recovery in 40 s (Fig. 6A). The fact that this recovery rate is significantly slower than that measured for YFP or TAF10 alone (3 s versus 250 ms) (Fig. 6A) is consistent with the notion that the slower mobility of TAF10 in the nucleus is a result of the binding of TAF10 to a nuclear structure, such as chromatin, possibly after an interaction with TAF8, and possible subsequent incorporation into complexes such as TFIID/TFTC. Note that comparable t50s (between 3 and 8 s) were observed for other chromatin-associated proteins, such as BRG1, PCAF, and XPB (39). Importantly, a similar slow recovery kinetic was observed in the nucleus for YFP-TAF10-NLSTAF8, which contains a heterologous NLS sequence (Fig. 6A). These data together support the idea that when TAF10 is in the nucleus it can bind to similar nuclear proteins and structures, as YFP-TAF10 cotranslocated with TAF8, further indicating that the interaction partners of TAF10 play a role primarily in the nuclear accumulation of the factor but that once it is in the nucleus it can dynamically incorporate into TAF-containing complexes and/or bind to nuclear structures.
FIG. 6.
YFP-TAF10 diffuses freely within the cytoplasm but binds to structures in the nucleus. (A) Quantitative FRAP analysis of YFP, YFP-TAF10 in the cytoplasm, nuclear YFP-TAF10-NLSTAF8, and nuclear YFP-TAF10 after coexpression with TAF8. YFP and cytoplasmic YFP-TAF10 show similar, very fast recovery kinetics in the cytoplasm (250 ms), whereas nuclear YFP-TAF10 shows much slower recovery kinetics (3 s) than its cytoplasmic counterpart. (B) Confocal series of pictures of transfected cells before and after photobleaching of the nuclear fluorescent pool of either YFP or YFP-TAF10. YFP shows fast and almost complete recovery within the first 5 min after bleaching. YFP-TAF10 exhibits only minimal recovery even after 30 min.
To further investigate whether the weak nuclear pool of overexpressed TAF10 is a result of its limited nuclear translocation by endogenous TAF8, TAF3, and/or SPT7L and to exclude a pure diffusion event, we eliminated the fluorescent nuclear pool of YFP-TAF10 by photobleaching. To this end, the loss of cytoplasmic fluorescence and the recovery of fluorescence in the nucleus were measured and normalized by the general loss of fluorescence due to bleaching during the imaging process. Figure 6B shows that 30 min after photobleaching of nuclear YFP-TAF10 there was still no, or only minimal, recovery in the nucleus, in contrast to YFP alone for which we observed a substantial amount of fluorescence recovery during the first 2 min following photobleaching. Similar results were obtained when we used the YFP-tet protein, comparable in size to YFP-TAF10, which also diffuses between the two subcellular compartments (data not shown). These data suggest that YFP-TAF10 cannot freely diffuse from the cytoplasm to the nucleus, as does YFP alone, and that the weak nuclear pool of overexpressed TAF10 is due to its transport to the nucleus by the limited pool of its endogenous partners.
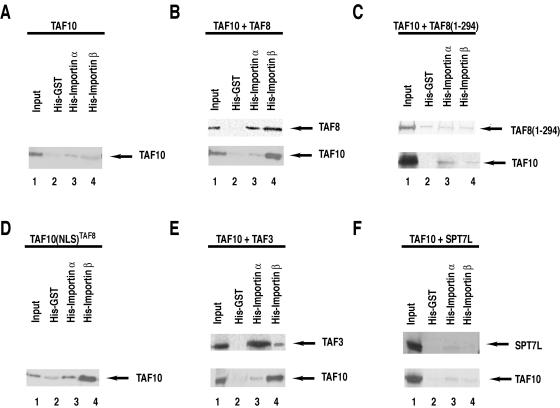
In contrast to TAF10 alone, TAF10/TAF8 and the TAF10/TAF3 HF-containing pairs bind to importin β in vitro.
