Abstract
Single-cell RNA sequencing (scRNA-seq) has allowed for the profiling of host and virus transcripts and host-virus interactions at single-cell resolution. This review summarizes the existing scRNA-seq technologies together with their strengths and weaknesses. The applications of scRNA-seq in various virological studies are discussed in depth, which broaden the understanding of the immune atlas, host-virus interactions, and immune repertoire. scRNA-seq can be widely used for virology in the near future to better understand the pathogenic mechanisms and discover more effective therapeutic strategies.
Keywords: Single-cell RNA sequencing (scRNA-seq), Host-virus interaction, COVID-19, Flavivirus
Highlights
-
•
The existing scRNA-seq technologies together with their strengths and weaknesses are summarized.
-
•
Applications of scRNA-seq in SARS-CoV-2 and flavivirus studies are extensively discussed.
-
•
Applications of scRNA-seq in studying immune atlas, host-virus interactions, and immune repertoire are further explored.
1. Introduction
The transcriptome (the collection of all mRNAs in cells) plays a crucial role in comprehensively understanding gene expression due to its essential connections to the genome and proteome and its ability to transmit biological genetic information. High-throughput sequencing makes it easier to obtain transcriptomes of specific tissues in a certain state. Since the 21st century, the advent of next-generation sequencing (NGS) and third-generation sequencing has made transcriptome sequencing cheaper and faster (Dijk et al., 2018).
The development of high-throughput sequencing technologies paved the way for further optimization down to the single-cell level. In recent years, the emergence of single-cell RNA sequencing (scRNA-seq) has allowed for analyzing transcriptomes for individual cells (Dai et al., 2022). Compared with bulk RNA sequencing, scRNA-seq facilitates an exploration of intracellular gene expression and the gene regulatory network at the resolution of the individual cell (Choi et al., 2020). With broad research prospects, scRNA-seq has been adopted to identify rare cell populations, screen abnormal cells, and reconstruct developmental trajectories (Hwang et al., 2018).
More efficient scRNA-seq technologies have been widely used in virology, playing a crucial role in interrogating the mechanisms of virus-host interactions and antiviral immune responses and assessing the efficacy of vaccinations (Luo et al., 2020). In this review, we present an overview of existing scRNA-seq technologies and their strengths and weaknesses, discuss how these approaches can improve our understanding of viral pathogenesis and immunological responses, and predict cutting-edge trends in this field. This review provides a reference for researchers who want to interrogate virus-host interactions by scRNA-seq. Readers may choose the appropriate scRNA-seq method based on specific scientific questions.
2. ScRNA-seq technologies
Although population-level transcriptomics screens have existed, they obscured the cell heterogeneity. Therefore, it is necessary to popularize scRNA-seq for biomedical research. In this section, we will briefly summarize the different methods and analysis workflow of scRNA-seq.
2.1. Library construction and sequencing
With a large number of available scRNA-seq technologies, each has distinct technical methods. Most scRNA-seq technologies are based on the same workflow (Fig. 1): (1) single-cell isolation; (2) cell capture and lysis to release mRNA and add unique nucleotide barcodes; (3) reverse transcription to cDNA; (4) cDNA amplification; (5) cDNA library preparation; and (6) sequencing.
Fig. 1.
scRNA-seq analysis is based on the workflow: (1) single cell isolation; (2) cells capture and adding barcode; (3) reverse transcription; (4) cDNA amplification; (5) cDNA library preparation; and (6) sequencing.
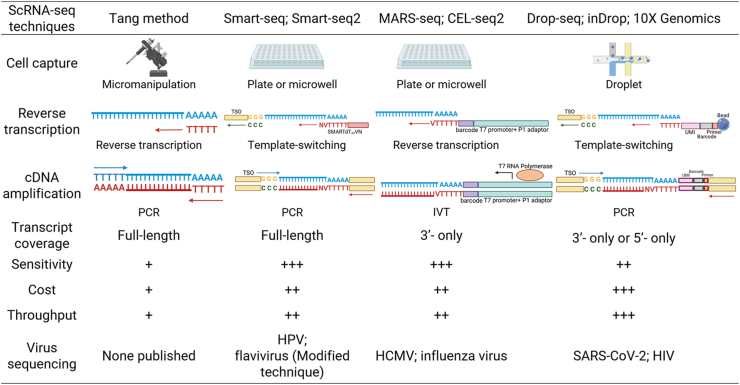
Diversification of scRNA-seq protocols, each with characteristic pros and cons, requires researchers to make suitable options according to the experimental purpose and funding. Here, we list several available scRNA-seq methods with their strengths, weaknesses, and applications in different viral studies (Fig. 2).
Fig. 2.
scRNA-seq technologies and their applications in virus sequencing. Smart-seq, switching mechanism at the 5′ end of the RNA template sequencing; MARS-seq, massively parallel single-cell RNA-sequencing; CEL-seq, cell expression by linear amplification and sequencing; TSO, template switch oligo; PCR, polymerase chain reaction; IVT, in-vitro transcription; HPV, human papillomavirus; HCMV, human cytomegalovirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; HIV, human immunodeficiency virus.
More specifically, in the first step of scRNA-seq, it is critical to select the proper single-cell sample protocol based on cell types, culture conditions, matrix content, and cell viability (Picelli, 2017). Then, individual cells in suspension are captured. Each capture method has characteristic strengths and weaknesses. Micromanipulation, the earliest cell capture method, was used to capture mouse oocytes by the Tang method (Tang et al., 2009). Although the method could observe the cells microscopically and generated 3 kb cDNA molecules, it was time-consuming and inefficient. With technological advances, individual cells can be captured to plates or parallel microfluidic channels, in which the reverse transcription of mRNA is performed and unique nucleotide code barcodes are added to each single cell. The methods based on well plates (typically 96- or 396-well plates), for example MARS-seq, allow presorting of cells by fluorescence-activated cell sorting (FACS) (Luo et al., 2020), which allows for analyzing rare cell populations with high sensitivity, but their throughput is limited by the well plates. Generally, plate-based platforms can reliably quantify up to 10,000 genes per cell (Paik et al., 2020). In addition, methods based on microfluidic mechanisms increase the throughput significantly by encapsulating single cells in oil droplets, which allows expression profiling for thousands of cells and ranging from 104 to 105 reads per cell, but their cost is high, and the capture cell count is unstable (Wang et al., 2021).
Reverse transcription is also continuously improving. The Tang method reverse transcribed polyadenylated RNA into cDNA using an oligo-dT primer carrying a specific anchor sequence, which has weaknesses of a 3′ end bias of transcripts and low accuracy (Zhang et al., 2021). Later experimental methods, such as Smart-Seq and Smart-Seq2, have made use of Moloney mouse leukemia virus (MMLV) template switching (TS) for reverse transcription, which increases transcription efficiency and sensitivity but has a lower read coverage toward the 5′-end of the transcripts (Picelli et al., 2014; Ramsköld et al., 2012). Ziegenhain et al. proved that compared with CEL-seq2 Drop-seq and MARS-seq, Smart-seq2 has the most even coverage across transcripts (Ziegenhain et al., 2017). After cDNA generation by reverse transcription, amplification of cDNA should be performed with either PCR or in vitro transcription (IVT). MARS-seq and CEL-seq make use of IVT to linearly amplify the RNA before reverse transcription (Hashimshony et al., 2012). As more than 98% of exonic reads come from the sense strand, whose sequence is identical to mRNA (except that T is replaced by U), IVT has high strand specificity. Because transcription only starts from the end with poly(T), there is a large 3′ bias compared with PCR (Picelli, 2017).
