Abstract
The mitogen-activated protein kinase extracellular signal-regulated kinase (ERK) is activated following engagement of the T-cell receptor and is required for interleukin 2 (IL-2) production and T-cell proliferation. This activation is enhanced by stimulation of the coreceptor CD28 and inhibited by the coreceptor CTLA-4. We show that the small G protein Rap1 is regulated in the opposite manner; it is inhibited by CD28 and activated by CTLA-4. Together, CD3 and CTLA-4 activate Rap1 in a sustained manner. To delineate T-cell function in the absence of Rap1 activity, we generated transgenic mice expressing Rap1GAP1, a Rap1-specific GTPase-activating protein. Transgenic mice showed lymphadenopathy, and transgenic T cells displayed increased ERK activation, proliferation, and IL-2 production. More significantly, the inhibitory effect of CTLA-4 on T-cell function in Rap1GAP1-transgenic T cells was reduced. We demonstrate that CTLA-4 activates Rap1, and we propose that intracellular signals from CTLA-4 antagonize CD28, at least in part, at the level of Rap1.
The activation of T cells requires antigen recognition through the T-cell receptor (TCR) and costimulation via the CD28 coreceptor, which binds the family of B7 ligands (B7-1 and B7-2) expressed on antigen-presenting cells (APCs) (35). Additional complexity comes with another coreceptor, CTLA-4, that also binds B7 and can antagonize the ability of CD28 to stimulate T-cell activation and interleukin 2 (IL-2) accumulation (25, 27, 28, 42, 44). Recent studies have suggested that inhibition of extracellular signal-regulated kinases (ERKs) may contribute to this action (6, 9, 38). ERKs are activated following costimulation by TCR/CD28 and are required for AP-1-dependent transcription of IL-2, a cytokine that is essential for T-cell proliferation (32, 46). Cross-linking anti-CTLA-4 antibodies (that activate CTLA-4) inhibit both ERK activation and IL-2 production following costimulation by TCR/CD28 (6, 9). The mechanism by which CTLA-4 inhibits ERK activation is not known.
The small G protein Ras mediates TCR activation of ERKs (7). TCR stimulation triggers the activation of Ras via the Ras guanine nucleotide exchange factors (GEF) Cal DAG-GEF II (also called Ras-GRP1) (17) and SOS (47). Ras is a positive activator of the mitogen-activated protein (MAP) kinase kinase kinase Raf-1 that can phosphorylate and activate the MAP kinase kinase MEK, which, in turn, can activate ERK. Potential negative regulatory molecules are also activated by TCR stimulation. One of these is the small G protein Rap1 that, like Ras, can be inhibited by specific GTPase-activating proteins (Rap1GAPs). Rap1 is activated following stimulation of the TCR (1, 8) and, like Ras, is rapidly recruited to the plasma membrane upon activation (3). During CD28 costimulation, Rap1 activation is inhibited (8, 22, 33) and ERK signaling is enhanced (8). Conversely, T lymphocytes lacking the Rap1GAP Spa-1 show constitutively elevated Rap1 activity, diminished ERK activation, and a decreased response to TCR stimulation (21). Models of T-cell anergy induced by lack of CD28 costimulation also show constitutively elevated Rap1 activity, diminished ERK activation, and decreased response to TCR stimulation (4). These correlations support the idea that Rap1 is a negative regulator of T-cell activation. In contrast, Rap1 has been proposed to augment T-cell activation via its enhancement of integrin-dependent adhesion (22, 39). Most models for the study of Rap1 function in T cells have examined constitutive Rap1 activation and, therefore, do not directly test the physiological role of Rap1 (21, 39). To better address the function of Rap1 in T-cell activation, we expressed Rap1GAP1 in T cells. This study describes the effects of ectopic Rap1GAP1 expression in peripheral T cells.
Rap1GAP1-expressing T cells were inhibited in their TCR-dependent activation of Rap1 and showed enhanced T-cell activation in vitro. Rap1GAP1-transgenic mice showed lymph nodes that were hypercellular and characterized by the presence of activated T cells. The enhanced activation in the Rap1GAP1-expressing T cells was associated with the reduced ability of CTLA-4 to down-regulate T-cell function. These studies suggest strongly that activation of Rap1 by CTLA-4 was required for CTLA-4 to block ERK signaling through the TCR and inhibit IL-2 production. We propose that Rap1 activation is dynamically regulated by CTLA-4 and CD28 to modulate T-cell responses.
MATERIALS AND METHODS
Cell lines and antibodies.
CH27 B-lymphoblastoid cells were maintained in RPMI 1640 (Life Technologies, Bethesda, MD) containing 10% fetal calf serum (HyClone, Logan, UT) and supplemented with 1 mM l-glutamine, sodium pyruvate (100 mg/ml), 5 × 10−5 M β-mercaptoethanol, essential and nonessential amino acids (Life Technologies, Bethesda, MD), 100 U/ml penicillin G, 100 U/ml streptomycin, and 50 μg/ml gentamicin.
The following antibodies were purchased from BD Pharmingen, San Diego, CA: Fluorescein (FITC)- and phycoerythrin (PE)-labeled anti-CD8 monoclonal antibodies (53-6.7), Cy-Chrome-labeled anti-CD4 monoclonal antibodies (RM4-5), FITC- and biotin-labeled anti-CD69 (H1.2F3), biotin-labeled anti-CD44 (IM7), PE-labeled anti-Vα11 (RR8-1), FITC-labeled anti-Vβ3 (KJ125), unconjugated and PE-labeled anti-CTLA4 (9H10), anti-CD3 (145-2C11), and anti-CD28 (37.51). Streptavidin-APC was also purchased from BD Pharmingen, San Diego, CA. Anti-Rap1(121), anti-PLCγ1(530), and anti-ERK2 (C14) were purchased from Santa Cruz Biotechnology, Inc., Santa Cruz, CA. Anti-phospho-p44/42 (ERK1 and ERK2) MAP kinase and anti-phospho-PLCγ1 (Tyr 771) antibodies were purchased from Cell Signaling Technology, Beverly, MA. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit were purchased from Amersham, Piscataway, NJ.
Rap1GAP1-transgenic mice.
The cDNA encoding Flag-Rap1GAP1 was inserted downstream of the β-actin promoter. This plasmid was used to generate transgenic mouse lines in C57BL/6-JxDBA mice by standard procedures. Founders were genotyped by PCR analysis using vector-specific primers (5′ Flag primer: TACAAGGACGACGATGACAAG; 3′ Rap1Gap1 primer: TCTTCACACACCAACTTTGC), and transgene-expressing founders were backcrossed with wild-type C57BL/6-J mice. Two lines for this transgene were established and shown to have similar amounts of Flag-Rag1GAP1 expression in thymocytes and similar levels of inhibition of Rap1 activity. One Flag-Rap1GAP1-expressing line was chosen to cross to the AD10 TCR-transgenic line maintained on the B10.BR background (23), and F1 mice were typed and studied. Experiments were performed with adult mice, except for thymocyte staining, which was performed with thymocytes from 4-week-old mice. In all animal studies, experiments were performed in compliance with the relevant laws and institutional guidelines and were approved by the Institutional Animal Care and Use Committee.
Flow cytometric analysis.
Thymocytes and lymph node cells were stained for surface expression of CD3, CD8, CD4, CD25 and CD44, and CD69. To confirm the phenotype of the AD10-transgenic TCR, the surface expression of Vα11 and Vβ3 was determined. CTLA-4 expression was determined following fixation and permeabilization with Cytofix/Cytoperm (BD Pharmingen, San Diego, CA) according to the manufacturer's instructions. Cells were analyzed by using a FACSCalibur flow cytometer (Becton Dickinson).
In vitro T-cell priming and stimulation.