Different NLSs have been reported to interact with distinct importins (16). The best-studied interactions involve the import receptor for classical NLS-containing proteins, consisting of an importin β/importin α dimer. Importin α recognizes the cargo NLS but requires importin β to cross the nuclear pore complex and to deliver the cargo protein inside the nucleus (16). To further investigate the mechanism of the translocation of the TAF10/TAF3, TAF10/TAF8, and TAF10/SPT7L heterodimers into the cell nucleus, we performed in vitro pull-down experiments. To this end, Ni-NTA resin-bound six-His-tagged GST (as a control), six-His-tagged importin α (Rch-1), and six-His-tagged importin β (15) were incubated with extracts from HeLa cells transfected with either a TAF10 expression vector alone or combinations of expression vectors expressing TAF10 with each of its interacting partners (Fig. 7). When extracts from TAF10-transfected cells, in which TAF10 is mostly cytoplasmic, were analyzed, a very weak interaction with importin α or importin β was observed (Fig. 7A). On the other hand, when extracts containing coexpressed TAF10/TAF8 or TAF10/TAF3 were incubated with the different importin-containing beads, we observed efficient binding of TAF10 to importin β (Fig. 7B and E, lower panels). Control experiments showed that YFP-TAF8 and Flag-TAF3, when used individually for transfection, interacted with both importins but predominantly with importin α. The above-described results suggest that TAF10 alone does not bind to either importin α or β but that it can bind to importin β through its dimerization with either TAF3 or TAF8. These findings are in good agreement with our in vivo data and further show that the importin α/β-dependent nuclear translocation process of TAF10 involves its dimerization with either TAF3 or TAF8.
FIG. 7.
In vitro TAF10 binding to importin β is dependent on the coexpression of either TAF8 or TAF3, but not SPT7L. (A to F) Six-His-tagged GST, six-His-tagged importin α, and six-His-tagged importin β were expressed in Escherichia coli and bound to Ni-NTA beads that were then extensively washed. HeLa cells were cotransfected with CFP- and YFP- or Flag-containing expression vectors (as indicated at the top of each panel); 48 h after transfection, WCEs were prepared and the extracts were incubated with the control His-tagged GST- as well as the importin α- and importin β-containing beads. Beads were then washed and bound proteins analyzed by Western blotting with an anti-GFP antibody. A small amount (0.1%) of the input extract was also analyzed on the same blots.
To further investigate in vitro the NLS requirement of TAF10 for binding to importin α or β, we repeated the previous experiments with extracts from HeLa cells either cotransfected with TAF10 and the NLS-lacking TAF8(1-294), which both remain in the cytoplasm, or transfected with the TAF10-NLSTAF8 expression vector, which is exclusively nuclear. When TAF8 lacks its NLS, as in the TAF8(1-294) mutant, it cannot bind to either importin α or importin β and consequently TAF10 does not interact with importin β (Fig. 7C). However, when TAF10 protein contains a heterologous NLS, it can efficiently interact with importin α and mainly with importin β (Fig. 7D). The data given above together indicate that TAF10 needs to have an NLS-containing partner, or a heterologous NLS fused to it, to be able to bind to importin β.
YFP-SPT7L was not found to bind to either importin α or importin β, and coexpression with TAF10 did not change the affinity of either protein for the importins (Fig. 7F). Thus, our results also suggest that for nuclear translocation the TAF10/SPT7L complex uses a different pathway than importin α/β. These in vitro interaction studies further show that the nuclear import of TAF10 is regulated through its interaction partners.
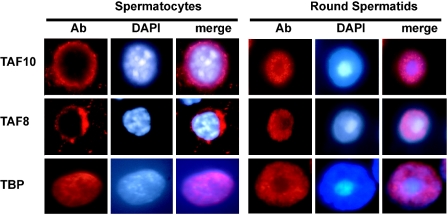
Endogenous TAF10 and TAF8 are cytoplasmic in primary adult spermatocytes.
In order to gain further insight into the nuclear transport of TAF10 in a natural setting, we have investigated its intracellular localization at various stages of the differentiation process of adult male germ cells. Both TAF8 and TAF10 are expressed in these cells and are likely to participate in the complex transcriptional program which takes place postmeiotically (44). Our analysis reveals that the proteins display highly similar distribution patterns, being cytoplasmic in primary adult spermatocytes and then nuclear in round spermatids. Notably, both TAF8 and TAF10 localize in the perinuclear cytoplasmic region in spermatocytes but are excluded from the transcriptionally inactive, highly heterochromatic chromocenter in the round spermatids (Fig. 8). TBP, in contrast, remains nuclear at all times during these differentiation steps (Fig. 8) (27). Thus, the processes of nuclear transport of TAF8 and TAF10 appear to occur in concert, possibly to allow the proper activation of the postmeiotic transcriptional program.
