Abstract
The mortality of patients with severe pneumonia caused by H1N1 infection is closely related to viral replication and cytokine storm. However, the specific mechanisms triggering virus replication and cytokine storm are still not fully elucidated. Here, we identified hypoxia inducible factor-1α (HIF-1α) as one of the major host molecules that facilitates H1N1 virus replication followed by cytokine storm in alveolar epithelial cells. Specifically, HIF-1α protein expression is upregulated after H1N1 infection. Deficiency of HIF-1α attenuates pulmonary injury, viral replication and cytokine storm in vivo. In addition, viral replication and cytokine storm were inhibited after HIF-1α knockdown in vitro. Mechanistically, the invasion of H1N1 virus into alveolar epithelial cells leads to a shift in glucose metabolism to glycolysis, with rapid production of ATP and lactate. Inhibition of glycolysis significantly suppresses viral replication and inflammatory responses. Further analysis revealed that H1N1-induced HIF-1α can promote the expression of hexokinase 2 (HK2), the key enzyme of glycolysis, and then not only provide energy for the rapid replication of H1N1 virus but also produce lactate, which reduces the accumulation of the MAVS/RIG-I complex and inhibits IFN-α/β production. In conclusion, this study demonstrated that the upregulation of HIF-1α by H1N1 infection augments viral replication and cytokine storm by cellular metabolic reprogramming toward glycolysis mainly through upregulation of HK2, providing a theoretical basis for finding potential targets for the treatment of severe pneumonia caused by H1N1 infection.
Keywords: H1N1, Severe pneumonia, Virus replication, Hypoxia inducible factor-1α, Glycolysis
Highlights
-
•
HIF-1α protein expression is upregulated after H1N1 infection.
-
•
HIF-1α is one of the major host molecules that facilitates H1N1 replication and promotes cytokine storm.
-
•
Mortality was decreased in HIF-1α knockdown mice infected with H1N1 compared to wild-type H1N1-infected mice.
-
•
This study explores the mechanism of HIF-1α, which regulates glucose metabolism reprogramming in alveolar epithelial cells.
1. Introduction
The avian influenza H1N1 virus has overcome species barriers and causes severe infections in humans. H1N1 infection usually presents with severe pneumonia, often complicated by acute respiratory distress syndrome and multiorgan failure (Gran et al., 2013; Pleschka, 2013; Iuliano et al., 2018). More importantly, in certain scenarios, H1N1 has been found to be resistant to currently approved anti-influenza drugs (such as neuraminidase inhibitors) (Li et al., 2015; Zumla et al., 2016). Therefore, an in-depth study of the mechanism by which H1N1 induces severe pneumonia would provide a key theoretical basis for identifying new targets for the treatment of H1N1 infection.
Current studies have shown that H1N1 first utilizes its surface protein hemagglutinin (HA) to bind to sialic acid residues on the alveolar epithelial cell membrane (Herold et al., 2015), after which the virus invades the alveolar epithelial cell through endocytosis and robustly replicates. H1N1 virus replication is the key initiating factor of severe pneumonia (Short et al., 2016). Specifically, H1N1 virions undergo uncoating, biosynthesis, assembly and release from host cells to complete their process of self-replication after infecting alveolar epithelial cells (Shapira et al., 2009). H1N1 viral RNA can activate host immune response pathways such as the retinoic acid-inducible gene I (RIG-I), Toll-like receptor (mainly TLR3 and TLR7) and the NLRP3 inflammasome pathways, which is followed by the production of proinflammatory factors, chemokines and type I interferons (IFN-α/β) (Murdaca et al., 2021). However, damage to the alveolar epithelial–endothelial barrier is often caused by a continuous and uncontrollable storm of inflammatory factors, which results in abnormal accumulation of proteinaceous edema fluid containing fibrin, red blood cells and inflammatory cells in the alveolar cavity, accompanied by reduced alveolar gas exchange, which eventually causes serious respiratory failure and progresses to severe pneumonia (Iwasaki and Pillai, 2014; Beljanski et al., 2015).
Severe pneumonia induced by H1N1 infection can cause severe breathing difficulties and refractory hypoxemia (Sarda et al., 2019). Studies have found that hypoxia inducible factor-1α (HIF-1α) is an important transcription factor activated in cells to adapt to hypoxic conditions (Akman et al., 2021). Under normoxic conditions, HIF-1α is quickly hydroxylated by oxygen-dependent prolyl hydroxylase (PHD) and further degraded in the proteasome after being ubiquitinated. Under hypoxic conditions, HIF-1α cannot be hydroxylated due to the reduced activity of PHD. The stabilized HIF-1α protein is then transferred to the nucleus, where it associates with HIF-1β and the cofactor P300/CBP to promote the transcription of downstream genes (Triner and Shah, 2016). The activity of HIF-1α is also affected by hypoxia-inducible factor inhibitor-1 (FIH-1). FIH-1 can hydroxylate the asparagine residue at position 803 of HIF-1α and then weaken the binding of HIF-1α to the cofactor P300/CBP. Therefore, the activity of HIF-1α is inhibited. Notably, multiple studies have shown that the expression of HIF-1α can also be upregulated under normoxic conditions (Koyasu et al., 2018). A host-based whole-genome sequencing study identified the potential involvement of the HIF-1 transcription factor and the IFN-γ pathway in the pathogenesis of severe influenza A (H1N1) infection (Li et al., 2021). H1N1 infection can cause lung and intestinal injury and imbalance of the intestinal flora. Houttuynia cordata polysaccharide, an important herbal medicine extracted from Houttuynia cordata, was found to significantly inhibit the expression of HIF-1α, reduce the quantity of mucosubstances in goblet cells and restore the level of zonula occludens-1 in the intestine (Chen et al., 2019). It was found that mortality in H1N1-infected AANAT−/− melatonin-deficient mice was significantly higher than that in wild-type mice. Additionally, the administration of melatonin significantly reduced mortality caused by H1N1 infection. These results suggested a protective effect of melatonin against H1N1 infection. Further study revealed that melatonin downregulated gene expression in the HIF-1 pathway and inhibited the H1N1 infection-induced release of proinflammatory cytokines from mast cells (Huo et al., 2023). HIF-1α was found to be responsible for the glycolysis and oxidative phosphorylation-induced germinal center response and alterations in follicular helper T-cell differentiation after H1N1 infection. Blocking glycolysis and upregulating oxidative phosphorylation activity significantly restored follicular helper T-cell differentiation after H1N1 infection and ameliorated inflammatory damage in mice (Dong et al., 2019). In our previous studies, we found that H1N1 infection could reduce the degradation of HIF-1α by inhibiting the function of the host cell proteasome and the expression of FIH-1 (Guo et al., 2017; Ren et al., 2019). However, the mechanism of action of HIF-1α in H1N1-infected alveolar epithelial cells is still unclear.
In this study, we found that HIF-1α promotes the expression of key glycolytic enzymes, which induces a shift in host cell glucose metabolism to glycolysis. As a result, on the one hand, this shift provides energy for viral replication. On the other hand, the lactate produced via glycolysis directly targets and binds to MAVS, abolishing the mitochondrial localization of MAVS and the formation of the MAVS/RIG-I complex, thereby inhibiting the production of downstream IFN-α/β and blocking the host cell innate immune response, which together increase viral replication and pathogenicity. Consequently, the continuous uncontrollable cytokine storm aggravates lung damage, leading to severe pneumonia.
2. Materials and methods
2.1. Animals and treatment
Surfactant protein C (SPC) is mainly expressed in type II alveolar epithelial cells. Spc-Cre mice and HIF-1αfl/fl mice were generated on a C57BL/6 background. Spc-Cre mice and HIF-1αfl/fl mice were purchased from Shanghai Model Organisms Center, Inc. The sequences of the HIF-1αfl/fl genotyping primers were 5′-GAAGTAAGCACCTGGAAGTAGTAG-3′ (forward) and 5′-CAATCATTCCAAAGTATCTCAGTA-3′ (reverse). The sequences of the Spc-Cre genotyping primers were 5′-TTGGCCTTGGCTGAGCTTAGACAT-3′ (forward) and 5′-CCTTCACGACATTCAACAGACCTT-3′ (reverse). SPF-grade 6- to 8-week-old male C57BL/6 mice (weighing 18–20 g) were purchased from Shanghai Sippr-BK Laboratory Animal Corp., Ltd. The mouse HIF-1α shRNA AAV6 was constructed by Shandong ViGene Biosciences (Jinan, Shandong, China). The negative control was AAV6-U6-GFP with insertion of a nonsense sequence. C57BL/6 mice were anesthetized and administered HIF-1α shRNA AAV6 or the negative control by nasal drops (30 μL per nostril, 1.0 × 1012 vg/mL). Changes in HIF-1α expression in lung tissues were detected after continued feeding for approximately two weeks to verify the efficiency of HIF-1α inhibition. All animals were maintained in the Animal Experimental Center of the Shanghai General Hospital Affiliated with Shanghai Jiaotong University. All procedures in this investigation were approved by the Animal Care and Use Committee of the Research Institute at the Shanghai General Hospital Affiliated with Shanghai Jiaotong University.