Single-cell suspensions of splenocytes from AD10 TCR-transgenic and AD10xRap1GAP1-transgenic mice were primed in vitro with 2.5 μM moth cytochrome c (MCC) peptide for 40 h. Lymphocytes were isolated from primary cultures by density centrifugation using Lympholyte M (Cedarlane, Hornby, Ontario, Canada). T cells were rested for 5 h in complete T-cell medium at 37°C and then restimulated in wells coated with plate-bound antibodies (immobilized antibodies used were 0.5 μg/ml anti-CD3, 10 μg/ml anti-CD28, and 10 μg/ml anti-CTLA-4, as indicated in the figures) overnight at 4°C. Immobilized CTLA-4 antibody stimulates the CTLA-4 coreceptor to inhibit T-cell function (13, 24, 37). Ten million T cells were added to antibody-coated wells and centrifuged for 30 s, incubated for the times indicated in the figures at 37°C, and then lysed with 2× lysis buffer. Alternatively, naïve T cells were stimulated with soluble anti-CD3 as previously described (16). For experiments designed to interfere with normal CTLA-4 function, T-cell blasts were preincubated with soluble anti-CTLA-4 antibody (10 μg/ml), which has been shown to block CTLA-4 engagement by B7 and inhibits the negative regulation of T cells by CTLA-4 (24). For Rap1 activation by APCs, 107 rested T-cell blasts, preincubated with or without soluble anti-CTLA-4 antibody, were incubated in a pellet with equal numbers of irradiated B10.BR splenocytes, preloaded with or without 5 μΜ moth cytochrome C (ΜCC; 1 h at 37°C). Following stimulation, cells were lysed and Rap1, Ras, PLCγ1, and ERK activation was determined as described below.
In vitro Rap1 and Ras activation assay.
The Rap1 assay was performed as previously described (8, 16). The glutathione S-transferase (GST)-Ral-GDS-Ras binding domain (RBD) fusion protein was a gift of J. L. Bos, Utrecht University, Utrecht, The Netherlands. Ras assays were performed using the Ras activation kit using GST-Raf-1-RBD (Upstate Biotechnology, Inc., Lake Placid, NY) according to the manufacturer's instructions.
Western blotting.
Proteins were separated in a 7.5 to 12% gel, followed by transfer to a polyvinylidene difluoride membrane. Phosphorylation of ERK1, ERK2, and PLCγ1 was detected in total-cell lysate by immunoblotting with phosphospecific MAP kinase and PLCγ1 antibodies. ERK2, PLCγ1, Rap1, and Ras levels were detected by immunoblotting with anti-ERK2, anti-PLCγ1, anti-Rap1, and anti-Ras antibodies, respectively. Proteins were detected using horseradish peroxidase-conjugated secondary antibodies followed by enhanced chemiluminescence.
T-cell proliferation assay.
T-cell proliferation was assayed as previously described (16). Splenocytes were assayed at a concentration of 106 cells/ml in the presence of 0.25 μg/ml soluble anti-CD3 antibody or 0.5 μM MCC peptide.
IL-2 ELISA.
The amount of IL-2 in culture supernatants (100 μl) was measured at 12 to 16 h for rested T-cell blasts incubated with irradiated B10.BR splenocytes and at 24 h for naïve AD10 splenocytes incubated with MCC peptide, by enzyme-linked immunosorbent assay (ELISA; eBioscience, San Diego, CA) according to the manufacturer's instructions.
Fluorescence-activated cell sorter (FACS) conjugate assay.
CH27 B-lymphoblastoid cells (APCs) were labeled with 1 μM DiI (Molecular Probes, Eugene OR) for 10 min at 37°C and then incubated overnight with 10 μM MCC. T-cell blasts were labeled with 0.5 μM 5,6-carboxyfluorescein diacetate (Molecular Probes, Eugene OR) for 30 min at 37°C and rested overnight. Equal numbers of APCs and T cells (105 cells) were incubated for 30 min at 37°C. Nonspecific aggregates were disrupted by vortexing, and the samples were analyzed with a FACSCalibur flow cytometer (Becton Dickinson, San Diego, CA). The percentage of conjugates (defined as the number of live-gated, double-positive events [upper right quadrant] divided by the total number of live green T cells) was determined for each sample.
Live cell imaging.
Live T-cell interactions were imaged as described previously (45). One day prior to the experiment, 2.5 × 105 fibroblasts were seeded onto 0.17-mm Delta T-cell culture dishes (Bioptechs, Butler, PA) in 1 ml of complete Dulbecco's modified Eagle medium. The next day, T-cell blasts were resuspended at 2 × 106/ml in phosphate-buffered saline containing 10% fetal bovine serum, 1 mM CaCl2, and 0.5 mM MgCl2 and incubated with 1 μM fura-2-acetoxymethyl ester (fura-2; Molecular Probes, Eugene, OR) for 30 min at 37°C in the dark. Cells were washed and resuspended at 107/ml in phosphate-buffered saline containing 1 mM CaCl2 and 0.5 mM Mg2+ and incubated at room temperature for 15 min to allow for further hydrolysis of the dye. Cells were stored on ice until use. After the addition of 2.5 × 105 T cells to the culture dish, alternating 600× green fluorescent (528 nm), differential interference contrast, and, for fura-2 analysis, excitation at 340 nm and 380 nm, images were captured every 8 to 12 s with an Applied Precision Instruments DeltaVision image restoration system (Issaquah, WA).
We have defined conjugates as a functional interaction between a T cell and an APC. A functional interaction was distinguished microscopically from passive contact between T cells and APCs by three criteria, namely: (i) the T cell underwent a morphological change as it flattened out against the APC, (ii) the T cell fluxed calcium, and (iii) the T cell caused a redistribution of MCC:I-Ek:green fluorescent protein (GFP) to the APC/T-cell interface. For AD10 T cells, we examined 161 T-cell interactions with 16 APCs. For AD10xRap1GAP1 T cells, we examined 224 T-cell interactions with 25 APCs. The percentage of conjugates was calculated as the number of conjugates formed per hundred T cells that contacted APCs. Statistical significance was determined by two-tailed Student's t test.
RESULTS
CTLA-4 inhibits ERKs and activates Rap1.
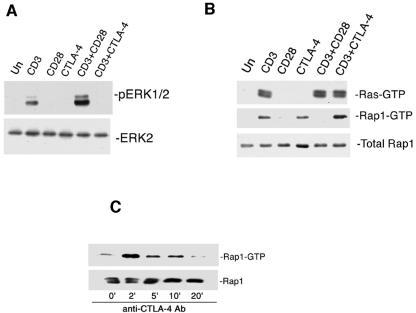
CTLA-4 is a well-described inhibitor of T-cell function (12). AD10 T cells were primed in vitro for 40 h to induce CTLA-4 expression. Engagement of CD3 on AD10 T-cell blasts activated ERKs, and this was enhanced by CD28 costimulation (Fig. 1A). In contrast, CTLA-4 costimulation blocked CD3 activation of ERKs in AD10 T-cell blasts (Fig. 1A). Neither CD28 nor CTLA-4 regulated ERKs in the absence of CD3 activation.
FIG. 1.
CD28 and CTLA-4 have opposing actions on ERKs and Rap1. (A) In vitro-primed AD10 T cells were left untreated (Un) or treated for 5 min with immobilized anti-CD3 (CD3), anti-CD28 (CD28), and/or anti-CTLA-4 (CTLA-4) as indicated. Cells extracts were analyzed by immunoblotting with antibodies specific for phosphorylated ERK1/2 (pERK1/2). The levels of total ERK2 are indicated as a loading control. (B) In vitro-primed AD10 T cells were left untreated (Un) or treated for 5 min with immobilized anti-CD3 (CD3), anti-CD28 (CD28), or anti-CTLA-4 (CTLA-4). Activated Rap1 (Rap1-GTP) and Ras (Ras-GTP) were precipitated using GST-RalGDS and GST-Raf-1GDS, respectively, and analyzed by immunoblotting with Rap1- or Ras-specific antibodies. Total levels of Rap1 are indicated as loading controls for both assays. (C) In vitro-primed AD10 T cells were left untreated (Un) or treated for the indicated times with immobilized anti-CTLA-4 (CTLA-4). Activated Rap1 (Rap1-GTP) was pulled down using GST-RalGDS and analyzed by immunoblotting with Rap1-specific antibodies (Ab). Total levels of Rap1 are indicated as a loading control.