FIG. 8.
Endogenous TAF10 and TAF8 are cytoplasmic in primary adult spermatocytes but not in round spermatids. Squash preparations from segments of defined stages of the mouse adult seminiferous tubuli were obtained by transillumination-assisted microdissection and probed with anti-TAF10, anti-TAF8, or anti-TBP antibody (left panels of each series). The middle and right panels show the corresponding DAPI-stained DNA and merged images, respectively. Magnification, ×100. Ab, antibody.
DISCUSSION
In recent years, a significant amount of effort has been put into understanding the structure, assembly, and diverse functions of TFIID and other TAF-containing complexes. The mostly biochemical experiments have shown that HF-containing TAFs mediate many of the subunit interactions within the TFIID and STAGA-/TFTC-type complexes (through their HF motifs), assuring the integrity of the complexes. They also play a key role in promoter recognition, exert coactivator functions, and by interacting with components of the general transcription machinery facilitate preinitiation complex recruitment (11, 24, 25, 41).
Here we found a new role for the HF-containing TAFs by investigating the regulation of TAF10 nuclear localization by its three HF-containing interaction partners. We observed that overexpressed TAF10, in contrast to the native protein, is more abundant in the cytoplasm of HeLa cells than in the nucleus and can be transferred to the nucleus upon overexpression of one of its partners. In agreement with the in vitro data, this result indicates that TAF10 interacts with its three HF-containing partners, TAF3, TAF8, and SPT7L, also in vivo. Moreover, we observed that if we eliminate the weak fluorescent nuclear pool of TAF10 by photobleaching, the remaining cytoplasmic TAF10 pool does not translocate to the nucleus through a passive diffusion mechanism. In contrast, YFP and YFP-tet proteins freely diffuse through the two subcellular compartments. These data together with previous observations with Drosophila cells (14) suggest that the expression levels of endogenous partners of TAF10 (TAF3, TAF8, and SPT7L) can be limited in the cells and that “free” TAF10, which is not part of any histone fold dimer combination, remains cytoplasmic (see also below). We investigated two different possibilities for the failure of the “free” or overexpressed pool of TAF10 to localize to the nucleus: (i) the overexpressed protein cannot be retained in the nucleus and is rapidly exported to the cytoplasm due to the limited amounts of the endogenous retention partner in the nucleus or (ii) TAF10, due to the lack of a functional NLS or because a cytoplasmic protein masks its NLS, can be imported into the nucleus only as a dimer with one of its NLS-containing partners and thus the “free” TAF10 protein would remain cytoplasmic. Several lines of evidence confirmed the second possibility. LMB treatment did not change the cytoplasmic localization of TAF10, suggesting that it is not exported into the cytoplasm through a CRM1-dependent pathway. Interestingly, the nuclear export of another TAF, TAF4b (formerly TAFII105), is mediated by a composite nuclear export signal and thus the nuclear export of TAF4b was suggested to happen through a CRM1-independent nuclear export pathway (42). Nevertheless, the cytoplasmic localization of all the truncated derivatives of TAF10 excluded the existence of an export signal insensitive to LMB and rather indicated the absence of a domain that can function as a nuclear localization signal. This conclusion was also supported by the observation that a heterologous NLS can drive TAF10 into the nucleus. The observation that the overexpressed CFP-TAF10-NLSSV40 and CFP-TAF10-NLSTAF8 fusion proteins remain nuclear excludes export or cytoplasmic retention as a mechanism for the cytoplasmic accumulation of the exogenously expressed TAF10 protein. The above-mentioned conclusion is in complete agreement with the fluorescent recovery kinetics after photobleaching measurements. The diffusion is measured in FRAP experiments over a micrometer range, and the transient interaction of a protein with subcellular structures is rate limiting and slows down the overall recovery rate of a given protein in a FRAP assay. As the t50 measured for TAF10 in the cytoplasm ranges from 10 to 100 ms, similar to the rate of free diffusion of YFP or CFP itself, our data suggest that TAF10, when excluded from the nucleus, moves like a “free” protein without binding to any partner or cytoplasmic subcellular structures. Although YFP-TAF10 does not seem to bind tightly to any cytoplasmic structure or partner, we cannot exclude the possibility of transient association of TAF10 with a cytoplasmic partner that would somehow counteract the passive diffusion of YFP-TAF10 to the nucleus. Alternatively, it could be possible that TAF10 in the cytoplasm forms dimers or oligomers (as suggested earlier [21]), which then would not be able to diffuse passively through the nuclear pore.