The H1N1 influenza virus strain A/PR/8/34 was a gift from Prof. Haikun Wang of the Shanghai Pasteur Institute of the Chinese Academy of Sciences. The H1N1 virus was subcultured in chicken embryos for two passages. The allantoic fluid then was filtered and stored at −80 °C for later use. The mice were divided into a blank control group, oseltamivir (OST; YiChang HEC ChangJiang Pharmaceutical Co., Ltd.) groups (OST L, 30 mg/kg, intragastric; OST H, 60 mg/kg, intragastric), shHIF-1α group, and 2-DG (Sigma–Aldrich) group (500 mg/kg, intraperitoneal) (n = 5 mice per group). After anesthetization by intraperitoneal injection of Zoletil 50 (Shanghai, China), the mice were administered the corresponding dose (1000 pfu) of H1N1 virus via nasal drops. The mice were then placed in an isolation cage with an independent air supply, reared normally, and sacrificed at the indicated time. The lung tissues, bronchoalveolar lavage fluid and serum of the mice were collected and analyzed.
2.2. Cell culture and treatment
The human lung cancer cell line (A549) was purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences. Although A549 cells are a type of cancer cell, studies have shown that A549 cells still retain the characteristics of alveolar epithelial cells. In addition, A549 cells are widely used in lung disease research (Foster et al., 1998; Hittinger et al., 2015; Zhou et al., 2015). Thus, we used A549 cells as alveolar epithelial cells in this study. The cells were cultured in F-12K medium containing 10% fetal bovine serum (Gibco, Grand Island, NY, USA) with 5% CO2 at 37 °C. For H1N1 virus infection, the experiments were carried out in the biological safety cabinet of the BSL-2 laboratory. Briefly, A549 cells were plated in a 6-well plate and incubated overnight. The cells were then incubated with H1N1 virus (MOI = 1) for 2 h with shaking of the plate once every 20–30 min, washed 2 times with PBS, treated with virus maintenance solution (F-12K + 25 mmol/L HEPES + 1% double antibiotics + 0.25 μg/mL TPCK-treated trypsin) and incubated for an additional 24 h. The infected cells were then ready for further analysis. Sodium l-lactate was purchased from Sigma–Aldrich (St. Louis, MO, USA). The lactate group was incubated with 10 mmol/L lactate for 24 h.
2.3. Cell transfection
siRNAs targeting HIF-1α or HK2 were generated by GenePharma (Shanghai, China). The HK2 plasmid was obtained from Genechem (Shanghai, China). The vector used to construct the HK2 plasmid was GV230 containing the XhoI/KpnI restriction site (Genechem, Shanghai, China). The negative control vector was CON083 (CMV-MCS-EGFP-SV40-Neomycin). For transfection, A549 cells were plated in 6-well plates at a concentration of 2 × 105 cells/well. After the cell confluence reached approximately 70%, HIF-1α siRNA (100 pmol per well), HK2 siRNA (100 pmol per well) or HK2 plasmid (2 μg per well), as indicated, were transfected with Lipofectamine 2000 (Thermo Fisher Scientific, Grand Island, NY, USA) according to the manufacturer's instructions. After 24 h of transfection, the cells were incubated with H1N1 virus (MOI = 1) for 24 h. The sequences of the HIF-1α siRNA, HK2 siRNA and HK2 plasmid can be found in Supplementary Table S1.
2.4. Real-time quantitative PCR
Mouse lung tissues and A549 cells were lysed, and total RNA was extracted with TRIzol (Thermo Fisher Scientific, Grand Island, NY, USA). The extracted RNA was reverse-transcribed into cDNA by using Prime Script™ RT Master Mix (TAKARA, Dalian, Liaoning, China). Real-time PCR was performed with a SYBR® Premix Ex Taq™ Kit (TAKARA, Dalian, Liaoning, China) for measurement of the mRNA expression levels of HIF-1α, HK2, M, IFN-α and IFN-β in an Applied Biosystems QuantStudio 6 Flex system by Life Technologies. β-Actin was used as the internal control. Relative expression levels were calculated by the 2–▵▵Ct method and normalized to β-actin expression levels. The primer sequences for amplification of each target gene and the internal reference gene are shown in Supplementary Table S2.
2.5. Western blotting
The lung tissues were ground into powder in liquid nitrogen. The lung tissue powder and A549 cells were lysed in RIPA buffer (Beyotime Biotechnology, Shanghai, China) supplemented with protease inhibitors and were then stored at −80 °C. Nuclear and cytoplasmic proteins were extracted with NE-PER nuclear and cytoplasmic extraction reagents (Thermo Fisher Scientific Inc., Grand Island, NY, USA) according to the instructions. The concentration of the lysates was measured with a BCA Protein Assay Kit (Beyotime Biotechnology, Shanghai, China), and 30 μg of protein from each lysate sample was separated by SDS‒PAGE (Beyotime Biotechnology, Shanghai, China). The membranes were blocked in 5% nonfat milk at 25 °C for 1 h and incubated with primary antibodies targeting HIF-1α (1:500, Source: rabbit, Catalog: BS3514, Bioworld Technology, Inc., St. Louis Park, MN, USA), HK2 (1:1000, Source: rabbit, Catalog: ab209847, Abcam, Cambridge, MA, USA), NP (1:1000, Source: rabbit, Catalog: GTX125989, GeneTex, San Antonio, TX, USA), IFN-α (1:500, Novusbio, Source: rabbit, Catalog: NBP2-75930, Littleton, CO, USA), IFN-β (1:500, Thermo Fisher Scientific, Source: rabbit, Catalog: PA5-20390, Grand Island, NY, USA), MAVS (1:1000, Source: rabbit, Catalog: ab31334, Abcam, Cambridge, MA, USA), RIG-I (1:500, Source: mouse, Catalog: sc-376845, Santa Cruz, CA, USA), Lamin B1 (1:1000, Source: mouse, Catalog: 33-2000, Thermo Fisher Scientific Inc., Grand Island, NY, USA) or β-actin (1:1000, Source: mouse, Catalog: AF0003, Beyotime Biotechnology, Shanghai, China), respectively, overnight at 4 °C. The next day, the membranes were washed and incubated with a horseradish peroxidase-conjugated secondary antibody (1:2000, Beyotime Biotechnology, Shanghai, China) for an additional 1 h at 25 °C. The signals of each protein were visualized by ECL (Millipore, Billerica, MA, USA) with a Tanon 5200 imaging system. β-Actin was used as an internal control.
2.6. Immunofluorescence analysis
A549 cells were plated in a glass-bottom culture dish and treated as indicated. The treated cells were rinsed with PBS 3 times, fixed with 4% paraformaldehyde for 10 min at 25 °C, rinsed with PBS 3 times, permeabilized with 0.1% Triton X-100 (Sigma–Aldrich, St. Louis, MO, USA) for 15 min and blocked with 5% BSA blocking solution (Beyotime Biotechnology, Shanghai, China) for 60 min at 25 °C. The cells were then incubated with a primary antibody (anti-HIF-1α, 1:100; anti-NP, 1:100; anti-MAVS, 1:100; anti-RIG-I, 1:100) diluted with Immunol staining primary antibody dilution buffer (Beyotime Biotechnology) overnight at 4 °C. The incubated cells were rinsed with PBS three times and stained with a diluted fluorescently labeled secondary antibody (1:200) at 25 °C for 2 h. Nuclei were stained with diluted DAPI (Beyotime Biotechnology, Shanghai, China) for 5 min. The stained cells were then observed and imaged by using a confocal microscope (Leica TCS SP8, Leica). For mouse lung tissues, frozen sections were prepared and subjected to immunofluorescence staining using procedures similar to those mentioned above.
2.7. Enzyme-linked immunosorbent assay (ELISA)
ELISA kits for human or mouse IL-1β, IL-6 and TNF-α were purchased from Anogen (Beijing, China). An ELISA kit for human HIF-1α was purchased from Sigma–Aldrich (St. Louis, MO, USA). Mouse IFN-α and IFN-β ELISA kits were purchased from Neobioscience. Serum, alveolar lavage fluid, and A549 cell culture supernatant were prepared and subjected to ELISA according to the manufacturer's instructions.
2.8. Hematoxylin–eosin (HE) staining
The left lower lung lobes were isolated and fixed with 4% paraformaldehyde (Servicebio, Wuhan, China). The fixed tissues were then dehydrated with 20% sucrose overnight. The lung tissues were then embedded in paraffin, sliced into 4 μm sections, and subjected to HE staining. The stained sections were observed and imaged with a 100× optical microscope (Leica DM5500 B).
2.9. Lactate and ATP measurement
The lactate concentration in mouse serum and A549 cell culture supernatant was determined using a lactic acid assay kit (Nanjing Jiancheng Bioengineering Institute, Nanjing, China). Mouse lung tissues and A549 cells were lysed according to the manufacturer's instructions for ATP measurement using an ATP assay kit (Beyotime Biotechnology, Shanghai, China). Each sample was measured in technical triplicate.