TCR activation of ERKs requires the small G protein Ras (7), and Ras was activated in T-cell blasts by engagement of the TCR by immobilized anti-CD3 (Fig. 1B, upper panel). However, costimulation via CD28 or CTLA-4 had no effect on Ras activation induced by CD3 engagement (Fig. 1B, upper panel). Another small G protein that has been proposed to regulate ERK activation in T cells is Rap1. Engagement of CD3 on AD10 T-cell blasts activated the small G protein Rap1, and this activation by CD3 was blocked by costimulation of CD28 (Fig. 1B, middle panel). Interestingly, engagement of CTLA-4 also was able to activate Rap1 in T-cell blasts, and engagement of CD3 and CTLA-4 showed an additive effect on the activation of Rap1 (Fig. 1B, middle panel).
The time course of Rap1 activation following engagement of CTLA-4 is shown in Fig. 1C. CTLA-4 engagement induced a rapid robust activation of Rap1 that was maximal at 2 min and remained elevated through 10 min (Fig. 1C). APCs also had the ability to activate Rap1 in AD10 T-cell blasts (Fig. 2A and B). Without antigen, APCs induced a weak transient activation of Rap1 that could be inhibited by blocking CTLA-4 on the T cells with soluble antibodies (26) (Fig. 2A and B). In the presence of 5 μM MCC peptide as an antigen, APCs induced an activation of Rap1 for up to 20 min. Five minutes after peptide stimulation, CTLA-4-blocking antibodies had no detectable effect on peptide-induced Rap1 activation. However, by 10 and 20 min after peptide stimulation, blocking CTLA-4 with soluble antibody resulted in diminished Rap1 activity (Fig. 2A and B). This is consistent with the idea that Rap1 is rapidly activated by receptors other than CTLA-4, including the TCR itself (8, 16, 39, 40). Indeed, the kinetics of the residual Rap1 activation in the absence of CTLA-4 mirrors that seen by stimulation of the TCR alone (39). In the same experiment, we examined whether blocking CTLA-4 might enhance Ras activation. In the absence of antigen, APCs could not activate Ras. In the presence of antigen, Ras was rapidly activated, but it was not further enhanced following blockade of CTLA-4 at 5, 10, or 20 min (Fig. 2C and D). These results and the previous data suggest that the ability of CTLA-4 to limit ERK activation is not mediated by the inhibition of Ras but may be mediated by the activation of Rap1.
FIG. 2.
Activation of Rap1 and Ras in T cells by APCs. (A) In vitro-primed AD10 T cells were incubated for the indicated times with APCs with or without 5 μM MCC peptide and soluble (blocking) Ab to CTLA-4. Active Rap1 (Rap1-GTP) was pulled down from cell extracts and analyzed by immunoblotting with Rap1-specific antibodies. Total Rap1 in cell extracts is indicated as a loading control. (B) Quantitation of Rap1 activation using data obtained from three independent experiments, normalized to the values seen at 0 min and expressed as means ± standard errors (SE). (C) In vitro-primed AD10 T cells were incubated for the indicated times with APCs with or without 5 μM MCC peptide and soluble (blocking) Ab to CTLA-4, as for panel A. Active Ras (Ras-GTP) was pulled down from cell extracts and analyzed by immunoblotting with Ras-specific antibodies. Total Ras in cell extracts is indicated as a loading control. (D) Quantitation of Ras activation using data obtained from three independent experiments, normalized to the values seen at 0 min and expressed as mean ± SE.
Transgenic expression and function of Rap1GAP1.
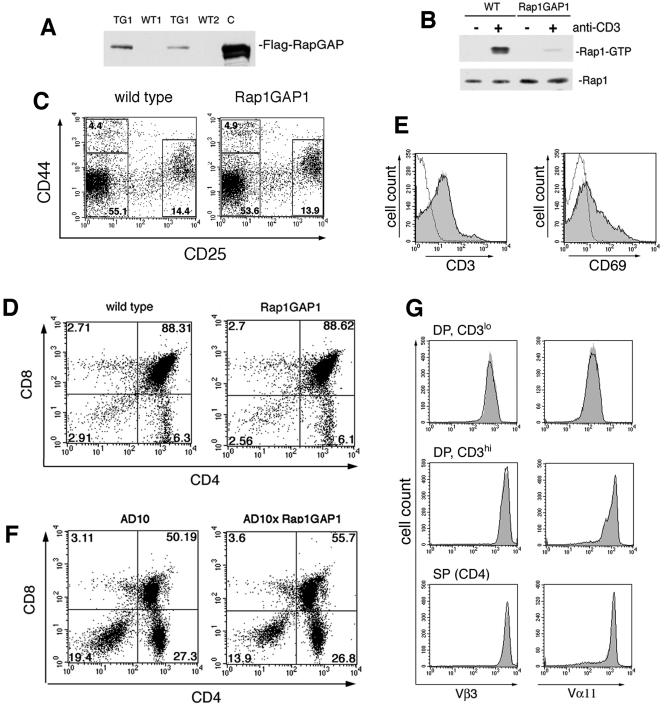
To examine the role of endogenous Rap1 activation on CTLA-4 function, we generated transgenic mice that overexpress the Rap1-GTPase Rap1GAP1 (tagged with a FLAG epitope) under control of the human β-actin promoter. Tagged Rap1GAP1 in the T cells of transgenic mice was detected by Western blotting using the FLAG monoclonal antibody (Fig. 3A). Transgenic mice generated from multiple lines displayed expression of Flag-Rap1GAP1. One of these lines (TG1) was used for subsequent studies in T cells.
FIG. 3.
Expression and function of Flag-Rap1GAP1 in murine T cells. (A) Immunoblot detection of the Flag-Rap1GAP1 transgene in murine T cells. Cell extracts from equal numbers of thymocytes from wild-type (WT1 and WT2) and transgenic littermates (TG1 and TG2) were analyzed by immunoblotting with Flag-specific antibodies. Lysates from Jurkat cells transfected with the Flag-Rap1GAP1 construct were included as a control (C). (B). Rap1 activation is blocked in Rap1GAP1-transgenic T cells. Following stimulation of wild-type (WT) and Rap1GAP1-transgenic (Rap1GAP1) T cells by anti-CD3 cross-linking, active Rap1 (Rap1-GTP) was pulled down from cell extracts and analyzed by immunoblotting with Rap1-specific antibodies. Total Rap1 in cell extracts is indicated as a loading control. (C) DN thymocyte maturation is normal in Rap1GAP1-transgenic mice. Thymocytes from wild-type and Rap1GAP1-expressing mice were analyzed for expression of CD44 and CD25. Cells were gated on the CD4−CD8− DN population, and the numbers represent percentages of the gated thymocyte subsets. (D) Normal distribution of CD4 and CD8 subgroups in Rap1GAP1-transgenic mice. Thymocytes from wild-type and Rap1GAP1-transgenic mice were stained to show expression of CD4 and CD8. The percentage of T-cell subsets is shown in each quadrant. (E) CD3 and CD69 expression is normal in the Rap1GAP1-transgenic mice. Overlay histograms show CD3 (left panel) and CD69 (right panel) expression on DP thymocytes from wild-type (black line) and Rap1GAP1-transgenic (gray fill) mice compared to control staining (dotted line). Data are representative of results of three experiments. (F) Normal distribution of CD4 and CD8 DP and SP subgroups in AD10 and AD10xRap1GAP1 mice. Thymocytes were stained to show expression of CD4 and CD8. The percentage of T-cell subsets is shown in each quadrant. (G) Normal expression of TCR in AD10 and AD10xRap1GAP1 thymocytes. Overlay histograms showing expression of Vβ3 (left panels) and Vα11 (right panels) on thymocytes from AD10 (black line) and AD10xRap1GAP1 (gray fill).
Rap1GAP1 is constitutively active when ectopically expressed and has been used to selectively inhibit Rap1 in a variety of cells (8, 36). Rap1GAP1 lacks the catalytic arginine, present in other GAPs, that is required for GAP activity on Ras, Rho, and Rab (14). In contrast, Rap1GAP has been shown to utilize a unique catalytic asparagine to stimulate GTP hydrolysis, which provides specificity for Rap (14). In addition, the presence of two lysines within the catalytic region of Rap1 and not present in Ras, Rho, and Rab (14) further ensures the specificity of the catalytic asparagine in Rap1 to function in GTP hydrolysis (5, 14). In peripheral T cells expressing transgenic Rap1GAP1, the activation of Rap1 by soluble anti-CD3 cross-linking was significantly reduced (Fig. 3B), establishing that Rap1GAP1 expression inhibited Rap1 activation in these cells.