Sequencing is usually followed by library preparation of the cDNA (Paik et al., 2020). At present, most studies use the Nextera kit for library preparation and the NGS platform (such as Illumina) for sequencing. Although new sequencing platforms have been constantly developed, the Illumina HiSeq X system remains the highest output platform due to its highly accurate data and low cost (Caruccio, 2011; Levy and Boone, 2019).
Considering the efficiency, sensitivity and cost of different scRNA-seq methods, we have the following suggestions for investigators intending to perform virological studies by scRNA-seq. Studies that aim to investigate the response of different cells to viruses and the interaction between cells in organs and tissues, which require sequencing a mass of cells, are most suitable for microfluidic-based methods, including Drop-seq, inDrop and 10X Genomics. In contrast, if researchers want to investigate the role of a particular or rare group of cells (e.g., memory B cells, effector T cells, etc.) in viral infections, it is rational to choose plate-based protocols and presort the cells by FACS before capturing cells to well plates. We believe that with the continuous innovation of equipment, the scRNA-seq technology mentioned above will become cheaper and more efficient in the future, providing new tools for researchers to explore the interaction between viruses and hosts.
2.2. Analysis workfolws and software
After the scRNA-seq workflow is completed, bioinformatics analysis of single-cell datasets must be performed, which also determines the accuracy and significance of the results. To date, various scRNA-seq analysis methods have been developed successively, which have expanded the application of scRNA-seq. The classical scRNA-seq analytic workflow includes read mapping, quality control, gene expression quantification, normalization, feature selection, cell clustering, dimensionality reduction, differential expression, and trajectory inference (Waickman et al., 2021). R or Python with various software packages is generally used to complete scRNA-seq analysis. We list the common scRNA-seq analysis methods, software packages, and their functions (Table 1). While the comparison and instruction of these packages will not be discussed here in detail and the interested reader is referred to some excellent reviews in Table 1 or published elsewhere (Balzer et al., 2021; Grün and van Oudenaarden, 2015).
Table 1.
scRNA-seq analytical workflows and software.
| Method | Software package (programming language) | Function | Key references |
|---|---|---|---|
| Read mapping | Cellranger (Linux); UMI-tools (Python); kallisto (Python); STAR (Python) | Data matrix generation | (Bray et al., 2016; Dobin et al., 2013; Smith et al., 2017) |
| Quality control | DoubletDecon (R); Scrublet (Python); SoupX (R); CellBender (Python) | Filter out cells by doublet detection tools and control the ambient RNA contamination | (DePasquale et al., 2019; Wolock et al., 2019; Young and Behjati, 2020) |
| Data integration; normalization; visualization and clustering; differential expression genes | Seurat (R); Scater (R); Scanpy (Python); Garnett (R) | Normalize data; visualize and cluster cells by nonlinear dimension-reduction methods such as t-SNE, UMAP, and Louvain and linear methods like PCA; gene differential expression analysis | (Hie et al., 2019; Lun et al., 2016; Michielsen et al., 2021; Stuart and Satija, 2019) |
| Enrichment analysis | clusterProfiler (R); singleseqgset (R); topGO (R) | Label genes based on involvement in common biologic processes, cellular components, and molecular function by reference to MSigDB, GO, KEGG, GSEA, etc. databases | (The Gene Ontology Consortium, 2017; Kanehisa et al., 2017; Liberzon et al., 2011) |
| Cell type annotation | SingleR (R); CHETAH (R); SCINA (R) | Annotate cell clusters based on the external dataset or markers (Classic or automated) | (Aran et al., 2019; de Kanter et al., 2019; Zhang et al., 2019) |
| Trajectory analysis | Monocle (R); TradeSeq (R) | Build the dynamic models of gene expression and observe the continuous heterogeneity of cells | (Trapnell et al., 2014; Van den Berge et al., 2020) |
| Gene regulatory network | CellPhoneDB (Python); CellChat (R); Celltalker (R); SCENIC (R) | Perform ligand-receptor analysis and infer the interaction between cell clusters by reference to CellPhoneDB, Connectome, RcisTarget, etc. databases | (Aibar et al., 2017; Raredon et al., 2022; Vento-Tormo et al., 2018) |
UMI, unique molecular identifier; t-SNE, t-distributed stochastic neighbor embedding; UMAP, Uniform manifold approximation, and projection; PCA, principal component analysis; MSigDB, Molecular Signatures Database; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, gene set enrichment analysis.
3. Applications of scRNA-seq in virology
ScRNA-seq is now a routine tool in biomedical research and has been widely used in oncology (Levitin et al., 2018), neuroscience (Ofengeim et al., 2017), genetics (Zhang and Liu, 2019), and virology (Qu et al., 2020). With the increasing demands, various scRNA-seq sequencing technologies (such as viral sequencing genomes and T-cell receptor or B-cell receptor sequencing genomes) and analytical methods (such as gene enrichment analysis, developmental trajectory analysis, and cell‒cell communication analysis) have been developed. ScRNA-seq provides an understanding of the mechanisms regulating particular genes in a specific cell type and their response to a particular environmental challenge with single-cell resolution. When these methods are used in virological studies, they can comprehensively reveal viral infection and antiviral mechanisms, providing insight into viral pathogenesis (Fig. 3). We present an overview of studies that have used scRNA-seq to characterize host-virus interactions, including SARS-CoV-2, flaviviruses, and other viruses.
Fig. 3.
scRNA-seq technologies reveal the host-virus interaction. With technological innovation, variety of analysis of scRNA-seq has been available: viral genome sequencing, gene regulatory network, T-cell receptor or B-cell receptor sequencing and cell clusters comparison. These effective scRNA-seq analysis has been applied to reveal the host-virus interaction including the viral burden detection, viral pathogenesis, host immune atlas and host immune repertoire.
3.1. Applications in SARS-CoV-2
As of November 2023, there have been more than 771 million cumulative confirmed cases and 6.98 million cumulative deaths worldwide due to coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 infection (WHO, 2023). The worldwide public health emergency has driven researchers to conduct a multitude of studies on SARS-CoV-2. Since scRNA-seq allows the assessment of cellular heterogeneity, identification of cellular states, and evaluation of dynamic cellular transitions at single-cell resolution and accuracy, it has played a key element in profiling the immune atlas after SARS-CoV-2 infection, revealing the virus-host interaction, and assessing the effectiveness of the vaccine.