We also investigated the possible mechanism by which TAF10 enters into the nucleus. The lack of nuclear localization of TAF10 by its truncated partners, from which the NLSs have been deleted, suggested that TAF10 does not have an NLS and enters the nucleus only with its NLS-containing partners. Moreover, we showed that TAF10 could not efficiently interact with importin α and β when overexpressed alone. On the other hand, TAF3 and TAF8 interact efficiently with importin α, probably through their NLSs, and by dimerizing with TAF10 through their histone fold domains can bring TAF10 to importin β. Transport receptors cannot always bind their cargoes directly; in some cases they need an adapter molecule, which complicates the transport cycles considerably. The best-studied adapter is importin α, which mediates import of proteins that carry mostly classical NLSs. Importin α binds both the cargo and importin β in the cytoplasm. Importin β accounts for contacts with the nuclear pore complex (8, 16) and mediates translocation of the trimeric NLS-containing factor-importin α/β complex from the cytoplasmic to the nuclear side of the nuclear pore complex. TAF10 lacks a functional NLS; thus, it does not bind to importin α and cannot subsequently bind to importin β. TAF8 or TAF3 alone is able to bind to importin α, but in the presence of TAF10, TAF8 or TAF3 binds predominantly to importin β. It is possible that in the TAF8-TAF10 (or TAF3-TAF10) heterodimer, the conformation of TAF8 (or TAF3) is changed and thus the dimer has a higher affinity for importin β. The fact that the third interaction partner of TAF10, SPT7L, uses an import pathway other than the importin α/β complex to achieve the nuclear import of the corresponding dimer suggests a different regulatory mechanism for the nuclear import and the consequent incorporation of the SPT7L-TAF10 dimer into the TFTC complex.
Recent development in in vivo imaging technologies allowed us to start to investigate fundamental mechanistic questions about how transcription complexes assemble and thus to better understand how they regulate transcription in the cells. In this respect, the important questions to answer are whether the different complexes, which have been purified biochemically as intact entities and which have been suggested to be recruited to the preinitiation complex as multisubunit complexes, are (i) assembled as subcomplexes or whole complexes in the cytoplasm, (ii) assembled as subcomplexes or whole complexes in the nucleus, or (iii) assembled within the nucleoplasm before DNA binding or on the promoter of a given gene. The TAF-containing complexes TFIID and TFTC have a unique characteristic because most of their components form dimers through their histone fold domains (11, 25). The present study on the nuclear localization of TAF10 reveals a new regulatory mechanism in the assembly of the TAF-containing complexes. We show that the first step of the assembly of these complexes takes place in the cytoplasm by the formation of histone fold dimers and is followed by coimport into the nucleus. Our biochemical and in vivo data together show that once TAF10 is imported into the nucleus, it presumably incorporates efficiently into the complexes and binds to chromatin, suggesting that in the case of TAF10 this first step of assembly in the cytoplasm is fundamentally important for the completion of the whole process.
Drosophila has two distinct homologues of human TAF10, D. melanogaster TAF10 and TAF10b, which are encoded by two paralogue genes. It has been shown that the subcellular localization of both D. melanogaster TAF10 and TAF10b proteins changes during development and can also depend on the cell type (14). Here we report that the intracellular localization of mouse TAF10 during adult spermatogenesis is regulated in concert with one of its partners, TAF8. Because of the lack of antibodies against TAF3 and SPT7L, which would display a specific signal in male germ cells, we cannot exclude the possibility that these two other partners may be regulated in a similar manner. We have found that TAF10 is cytoplasmic in pachytene spermatocytes and nuclear in round spermatids following the localization of TAF8 in the same cell types. This observation reinforces the notion that the concerted nuclear transport of TAF10 and TAF8 is indeed a physiologically relevant event, as it coincides with the postmeiotic wave of transcriptional activation (44). Importantly, other TAFs, such as TAF7L, show this developmentally regulated intracellular localization as well (40), suggesting a general control mechanism of the composition of TFIID and other TAF-containing complexes during spermatogenesis.