Thymocyte development in the Rap1GAP1-transgenic mice was grossly normal. We examined the CD4−CD8− double negative (DN) population by staining for the maturation markers CD44 and CD25. Both the wild-type and Rap1GAP1-expressing thymocytes showed normal distributions of the DN subpopulations (Fig. 3C). Rap1GAP1-transgenic mice also showed a normal distribution of CD4+CD8+ double positive (DP), CD4+ single positive (SP), and CD8+ SP thymocytes (Fig. 3D). The progression of thymocytes from DP to SP can be evaluated by examining the surface expression of CD3 and CD69. Expression profiles of CD3 and CD69 in the DP thymocytes from Rap1GAP1-expressing mice were the same as the wild type, suggesting grossly normal positive selection (Fig. 3E).
Thymic development in mice with a polyclonal TCR repertoire is difficult to evaluate. Therefore, we evaluated the Rap1GAP1-transgenic mice crossed to the TCR-transgenic line, AD10. The AD10 TCR-transgenic mice express Vβ3 and Vα11 specific for MCC. The AD10 TCR-transgenic thymocyte populations are skewed towards the CD4 lineage, which reflected MHC class II positive selection (Fig. 3F). We noted a decrease in the percentage of DN thymocytes in the AD10xRap1GAP1 mice that was consistent among multiple sibling pairs; however, this did not appear to influence the percentages of cells within DP and SP subsets. As DP thymocytes undergo positive selection, they up-regulate CD3 expression from low (CD3lo) to high (CD3hi). In the DP population, the majority of CD3lo thymocytes expressed the transgenic TCR, as indicated by the expression of Vβ3 and Vα11 (Fig. 3G, top panels). The expression of Vβ3 and Vα11 was increased in the CD3hi DP population (Fig. 3G, middle panels), indicating that the TCR-transgenic thymocytes were being positively selected. This high expression of CD3, Vβ3, and Vα11 was also maintained in the CD4 SP thymocytes that had undergone positive selection (Fig. 3G, bottom panels). This phenotype was the same in the Rap1GAP1-expressing thymocytes, both in the percentage of CD4+ and the expression of Vβ3 and Vα11 (Fig. 3G). Therefore, expression of Rap1GAP1 did not prevent positive selection of AD10 thymocytes.
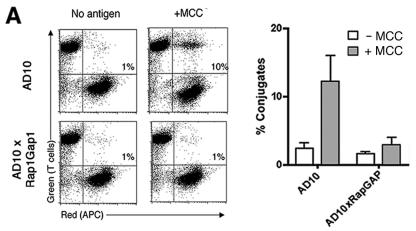
Rap1 activation has been shown to enhance integrin-mediated adhesion in T cells, as measured by conjugate formation with APCs (15, 22). By FACS analysis, AD10 T cells were shown to form strong conjugates with APCs in an antigen-dependent manner (Fig. 4A). In Rap1GAP1-expressing T cells, the antigen-dependent induction of strong conjugates was blocked (Fig. 4A). These data support previous data implicating Rap1 in T-cell adhesion (15, 22, 39). T cells need to interact with APCs to become activated; therefore, we examined microscopically the ability of Rap1GAP1-expressing T cells to make conjugates. Both AD10 and AD10xRap1GAP1 T cells flattened out against the APC, fluxed calcium, and were able to redistribute MCC:I-Ek:GFP to the interface of T-cell/APC contact (Fig. 4B). These data demonstrate that T cells were capable of making conjugates in the absence of Rap1 activation even though they were incapable of making the strong conjugates that withstand the shear forces generated in the FACS assay. The percentage of Rap1GAP1-expressing T cells forming conjugates with APCs was 34.5 ± 4.0% and was not significantly different from the percentage of the AD10 T cells that made conjugates (36.8 ± 5.1%). These data support a model that Rap1-dependent enhancement of adhesion was not required for antigen-dependent T-cell activation under the conditions investigated here. Importantly, the absence of Rap1-dependent adhesion was not limiting T-cell/APC conjugates measured microscopically in this model.
FIG. 4.
Rap1 activation regulates T-cell adhesion. (A) Rap1 activation induces strong T-cell conjugate formation with APCs. Conjugate formation of AD10 and AD10xRap1GAP1 T-cell blasts (green) and CH27 B cells as APCs (red) that were loaded with 10 μM MCC (+MCC) or without antigen (−MCC). Following a 30-min incubation of equal numbers of T cells and APCs, conjugate formation was analyzed by flow cytometry. The percentage of T cells forming conjugates is indicated in each upper right quadrant (left panel). Quantification of conjugate formation by AD10 and AD10xRap1GAP1 T cells is shown in the right panel. Data are expressed as means ± standard deviations of results of three independent experiments. (B) Rap1GAP1-transgenic T cells that formed conjugates with APCs fluxed calcium (middle panels) and flattened out against the APCs and redistributed MHC:I-Ek:GFP at the T-cell-APC interface (right panels). Resting T cells were loaded with fura-2 before being introduced into wells containing transfected fibroblast APCs expressing MCC:I-Ek:GFP. For both AD10 controls (top panels) and AD10xRap1GAP1 (bottom panels), a representative series of pictures is shown. Blue represents differential interference contrast and green represents MCC:I-Ek:GFP. Left panels, images of the T cell approaching the APC just prior to contact. Middle panels, images of the T cell making contact with the APC, showing the level of intracellular calcium within the T cell (shown in red). Right panels, images of the T cell subsequent to calcium flux, showing the T cell flattened against the APC, with MCC:I-Ek:GFP redistributed to the interface between the T cell and the APC. Bars, 15 microns.
T cells are activated in the Rap1GAP1-expressing mice.
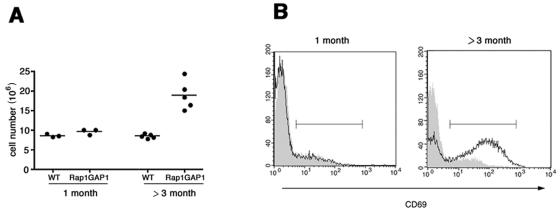
Overexpression of Rap1GAP1 in T cells yielded a phenotype of lymphadenopathy in adult mice. One-month-old Rap1GAP1-transgenic mice showed the same numbers of cells in their inguinal lymph nodes as age-matched control animals (Fig. 5A). However, adult Rap1GAP1-transgenic mice (over 3 months old) showed hypercellularity of lymph nodes, with cellularity twofold higher than those for age-matched wild-type controls (Fig. 5A).
FIG. 5.
Accumulation of activated T cells in the lymph nodes of Rap1GAP1-transgenic animals. (A) Inguinal lymph nodes were isolated from wild-type (WT) and Rap1GAP1-transgenic animals at the indicated ages, and the cell numbers per animal were determined. The bars denote the means for each group. (B) Inguinal lymph node cells from wild-type (gray shading) and Rap1Gap1-transgenic (black line) animals were stained for CD3 and the activation marker CD69 and were gated on CD3. The bar denotes the gate used to evaluate CD69 levels.
In the lymph nodes of the adult Rap1GAP1-transgenic mice (over 3 months old), there was an increase in CD4+ cells compared to age-matched wild-type controls. The mean percentages (± standard errors) of CD4+ and CD8+ T cells within wild-type lymph nodes were 26.5 ± 0.9, and 24.2 ± 1.7, respectively, whereas for the Rap1GAP1-positive lymph nodes they were 30.5 ± 2.5 and 16.4 ± 2.0, respectively (n = 3).
To examine whether Rap1GAP1-expressing mice accumulated activated T cells in their lymph nodes, cells were stained for CD3 and the activation marker CD69. T cells from 1-month-old wild-type and Rap1GAP1-transgenic mice showed a basal level of CD69-positive cells of 11.6 ± 0.6% and 15.3 ± 3.4%, respectively (n = 3) (Fig. 5B). In contrast, lymph node cells from adult Rap1GAP1-expressing mice (over 3 months old) showed a significant increase in the percentage of T cells expressing CD69 (63.8 ± 7.2%; n = 6) compared to levels on T cells from age-matched wild-type animals (6.7 ± 1.8%; n = 6) (Fig. 5B). Despite the presence of large numbers of activated T cells in the adult Rap1GAP1-transgenic mice, there was no evidence of infiltrate into the heart, lung, gut, or liver (data not shown). When crossed to the TCR-transgenic line, AD10, the adult Rap1GAP1-expressing mice, no longer showed the lymphadenopathy and did not accumulate activated T cells within inguinal lymph nodes (data not shown).