3.1.1. Immune atlas study
ScRNA-seq allows for the identification of cell populations and the comparison of cell type proportions in different conditions. An increasing proportion of certain cells usually indicates stronger local cell growth, the transition from other cell types, or the recruitment of cells from surrounding tissues. In contrast, a decreasing proportion of certain cells usually means more cell death, transition to other cell types, or migration outward (Liu et al., 2022). When the immune system is confronted with infectious pathogens, heterogeneous immune cells are usually recruited to infected organs and participate in various critical biological processes, including pathogen recognition, antigen presentation, and cell killing. Many studies have revealed the pathogenesis of SARS-CoV-2 infection by comparing the proportion alteration of immune cell types in COVID-19 patients of different severities and healthy controls.
Most studies have used peripheral blood mononuclear cell (PBMC) samples from subjects to profile peripheral immune responses after COVID-19 infection. Wilk et al. (2020) defined a peripheral immune profile of severe COVID-19 patients by scRNA-seq. COVID-19 patients had increased developing neutrophils and CD14+ monocytes but depletion of several innate immune cells, including γδ T cells, plasmacytoid dendritic cells (pDCs), conventional dendritic cells (DCs), CD16+ monocytes, and natural killer (NK) cells. Except for severe COVID-19 patients, patients with a long duration of viral shedding also have significant alterations in immune cell composition and phenotype compared with healthy donors. Yang et al. (2021) found that the decreasing trend of NK cells and the increased regulatory T cells may be associated with the long duration of viral shedding.
ScRNA-seq can also be used to investigate the changes in cell types in infected tissues. Delorey et al. (2021) established single-cell atlases of the lung, kidney, liver, and heart for COVID-19 patients via scRNA-seq. The lungs of COVID-19 patients showed significantly reduced type II alveolar cells (AT2) and increased dendritic cells, macrophages, NK cells, fibroblasts, lymphatic endothelial cells, and vascular endothelial cells in comparison to normal lungs. For the heart, reduced cardiomyocytes and pericytes and increased vascular endothelial cells were observed in COVID-19 patients.
ScRNA-seq performed on COVID-19 patient tissue (PBMCs or target organs) is the most direct way to reflect viral infection. It is not necessary to worry about unavailable patient samples, as an increasing number of publicly accessible scRNA-seq datasets are currently available for analysis (Das et al., 2018; Wang et al., 2018). However, an immune atlas or cell profiles of virus-infected organs only provide an inexhaustive view of infection, which requires further analysis of particular cell types or gene expression to reveal the mechanism of viral infection.
3.1.2. Host-virus interactions study
Another critical function of scRNA-seq is to characterize the transcriptome of various cell types in response to SARS-CoV-2 infection. Differential gene expression analysis is a common approach to obtaining gene expression profiles comparatively between COVID-19 patients and healthy controls, which is significant for screening receptors and understanding pathogenesis. For differentially expressed genes, gene enrichment analysis is often performed to identify the activated and deactivated pathways and biological processes that are most closely related to SARS-CoV-2 infection (Qu et al., 2020).
ScRNA-seq can identify susceptible cells by examining individual cell transcriptomes (including viral transcriptome and host transcriptome). Cells with a high viral load or active proliferation may be target cells attacked by the virus or play a key role in SARS-CoV-2 colonization and spreading. Ren et al. (2021) performed scRNA-seq analysis on samples from the respiratory system and indicated the presence of viral RNAs of SARS-CoV-2 in epithelial cells and immune cells, including neutrophils, macrophages, plasma B cells, T cells, and NK cells. Intriguingly, only a subset of epithelial cells expresses angiotensin I converting enzyme 2 (ACE2), which plays a critical role in mediating SARS-CoV-2 entry (Hoffmann et al., 2020). Therefore, it would be interesting to investigate how SARS-CoV-2 enters these cells. Are there still unknown receptors that help SARS-CoV-2 infect cells?
ACE2 is the host receptor of SARS-CoV-2 to infect human cells, while RNA-seq-derived datasets from major human physiological systems reveal that ACE2 is not highly expressed in some target cells of SARS-CoV-2 (Zou et al., 2020). Qi et al. (2020) assumed that SARS-CoV-2 may depend on a co-receptor or other auxiliary membrane proteins to facilitate its binding and entry. They collected single-cell gene expression data from 13 tissues, including the liver, lung, colon, spleen, kidney, etc., and explored the expression spectrum of ACE2, single-stranded RNA (ssRNA) viral receptors and other membrane proteins in human tissues to elaborate the potential relationship between ACE2 and other membrane proteins or viral receptors. Among the 94 genes that were significantly associated with ACE2, ANPEP, ENPEP, and DPP4 were the top three most associated. ANPEP and DPP4 are the known receptors for coronavirus, suggesting that ENPEP may be another potential receptor for coronavirus that needs further experimental validation.
It is a valid analytical strategy to screen out the possible co-receptors by analyzing scRNA-seq data based on existing receptor databases, which can help researchers select the particular genes that may be most associated with viral infection among hundreds or thousands of genes, especially saving manpower and material resources. However, this strategy will be limited by the scRNA-seq data and receptor datasets, which means researchers cannot obtain new findings outside the data or datasets, and it should be particularly noted that experimental validation of the scRNA-seq analysis is essential.
In addition to screening receptors, scRNA-seq analysis of immune cells can elucidate the immunoregulatory mechanisms of the host after SARS-CoV-2 infection. The expression of type I/III interferon response genes, which plays a significant role in innate immune responses to viral infection, was elevated in COVID-19 patients compared to healthy controls. An increase in cytokines such as CCL4, CXCL10, interleukin (IL)-7 and IL-1α in patient serum is associated with severe and critical COVID-19, suggesting that the recruitment of monocytes and NK lymphocytes and the activity of T cells affect SARS-CoV-2 progression. For T cells, differential gene expression analysis and gene-set enrichment analysis identified pathways associated with inflammation and T-cell activation, including IL-2-STAT5 signaling, mTORC1 signaling, inflammatory response, IFNγ response, and IL-6– JAK–STAT3 signaling (Stephenson et al., 2021).
Most scRNA-seq studies analyze the differential gene expression between virus-infected and control samples after cell cluster definition to investigate the regulatory mechanisms at the gene level after infection. The analysis of immune cells provides a view of the immune responses of the host, paving the way for the analysis of cell-cell interactions.
3.1.3. Immune repertoire study
The immune repertoire refers to the sum of B cells and T cells with distinct antigen specificity at any given time, which mediate antigen-specific adaptive immunity by recognizing and eliminating antigens originating from infection (Liu and Wu, 2018). Immune repertoire studies play an important role in identifying neutralizing antibodies and testing vaccination efficacy.
The unique TCR or BCR on the surface of each T-cell or B-cell is critical for specific antigen recognition. At the early stage, asymptomatic COVID-19 patients have more diverse TCRs than moderate or severe patients, indicating that effective TCR diversification on CD4+ or CD8+ T cells may contribute to the immune control and outcome of COVID-19 (Zhao et al., 2021). In addition, scRNA-seq for B cells can identify neutralizing antibodies that have strong therapeutic and prophylactic efficacy. Cao et al. (2020) performed scRNA-seq and BCR sequencing of antigen-binding B cells from the plasma of convalescent COVID-19 patients and identified 14 highly active SARS-CoV-2-neutralizing antibodies.
scRNA-seq is also applied to assess vaccination efficacy by monitoring T-cell responses and antibody generation after vaccine injection. Sureshchandra et al. (2021) measured humoral and cellular responses to mRNA vaccination in four volunteers and compared their immune repertoire changes with those in three convalescent individuals who experienced asymptomatic COVID-19. They found activated CD4+ T cells associated with Th1 and Th17 cells and expanded memory clone CD8+ T cells in vaccinated individuals. Although neutralizing titers in the vaccinated group were comparable to those in recovered individuals after the first dose, the levels were much greater after the booster. In addition to antibody titers, the evolution of the SARS-CoV-2-specific B-cell repertoire can also be characterized by analyzing the blood sample of a healthy vaccinated participant at multiple time points via scRNA-seq (Kramer et al., 2022).