Similar regulatory interactions that specifically affect the subcellular localization of proteins have been described (4). In particular, the nuclear localization of the extradenticle and PBX1 proteins is regionally restricted during Drosophila and mammalian development and depends on the nuclear localization of their interacting proteins HTH and PREP1 (4).
Here we describe a novel mechanism in which HF-containing proteins can regulate the local action of their interacting partners by facilitating their import into the nucleus. This also suggests that in its cytoplasm, depending on with which interaction partner TAF10 heterodimerizes, the cell determines into which TAF-containing complex (TFIID or TFTC) TAF10 will be incorporated once transported into the nucleus. Thus, in this study we have unraveled a novel regulatory role for the three histone fold-containing interacting partners of TAF10. The question of whether this is a general role for most of the known histone fold-containing TAFs or other proteins during cellular differentiation or metazoan development remains to be answered.
Acknowledgments
We are grateful to T. Misteli for helpful discussions, advice, and support; to the NCI and the IGBMC imaging facilities; to A. Jànoshàzi and T. Karpova for imaging training; to M. Oulad-Abdelghani for generating antibodies; to I. Davidson and G. Gangloff for TAF3 expression plasmids; to D. Gorlich, A. Kretsovali, and J. Papamatheakis for the importin plasmids; to S. Gorski and E. Meshorer for advice in FRAP experiments; and to M. Frontini, A. Magklara, E. Martinez, and S. John for critically reading the manuscript. We also thank the IGBMC cell culture facility for providing cells and I. Kolb-Cheynel for help with the recombinant baculoviruses.
E.S. was supported by an EMBO long-term fellowship, M.A.D. by a fellowship from the European Community (grant HPRN-CT-2000-00088), and G.F. by a fellowship of the Fondation de la Recherche Médicale. This work was supported by funds from INSERM, CNRS, Hôpital Universitaire de Strasbourg, Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, and the Front Nationale de la Science ACI and by European Community RTN (HPRN-CT-2000-00087, HPRN-CT-2000-00088, and HPRN-CT 00504228), STREP (LSHG-CT-2004-502950), and AICR (03-084) grants.
REFERENCES
- 1.Albright, S. R., and R. Tjian. 2000. TAFs revisited: more data reveal new twists and confirm old ideas. Gene 242:1-13. [DOI] [PubMed] [Google Scholar]
- 2.Bell, B., E. Scheer, and L. Tora. 2001. Identification of hTAF(II)80 delta links apoptotic signaling pathways to transcription factor TFIID function. Mol. Cell 8:591-600. [DOI] [PubMed] [Google Scholar]
- 3.Bell, B., and L. Tora. 1999. Regulation of gene expression by multiple forms of TFIID and other novel TAFII-containing complexes. Exp. Cell Res. 246:11-19. [DOI] [PubMed] [Google Scholar]
- 4.Berthelsen, J., C. Kilstrup-Nielsen, F. Blasi, F. Mavilio, and V. Zappavigna. 1999. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 13:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brand, M., C. Leurent, V. Mallouh, L. Tora, and P. Schultz. 1999. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science 286:2151-2153. [DOI] [PubMed] [Google Scholar]
- 6.Brou, C., S. Chaudhary, I. Davidson, Y. Lutz, J. Wu, J. M. Egly, L. Tora, and P. Chambon. 1993. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatton, B., A. Bahr, J. Acker, and C. Kedinger. 1995. Eukaryotic GST fusion vector for the study of protein-protein associations in vivo: application to interaction of ATFa with Jun and Fos. BioTechniques 18:142-145. [PubMed] [Google Scholar]
- 8.Chook, Y. M., and G. Blobel. 2001. Karyopherins and nuclear import. Curr. Opin. Struct. Biol. 11:703-715. [DOI] [PubMed] [Google Scholar]
- 9.Dubrovskaya, V., A.-C. Lavigne, I. Davidson, J. Acker, A. Staub, and L. Tora. 1996. Distinct domains of hTAFII100 are required for functional interaction with transcription factor TFIIFβ (RAP30) and incorporation into the TFIID complex. EMBO J. 15:3702-3712. [PMC free article] [PubMed] [Google Scholar]