Rap1GAP1-expression enhances ERK activation.
ERK activation following cross-linking of soluble anti-CD3 in the wild-type and Rap1GAP1-transgenic T cells was measured (Fig. 6A). In wild-type T cells, stimulation of ERK activity was evident at 0.5 μg/ml antibody and increased in a dose-dependent manner. In the Rap1GAP1-expressing T cells, ERK activity was detectable at 0.25 μg/ml and was increased over that of wild-type T cells at low concentrations of anti-CD3 antibody in multiple experiments (Fig. 6A). We propose that Rap1 activation following limiting CD3 cross-linking may increase the threshold for ERK activation.
FIG. 6.
ERK activation is enhanced in Rap1GAP1-transgenic T cells. (A) Lowered threshold for ERK activation in Rap1GAP1-transgeneic T cells. Naïve Rap1GAP1-transgenic or wild-type T cells were stimulated for 5 min with soluble cross-linked anti-CD3 at the indicated concentrations. Cell extracts were analyzed by immunoblotting with phospho-ERK1/2 antibodies (pERK1/2) (left panel). The levels of total ERK2 are indicated as a loading control (lower panel). Data were obtained from three independent experiments, normalized to wild-type ERK activity at 1.0 μg/ml, and expressed as means ± SE (right panel). (B) Loss of ERK inhibition by CTLA-4 in Rap1GAP1-expressing T cells. In vitro-primed AD10 and AD10xRap1GAP1-transgenic T cells were left untreated (UT) or stimulated for 5 min with immobilized anti-CD3 (CD3), anti-CD28 (CD28), or anti-CTLA-4 (CTLA-4). Cells extracts were analyzed by immunoblotting with antibodies specific for phosphorylated ERK1/2 (pERK1/2) (left panel). The levels of total ERK2 are indicated as a loading control (lower panel). Data were obtained from three independent experiments, normalized to wild-type ERK activity in response to CD3+CD28, and expressed as means ± SE (right panel). (C) Loss of Rap1 activation by CD3 and CTLA-4 in AD10xRap1GAP1-transgenic T cells. In vitro-primed AD10- and AD10xRap1GAP1-transgenic T-cell blasts were left untreated (Un) or treated for 5 min with immobilized anti-CD3 (CD3), anti-CD28 (CD28), or anti-CTLA-4 (CTLA-4). Activated Rap1 (Rap1-GTP) was precipitated using GST-RalGDS and was analyzed by immunoblotting with Rap1-specific antibodies (left panel). Total levels of Rap1 are indicated as loading controls (lower panel). Data were obtained from three independent experiments, normalized to wild-type Rap1 activity in response to CD3, and expressed as means ± SE (right panel).
We also tested the impact of Rap1GAP1 expression on the ability of CTLA-4 to inhibit ERKs in rested T-cell blasts. The kinetics and levels of CTLA-4 expression on the surface of AD10xRap1GAP1 T-cell blasts were the same as for AD10 T-cell blasts (data not shown). In wild-type AD10 T-cell blasts, ERK activation via immobilized antibodies to CD3 and/or CD28 was blocked by CTLA-4 engagement (Fig. 6B). These data demonstrate that CTLA-4 can block ERK activation by TCR even when CD28 is engaged. However, this block was largely absent in the Rap1GAP1-expressing AD10 T-cell blasts (Fig. 6B), suggesting that Rap1 contributes to signals from CTLA-4 to inhibit ERK activity.
The effect of activation of CD3, CD28, and CTLA-4 on Rap1 activation was examined also with AD10 and AD10xRap1GAP1 T cells. As shown previously, CD3 activation of Rap1 was blocked by CD28. However, Rap1 inhibition by CD28 was largely reversed by CTLA-4 (Fig. 6C) demonstrating that CTLA-4 can activate Rap1 even in the presence of signals from CD28. In the AD10xRap1GAP1 T cells, Rap1 activation by CD3 and CTLA-4 was completely inhibited (Fig. 6C). Taken together, these data suggest that in the absence of Rap1 activation, the ability of CTLA-4 to inhibit ERKs is dramatically diminished.
Rap1GAP1-expression enhances T-cell function.
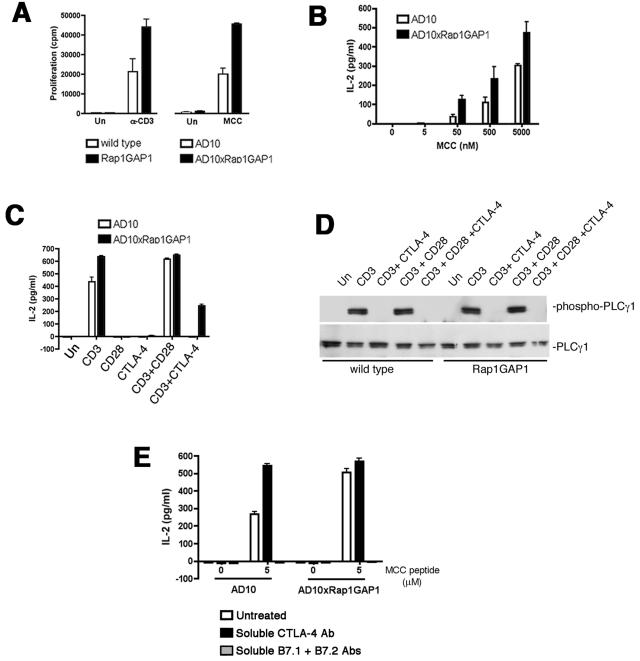
To examine the role of Rap1 on T-cell function, we assayed proliferation and IL-2 production for wild-type and Rap1GAP1-transgenic T cells. For proliferation assays, splenocytes from Rap1GAP1 and AD10xRap1GAP1-transgenic mice were stimulated with soluble anti-CD3 or MCC peptide, respectively. In both cases, T cells that expressed the Rap1GAP1 transgene showed increased proliferation in response to both anti-CD3 and MCC peptide compared to T cells from the respective littermate controls (Fig. 7A). AD10 and AD10xRap1GAP1 T cells both made IL-2 in response to MCC peptide in a dose-dependent manner (Fig. 7B). However, AD10xRap1GAP1 T cells made more IL-2 than AD10 T cells at all concentrations of MCC peptide.
FIG. 7.
T-cell activation is enhanced in Rap1GAP1-transgenic T cells. (A) Increased proliferation of Rap1GAP1-transgenic T cells. Wild-type and Rap1GAP1-transgenic splenocytes were left untreated (Un) or treated with 0.25 μg/ml cross-linked anti-CD3 (α-CD3) (left panel). AD10 and AD10xRap1GAP1 splenocytes were left untreated (Un) or treated with 0.5 μM MCC peptide (MCC) (right panel). Proliferation was quantified via [3H]thymidine incorporation. Data are the means ± SE (n = 3). (B) Increase in IL-2 production in GAP1-transgenic T cells. AD10 and AD10xRap1GAP1 splenocytes were cultured with MCC peptide at the indicated concentrations. Supernatants were harvested for IL-2 measurement by ELISA. The data are the means ± SE (n = 3). (C) CTLA-4 cannot inhibit IL-2 production in Rap1GAP1-transgenic T cells. In vitro-primed AD10 and AD10xRap1GAP1-expressing T cells were left untreated (Un) or treated for 5 min with immobilized anti-CD3 (CD3), anti-CD28 (CD28), and/or anti-CTLA-4 (CTLA-4). Supernatants were harvested for IL-2 measurement by ELISA. The data are the means ± SE (n = 3). (D) CTLA-4 inhibits PLCγ1 phosphorylation in Rap1GAP1-expressing T cells. In vitro-primed AD10 and AD10xRap1GAP1-expressing T cells were left untreated (Un) or treated for 5 min with immobilized anti-CD3 (CD3), anti-CD28 (CD28), and anti-CTLA-4 (CTLA-4) as indicated. Cells extracts were analyzed by immunoblotting with antibodies specific for phosphorylated PLCγ1 (phospho-PLCγ1) (upper panel). The levels of total PLCγ1 are indicated as a loading control (lower panel). (E) In vitro-primed AD10 and AD10xRap1GAP1-expressing T cells were incubated with irradiated B10.BR splenocytes in the presence or absence of 5 μM MCC peptide and/or soluble CTLA-4 Ab or soluble B7-1 and B7-2 Ab. Supernatants were harvested for IL-2 measurement by ELISA. The data are the means ± SE (n = 3).