To date, many immune repertoire studies have used scRNA-seq, which is inseparable from the development of TCR and BCR sequencing technologies. Because microfluidic-based methods allow customized TCR/BCR transcript capture, for example, DART-seq or RAGE-Seq (Singh et al., 2019), TCR/BCR sequencing is most commonly combined with microfluidic-based platforms, such as 10X Genomics. However, if rare immune cell clusters are to be investigated, as mentioned earlier, the 10× platform may be limited. Huang et al. (2019) modified the Smart-Seq2 protocol and developed a technology (SELECT-seq) to isolate rare T cells and to analyze their transcriptome and TCR composition.
3.2. Applications in flaviviruses
The Flavivirus genus encompasses more than 75 positive-stranded enveloped RNA viruses transmitted by arthropods (primarily ticks and mosquitoes). Important flaviviruses that infect humans include Zika virus (ZIKV), dengue virus (DENV), West Nile virus (WNV), Japanese encephalitis virus (JEV) and yellow fever virus (YFV), which may cause severe outbreaks and pose a substantial danger to human health worldwide (Zhao et al., 2021). To date, flavivirus-related scRNA-seq analysis is still inadequate, and further studies are needed.
3.2.1. Host-virus interaction study
One of the limitations of scRNA-seq in the application of flavivirus-related studies is that many scRNA-seq platforms cannot capture nonpolyadenylated transcripts (Salmen et al., 2022). Viruses in the Flaviviridae family produce nonpolyadenylated transcripts during viral multiplication (Brinton and Basu, 2015). Therefore, it is important to develop a scRNA-seq method to capture the nonpolyadenylated transcriptome in cells to study viral infection dynamics. Based on Smart-seq2, Zanini et al. (2018) developed a new scRNA-seq technology, named virus-inclusive single-cell RNA-Seq (viscRNA-Seq), to sequence and quantify the whole transcriptome and the viral RNA of single cells. They have applied this technology to analyze the transcriptome dynamics of ZIKV and DENV infection in cultured human hepatoma (Huh7) cells. After demonstrating the effectiveness of the technique, they combined fluorescence-activated cell sorting (FACS) with viscRNA-Seq to profile the host and viral transcriptomes in PBMCs collected from DENV-infected individuals. For individuals with severe dengue fever, many antiviral IFN response genes were strongly upregulated in a cell type-specific manner, which indicates that distinct cell populations respond differently to the same viral infection. In severe dengue subjects, DENV RNA was enriched in IgM-positive B cells and some naive B cells. In addition, B-cell activation and tissue-specific homing signals were upregulated (Zanini et al., 2018). The above results highlight the utility of viscRNA-Seq as a powerful tool to explore host-virus interactions at the single-cell level.
Waickman et al. (2021) also utilized scRNA-seq to analyze PBMCs from DENV-infected individuals and revealed differences in the immune response and cellular transcriptional profiles between experimental and natural primary DENV-1 infections. Two datasets revealed that both experimental and natural primary DENV-1 induced the activation and expansion of lymphocyte populations, but natural infection had a more pronounced pattern of inflammatory gene upregulation.
In addition to DENV, O'Neal et al. (O'Neal et al., 2019) developed a WNV infection model in murine fibroblast L929 cells and performed gene ontology (GO) enrichment analysis to identify the pathways that might be linked to infection. The top pathways positively correlated with WNV RNA included transcriptional regulation, amino acid transport and rRNA processing, and 124 IFN-stimulated genes were negatively correlated with intracellular viral abundance.
In 2023, studies on flaviviruses using scRNA-seq have increased, especially regarding ZIKV. Considering the harmful effects of ZIKV on the fetus during pregnancy, Wu et al. established ZIKV infection in human trophoblast stem cells (hTSCs)-derived trophoblast organoid. Approaches including gene knockout and bulk transcriptome sequencing provide important insights for susceptibility to ZIKV and structural disruption in organoids, results of scRNA-seq further explain the pathogenic mechanism, revealing that ZIKV disrupted the stemness of hTSCs and the proliferation of cytotrophoblast cells (Wu et al., 2023). Obviously, scRNA-seq has the advantage over other traditional technologies, which it can uncover pathological mechanisms in detail and differentiation trajectories of infected cells. In addition, Yang et al. focused on the potential threat of ZIKV to the male reproductive system. Using scRNA-seq, they proved that mouse testicular spermatogenic cells were susceptible to ZIKV, but there was no obvious apoptosis, and the classical pathway of complement in monocyte/macrophage was activated after infection. Subsequently, Yang et al. performed traditional experiments such as RT-qPCR, ELISA and immunofluorescence staining to verify the results of scRNA-seq and confirmed that S100A4+ monocytes activated the complement and then resulted in the damage of spermatogenic cells in testes after ZIKV infection (Yang et al., 2023). Wu and Yang et al. show another strategy of scRNA-seq research, which make use of traditional virological techniques to verify the mechanisms provided by scRNA-seq.
Although the availability of viscRNA-Seq has been confirmed in detecting intracellular ZIKV and DENV RNA, its application is not yet widespread. Understanding the pathogenic mechanisms of viruses in the Flaviviviridae family relies on scRNA-seq methods that have been developed to measure nonpolyadenylated transcripts. Considering the application of scRNA-seq in ZIKV studies in recent years, we anticipate that more protocols will be available in the future to profile intracellular flavivirus RNA sequences, especially methods suitable for microfluidic mechanisms.
3.2.2. Study of the immune repertoire
The development of the DENV vaccine has been a challenge because of the antibody-dependent enhancement (ADE) effect caused by its four serotypes. ADE means that cross-reactive antibodies from the first infection will bind to virus particles during the secondary infection and enter FcγR-bearing cells to aggravate the disease (Acosta and Bartenschlager, 2016). As a result, it is necessary to investigate whether or how memory B cells (MBCs) generate antibodies that respond to neutralizing conserved epitopes of a subsequent heterologous vaccine or infection. Wong et al. (2020) used scRNA-seq to detect the bone marrow of mice at different time points after vaccination and found an extensive clonal overlap of complementarity determining region 3 (CDR3) regions of both IgH and Igκ between pre-MBCs, mature MBCs, and germinal center B cells, which indicates that MBCs hardly generate new specificities after vaccination. Therefore, limiting MBC diversity is an effective strategy to reduce ADE for DENV vaccines.