2.10. Coimmunoprecipitation (Co-IP)
Mouse lung tissues and A549 cells were lysed in RIPA buffer as mentioned for Western blotting. The lysates were centrifuged at 12,000×g and 4 °C for 15 min. Supernatants were collected, and 400 μg lysate samples were incubated with 2 μg of an anti-MAVS primary antibody overnight at 4 °C in a rotator. Then, 20 μL Protein A/G agarose (Beyotime Biotechnology, Shanghai, China) was added and incubated for an additional 3 h at 4 °C, after which the lysates were washed 5 times with PBS and subjected to Western blot analysis with primary antibodies specific for MAVS (Abcam, Source: rabbit, Catalog: ab31334, Abcam, Cambridge, MA, USA) and RIG-I (Santa Cruz, Source: mouse, Catalog: sc-376845, Santa Cruz, CA, USA).
2.11. Statistical analysis
SPSS 22.0 and GraphPad Prism 8.0 software were used for statistical analysis. Each experiment was repeated at least three times independently. All data are presented as the means ± standard deviations. Student's t-test and one-way analysis of variance (ANOVA) were performed, and P < 0.05 was considered to indicate a statistically significant difference.
3. Results
3.1. HIF-1α knockdown attenuates virus replication and cytokine storm in H1N1-infected mice
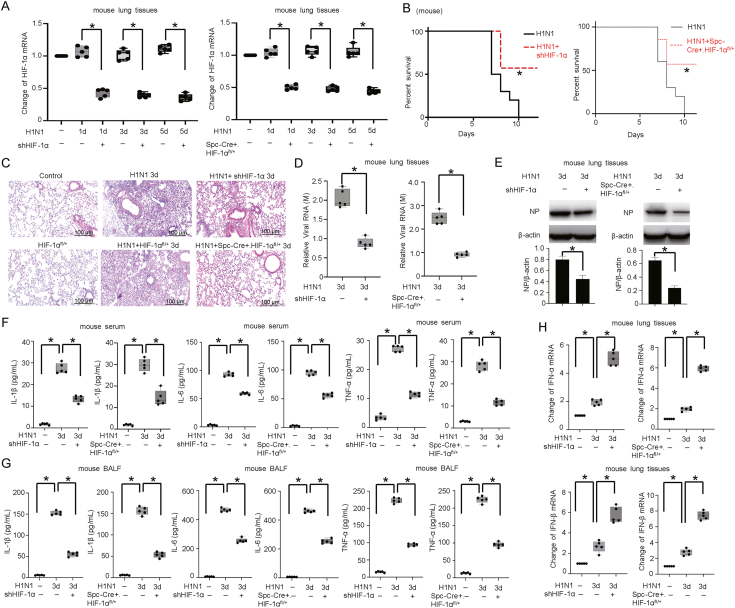
To understand the correlation between H1N1 infection and infection-induced hypoxic conditions, we utilized the H1N1 infection mouse model. The lung tissues of infected mice showed obvious inflammatory cell infiltration, pulmonary interstitial edema and increased alveolar exudation, which peaked on the 3rd day after H1N1 infection (Supplementary Fig. S1A). No significant changes were observed in the mRNA expression of HIF-1α after H1N1 infection in either lung tissue or A549 cells (Supplementary Fig. S1B). HIF-1α protein exhibited significantly increased expression and accumulated in the nucleus after H1N1 infection in A549 alveolar epithelial cells (Supplementary Fig. S1C, S1D). To explore the role of HIF-1α in H1N1 virus infection, we employed Spc-Cre+HIF-1αfl/+ C57BL/6 mice and adeno-associated viral shRNA transduction for HIF-1α silencing (Fig. 1A, Supplementary Fig. S2). HIF-1α knockdown mice showed decreased mortality compared to wild-type H1N1-infected mice after H1N1 infection (Fig. 1B). HIF-1α knockdown also attenuated inflammatory cell infiltration and tissue edema in H1N1-infected mice, as shown by histopathological analysis (Fig. 1C). These results suggest that knockdown of HIF-1α ameliorates H1N1-induced pulmonary injury.
Fig. 1.
HIF-1α knockdown attenuates viral replication and cytokine storm in H1N1-infected mice. A The expression of HIF-1α mRNA after H1N1 infection (1000 pfu per mouse) in lung tissues of mice with or without HIF-1α knockdown (n = 5) was measured by qPCR. B The survival of H1N1 (1000 pfu per mouse, n = 10)-infected mice with or without HIF-1α knockdown. C HE staining of lung tissue from H1N1-infected (1000 pfu per mouse) mice with or without HIF-1α knockdown on Day 3 after infection (scale bar = 100 μm) (n = 5). D, E The expression of viral RNA (M) and NP protein in the lung tissues was measured by qPCR and Western blotting on Day 3 after infection (n = 5). β-Actin served as the loading control. F, G The concentrations of IL-1β, IL-6 and TNF-α in mouse serum and BALF on Day 3 (n = 5) were measured by ELISA. H, I The amounts of IFN-α/β in lung tissues and serum on Day 3 were measured by qPCR and ELISA (n = 5). J The levels of IFN-α/β in lung tissues from the Day 1, 3, and 5 groups or Day 3 group were measured by Western blotting (n = 5). β-Actin served as the loading control. The data are presented as the means with SDs. Statistical analysis was performed by Student's t-test and one-way ANOVA. ∗P < 0.05.
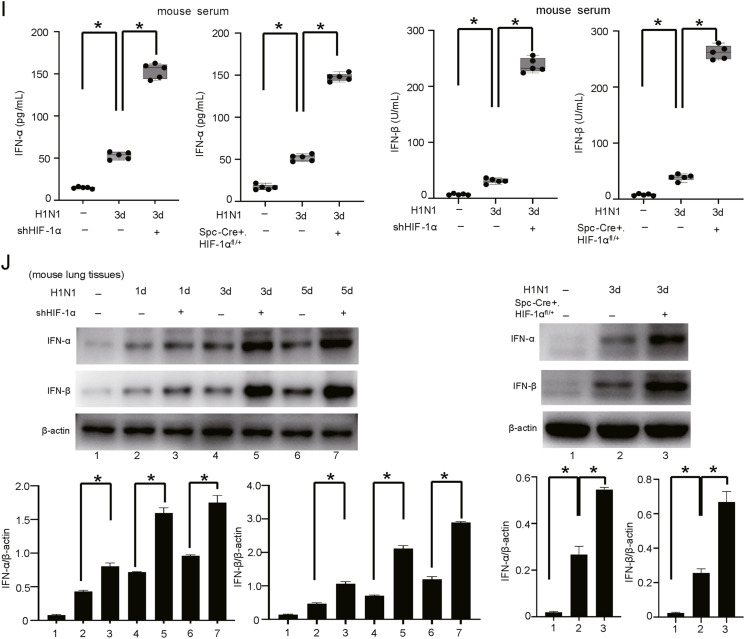
The mortality of patients with severe pneumonia caused by H1N1 virus is closely related to viral replication and cytokine storm (Herold et al., 2015; Murdaca et al., 2021). We observed that HIF-1α knockdown significantly reduced the expression of viral RNA (M gene) and the nucleocapsid (NP) protein (Fig. 1D and E) in vivo. We then investigated the levels of IL-1β, IL-6 and TNF-α in H1N1-infected mice. Our results showed that the concentrations of proinflammatory cytokines (IL-1β, IL-6 and TNF-α) were significantly increased in the serum and bronchoalveolar lavage fluid (BALF) of wild-type H1N1-infected mice. Knockdown of HIF-1α significantly decreased the concentration of proinflammatory cytokines after infection (Fig. 1F and G). As innate immunity is the host's first line of defense against viral infection, and type I interferon (IFN-I) is an important effector molecule involved in the antiviral immune response (Samuel, 2001; Haller and Kochs, 2002; Krug et al., 2003), we further monitored the expression of IFN-α/β, the major cytokines in the defense against viral infection. In response to H1N1 infection, the levels of IFN-α/β in mouse lung tissues and serum were increased, and the degree of IFN-α/β secretion was further significantly increased in mice with HIF-1α knockdown (Fig. 1H–J). These results demonstrated that HIF-1α is important for H1N1 replication and cytokine storm induced by H1N1 infection in mice.