Rap1GAP1 expression also reduced the antagonism of IL-2 production by CTLA-4 in T-cell blasts. AD10 T-cell blasts produced IL-2 in response to anti-CD3, and CTLA-4 significantly blocked this production (Fig. 7C). The AD10xRap1GAP1 T-cell blasts made more IL-2 than the AD10 T cells in response to anti-CD3; however, the ability of CTLA-4 costimulation to down-regulate IL-2 production in response to anti-CD3 was significantly reduced (Fig. 7C). Therefore, the inhibition of IL-2 production by CTLA-4 was mediated, in part, via Rap1. It is well known that CTLA-4 has multiple mechanisms to inhibit T-cell function (12, 34), and the data suggest that Rap1 mediates the inhibition of ERKs by CTLA-4 but does not inhibit other known pathways of CTLA-4 inhibition of IL-2. Some of these pathways include inhibition of PLCγ, the recruitment of the SHP-2 phosphatase (10), and regulation of ERK-independent transcription factors that remain inhibited by CTLA-4 (18). To confirm that ERK-independent actions of CTLA-4 were retained in the Rap1GAP1-transgenic T cells, we examined TCR-dependent tyrosine phosphorylation of PLCγ1 (19). CTLA-4 inhibited the phosphorylation of PLCγ1 in Rap1GAP1-expressing as well as wild-type T cells (Fig. 7D).
Antigen-dependent IL-2 production in AD10 T-cell blasts is enhanced by blocking CTLA-4 with soluble antibodies (Fig. 7E). This is consistent with the function of CTLA-4 in limiting IL-2 production. AD10xRap1GAP1 T-cell blasts produce more IL-2 than AD10 T-cell blasts in response to peptide. These levels were similar to that seen with AD10 blasts after the blocking of CTLA-4 (Fig. 7E). Blocking antibodies to CTLA-4 did not further enhance IL-2 production in AD10xRap1GAP1 T-cell blasts, suggesting that signals from CTLA-4 were not inhibiting IL-2 production in these cells (Fig. 7E). As expected, the enhancement of IL-2 production was completely inhibited by blocking signals through B7.
DISCUSSION
CTLA-4/B7 interactions are a well-established mechanism for inhibiting signals generated downstream of the TCR. We have shown that CTLA-4 activation, both in the presence and absence of TCR stimulation, activates the small G protein Rap1. This ability of CTLA-4 to activate Rap1 was established using immobilized (activating) anti-CTLA-4 as well as soluble antibodies to CTLA-4, which inhibit CTLA-4 engagement by B7 molecules expressed on APCs (44). Using T cells deficient in Rap1 activation, which were isolated from Rap1GAP1-transgenic mice, we have also shown that the consequence of the activation of Rap1 by CTLA-4 is to limit TCR-dependent signals via Ras, and we propose that this is one of many mechanisms by which CTLA-4 inhibits T-cell function.
Rap1GAP1 has unique structural features compared to the GAPs of Ras, Rho, Arf, Rab, or Ran (5, 14) that dictate its specificity towards Rap. Unlike other GAPs, Rap1GAP1 uses a catalytic asparagine to stimulate GTP hydrolysis, which targets interactions with Rap and limits its interactions with other small G proteins (5). While the main function of Rap1Gap1 is thought to be the down-regulation of Rap activity by GTP hydrolysis, it is possible that other functions exist. For example, Rap1GAPs have been shown to associate with selected G protein alpha subunits (30). Although there is no evidence that G protein alpha subunits participate in CTLA-4 function, there remains the possibility that there are other protein interactions of Rap1GAP1 that regulate signaling pathways via mechanisms that are distinct from its GAP activity towards Rap. Our data show that the inhibition of Rap1 activation via Rap1GAP1 expression enhanced TCR-dependent ERK activation, IL-2 production, proliferation, and CD69 expression, consistent with a role of endogenous Rap1 to limit T-cell activation. These data complement nicely those results seen with peripheral T cells from Spa-1-null mice, which displayed decreased ERK activity and developed anergy associated with constitutive Rap1 activation (21). We also demonstrate that Rap1 activation by CTLA-4 antagonized TCR-dependent ERK activation, both in the presence and absence of CD28, and limited IL-2 production. Immobilized anti-CTLA-4 antibody in the absence of additional stimuli was sufficient to activate Rap1, demonstrating that CTLA-4 can signal independently of TCR. We propose that Rap1 activation by CTLA-4 may contribute to its antagonism of both TCR and CD28 signaling.
The data suggest that Rap1 mediates the inhibition of ERKs by CTLA-4. While limiting Rap1 activation completely reversed the inhibition of ERKs by CTLA-4, the same conditions did not completely restore IL-2 production in these T cells. These data are consistent with the existence of multiple mechanisms by which CTLA-4 is known to inhibit IL-2, including the recruitment of the SHP-2 phosphatase (12). Additionally, while increased ERK activity may activate Elk-1 and induce Fos transcription required to drive the IL-2 promoter, inhibition of IL-2 transcription by CTLA-4 may not be fully reversed by blocking Rap1 activation, as signaling to other relevant transcription factors may remain inhibited by CTLA-4 (18). We asked whether additional inhibitory mechanisms were still functioning in Rap1GAP1-expressing T cells by examining TCR-dependent tyrosine phosphorylation of PLCγ1 (19). CTLA-4 inhibited the phosphorylation of PLCγ1 in Rap1GAP1-expressing as well as wild-type T cells. Moreover, the inhibitory actions of Rap1 appear to be selective, as the inhibition of T-cell function by IL-10 was not blocked in Rap1GAP1-expressing T cells (data not shown).
Rap1GAP1-transgenic mice displayed a marked lymphadenopathy that manifested after they reached 3 months of age. The phenotype of the Rap1GAP1 animals was milder than that seen for animals lacking CTLA-4 (11, 41). Rap1GAP1 mice did not suffer increased mortality or show the overt autoimmune disease or lymphocytic infiltrate that was seen with CTLA-4-null mice (data not shown). Rather, the phenotype of the Rap1GAP1-transgenic mice was similar to that seen with mice expressing mutant CTLA-4 that lacks its cytoplasmic tail. Both mice display hyperactivity of peripheral T cells and lymphadenopathy, without increased mortality or infiltrate (29). The modest phenotype of both mice is consistent with the notion that the full CTLA-4 molecule provides multiple mechanisms of CTLA-4 inhibition, involving both inhibitory signals generated from the cytoplasmic domain and antagonism of B7 signaling via the extracellular domain (9, 31, 43). We suggest that Rap1 participates in the inhibitory signals that are generated from the cytoplasmic domain of CTLA-4.
The increase in CD4+ T cells in the periphery does not appear to be due to an effect of Rap1GAP1 in the thymus. In the Rap1GAP1 and the AD10xRap1GAP1-transgenic animals, the ectopic expression of Rap1GAP1 did not grossly affect the thymocyte development. The absence of effects of Rap1 on T-cell development has also been reported for thymocytes that have heightened Rap1 activity from mice deficient in the endogenous T-cell Rap1GAP Spa-1 (21) and for the P14 TCR-transgenic mouse expressing constitutively active RapV12 (39). However, RapV12 expression enhanced positive selection in a low-affinity selection model, the HY-TCR-transgenic model (39). This enhancement of positive selection was due to the promotion of T-cell adhesion to thymic epithelial cells.
Rap1 activation of LFA-dependent adhesion has been shown to enhance T-cell function (39). However, potential inhibitory pathways of Rap1 also exist (8, 22). This has been formally tested in experiments shown in the present study employing antibody stimulation of T cells in the absence of APCs that provide an opportunity to examine Rap1 functions under conditions under which Rap1-dependent adhesion cannot occur. Under these conditions, Rap1 activation can limit signaling to ERKs. Our study supports a role for Rap1 in antagonizing T-cell function that is distinct from the actions of Rap1 to enhance integrin-dependent adhesion (39).