- 10.Frontini, M., C. Imbriano, A. diSilvio, B. Bell, A. Bogni, C. Romier, D. Moras, L. Tora, I. Davidson, and R. Mantovani. 2002. NF-Y recruitment of TFIID, multiple interactions with histone fold TAF(II)s. J. Biol. Chem. 277:5841-5848. [DOI] [PubMed] [Google Scholar]
- 11.Gangloff, Y., C. Romier, S. Thuault, S. Werten, and I. Davidson. 2001. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 26:250-257. [DOI] [PubMed] [Google Scholar]
- 12.Gangloff, Y. G., J. C. Pointud, S. Thuault, L. Carre, C. Romier, S. Muratoglu, M. Brand, L. Tora, J. L. Couderc, and I. Davidson. 2001. The TFIID components human TAF(II)140 and Drosophila BIP2 [TAF(II)155] are novel metazoan homologues of yeast TAF(II)47 containing a histone fold and a PHD finger. Mol. Cell. Biol. 21:5109-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gangloff, Y. G., S. L. Sanders, C. Romier, D. Kirschner, P. A. Weil, L. Tora, and I. Davidson. 2001. Histone folds mediate selective heterodimerization of yeast TAF(II)25 with TFIID components yTAF(II)47 and yTAF(II)65 and with SAGA component ySPT7. Mol. Cell. Biol. 21:1841-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgieva, S., D. B. Kirschner, T. Jagla, E. Nabirochkina, S. Hanke, H. Schenkel, C. de Lorenzo, P. Sinha, K. Jagla, B. Mechler, and L. Tora. 2000. Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol. Cell. Biol. 20:1639-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorlich, D., P. Henklein, R. A. Laskey, and E. Hartmann. 1996. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15:1810-1817. [PMC free article] [PubMed] [Google Scholar]
- 16.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 17.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 18.Guermah, M., K. Ge, C. M. Chiang, and R. G. Roeder. 2003. The TBN protein, which is essential for early embryonic mouse development, is an inducible TAFII implicated in adipogenesis. Mol. Cell 12:991-1001. [DOI] [PubMed] [Google Scholar]
- 19.Helmlinger, D., S. Hardy, S. Sasorith, F. Klein, F. Robert, C. Weber, L. Miguet, N. Potier, A. Van-Dorsselaer, J. M. Wurtz, J. L. Mandel, L. Tora, and D. Devys. 2004. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 13:1257-1265. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez-Hernandez, A., and A. Ferrus. 2001. Prodos is a conserved transcriptional regulator that interacts with dTAF(II)16 in Drosophila melanogaster. Mol. Cell. Biol. 21:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacq, X., C. Brou, Y. Lutz, I. Davidson, P. Chambon, and L. Tora. 1994. Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell 79:107-117. [DOI] [PubMed] [Google Scholar]
- 22.Kirchner, J., S. L. Sanders, E. Klebanow, and P. A. Weil. 2001. Molecular genetic dissection of TAF25, an essential yeast gene encoding a subunit shared by TFIID and SAGA multiprotein transcription factors. Mol. Cell. Biol. 21:6668-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirschner, D. B., E. vom Baur, C. Thibault, S. L. Sanders, Y. G. Gangloff, I. Davidson, P. A. Weil, and L. Tora. 2002. Distinct mutations in yeast TAF(II)25 differentially affect the composition of TFIID and SAGA complexes as well as global gene expression patterns. Mol. Cell. Biol. 22:3178-3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kouskouti, A., E. Scheer, A. Staub, L. Tora, and I. Talianidis. 2004. Gene-specific modulation of TAF10 function by SET9-mediated methylation. Mol. Cell 14:175-182. [DOI] [PubMed] [Google Scholar]
- 25.Leurent, C., S. Sanders, C. Ruhlmann, V. Mallouh, P. A. Weil, D. B. Kirschner, L. Tora, and P. Schultz. 2002. Mapping histone fold TAFs within yeast TFIID. EMBO J. 21:3424-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leurent, C., S. L. Sanders, M. A. Demeny, K. A. Garbett, C. Ruhlmann, P. A. Weil, L. Tora, and P. Schultz. 2004. Mapping key functional sites within yeast TFIID. EMBO J. 23:719-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martianov, I., S. Brancorsini, A. Gansmuller, M. Parvinen, I. Davidson, and P. Sassone-Corsi. 2002. Distinct functions of TBP and TLF/TRF2 during spermatogenesis: requirement of TLF for heterochromatic chromocenter formation in haploid round spermatids. Development 129:945-955. [DOI] [PubMed] [Google Scholar]