3.2. HIF-1α regulates H1N1 replication and the host inflammatory response in vitro
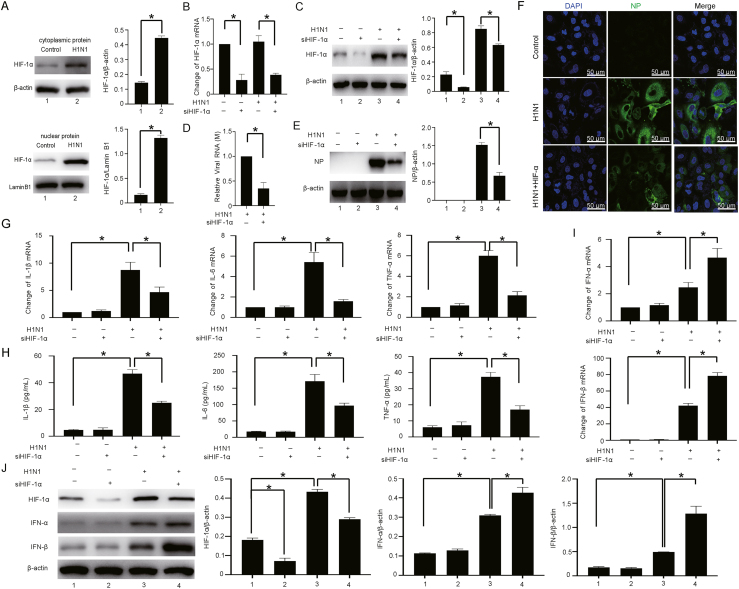
In this research, we also confirmed that H1N1 virus infection upregulates the expression of HIF-1α and promotes HIF-1α translocation to the nucleus in A549 cells (Fig. 2A), as shown in our previous studies (Guo et al., 2017). To verify the potential roles of HIF-1α in the modulation of H1N1 replication and the host inflammatory response in vitro, siRNA targeting HIF-1α was used to knock down the expression of HIF-1α in A549 cells (Fig. 2B and C). We found that HIF-1α silencing significantly reduced the expression of the viral M gene (Fig. 2D). Western blot and immunofluorescence analyses further revealed that HIF-1α silencing significantly decreased the protein level of viral NP in H1N1-infected A549 cells (Fig. 2E and F). The mRNA levels of proinflammatory factors, including IL-1β, IL-6 and TNF-α, were significantly increased after H1N1 infection. However, the mRNA levels of these proinflammatory factors (IL-1β, IL-6 and TNF-α) were strikingly reduced in HIF-1α deficient cells (Fig. 2G). A similar result was observed in the cell culture supernatant by ELISA (Fig. 2H). We also found that the mRNA and protein levels of IFN-α/β were significantly increased after H1N1 virus infection, while the increases in the IFN-α/β levels were further enhanced by HIF-1α inhibition (Fig. 2I and J). Taken together, these results suggest that HIF-1α plays a role in viral replication and the host inflammatory response in A549 alveolar epithelial cells.
Fig. 2.
HIF-1α regulates H1N1 replication and the host inflammatory response in vitro. A The level of HIF-1α protein in the cytosolic and nuclear fractions after H1N1 virus infection. A549 cells were treated with H1N1 virus (MOI = 1) for 24 h. Then, the cytosolic and nuclear fractions were extracted separately. The level of HIF-1α protein in the cytosolic and nuclear fractions was measured by Western blotting. β-Actin and Lamin B1 served as the loading controls. B, C The expression levels of HIF-1α mRNA and protein in H1N1-infected A549 cells (MOI = 1 for 24 h) transfected with or without siRNA targeting HIF-1α were measured by qPCR and Western blotting. β-Actin served as the loading control. D, E The expression of viral M mRNA and NP protein in H1N1-infected A549 cells (MOI = 1 for 24 h) transfected with or without siRNA targeting HIF-1α were measured by qPCR and Western blotting. β-Actin served as the loading control. F Immunofluorescence analysis of NP protein in H1N1-infected A549 cells (MOI = 1 for 24 h) transfected with or without siRNA targeting HIF-1α (scale bar = 50 μm). G The expression of IL-1β, IL-6 and TNF-α mRNA in H1N1-infected A549 cells (MOI = 1 for 24 h) transfected with or without siRNA targeting HIF-1α was measured by qPCR. H The levels of IL-1β, IL-6 and TNF-α in the culture supernatant of H1N1-infected A549 cells (MOI = 1 for 24 h) after transfection with or without siRNA targeting HIF-1α were measured by ELISA. I, J The protein expression of IFN-α/β and HIF-1α in H1N1-infected A549 cells (MOI = 1 for 24 h) after transfection with or without siRNA targeting HIF-1α was measured by Western blotting. β-Actin served as the loading control. The data are presented as the means with SDs. Statistical analysis was performed by Student's t-test and one-way ANOVA. ∗P < 0.05.
3.3. HIF-1α promotes H1N1 viral replication and the host inflammatory response by regulating HK2-mediated glycolysis
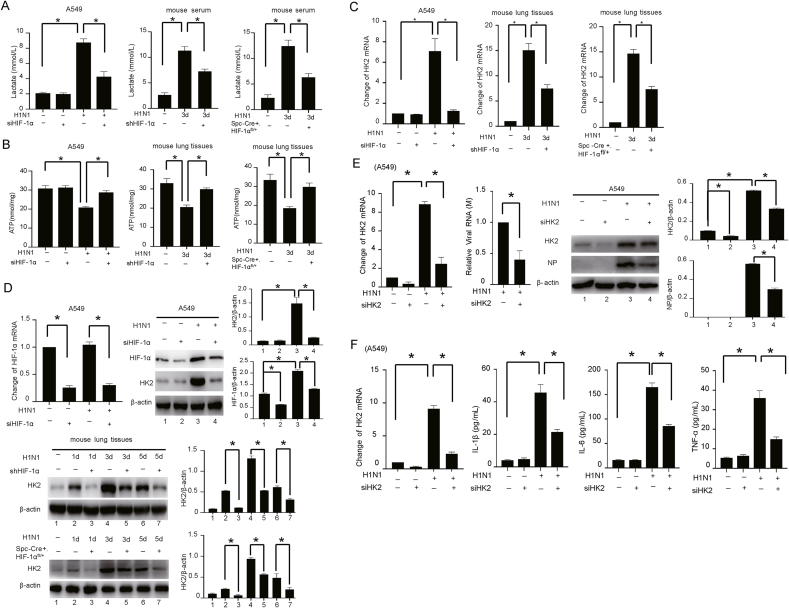
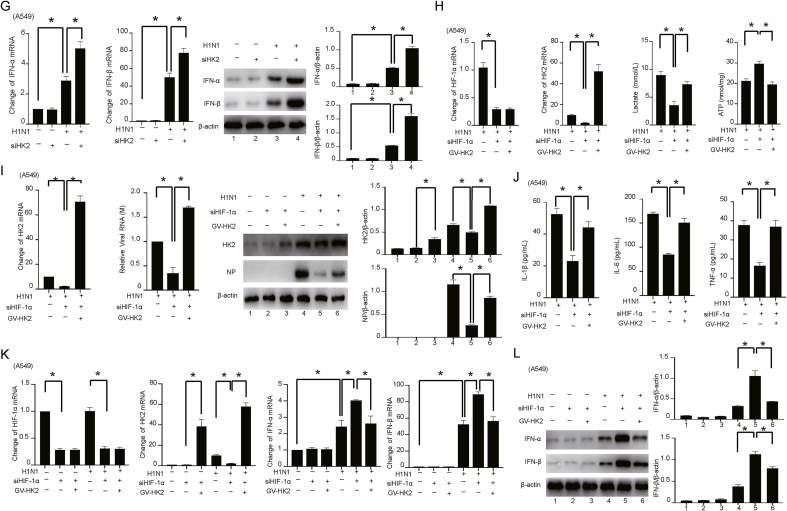
Metabolic reprogramming is an important mechanism for the regulation of viral replication (Fontaine et al., 2015; Chuang et al., 2017; Passalacqua et al., 2019). The rapid replication of H1N1 virus depends on the organelles, energy supply and biosynthetic materials of the host cells (Watanabe et al., 2010). Studies have shown that HIF-1α plays an important role in regulating glycolysis (Papandreou et al., 2006; Zhang et al., 2016). However, the role and mechanism of HIF-1α in H1N1-induced glycolysis are still unclear. We found that HIF-1α silencing attenuated the H1N1-induced lactate production and increased ATP level in vivo and in vitro (Fig. 3A and B). Similar patterns of changes in ATP, ADP and AMP levels were also detected by HPLC (Supplementary Fig. S3). To identify the potential critical enzymes during H1N1-induced glycolysis mediated by HIF-1α, we used qPCR to determine the expression profiles of glycolytic enzymes in cells with or without HIF-1α silencing. Our study showed that the expression of multiple metabolic enzymes, especially HK2, was increased after H1N1 infection (Supplementary Fig. S4). Importantly, knockdown of HIF-1α reversed the increases in the mRNA and protein levels of HK2 induced by H1N1 (Fig. 3C and D).
Fig. 3.
HIF-1α promotes H1N1 viral replication and the host inflammatory response by regulating HK2-mediated glycolysis. A The levels of lactate in H1N1-infected mouse serum (n = 5, 1000 pfu per mouse) and culture supernatant of A549 cells (MOI = 1 for 24 h) with or without HIF-1α knockdown were measured by colorimetry according to the instructions of the kit. B The levels of ATP in H1N1-infected mouse lung tissues (n = 5, 1000 pfu per mouse) or A549 cells (MOI = 1 for 24 h) with or without HIF-1α knockdown were measured by colorimetry according to the instructions of the kit. C, D The mRNA and protein expression of HIF-1α and HK2 in H1N1-infected mouse lung tissues (n = 5, 1000 pfu per mouse) or A549 cells (MOI = 1 for 24 h) with or without HIF-1α knockdown were detected by qPCR and Western blotting. β-Actin served as the loading control. E The expression of HK2 and viral M and NP in H1N1-infected A549 cells (MOI = 1 for 24 h) transfected with siRNA targeting HK2 were measured by qPCR and Western blotting. F The expression of HK2 mRNA in A549 cells transfected with or without siRNA targeting HK2 was measured by qPCR. The concentrations of IL-1β, IL-6 and TNF-α in the culture supernatant of A549 cells transfected with or without siRNA targeting HK2 were measured by ELISA. G The expression levels of IFN-α/β mRNA and protein in H1N1-infected A549 cells (MOI = 1 for 24 h) transfected with or without siRNA targeting HK2 were measured by qPCR and Western blotting. β-Actin served as the loading control. H The levels of HIF-1α mRNA, HK2 mRNA and ATP in H1N1-infected A549 cells (MOI = 1 for 24 h) and lactate in the culture supernatant were measured by qPCR or colorimetry according to the kit instructions after transfection of siRNA targeting HIF-1α together with ectopic expression of HK2 as indicated. I The expression levels of HK2 mRNA, viral M mRNA and NP protein in H1N1-infected A549 cells (MOI = 1 for 24 h) with transfection of siRNA targeting HIF-1α alone or together with ectopic expression of HK2, as indicated, were measured by qPCR and Western blotting. J The concentrations of IL-1β, IL-6 and TNF-α in the culture supernatant of H1N1-infected A549 cells (MOI = 1 for 24 h) with transfection of siRNA targeting HIF-1α alone or together with ectopic expression of HK2, as indicated, were measured by ELISA. K, L The expression levels of HIF-1α mRNA, HK2 mRNA, IFN-α/β mRNA and protein in H1N1-infected A549 cells (MOI = 1 for 24 h) with transfection of siRNA targeting HIF-1α alone or together with ectopic expression of HK2, as indicated, were measured by qPCR and Western blotting. β-Actin served as the loading control. The data are presented as the means with SDs. Statistical analysis was performed by Student's t-test and one-way ANOVA. ∗P < 0.05.