Previous studies using RapV12 have demonstrated that enhanced Rap1-dependent adhesion could potentiate T-cell activation. However, the loss of Rap1-dependent adhesion may not be limiting for all functional T-cell interactions. In the present study, although we demonstrate the ability of Rap1 to enhance adhesion, this effect does not overcome the actions of Rap1 to inhibit ERKs. We determined that Rap1GAP1-expressing T cells were able to form conjugates (as evaluated microscopically) at the same frequency as wild-type T cells. It is likely that the conjugates made by the Rap1GAP1-expressing T cells may not be as strong as those made by wild-type T cells, as we were able to detect differences in adhesion when we evaluated conjugates by FACS analysis. However, the data suggest strongly that the absence of Rap1-dependent adhesion does not limit T-cell activation in this model.
The differences between this study and that of others (39) may also reflect differences in the T cells used in each study. In a previous study, Rap1-dependent adhesion was shown to enhance thymocyte and naïve T-cell responses (39). In this study, we have used rested in vitro T-cell blasts, which have lower activation thresholds than naïve T cells (20) and therefore may be less dependent on Rap1-dependent adhesion to become activated.
In this study, we used the AD10 TCR-transgenic model. In other studies, the use of different TCR-transgenic systems may contribute to the distinct roles of Rap1 in T-cell responses. For example, constitutive activation of Rap1 enhanced positive selection in the HY-TCR-transgenic model (39) but not in the P14 TCR-transgenic model (39) or the AD10 TCR-transgenic mouse expressing Rap1GAP1 shown in the present study. In addition, the requirement for integrin-dependent adhesion in T-cell activation can be influenced by the affinity of the transgenic TCR for the antigen peptide used. LFA-1/ICAM interactions are essential for T-cell responses by low-affinity peptide (2), but are not required for high-affinity interactions (2). This suggests that Rap1-dependent effects of adhesion may also be affinity dependent. Indeed, Rap1 enhancement of TCR function in the P14 TCR model was more evident with low-affinity peptide (39) but was not evident with the high-affinity model used in this study. Therefore, under the conditions of this study, we believe that the antagonism of ERKs by Rap1 was favored over its enhancement of adhesion in regulating T-cell activation. We suggest that Rap1-dependent LFA activation is not the dominant action of Rap1 in T-cell responses to high-affinity ligands.
We demonstrate here that CD28 inhibits Rap1 to augment T-cell function and that CTLA-4 activates Rap1 to antagonize T-cell function. In this study, we show that CTLA-4 activates Rap1 to antagonize ERK signaling that, in turn, contributes to the down-regulation of IL-2 production and proliferation. We propose that this negative regulation of T-cell function by Rap1 is independent of its role in augmenting T-cell function through integrin-mediated adhesion. The coordinated functions of TCR engagement and costimulation by CD28 and CTLA-4 utilize Rap1 to regulate the outcome of T-cell function following T-cell/APC interactions.
Acknowledgments
We thank M. Findlay, C. Huddleston, and B. Hunter for technical assistance.
This work was funded by NIH NIAID grants AI047337 (P.J.S.S.) and AI50823 (D.C.P.).
REFERENCES
- 1.Amsen, D., A. Kruisbeek, J. L. Bos, and K. Reedquist. 2000. Activation of the Ras-related GTPase Rap1 by thymocyte TCR engagement and during selection. Eur. J. Immunol. 30:2832-2841. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann, M. F., A. Gallimore, E. Jones, B. Ecabert, H. Acha-Orbea, and M. Kopf. 2001. Normal pathogen-specific immune responses mounted by CTLA-4-deficient T cells: a paradigm reconsidered. Eur. J. Immunol. 31:450-458. [DOI] [PubMed] [Google Scholar]
- 3.Bivona, T. G., H. H. Wiener, I. M. Ahearn, J. Silletti, V. K. Chiu, and M. R. Philips. 2004. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 164:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boussiotis, V. A., G. J. Freeman, A. Berezovskaya, D. L. Barber, and L. M. Nadler. 1997. Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science 278:124-128. [DOI] [PubMed] [Google Scholar]
- 5.Brinkmann, T., O. Daumke, U. Herbrand, D. Kuhlmann, P. Stege, M. R. Ahmadian, and A. Wittinghofer. 2002. Rap-specific GTPase activating protein follows an alternative mechanism. J. Biol. Chem. 277:12525-12531. [DOI] [PubMed] [Google Scholar]
- 6.Calvo, C. R., D. Amsen, and A. M. Kruisbeek. 1997. Cytotoxic T lymphocyte antigen 4 (CTLA-4) interferes with extracellular signal-regulated kinase (ERK) and Jun NH2-terminal kinase (JNK) activation, but does not affect phosphorylation of T cell receptor ζ and ZAP70. J. Exp. Med. 186:1645-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantrell, D. A. 2003. GTPases and T cell activation. Immunol. Rev. 192:122-130. [DOI] [PubMed] [Google Scholar]
- 8.Carey, K. D., T. J. Dillon, J. M. Schmitt, A. M. Baird, A. D. Holdorf, D. B. Straus, A. S. Shaw, and P. J. Stork. 2000. CD28 and the tyrosine kinase lck stimulate mitogen-activated protein kinase activity in T cells via inhibition of the small G protein Rap1. Mol. Cell. Biol. 20:8409-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carreno, B. M., F. Bennett, T. A. Chau, V. Ling, D. Luxenberg, J. Jussif, M. L. Baroja, and J. Madrenas. 2000. CTLA-4 (CD152) can inhibit T cell activation by two different mechanisms depending on its level of cell surface expression. J. Immunol. 165:1352-1356. [DOI] [PubMed] [Google Scholar]
- 10.Chambers, C. A. 2001. The expanding world of co-stimulation: the two-signal model revisited. Trends Immunol. 22:217-223. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, C. A., D. Cado, T. Truong, and J. P. Allison. 1997. Thymocyte development is normal in CTLA-4-deficient mice. Proc. Natl. Acad. Sci. USA 94:9296-9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers, C. A., M. S. Kuhns, J. G. Egen, and J. P. Allison. 2001. CTLA-4-mediated inhibition in regulation of T cell responses: mechanisms and manipulation in tumor immunotherapy. Annu. Rev. Immunol. 19:565-594. [DOI] [PubMed] [Google Scholar]
- 13.da Rocha Dias, S., and C. E. Rudd. 2001. CTLA-4 blockade of antigen-induced cell death. Blood 97:1134-1137. [DOI] [PubMed] [Google Scholar]
- 14.Daumke, O., M. Weyand, P. P. Chakrabarti, I. R. Vetter, and A. Wittinghofer. 2004. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429:197-201. [DOI] [PubMed] [Google Scholar]
- 15.de Bruyn, K. M., S. Rangarajan, K. A. Reedquist, C. G. Figdor, and J. L. Bos. 2002. The small GTPase Rap1 is required for Mn2+- and antibody-induced LFA-1- and VLA-4-mediated cell adhesion. J. Biol. Chem. 277:29468-29476. [DOI] [PubMed] [Google Scholar]
- 16.Dillon, T. J., V. Karpitski, S. A. Wetzel, D. C. Parker, A. S. Shaw, and P. J. Stork. 2003. Ectopic B-Raf expression enhances extracellular signal-regulated kinase (ERK) signaling in T cells and prevents antigen-presenting cell-induced anergy. J. Biol. Chem. 278:35940-35949. [DOI] [PubMed] [Google Scholar]
- 17.Ebinu, J. O., S. L. Stang, C. Teixeira, D. A. Bottorff, J. Hooton, P. M. Blumberg, M. Barry, R. C. Bleakley, H. L. Ostergaard, and J. C. Stone. 2000. RasGRP links T-cell receptor signaling to Ras. Blood 95:3199-3203. [PubMed] [Google Scholar]