In addition, Afik et al. (2017) developed software, ‘TCR Reconstruction Algorithm for Paired-End Single cell’ (TRAPeS), to accurately reconstruct TCRs from TCR sequence information generated by scRNA-seq and relate specific TCR properties to the heterogeneity of the CD8+ T-cell state. This method is suitable for scRNA-seq with short read lengths and increased sensitivity of sequence reads. They collected and analyzed whole blood samples from a volunteer who was vaccinated with the 17D yellow fever vaccine and performed scRNA-seq. After identifying the subpopulations of CD8+ T cells by differentially expressed genes, they studied the TCR characteristics of different cells and indicated that naive CD8+ T cells had longer CDR3 regions than effector memory CD8+ T cells after vaccination.
Because high accuracy scRNA-seq data are required for immune repertoire studies, plate-based strategies are the common choice, such as Smart-seq2. Interestingly, in addition to customized TCR sequencing technology, it is also possible to reconstruct the TCR by a particular algorithm after sequencing. If more algorithms are applied in the future to profile TCR/BCR sequences of immune cells after viral infection, this will greatly reduce the cost of scRNA-seq.
3.3. Applications in other viruses
With the rapid development of scRNA-seq, many exciting viral studies have emerged, not just SARS-CoV-2 and flaviviruses. Here, we summarized the researches of scRNA-seq in influenza virus, human immunodeficiency virus (HIV), herpes simplex virus (HSV) for introduction, to provide a broader view from the RNA viruses with high variability, RNA viruses with latency and DNA viruses.
For influenza viruses, scRNA-seq can detect the viral load of each cell to investigate the heterogeneity of infection and characterize the antiviral response in various cell types (Steuerman et al., 2018). As only some cells induce IFN after influenza virus infection, Russell et al. (2019) explored how viral genetic diversity contributes to cell infection heterogeneity by establishing influenza A (H1N1) virus-infected A549 cells that carried IFN reporters and sequencing the infected IFN-positive cells on Illumina and PacBio (third generation sequencing) platforms. They found that defects of viruses may increase IFN induction, including the absence of the NS gene and amino acid mutations in nonstructural protein 1 or polymerase protein 1, and the viral defect may relate to innate immune induction of influenza but not a determining factor. The result also suggests that it is effective to perform scRNA-seq of the cell clusters of interest by creating pathway reporter cell lines. In this study, IFN-enriched infected cells were subjected to scRNA-seq by 10× Chromium after the cells were sorted by flow cytometry analyses.
The latent infection of HIV makes radical therapy difficult: Although antiretroviral treatment (ART) can effectively halt HIV disease progression and reduce plasma viremia to undetectable levels, it cannot eradicate the virus (Brandt et al., 2020). Understanding the characteristics of latently infected cells to eradicate them is thus the key to treating HIV patients, in which scRNA-seq is utilized to profile latent cells and investigate latency reactivation potential. Golumbeanu et al. (2018) identified 134 differentially expressed genes between resting-state cells and responsive-state cells and indicated that responsive-state cells display higher metabolic activity. Similar to Russell et al., the aim of Golumbeanu et al. was also to analyze cells of interest, while they chose to sort cells through FACS before performing sequencing.
HSV has gained considerable attention since it can cause life-threatening herpes simplex virus encephalitis (HSE). To characterize the role of microglia in HSE, Uyar et al. (2022) performed scRNA-seq on microglia and microglia-like cells isolated from mice infected with HSV-1. They revealed that microglia proliferated and highly expressed CD68 (a marker of phagolysosomal activity) after viral infection, suggesting that microglia participate in viral clearance by phagocytosis. In addition, a novel cell cluster associated with the severity of infection was observed and named “in transition” microglia/microglia-like cells, which increased NLRP3 inflammasome-mediated interleukin IL-1β production and promoted a proinflammatory response. Cell status defined by scRNA-seq algorithms may be a brand-new cell cluster, and profiling the viral gene expression dynamics and the immune responses of these cells can provide a deep view of host and viral interactions.
4. Conclusions and future directions
With the booming of scRNA-seq in recent years, more precise sequencing technology and greater diversity of analytical methods have continually emerged, providing researchers with a comprehensive research platform. There is no doubt that scRNA-seq breaks through some of the limitations of bulk RNA sequencing, the most important of which is that scRNA-seq reveals the heterogeneity of viral infections and cellular responses at the single-cell level. We anticipate that scRNA-seq will be combined with more bioinformatics approaches (e.g., spatial transcriptomics, proteomics, etc.) in the future, providing more information to understand the complexities of host viral interactions. The variety of scRNA-seq tools will allow us to document the expression of all genes in various cells with high temporal and spatial resolution, understand the changes in cellular functions and signaling pathways, and reveal the mechanisms of gene regulation and response to stimulation. In addition, the availability of more public scRNA-seq datasets and analytical methods will also constantly promote the credibility and diversity of scRNA-seq analysis.
Although scRNA-seq offers a broader application prospect in biomedical research, the high cost, low sequencing depth, and cell capture rate are urgent issues with scRNA-seq. As a result, future scRNA-seq development will focus on the advancement of library preparation, sequencing technology, and cell capture technology. We recommend that researchers should fully understand the advantages and limitations of different scRNA-seq technologies before conducting scRNA-seq studies to reveal host-virus interactions, considering the purpose and content of the experiment, of which virus type, cell type, cell number, budget and so on are all important factors. Additionally, noteworthy is that although scRNA-seq has shown powerful advantages in the field of virology and immunology, how to combine intracellular viral load with cell function requires more practical research, and the results of scRNA-seq also have to be validated by in vitro and in vivo experiments.
Conflict of interest
The authors declare that there are no conflicts of interest. Prof. Jing An is an editorial board member for Virologica Sinica and was not involved in the editorial review or the decision to publish this article.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2021YFC2300202); the National Natural Science Foundation of China (U1902210, 81871641, 81972979, 82172266, 82241071, and 81902048); the Support Project of High-level Teachers in Beijing Municipal Universities in the Period of 13th Five-year Plan (IDHT20190510); and the Beijing Key Laboratory of Emerging Infectious Diseases (DTKF202103).
Contributor Information
Pei-Gang Wang, Email: pgwang@ccmu.edu.cn.
Jing An, Email: anjing@ccmu.edu.cn.