To reveal whether HIF-1α activation promotes glycolysis by regulating HK2 during H1N1 infection, a siRNA targeting the backsplice sequence of HK2 (siHK2) was adopted. As expected, siHK2 transfection significantly downregulated HK2 expression and attenuated the elevation of the M gene mRNA and NP protein levels in H1N1-infected epithelial cells (Fig. 3E). Similarly, inhibition of HK2 also significantly suppressed H1N1-induced production of proinflammatory factors (IL-1β, IL-6 and TNF-α) (Fig. 3F). The mRNA and protein expression levels of IFN-α/β were significantly increased after HK2 inhibition (Fig. 3G). Collectively, these results indicate that HK2, a key enzyme of glycolysis, is involved in the pathogenic process of H1N1 infection.
To determine the role of the HIF-1α/HK2 axis in H1N1 infection, a plasmid for HK2 overexpression was used in HIF-1α-silenced A549 alveolar epithelial cells. There was no influence on cell viability after transfection with HIF-1α siRNA and the GV-HK2 plasmid (Supplementary Fig. S5). We found that HK2 overexpression significantly reversed the changes in lactate and ATP levels caused by HIF-1α silencing (Fig. 3H, Supplementary Fig. S6, S7A). Thus, the levels of viral RNA (M gene), the NP protein and proinflammatory factors (IL-1β, IL-6 and TNF-α), which were reduced by HIF-1α silencing, were increased after HK2 overexpression (Fig. 3I and J). HK2 overexpression significantly reduced the increase in IFN-α/β expression caused by inhibition of HIF-1α (Fig. 3K and L, Supplementary Fig. S7B). Thus, HIF-1α may promote a shift in glucose metabolism to glycolysis by regulating HK2, thereby increasing viral replication and enhancing the inflammatory response.
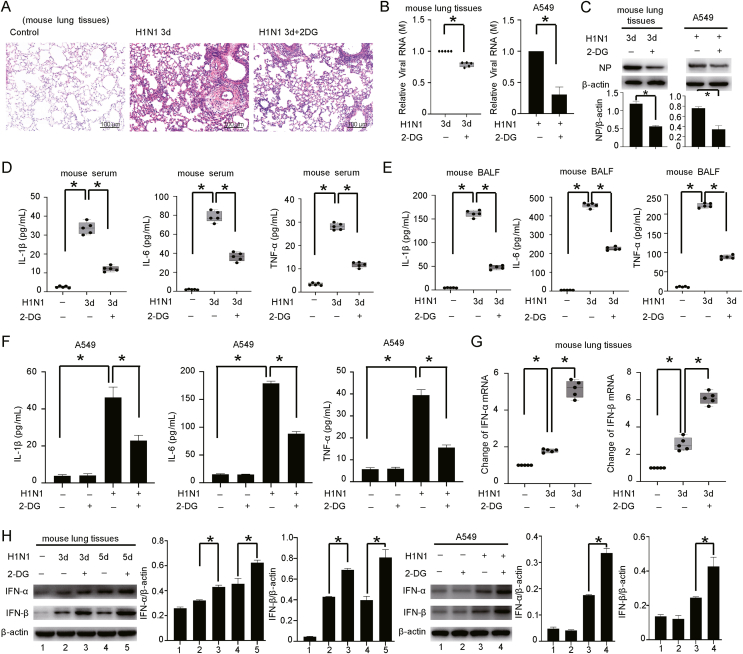
3.4. Inhibiting glycolysis reduces H1N1-induced lung injury
To verify the role of glycolysis in severe pneumonia caused by H1N1 infection, we used 2-DG to inhibit glycolysis and evaluated the changes in viral replication and the levels of host inflammatory factors. We first determined the minimum toxic concentration of 2-DG in A549 cells by a CCK-8 assay. The results showed that a minimum concentration of 20 mmol/L of 2-DG significantly decreased the viability of A549 cells after 24 and 48 h. Therefore, a concentration of 10 mmol/L 2-DG, which exhibited almost no toxicity to A549 cells over 48 h, was administrated in subsequent experiments (Supplementary Fig. S8). HE staining suggested that treatment with 2-DG significantly attenuated the lung injury (inflammatory cell infiltration and pulmonary interstitial edema) caused by H1N1 infection (Fig. 4A). Similar to the observation in our previous study (Ren et al., 2021), the addition of 2-DG to H1N1-infected A549 alveolar epithelial cells greatly reduced the levels of viral RNA (M gene) and the NP protein. Similar patterns were observed in vivo (Fig. 4B and C). In addition, administration of 2-DG significantly reversed the increases in the levels of proinflammatory factors (IL-1β, IL-6 and TNF-α) induced by H1N1 infection both in vivo (mouse serum and BALF) and in vitro (culture supernatant of A549 cells) (Fig. 4D–F). The expression levels of IFN-α/β were significantly increased by cotreatment with 2-DG (Fig. 4G and H, Supplementary Fig. S9). These results clearly suggest that glycolysis plays an important role in the replication of H1N1 virus and the host inflammatory response.
Fig. 4.
Inhibiting glycolysis attenuates H1N1-induced lung injury. A HE staining of lung tissues from H1N1-infected mice (n = 5, 1000 pfu per mouse) treated with or without 2-DG by intraperitoneal injection (500 mg/kg, once daily for 3 days). Scale bar = 100 μm. B, C The expression levels of M mRNA and NP protein in lung tissues (n = 5, 1000 pfu per mouse) and A549 cells (MOI = 1 for 24 h) infected with H1N1 virus and treated with or without 2-DG (10 mmol/L for 48 h for A549) were measured by qPCR and Western blotting. β-Actin served as the loading control. D, E The concentrations of IL-1β, IL-6 and TNF-α in the serum and BALF of H1N1-infected mice after treatment with or without 2-DG (n = 5, 1000 pfu per mouse) were measured by ELISA. F The concentrations of IL-1β, IL-6 and TNF-α in the culture supernatant of H1N1-infected A549 cells treated with or without 2-DG (10 mmol/L for 48 h) were measured by ELISA. G, H The expression of IFN-α/β in lung tissues of H1N1-infected mice (n = 5, 1000 pfu per mouse) or A549 cells (MOI = 1 for 24 h) treated with or without 2-DG (10 mmol/L for 48 h for A549) was evaluated by qPCR and Western blotting. β-Actin served as the loading control. The data are presented as the means with SDs. Statistical analysis was performed by Student's t-test and one-way ANOVA. ∗P < 0.05.
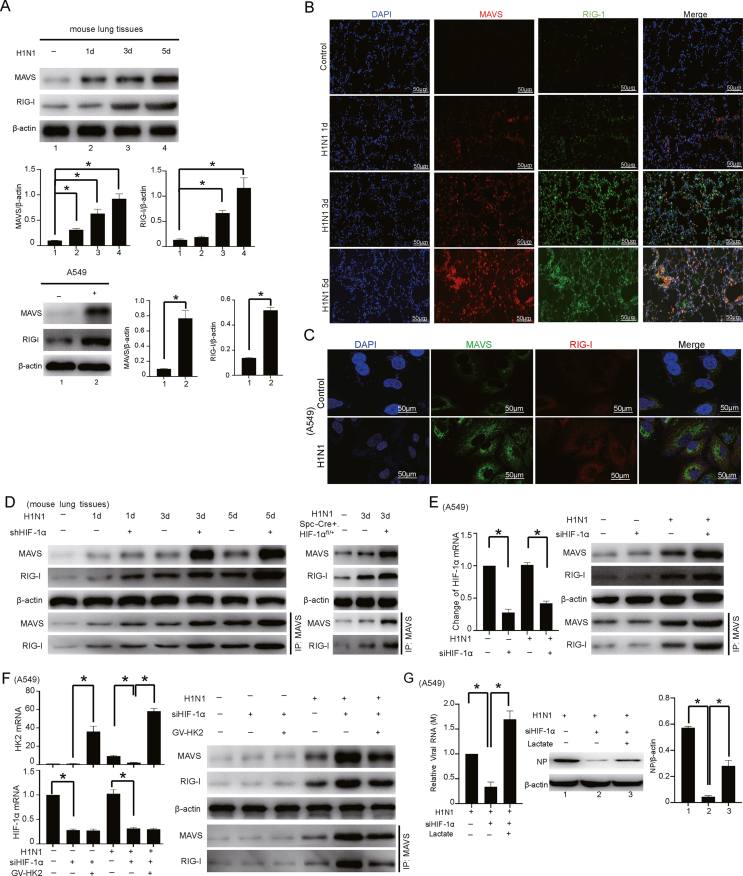
3.5. HIF-1α/HK2 axis-mediated lactate accumulation promotes H1N1 replication and the host inflammatory response through the MAVS/RIG-I signaling pathway
Lactate can directly target MAVS, which affects its correct mitochondrial localization (Thyrsted et al., 2021), further interfering with the formation of the complex between MAVS and RIG-I and ultimately preventing the activation of downstream pathways. Importantly, this effect is only stimulated by RNA viruses or intracellular RNA (Zhang et al., 2019). Our results showed that H1N1 infection activated the MAVS/RIG-I signaling pathway and that the MAVS and RIG-I protein levels increased gradually in a time-dependent manner after H1N1 infection (Fig. 5A). These two proteins shared similar patterns of abundance and subcellular localization in mouse lung tissues and in A549 cells after H1N1 infection (Fig. 5B and C).