- 28.Martinez, E. 2002. Multi-protein complexes in eukaryotic gene transcription. Plant Mol. Biol. 50:925-947. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, E., T. K. Kundu, J. Fu, and R. G. Roeder. 1998. A human SPT3-TAFII31-GCN5-L acetylase complex distinct from transcription factor IID. J. Biol. Chem. 273:23781-23785. [DOI] [PubMed] [Google Scholar]
- 30.Martinez, E., V. B. Palhan, A. Tjernberg, E. S. Lymar, A. M. Gamper, T. K. Kundu, B. T. Chait, and R. G. Roeder. 2001. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell. Biol. 21:6782-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metzger, D., E. Scheer, A. Soldatov, and L. Tora. 1999. Mammalian TAF(II)30 is required for cell cycle progression and specific cellular differentiation programmes. EMBO J. 18:4823-4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohan, I. W., E. Scheer, O. Wendling, D. Metzger, and L. Tora. 2003. TAF10 [TAF(II)30] is necessary for TFIID stability and early embryogenesis in mice. Mol. Cell. Biol. 23:4307-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moroianu, J. 1998. Distinct nuclear import and export pathways mediated by members of the karyopherin beta family. J. Cell. Biochem. 70:231-239. [PubMed] [Google Scholar]
- 34.Muller, F., and L. Tora. 2004. The multicoloured world of promoter recognition complexes. EMBO J. 23:2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muratoglu, S., S. Georgieva, G. Papai, E. Scheer, I. Enunlu, O. Komonyi, I. Cserpan, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schlitz, T. Howard, X. J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 37.Parvinen, M., and N. B. Hecht. 1981. Identification of living spermatogenic cells of the mouse by transillumination-phase contrast microscopic technique for ‘in situ’ analyses of DNA polymerase activities. Histochemistry 71:567-579. [DOI] [PubMed] [Google Scholar]
- 38.Phair, R. D., S. A. Gorski, and T. Misteli. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods Enzymol. 375:393-414. [DOI] [PubMed] [Google Scholar]
- 39.Phair, R. D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D. T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pointud, J. C., G. Mengus, S. Brancorsini, L. Monaco, M. Parvinen, P. Sassone-Corsi, and I. Davidson. 2003. The intracellular localisation of TAF7L, a paralogue of transcription factor TFIID subunit TAF7, is developmentally regulated during male germ-cell differentiation. J. Cell Sci. 116:1847-1858. [DOI] [PubMed] [Google Scholar]
- 41.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255:1-14. [DOI] [PubMed] [Google Scholar]
- 42.Rashevsky-Finkel, A., A. Silkov, and R. Dikstein. 2001. A composite nuclear export signal in the TBP-associated factor TAFII105. J. Biol. Chem. 276:44963-44969. [DOI] [PubMed] [Google Scholar]
- 43.Sanders, S. L., E. R. Klebanow, and P. A. Weil. 1999. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274:18847-18850. [DOI] [PubMed] [Google Scholar]
- 44.Sassone-Corsi, P. 2002. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science 296:2176-2178. [DOI] [PubMed] [Google Scholar]
- 45.Tora, L. 2002. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 16:673-675. [DOI] [PubMed] [Google Scholar]
- 46.Voss, A. K., T. Thomas, P. Petrou, K. Anastassiadis, H. Scholer, and P. Gruss. 2000. Taube nuss is a novel gene essential for the survival of pluripotent cells of early mouse embryos. Development 127:5449-5461. [DOI] [PubMed] [Google Scholar]
- 47.Wieczorek, E., M. Brand, X. Jacq, and L. Tora. 1998. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393:187-191. [DOI] [PubMed] [Google Scholar]
- 48.Wu, P. Y., C. Ruhlmann, F. Winston, and P. Schultz. 2004. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell 15:199-208. [DOI] [PubMed] [Google Scholar]
- 49.Wu, P. Y., and F. Winston. 2002. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22:5367-5379. [DOI] [PMC free article] [PubMed] [Google Scholar]