References
- Acosta E.G., Bartenschlager R. Paradoxical role of antibodies in dengue virus infections: considerations for prophylactic vaccine development. Expet Rev. Vaccine. 2016;15:467–482. doi: 10.1586/14760584.2016.1121814. [DOI] [PubMed] [Google Scholar]
- Afik S., Yates K.B., Bi K., Darko S., Godec J., Gerdemann U., Swadling L., Douek D.C., Klenerman P., Barnes E.J., Sharpe A.H., Haining W.N., Yosef N. Targeted reconstruction of T cell receptor sequence from single cell RNA-seq links CDR3 length to T cell differentiation state. Nucleic Acids Res. 2017;45:e148. doi: 10.1093/nar/gkx615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J., van den Oord J., Atak Z.K., Wouters J., Aerts S. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R., Butte A.J., Bhattacharya M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20:163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzer M.S., Ma Z., Zhou J., Abedini A., Susztak K. How to get started with single cell RNA sequencing data analysis. J. Am. Soc. Nephrol. 2021;32:1279–1292. doi: 10.1681/ASN.2020121742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt L., Cristinelli S., Ciuffi A. Single-cell analysis reveals heterogeneity of virus infection, pathogenicity, and host responses: HIV as a pioneering example. Annu Rev Virol. 2020;7:333–350. doi: 10.1146/annurev-virology-021820-102458. [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Brinton M.A., Basu M. Functions of the 3’ and 5’ genome RNA regions of members of the genus Flavivirus. Virus Res. 2015;206:108–119. doi: 10.1016/j.virusres.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., Chai X., He R., Li X., Lv Q., Zhu H., Deng W., Xu Y., Wang Y., Qiao L., Tan Y., Song L., Wang G., Du X., Gao N., Liu J., Xiao J., Su X., Du Z., Feng Y., Qin Chuan, Qin Chengfeng, Jin R., Xie X.S. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruccio N. Preparation of next-generation sequencing libraries using NexteraTM technology: simultaneous DNA fragmentation and adaptor tagging by in vitro transposition. Methods Mol. Biol. 2011;733:241–255. doi: 10.1007/978-1-61779-089-8_17. [DOI] [PubMed] [Google Scholar]
- Choi J.R., Yong K.W., Choi J.Y., Cowie A.C. Single-cell RNA sequencing and its combination with protein and DNA analyses. Cells. 2020;9:1130. doi: 10.3390/cells9051130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X., Cai L., He F. Single-cell sequencing: expansion, integration and translation. Briefings Funct. Genomics. 2022;21:280–295. doi: 10.1093/bfgp/elac011. [DOI] [PubMed] [Google Scholar]
- Das R., Bar N., Ferreira M., Newman A.M., Zhang L., Bailur J.K., Bacchiocchi A., Kluger H., Wei W., Halaban R., Sznol M., Dhodapkar M.V., Dhodapkar K.M. Early B cell changes predict autoimmunity following combination immune checkpoint blockade. J. Clin. Invest. 2018;128:715–720. doi: 10.1172/JCI96798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kanter J.K., Lijnzaad P., Candelli T., Margaritis T., Holstege F.C.P. CHETAH: a selective, hierarchical cell type identification method for single-cell RNA sequencing. Nucleic Acids Res. 2019;47:e95. doi: 10.1093/nar/gkz543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorey T.M., Ziegler C.G.K., Heimberg G., Normand R., Yang Y., Segerstolpe Å., Abbondanza D., Fleming S.J., Subramanian A., Montoro D.T., Jagadeesh K.A., Dey K.K., Sen P., Slyper M., Pita-Juárez Y.H., Phillips D., Biermann J., Bloom-Ackermann Z., Barkas N., Ganna A., Gomez J., Melms J.C., Katsyv I., Normandin E., Naderi P., Popov Y.V., Raju S.S., Niezen S., Tsai L.T.-Y., Siddle K.J., Sud M., Tran V.M., Vellarikkal S.K., Wang Y., Amir-Zilberstein L., Atri D.S., Beechem J., Brook O.R., Chen J., Divakar P., Dorceus P., Engreitz J.M., Essene A., Fitzgerald D.M., Fropf R., Gazal S., Gould J., Grzyb J., Harvey T., Hecht J., Hether T., Jané-Valbuena J., Leney-Greene M., Ma H., McCabe C., McLoughlin D.E., Miller E.M., Muus C., Niemi M., Padera R., Pan L., Pant D., Pe’er C., Pfiffner-Borges J., Pinto C.J., Plaisted J., Reeves J., Ross M., Rudy M., Rueckert E.H., Siciliano M., Sturm A., Todres E., Waghray A., Warren S., Zhang S., Zollinger D.R., Cosimi L., Gupta R.M., Hacohen N., Hibshoosh H., Hide W., Price A.L., Rajagopal J., Tata P.R., Riedel S., Szabo G., Tickle T.L., Ellinor P.T., Hung D., Sabeti P.C., Novak R., Rogers R., Ingber D.E., Jiang Z.G., Juric D., Babadi M., Farhi S.L., Izar B., Stone J.R., Vlachos I.S., Solomon I.H., Ashenberg O., Porter C.B.M., Li B., Shalek A.K., Villani A.-C., Rozenblatt-Rosen O., Regev A. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePasquale E.A.K., Schnell D.J., Van Camp P.-J., Valiente-Alandí Í., Blaxall B.C., Grimes H.L., Singh H., Salomonis N. DoubletDecon: deconvoluting doublets from single-cell RNA-sequencing data. Cell Rep. 2019;29:1718–1727.e8. doi: 10.1016/j.celrep.2019.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijk E.L. van, Jaszczyszyn Y., Naquin D., Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34:666–681. doi: 10.1016/j.tig.2018.05.008. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golumbeanu M., Cristinelli S., Rato S., Munoz M., Cavassini M., Beerenwinkel N., Ciuffi A. Single-cell RNA-seq reveals transcriptional heterogeneity in latent and reactivated HIV-infected cells. Cell Rep. 2018;23:942–950. doi: 10.1016/j.celrep.2018.03.102. [DOI] [PubMed] [Google Scholar]
- Grün D., van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163:799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- Hashimshony T., Wagner F., Sher N., Yanai I. CEL-seq: single-cell RNA-seq by multiplexed linear amplification. Cell Rep. 2012;2:666–673. doi: 10.1016/j.celrep.2012.08.003. [DOI] [PubMed] [Google Scholar]
- Hie B., Bryson B., Berger B. Efficient integration of heterogeneous single-cell transcriptomes using Scanorama. Nat. Biotechnol. 2019;37:685–691. doi: 10.1038/s41587-019-0113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Sikora M.J., Islam S., Chowdhury R.R., Chien Y., Scriba T.J., Davis M.M., Steinmetz L.M. Select sequencing of clonally expanded CD8+ T cells reveals limits to clonal expansion. Proc. Natl. Acad. Sci. U.S.A. 2019;116:8995–9001. doi: 10.1073/pnas.1902649116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B., Lee J.H., Bang D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018;50:96. doi: 10.1038/s12276-018-0071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer K.J., Wilfong E.M., Voss K., Barone S.M., Shiakolas A.R., Raju N., Roe C.E., Suryadevara N., Walker L.M., Wall S.C., Paulo A., Schaefer S., Dahunsi D., Westlake C.S., Crowe J.E., Jr., Carnahan R.H., Rathmell J.C., Bonami R.H., Georgiev I.S., Irish J.M. Single-cell profiling of the antigen-specific response to BNT162b2 SARS-CoV-2 RNA vaccine. Nat Commun. 2022;13:3466. doi: 10.1038/s41467-022-31142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin H.M., Yuan J., Sims P.A. Single-cell transcriptomic analysis of tumor heterogeneity. Trends Cancer. 2018;4:264–268. doi: 10.1016/j.trecan.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S.E., Boone B.E. Next-generation sequencing strategies. Cold Spring Harb Perspect Med. 2019;9:a025791. doi: 10.1101/cshperspect.a025791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Wu J. History, applications, and challenges of immune repertoire research. Cell Biol. Toxicol. 2018;34:441–457. doi: 10.1007/s10565-018-9426-0. [DOI] [PubMed] [Google Scholar]