Fig. 5.
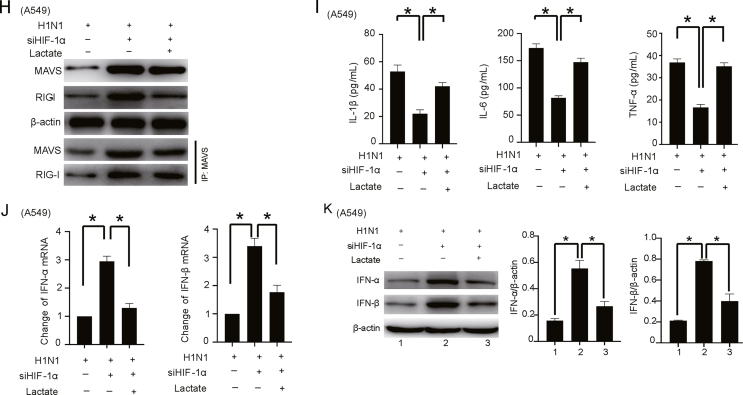
HIF-1α/HK2 axis-mediated lactate accumulation promotes H1N1 virus replication and the host inflammatory response through the MAVS/RIG-I signaling pathway. A The expression levels of MAVS and RIG-I in lung tissues (n = 5, 1000 pfu per mouse for Days 1, 3, and 5) and A549 cells (MOI = 1 for 24 h) with or without H1N1 virus infection as indicated were measured by Western blotting. B, C Immunofluorescence analysis of the expression and colocalization of MAVS and RIG-I in lung tissues of mice (n = 5, 1000 pfu per mouse, Days 1, 3, and 5) and A549 cells (MOI = 1 for 24 h) with or without H1N1 virus infection as indicated. Scale bar = 50 μm. D, E Co-IP analysis of the expression of and interaction between MAVS and RIG-I in H1N1-infected lung tissues of mice (n = 5, 1000 pfu per mouse for Days 1, 3, and 5) or A549 cells (MOI = 1 for 24 h) with or without HIF-1α knockdown. Changes in HIF-1α mRNA expression were detected with qPCR. F Co-IP analysis of the expression of and interaction between MAVS and RIG-I in H1N1-infected A549 cells (MOI = 1 for 24 h) with transfection of siRNA targeting HIF-1α alone or together with ectopic expression of HK2, as indicated. Changes in HIF-1α and HK2 mRNA were detected with qPCR. G The expression levels of M mRNA and NP protein in H1N1-infected A549 cells (MOI = 1 for 24 h) with transfection of siRNA targeting HIF-1α alone or together with exogenous lactate supplementation were measured by qPCR and Western blotting. H Co-IP analysis of the expression of and interaction between MAVS and RIG-I in H1N1-infected A549 cells (MOI = 1 for 24 h) with transfection of siRNA targeting HIF-1α alone or together with exogenous lactate supplementation. I The concentrations of IL-1β, IL-6 and TNF-α in the culture supernatant of H1N1-infected A549 cells (MOI = 1 for 24 h) with transfection of siRNA targeting HIF-1α alone or together with exogenous lactate supplementation were measured by ELISA. J, K The expression levels of IFN-α/β mRNA and protein in H1N1-infected A549 cells (MOI = 1 for 24 h) with transfection of siRNA targeting HIF-1α alone or together with exogenous lactate supplementation were measured by qPCR and Western blotting. β-Actin served as the loading control. The data are presented as the means with SDs. Statistical analysis was performed by Student's t-test and one-way ANOVA. ∗P < 0.05.
Given that the HIF-1α/HK2 axis is vital for H1N1 infection, we asked whether the increased expression of MAVS and RIG-I caused by H1N1 infection is correlated with this axis. Our results showed that H1N1 infection enhanced the interaction of MAVS and RIG-I and that knockdown of HIF-1α further enhanced this interaction in vivo and in vitro (Fig. 5D and E). To further confirm the role of HIF-1α in the interaction between MAVS and RIG-I after H1N1 infection, HEK293T cells were cotransfected with the MAVS, RIG-I or HIF-1α expression plasmid. The results shown in Supplementary Fig. S10 indicate that H1N1 infection can enhance the interaction of MAVS and RIG-I. In addition, overexpression of HIF-1α reduced the interaction of MAVS and RIG-I. Importantly, overexpression of HK2 reversed the increased interaction between MAVS and RIG-I, which was caused by the inhibition of HIF-1α during H1N1 infection (Fig. 5F). In H1N1-infected A549 cells, knockdown of HIF-1α inhibited viral replication, and the addition of exogenous lactate had the opposite effect (Fig. 5G). Silencing HIF-1α increased the expression of MAVS and RIG-I, and the effect was attenuated by adding exogenous lactate (Fig. 5H). Moreover, the levels of proinflammatory factors (IL-1β, IL-6 and TNF-α), which were reduced by HIF-1α knockdown, were increased by exogenous lactate supplementation (Fig. 5I). The mRNA and protein expression levels of IFN-α/β, which were increased by HIF-1α inhibition, were significantly reduced (Fig. 5J and K). These results suggest that HIF-1α/HK2 axis-mediated lactate accumulation promotes H1N1 replication and the host inflammatory response, probably through the MAVS/RIG-I signaling pathway.
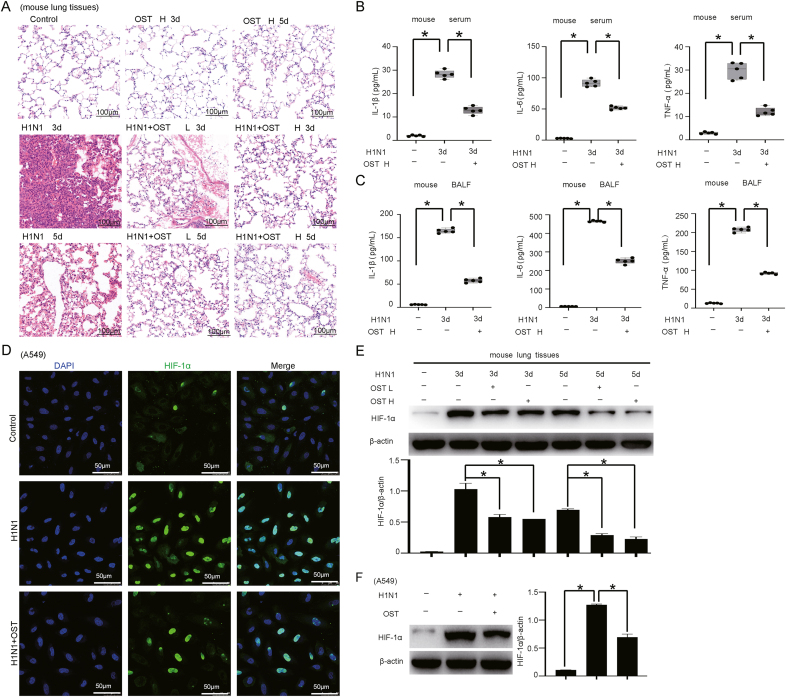
3.6. Oseltamivir inhibits the expression of HIF-1α during H1N1 infection
Neuraminidase inhibitors were identified as first-line drugs for the prevention and treatment of influenza, including oseltamivir. The metabolite of oseltamivir selectively binds to the active site of the influenza neuraminidase to block viral release from host cells (Uyeki, 2009). Our results showed that oseltamivir treatment ameliorated H1N1-induced lung injury (Fig. 6A) and reduced inflammatory cytokine (IL-1β, IL-6 and TNF-α) expression in vivo (Fig. 6B and C).
Fig. 6.