- 18.Fraser, J. H., M. Rincon, K. D. McCoy, and G. Le Gros. 1999. CTLA4 ligation attenuates AP-1, NFAT and NF-κB activity in activated T cells. Eur. J. Immunol. 29:838-844. [DOI] [PubMed] [Google Scholar]
- 19.Gatta, L., G. Calviello, F. Di Nicuolo, L. Pace, V. Ubaldi, G. Doria, and C. Pioli. 2002. Cytotoxic T lymphocyte-associated antigen-4 inhibits integrin-mediated stimulation. Immunology 107:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iezzi, G., K. Karjalainen, and A. Lanzavecchia. 1998. The duration of antigenic stimulation determines the fate of naive and effector T cells. Immunity 8:89-95. [DOI] [PubMed] [Google Scholar]
- 21.Ishida, D., H. Yang, K. Masuda, K. Uesugi, H. Kawamoto, M. Hattori, and N. Minato. 2003. Antigen-driven T cell anergy and defective memory T cell response via deregulated Rap1 activation in SPA-1-deficient mice. Proc. Natl. Acad. Sci. USA 100:10919-10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katagiri, K., M. Hattori, N. Minato, and T. Kinashi. 2002. Rap1 functions as a key regulator of T-cell and antigen-presenting cell interactions and modulates T-cell responses. Mol. Cell. Biol. 22:1001-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaye, J., N. J. Vasquez, and S. M. Hedrick. 1992. Involvement of the same region of the T cell antigen receptor in thymic selection and foreign peptide recognition. J. Immunol. 148:3342-3353. [PubMed] [Google Scholar]
- 24.Krummel, M. F., and J. P. Allison. 1995. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 182:459-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krummel, M. F., and J. P. Allison. 1996. CTLA-4 engagement inhibits IL-2 accumulation and cell cycle progression upon activation of resting T cells. J. Exp. Med. 183:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krummel, M. F., T. J. Sullivan, and J. P. Allison. 1996. Superantigen responses and co-stimulation: CD28 and CTLA-4 have opposing effects on T cell expansion in vitro and in vivo. Int. Immunol. 8:519-523. [DOI] [PubMed] [Google Scholar]
- 27.Lee, K. M., E. Chuang, M. Griffin, R. Khattri, D. K. Hong, W. Zhang, D. Straus, L. E. Samelson, C. B. Thompson, and J. A. Bluestone. 1998. Molecular basis of T cell inactivation by CTLA-4. Science 282:2263-2266. [DOI] [PubMed] [Google Scholar]
- 28.Mandelbrot, D. A., A. J. McAdam, and A. H. Sharpe. 1999. B7-1 or B7-2 is required to produce the lymphoproliferative phenotype in mice lacking cytotoxic T lymphocyte-associated antigen 4 (CTLA-4). J. Exp. Med. 189:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masteller, E. L., E. Chuang, A. C. Mullen, S. L. Reiner, and C. B. Thompson. 2000. Structural analysis of CTLA-4 function in vivo. J. Immunol. 164:5319-5327. [DOI] [PubMed] [Google Scholar]
- 30.Meng, J., J. L. Glick, P. Polakis, and P. J. Casey. 1999. Functional interaction between Gαgz and Rap1GAP suggests a novel form of cellular cross-talk. J. Biol. Chem. 274:36663-36669. [DOI] [PubMed] [Google Scholar]
- 31.Nakaseko, C., S. Miyatake, T. Iida, S. Hara, R. Abe, H. Ohno, Y. Saito, and T. Saito. 1999. Cytotoxic T lymphocyte antigen 4 (CTLA-4) engagement delivers an inhibitory signal through the membrane-proximal region in the absence of the tyrosine motif in the cytoplasmic tail. J. Exp. Med. 190:765-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okumura, N., M. Okada, and H. Nakagawa. 1994. Depolarization-induced tyrosine phosphorylation in PC12h cells. J. Biochem. 116:346-350. [DOI] [PubMed] [Google Scholar]
- 33.Reedquist, K. A., and J. L. Bos. 1998. Costimulation through CD28 suppresses T cell receptor-dependent activation of the Ras-like small GTPase Rap1 in human T lymphocytes. J. Biol. Chem. 273:4944-4949. [DOI] [PubMed] [Google Scholar]
- 34.Saito, T., and S. Yamasaki. 2003. Negative feedback of T cell activation through inhibitory adapters and costimulatory receptors. Immunol. Rev. 192:143-160. [DOI] [PubMed] [Google Scholar]
- 35.Salomon, B., and J. A. Bluestone. 2001. Complexities of CD28/B7: CTLA-4 costimulatory pathways in autoimmunity and transplantation. Annu. Rev. Immunol. 19:225-252. [DOI] [PubMed] [Google Scholar]
- 36.Schmitt, J. M., and P. J. S. Stork. 2000. β2-adrenergic receptor activates extracellular regulated kinases (ERKs) via the small G protein Rap1 and the serine/threonine kinase B-Raf. J. Biol. Chem. 275:25342-25350. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, H., S. D. Dias, H. Hu, and C. E. Rudd. 2001. A regulatory role for cytoplasmic YVKM motif in CTLA-4 inhibition of TCR signaling. Eur. J. Immunol. 31:2042-2050. [DOI] [PubMed] [Google Scholar]
- 38.Schneider, H., D. A. Mandelbrot, R. J. Greenwald, F. Ng, R. Lechler, A. H. Sharpe, and C. E. Rudd. 2002. Cutting edge: CTLA-4 (CD152) differentially regulates mitogen-activated protein kinases (extracellular signal-regulated kinase and c-Jun N-terminal kinase) in CD4+ T cells from receptor/ligand-deficient mice. J. Immunol. 169:3475-3479. [DOI] [PubMed] [Google Scholar]
- 39.Sebzda, E., M. Bracke, T. Tugal, N. Hogg, and D. A. Cantrell. 2002. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat. Immunol. 3:251-258. [DOI] [PubMed] [Google Scholar]
- 40.Suga, K., K. Katagiri, T. Kinashi, M. Harazaki, T. Iizuka, M. Hattori, and N. Minato. 2001. CD98 induces LFA-1-mediated cell adhesion in lymphoid cells via activation of Rap1. FEBS Lett. 489:249-253. [DOI] [PubMed] [Google Scholar]
- 41.Tivol, E. A., F. Borriello, A. N. Schweitzer, W. P. Lynch, J. A. Bluestone, and A. H. Sharpe. 1995. Loss of CTLA-4 leads to massive lymphoproliferation and fatal multiorgan tissue destruction, revealing a critical negative regulatory role of CTLA-4. Immunity 3:541-547. [DOI] [PubMed] [Google Scholar]
- 42.Tivol, E. A., S. D. Boyd, S. McKeon, F. Borriello, P. Nickerson, T. B. Strom, and A. H. Sharpe. 1997. CTLA4Ig prevents lymphoproliferation and fatal multiorgan tissue destruction in CTLA-4-deficient mice. J. Immunol. 158:5091-5094. [PubMed] [Google Scholar]
- 43.Vijayakrishnan, L., J. M. Slavik, Z. Illes, R. J. Greenwald, D. Rainbow, B. Greve, L. B. Peterson, D. A. Hafler, G. J. Freeman, A. H. Sharpe, L. S. Wicker, and V. K. Kuchroo. 2004. An autoimmune disease-associated CTLA-4 splice variant lacking the B7 binding domain signals negatively in T cells. Immunity 20:563-575. [DOI] [PubMed] [Google Scholar]
- 44.Walunas, T. L., D. J. Lenschow, C. Y. Bakker, P. S. Linsley, G. J. Freeman, J. M. Green, C. B. Thompson, and J. A. Bluestone. 1994. CTLA-4 can function as a negative regulator of T cell activation. Immunity 1:405-413. [DOI] [PubMed] [Google Scholar]
- 45.Wetzel, S. A., T. W. McKeithan, and D. C. Parker. 2002. Live cell dynamics and the role of costimulation in immunological synapse formation. J. Immunol. 169:6092-6101. [DOI] [PubMed] [Google Scholar]
- 46.Whitehurst, C. E., and T. D. Geppert. 1996. MEK1 and the extracellular signal-regulated kinases are required for the stimulation of IL-2 gene transcription in T cells. J. Immunol. 156:1020-1029. [PubMed] [Google Scholar]
- 47.Zhao, H., Y. Y. Li, R. V. Fucini, S. E. Ross, J. E. Pessin, and G. A. Koretzky. 1997. T cell receptor-induced phosphorylation of Sos requires activity of CD45, Lck, and protein kinase C, but not ERK. J. Biol. Chem. 272:21625-21634. [DOI] [PubMed] [Google Scholar]