- Liu W., Jia J., Dai Y., Chen W., Pei G., Yan Q., Zhao Z. Delineating COVID-19 immunological features using single-cell RNA sequencing. Innovation. 2022;3 doi: 10.1016/j.xinn.2022.100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lun A.T.L., Bach K., Marioni J.C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G., Gao Q., Zhang S., Yan B. Probing infectious disease by single-cell RNA sequencing: progresses and perspectives. Comput. Struct. Biotechnol. J. 2020;18:2962–2971. doi: 10.1016/j.csbj.2020.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michielsen L., Reinders M.J.T., Mahfouz A. Hierarchical progressive learning of cell identities in single-cell data. Nat. Commun. 2021;12:2799. doi: 10.1038/s41467-021-23196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengeim D., Giagtzoglou N., Huh D., Zou C., Yuan J. Single-cell RNA sequencing: unraveling the brain one cell at a time. Trends Mol. Med. 2017;23:563–576. doi: 10.1016/j.molmed.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal J.T., Upadhyay A.A., Wolabaugh A., Patel N.B., Bosinger S.E., Suthar M.S. West nile virus-inclusive single-cell RNA sequencing reveals heterogeneity in the type I interferon response within single cells. J Virol. 2019;93 doi: 10.1128/JVI.01778-18. -18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik D.T., Cho S., Tian L., Chang H.Y., Wu J.C. Single-cell RNA sequencing in cardiovascular development, disease and medicine. Nat. Rev. Cardiol. 2020;17:457–473. doi: 10.1038/s41569-020-0359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S. Single-cell RNA-sequencing: the future of genome biology is now. RNA Biol. 2017;14:637. doi: 10.1080/15476286.2016.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu L., Li S., Qiu H.J. Applications of single-cell RNA sequencing in virology. Yi Chuan. 2020;42:269–277. doi: 10.16288/j.yczz.19-223. [DOI] [PubMed] [Google Scholar]
- Ramsköld D., Luo S., Wang Y.-C., Li R., Deng Q., Faridani O.R., Daniels G.A., Khrebtukova I., Loring J.F., Laurent L.C., Schroth G.P., Sandberg R. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat. Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raredon M.S.B., Yang J., Garritano J., Wang M., Kushnir D., Schupp J.C., Adams T.S., Greaney A.M., Leiby K.L., Kaminski N., Kluger Y., Levchenko A., Niklason L.E. Computation and visualization of cell–cell signaling topologies in single-cell systems data using Connectome. Sci. Rep. 2022;12:4187. doi: 10.1038/s41598-022-07959-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wen W., Fan X., Hou W., Su Bin, Cai P., Li J., Liu Y., Tang F., Zhang F., Yang Y., He Jiangping, Ma W., He Jingjing, Wang P., Cao Q., Chen F., Chen Y., Cheng X., Deng G., Deng X., Ding W., Feng Y., Gan R., Guo C., Guo W., He S., Jiang C., Liang J., Li Y., Lin J., Ling Y., Liu H., Liu J., Liu N., Liu S.-Q., Luo M., Ma Q., Song Q., Sun W., Wang G., Wang F., Wang Y., Wen X., Wu Q., Xu G., Xie X., Xiong X., Xing X., Xu H., Yin C., Yu D., Yu K., Yuan J., Zhang B., Zhang P., Zhang T., Zhao J., Zhao Peidong, Zhou J., Zhou W., Zhong S., Zhong X., Zhang S., Zhu L., Zhu P., Zou B., Zou J., Zuo Z., Bai F., Huang X., Zhou P., Jiang Q., Huang Z., Bei J.X., Wei L., Bian X.W., Liu X., Cheng T., Li X., Zhao Pingsen, Wang F.S., Wang H., Su Bing, Zhang Zheng, Qu K., Wang X., Chen J., Jin R., Zhang Zemin. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell A.B., Elshina E., Kowalsky J.R., te Velthuis A.J.W., Bloom J.D. Single-cell virus sequencing of influenza infections that trigger innate immunity. J Virol. 2019;93:e00500–e00519. doi: 10.1128/JVI.00500-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmen F., De Jonghe J., Kaminski T.S., Alemany A., Parada G.E., Verity-Legg J., Yanagida A., Kohler T.N., Battich N., van den Brekel F., Ellermann A.L., Arias A.M., Nichols J., Hemberg M., Hollfelder F., van Oudenaarden A. High-throughput total RNA sequencing in single cells using VASA-seq. Nat. Biotechnol. 2022;40:1780–1793. doi: 10.1038/s41587-022-01361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M., Al-Eryani G., Carswell S., Ferguson J.M., Blackburn J., Barton K., Roden D., Luciani F., Giang Phan T., Junankar S., Jackson K., Goodnow C.C., Smith M.A., Swarbrick A. High-throughput targeted long-read single cell sequencing reveals the clonal and transcriptional landscape of lymphocytes. Nat. Commun. 2019;10:3120. doi: 10.1038/s41467-019-11049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson E., Reynolds G., Botting R.A., Calero-Nieto F.J., Morgan M.D., Tuong Z.K., Bach K., Sungnak W., Worlock K.B., Yoshida M., Kumasaka N., Kania K., Engelbert J., Olabi B., Spegarova J.S., Wilson N.K., Mende N., Jardine L., Gardner L.C.S., Goh I., Horsfall D., McGrath J., Webb S., Mather M.W., Lindeboom R.G.H., Dann E., Huang N., Polanski K., Prigmore E., Gothe F., Scott J., Payne R.P., Baker K.F., Hanrath A.T., Schim van der Loeff I.C.D., Barr A.S., Sanchez-Gonzalez A., Bergamaschi L., Mescia F., Barnes J.L., Kilich E., de Wilton A., Saigal A., Saleh A., Janes S.M., Smith C.M., Gopee N., Wilson C., Coupland P., Coxhead J.M., Kiselev V.Y., van Dongen S., Bacardit J., King H.W., Rostron A.J., Simpson A.J., Hambleton S., Laurenti E., Lyons P.A., Meyer K.B., Nikolić M.Z., Duncan C.J.A., Smith K.G.C., Teichmann S.A., Clatworthy M.R., Marioni J.C., Göttgens B., Haniffa M. Single-cell multi-omics analysis of the immune response in COVID-19. Nat. Med. 2021;27:904–916. doi: 10.1038/s41591-021-01329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerman Y., Cohen M., Peshes-Yaloz N., Valadarsky L., Cohn O., David E., Frishberg A., Mayo L., Bacharach E., Amit I., Gat-Viks I. Dissection of influenza infection in vivo by single-cell RNA sequencing. Cell Syst. 2018;6:679–691.e4. doi: 10.1016/j.cels.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- Sureshchandra S., Lewis S.A., Doratt B.M., Jankeel A., Coimbra Ibraim I., Messaoudi I. Single-cell profiling of T and B cell repertoires following SARS-CoV-2 mRNA vaccine. JCI Insight. 