Oseltamivir (OST) inhibits the expression of HIF-1α during H1N1 virus infection. A HE staining of lung tissue from H1N1-infected mice (n = 5, 1000 pfu per mouse, Days 3 and 5) treated with or without a low (L) (OST, 30 mg/kg, intragastric, once every 12 h for Days 3 and 5) or high (H) (OST, 60 mg/kg, intragastric, once every 12 h for Days 3 and 5) dose of oseltamivir. Scale bar = 100 μm. B, C The concentrations of IL-1β, IL-6 and TNF-α in serum and BALF from H1N1-infected mice (n = 5, 1000 pfu per mouse) after treatment with or without oseltamivir were measured by ELISA. D The distribution of HIF-1α protein in A549 cells after OST treatment (0.75 μg/mL for 48 h) was investigated by immunofluorescence staining (scale bar = 50 μm). E, F The protein expression level of HIF-1α in mouse lung tissues (n = 5, 1000 pfu per mouse for Days 3 and 5) and A549 cells (MOI = 1 for 24 h) after OST treatment was measured by Western blotting. β-Actin served as the loading control. The data are presented as the means with SDs. Statistical analysis was performed by one-way ANOVA. ∗P < 0.05.
Next, we asked whether HIF-1α expression is affected by oseltamivir treatment during H1N1 virus infection in vitro and in vivo. The IFA results showed that oseltamivir treatment suppressed the upregulation of HIF-1α expression induced by H1N1 infection in A549 cells (Fig. 6D). The HIF-1α protein level in mouse lung tissue was also significantly decreased in the oseltamivir-treated group compared to the non-treated group after H1N1 infection (Fig. 6E and F). Taken together, these results confirmed the positive role of HIF-1α in modulating H1N1 infection and the accompanying lung injury.
4. Discussion
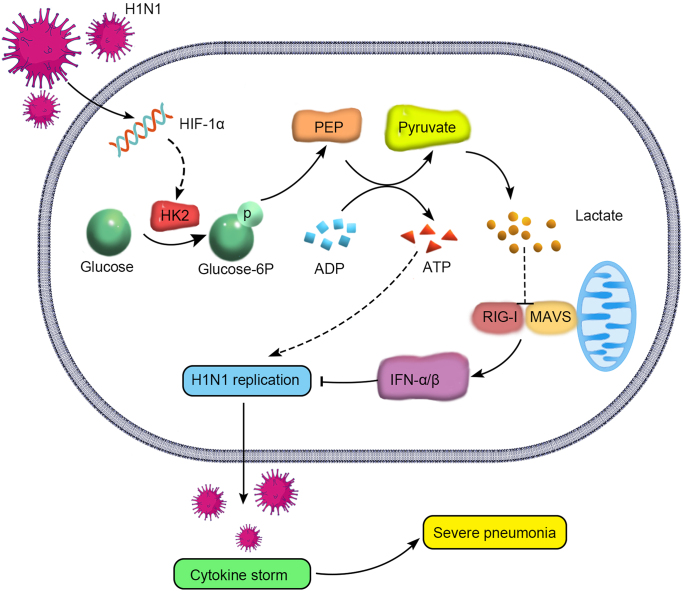
The mortality of patients with severe pneumonia caused by H1N1 virus is closely related to viral replication and cytokine storm (Matsuoka et al., 2016; Rondina et al., 2016). However, the specific mechanism is still unclear. In this study, we confirmed that HIF-1α is upregulated after H1N1 virus infection and found a new mechanism underlying the role of HIF-1α in H1N1-induced severe pneumonia. We showed that HIF-1α promotes glucose metabolism and a shift to glycolysis. Additional studies revealed that HIF-1α promotes the expression of HK2, the key enzyme of glycolysis. This not only rapidly provides energy for viral replication but also results in the production of lactate, which interacts with MAVS to prevent its mitochondrial localization and the formation of the MAVS/RIG-I complex; inhibits IFN-α/β production; and suppresses host cell innate immune responses, which further enhances virus replication and pathogenicity. As a result, the continuous uncontrollable cytokine storm aggravates lung damage, and severe pneumonia develops, increasing the mortality of patients (Fig. 7).
Fig. 7.
Diagram of the mechanism by which HIF-1α promotes glycolysis during H1N1-induced severe pneumonia. HIF-1α activation in alveolar epithelial cells promotes the expression of the key glycolytic enzyme HK2 after H1N1 infection and induces a shift in host cell glucose metabolism to glycolysis. On the one hand, this shift provides energy for viral replication. On the other hand, the lactate produced via glycolysis can directly target and bind to MAVS, abolish the mitochondrial localization of MAVS and the formation of the MAVS/RIG-I complex, inhibit downstream IFN-α/β production and block the host cell innate immune response, further increasing viral replication and pathogenicity. Finally, the continuous uncontrollable cytokine storm aggravates lung damage and leads to severe pneumonia.
It has been reported that the HIF-1α/HIF-1β heterodimer can upregulate the expression of DNASE1 and thus reduce HBV replication through catabolism of the viral DNA genome in the capsid (Hallez et al., 2019). However, HIF-1α is activated during dengue virus infection and promotes the replication of dengue virus (Frakolaki et al., 2018). Therefore, the role of HIF-1α in viral replication is context dependent. The function of HIF-1α in H1N1 infection is incompletely elucidated. It has been reported that total depletion of HIF-1α in alveolar epithelial cells leads to increased H1N1 virus replication and reduced survival in mice (Zhao et al., 2020). Our results also confirmed that HIF-1α knockout aggravated lung injury in mice infected with H1N1 (Supplementary Fig. S11). However, it has also been reported that the titer of anti-influenza A antibody is increased on Day 4 in virus-infected mice with alveolar epithelial cell-specific knockout of HIF-1α (Xi et al., 2017). Considering that HIF-1α might be an important factor in H1N1 infection, the role of HIF-1α in alveolar epithelial cells during H1N1 virus infection should be more precisely evaluated.
We therefore introduced knockdown but not knockout of HIF-1α to understand the role and mechanisms of HIF-1α in modulating H1N1 infection. Our preliminary study found that HIF-1α expression was significantly increased after H1N1 infection (Guo et al., 2017). In this study, we further clarified the mechanism of HIF-1α in H1N1 infection. HIF-1α inhibition greatly inhibits the expression of the viral M gene and NP protein during H1N1 infection in alveolar epithelial cells, indicating that HIF-1α is important for the replication of H1N1. Importantly, further analysis revealed that HK2, the key downstream target of HIF-1α, normalizes the H1N1 virus burden, which is reduced in alveolar epithelial cells with HIF-1α inhibition.
The inflammatory response usually results in an abnormal metabolic microenvironment (Aguilar-Cazares et al., 2022; Al Madhoun et al., 2022). Variations in the oxygen concentration, pH and immune metabolism regulate immune cells and inflammatory processes. HIF-1α directly mediates the overexpression of monocarboxylate transporter 4 (MCT-4) and aggravates inflammation in the context of arsenite-induced glycolysis (Luo et al., 2017). In macrophages, HIF-1α mainly induces the expression of inflammatory genes, such as IL-6, IL-1β and inducible nitric oxide synthase (iNOS), under conditions such as Helicobacter pylori infection (Matak et al., 2015). Studies have found that viral infection plays a role in promoting the stability of the HIF-1α protein in target cells, which is believed to be harmful to the host in some acute viral infections (Sharma et al., 2022; Zalpoor et al., 2022). In this research, we further explored the proinflammatory effects of HIF-1α after H1N1 infection, finding that HIF-1α knockdown reduced the production of proinflammatory factors (IL-1β, IL-6 and TNF-α) both in vitro and in vivo.
HIF-1α is an important modulator mediating a shift in cellular metabolism from oxidative phosphorylation to glycolysis by inducing the expression of glucose transporters, glycolytic enzymes and glycolysis-inducing factors (Sun et al., 2019). Recent studies have shown that respiratory syncytial virus (RSV) has the ability to promote the stability and accumulation of HIF-1α and reprogram cellular metabolism, specifically shifting it toward glycolysis and the pentose phosphate pathway (Morris et al., 2020). The protein and mRNA expression levels of HIF-1α and glycolytic enzymes in human liver tissue and hepatocytes were significantly higher after infection with hepatitis C virus than in the uninfected control group. These increases were accompanied by upregulation of serine biosynthetic enzymes, indicating a shift in cell metabolism toward the nucleotide synthesis necessary to promote HCV replication (Jung et al., 2016). In macrophages, as in tumor cells, HIF-1α gene expression and protein stability were increased even in aerobic environments. HIF-1α promotes glucose uptake by upregulating target genes such as those encoding glucose transporters and lactate dehydrogenase. HIF-1α gene-deficient mice show severe inflammation and develop sepsis under lipopolysaccharide stimulation (Tai et al., 2009; Mascanfroni et al., 2015; Miyasaka et al., 2015). Our research found that glucose metabolism shifts to glycolysis after H1N1 infection. By screening the expression of key glycolysis enzymes after H1N1 infection, we found that HK2 expression is increased after infection and is significantly reduced in HIF-1α-inhibited cells. HK2 has been reported to be a target gene of HIF-1α (Cao et al., 2020; Xu et al., 2020). In this study, we further revealed that HIF-1α upregulated HK2 expression in alveolar epithelial cells, which mediated H1N1-induced glycolytic reprogramming and thus drove H1N1 virus replication as well as the host inflammatory response.