2021;6 doi: 10.1172/jci.insight.153201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A., Lao K., Surani M.A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium Expansion of the gene ontology knowledgebase and resources. Nucleic Acids Res. 2017;45:D331–D338. doi: 10.1093/nar/gkw1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. Pseudo-temporal ordering of individual cells reveals dynamics and regulators of cell fate decisions. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyar O., Dominguez J.M., Bordeleau M., Lapeyre L., Ibáñez F.G., Vallières L., Tremblay M.-E., Corbeil J., Boivin G. Single-cell transcriptomics of the ventral posterolateral nucleus-enriched thalamic regions from HSV-1-infected mice reveal a novel microglia/microglia-like transcriptional response. J. Neuroinflammation. 2022;19:81. doi: 10.1186/s12974-022-02437-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berge K., Roux de Bézieux H., Street K., Saelens W., Cannoodt R., Saeys Y., Dudoit S., Clement L. Trajectory-based differential expression analysis for single-cell sequencing data. Nat. Commun. 2020;11:1201. doi: 10.1038/s41467-020-14766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R., Efremova M., Botting R.A., Turco M.Y., Vento-Tormo M., Meyer K.B., Park J.-E., Stephenson E., Polański K., Goncalves A., Gardner L., Holmqvist S., Henriksson J., Zou A., Sharkey A.M., Millar B., Innes B., Wood L., Wilbrey-Clark A., Payne R.P., Ivarsson M.A., Lisgo S., Filby A., Rowitch D.H., Bulmer J.N., Wright G.J., Stubbington M.J.T., Haniffa M., Moffett A., Teichmann S.A. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waickman A.T., Friberg H., Gromowski G.D., Rutvisuttinunt W., Li T., Siegfried H., Victor K., McCracken M.K., Fernandez S., Srikiatkhachorn A., Ellison D., Jarman R.G., Thomas S.J., Rothman A.L., Endy T., Currier J.R. Temporally integrated single cell RNA sequencing analysis of PBMC from experimental and natural primary human DENV-1 infections. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., de Mochel N.S.R., Christenson S.A., Cassandras M., Moon R., Brumwell A.N., Byrnes L.E., Li A., Yokosaki Y., Shan P., Sneddon J.B., Jablons D., Lee P.J., Matthay M.A., Chapman H.A., Peng T. Expansion of hedgehog disrupts mesenchymal identity and induces emphysema phenotype. J. Clin. Invest. 2018;128:4343–4358. doi: 10.1172/JCI99435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., He Y., Zhang Q., Ren X., Zhang Z. Direct comparative analyses of 10X Genomics Chromium and smart-seq2. Dev. Reprod. Biol. 2021;19:253–266. doi: 10.1016/j.gpb.2020.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2023. WHO Coronavirus (COVID-19) Dashboard.https://covid19.who.int (Accessed 14 November 2023) [Google Scholar]
- Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T., Simpson L.J., Grant P., Subramanian A., Rogers A.J., Blish C.A. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolock S.L., Lopez R., Klein A.M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 2019;8:281–291.e9. doi: 10.1016/j.cels.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong R., Belk J.A., Govero J., Uhrlaub J.L., Reinartz D., Zhao H., Errico J.M., D'Souza L., Ripperger T.J., Nikolich-Zugich J., Shlomchik M.J., Satpathy A.T., Fremont D.H., Diamond M.S., Bhattacharya D. Affinity-restricted memory B cells dominate recall responses to heterologous flaviviruses. Immunity. 2020;53:1078–1094.e7. doi: 10.1016/j.immuni.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Huang X.Y., Sun M.X., Wang Y., Zhou H.Y., Tian Y., He B., Li K., Li D.Y., Wu A.P., Wang H., Qin C.F. Zika virus targets human trophoblast stem cells and prevents syncytialization in placental trophoblast organoids. Nat. Commun. 2023;14:5541. doi: 10.1038/s41467-023-41158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Fan J., Huang J., Guo E., Fu Y., Liu S., Xiao R., Liu C., Lu F., Qin T., He C., Wang Z., Qin X., Hu D., You L., Li X., Wang T., Wu P., Chen G., Zhou J., Li K., Sun C. Clinical and molecular characteristics of COVID-19 patients with persistent SARS-CoV-2 infection. Nat. Commun. 2021;12:3501. doi: 10.1038/s41467-021-23621-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Liu L.B., Liu F.L., Wu Y.H., Zhen Z.D., Fan D.Y., Sheng Z.Y., Song Z.R., Chang J.T., Zheng Y.T., An J., Wang P.G. Single-cell RNA sequencing reveals the fragility of male spermatogenic cells to Zika virus-induced complement activation. Nat. Commun. 2023;14:2476. doi: 10.1038/s41467-023-38223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.D., Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience. 2020;9:giaa151. doi: 10.1093/gigascience/giaa151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanini F., Pu S.Y., Bekerman E., Einav S., Quake S.R. Single-cell transcriptional dynamics of flavivirus infection. Elife. 2018;7 doi: 10.7554/eLife.32942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanini F., Robinson M.L., Croote D., Sahoo M.K., Sanz A.M., Ortiz-Lasso E., Albornoz L.L., Rosso F., Montoya J.G., Goo L., Pinsky B.A., Quake S.R., Einav S. Virus-inclusive single-cell RNA sequencing reveals the molecular signature of progression to severe dengue. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E12363–E12369. doi: 10.1073/pnas.1813819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Liu L. Applications of single cell RNA sequencing to research of stem cells. World J. Stem Cell. 2019;11:722–728. doi: 10.4252/wjsc.v11.i10.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Luo D., Zhong X., Choi J.H., Ma Y., Wang S., Mahrt E., Guo W., Stawiski E.W., Modrusan Z., Seshagiri S., Kapur P., Hon G.C., Brugarolas J., Wang T. SCINA: a semi-supervised subtyping algorithm of single cells and bulk samples. Genes. 2019;10:E531. doi: 10.3390/genes10070531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang D., Peng M., Tang L., Ouyang J., Xiong F., Guo C., Tang Y., Zhou Y., Liao Q., Wu X., Wang H., Yu J., Li Y., Li X., Li G., Zeng Z., Tan Y., Xiong W. Single-cell RNA sequencing in cancer research. J. Exp. Clin. Cancer Res. 2021;40:81. doi: 10.1186/s13046-021-01874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Wang M., Cao Jing, Shen J., Zhou X., Wang D., Cao Jimin. Flavivirus: from structure to therapeutics development. Life. 2021;11:615. doi: 10.3390/life11070615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.N., You Y., Cui X.M., Gao H.X., Wang G.L., Zhang S.B., Yao L., Duan L.J., Zhu K.L., Wang Y.L., Li L., Lu J.H., Wang H.B., Fan J.F., Zheng H.W., Dai E.H., Tian L.Y., Ma M.J. Single-cell immune profiling reveals distinct immune response in asymptomatic COVID-19 patients. Signal Transduct. Targeted Ther. 2021;6:342. doi: 10.1038/s41392-021-00753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., Leonhardt H., Heyn H., Hellmann I., Enard W. Comparative analysis of single-cell RNA sequencing methods. Mol Cell. 2017;65:631–643.e4. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020;14:185–192. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]