Type I IFN plays an extremely important role in the host antiviral response (El-Baky et al., 2015; Soper et al., 2017). Increased levels of type I IFNs precede the host immune response, and the IFN response is the first defense system to respond to influenza virus infection (Samuel, 2001). RIG-I/MDA5 is a pattern recognition receptor (PRR) localized in the cytoplasm that recognizes double-stranded RNA (dsRNA) production and induces IFN synthesis in a TLR-independent manner. Excessive activation of the RIG-I signaling pathway leads to the upregulation of type I IFN expression (Song et al., 2022). Many viruses encode specific proteins that inhibit RIG-I-mediated expression of IFNs and thus evade elimination by the host immune system (Talon et al., 2000; Hayman et al., 2006).
Mitochondria were first confirmed to participate in innate immune regulation based on the discovery of mitochondrial antiviral-signaling protein (MAVS). MAVS is anchored to mitochondria and acts as a key linker protein in RIG-I signaling (Meylan et al., 2005; Seth et al., 2005). Specifically, after recognizing the virus, RIG-I undergoes conformational changes, with exposure of the CARD domain. The CARD domain then associates with the mitochondrial adaptor protein MAVS and induces the expression of type I IFN genes (Takeuchi and Akira, 2010). A recent study showed that lactate, a glycolytic metabolite, can directly target and bind to MAVS, disrupt its mitochondrial positioning, prevent it from forming a complex with RIG-I and then reduce the production of IFN-α/β (Zhang et al., 2019). Here, we found that H1N1 infection promotes the expression of MAVS and RIG-I and formation of the associated complex. HIF-1α inhibition or inhibition of glycolysis reduces the production of lactate, promotes the expression of MAVS and RIG-I and indirectly increases complex formation. Taken together, the above results suggest that H1N1 infection increases the HIF-1α protein level, promotes glycolysis to produce lactate, and thus inhibits the expression of MAVS and RIG-I and formation of the associated complex, leading to a reduction in IFN-α/β production, which ultimately facilitates the replication of H1N1 virus and aggravates lung injury through excessive inflammatory responses.
5. Conclusions
Overall, this study explored the potential molecular mechanism of HIF-1α and found that it regulates glucose metabolism reprogramming, which promotes H1N1 replication and host inflammatory responses. Therefore, this study provides a theoretical basis for identifying new molecular targets involved in metabolic disorders for the treatment of H1N1 infection to reduce the mortality of severe pneumonia associated with H1N1 infection.
Data availability
Data supporting the findings of this research are reported in the main text and figures and in the Supplementary Data. All reagents and materials generated in this study are available from the corresponding author.
Ethics statement
Animal experiments were conducted in accordance with the Chinese Laboratory Animal Welfare and Ethics Guidelines. The agreement was approved by the Ethics Committee of the Shanghai General Hospital affiliated with Shanghai Jiaotong University.
Author contributions
Xiaoxiao Meng: conceptualization, methodology, software, investigation, data curation, validation, writing-original draft, writing-review & editing. Yong Zhu: conceptualization, methodology, investigation, data curation, validation, writing-original draft, writing-review & editing. Wenyu Yang: methodology, investigation. Jiaxiang Zhang: methodology, investigation. Wei Jin: investigation, data curation. Rui Tian: investigation, data curation. Zhengfeng Yang: conceptualization, resources, project administration, supervision. Ruilan Wang: conceptualization, resources, project administration, supervision, funding acquisition.
Conflict of interest
The authors declare that there are no conflicts of interest.
Acknowledgements
This research was supported by a grant from the National Natural Science Foundation of China (No. 82072210); the Shanghai Municipal Science and Technology Commission, China (No. 20ZR1445200); the Chinese Federation of Public Health Foundation (GWLM202001); and the Three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai (No. GWV-10.1-XK25). The H1N1 influenza virus strain A/PR/8/34 was a gift from Professor Haikun Wang of the Shanghai Pasteur Institute of the Chinese Academy of Sciences.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virs.2023.11.010.
Contributor Information
Zhengfeng Yang, Email: Zhengfeng.yang@shgn.cn.
Ruilan Wang, Email: wangyusun@hotmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aguilar-Cazares D., Chavez-Dominguez R., Marroquin-Mucino M., Perez-Medina M., Benito-Lopez J.J., Camarena A., Rumbo-Nava U., Lopez-Gonzalez J.S. The systemic-level repercussions of cancer-associated inflammation mediators produced in the tumor microenvironment. Front. Endocrinol. (Lausanne) 2022;13 doi: 10.3389/fendo.2022.929572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akman M., Belisario D.C., Salaroglio I.C., Kopecka J., Donadelli M., De Smaele E., Riganti C. Hypoxia, endoplasmic reticulum stress and chemoresistance: dangerous liaisons. J. Exp. Clin. Cancer Res. 2021;40:28. doi: 10.1186/s13046-020-01824-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Madhoun A., Kochumon S., Al-Rashed F., Sindhu S., Thomas R., Miranda L., Al-Mulla F., Ahmad R. Dectin-1 as a potential inflammatory biomarker for metabolic inflammation in adipose tissue of individuals with obesity. Cells. 2022;11:2879. doi: 10.3390/cells11182879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beljanski V., Chiang C., Kirchenbaum G.A., Olagnier D., Bloom C.E., Wong T., Haddad E.K., Trautmann L., Ross T.M., Hiscott J. Enhanced influenza virus-like particle vaccination with a structurally optimized RIG-I agonist as adjuvant. J. Virol. 2015;89:10612–10624. doi: 10.1128/JVI.01526-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Wang M., Dong Y., Xu B., Chen J., Ding Y., Qiu S., Li L., Karamfilova Zaharieva E., Zhou X., Xu Y. Circular RNA circRNF20 promotes breast cancer tumorigenesis and warburg effect through miR-487a/HIF-1α/HK2. Cell Death Dis. 2020;11:145. doi: 10.1038/s41419-020-2336-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.Y., Li H., Lu X.X., Ling L.J., Weng H.B., Sun W., Chen D.F., Zhang Y.Y. Houttuynia cordata polysaccharide alleviated intestinal injury and modulated intestinal microbiota in H1N1 virus infected mice. Chin. J. Nat. Med. 2019;17:187–197. doi: 10.1016/S1875-5364(19)30021-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C., Prasanth K.R., Nagy P.D. The glycolytic pyruvate kinase is recruited directly into the viral replicase complex to generate ATP for RNA synthesis. Cell Host Microbe. 2017;22:639–652 e637. doi: 10.1016/j.chom.2017.10.004. [DOI] [PubMed] [Google Scholar]
- Dong L., He Y., Zhou S., Cao Y., Li Y., Bi Y., Liu G. HIF1α-dependent metabolic signals control the differentiation of follicular helper T cells. Cells. 2019;8:1450. doi: 10.3390/cells8111450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Baky N.A., Uversky V.N., Redwan E.M. Human consensus interferons: bridging the natural and artificial cytokines with intrinsic disorder. Cytokine Growth Factor Rev. 2015;26:637–645. doi: 10.1016/j.cytogfr.2015.07.012. [DOI] [PubMed] [Google Scholar]
- Fontaine K.A., Sanchez E.L., Camarda R., Lagunoff M. Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 2015;89:2358–2366. doi: 10.1128/JVI.02309-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster K.A., Oster C.G., Mayer M.M., Avery M.L., Audus K.L. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 1998;243:359–366. doi: 10.1006/excr.1998.4172. [DOI] [PubMed] [Google Scholar]
- Frakolaki E., Kaimou P., Moraiti M., Kalliampakou K.I., Karampetsou K., Dotsika E., Liakos P., Vassilacopoulou D., Mavromara P., Bartenschlager R., Vassilaki N. The role of tissue oxygen tension in dengue virus replication. Cells. 2018;7:241. doi: 10.3390/cells7120241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gran J.M., Kacelnik O., Grjibovski A.M., Aavitsland P., Iversen B.G. Counting pandemic deaths: comparing reported numbers of deaths from influenza a(H1N1)pdm09 with estimated excess mortality. Influenza Other Respir. Viruses. 2013;7:1370–1379. doi: 10.1111/irv.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Zhu Z., Zhang W., Meng X., Zhu Y., Han P., Zhou X., Hu Y., Wang R. Nuclear translocation of HIF-1α induced by influenza a (H1N1) infection is critical to the production of proinflammatory cytokines. Emerg. Microbes Infect. 2017;6:e39. doi: 10.1038/emi.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O., Kochs G. Interferon-induced mx proteins: dynamin-like GTPases with antiviral activity. Traffic. 2002;3:710–717. doi: 10.1034/j.1600-0854.2002.31003.x. [DOI] [PubMed] [Google Scholar]
- Hallez C., Li X., Suspene R., Thiers V., Bouzidi M.S., C M.D., Lucansky V., Wain-Hobson S., Gaudin R., Vartanian J.P. Hypoxia-induced human deoxyribonuclease I is a cellular restriction factor of hepatitis B virus. Nat. Microbiol. 2019;4:1196–1207. doi: 10.1038/s41564-019-0405-x. [DOI] [PubMed] [Google Scholar]
- Hayman A., Comely S., Lackenby A., Murphy S., McCauley J., Goodbourn S., Barclay W. Variation in the ability of human influenza A viruses to induce and inhibit the IFN-beta pathway. Virology. 2006;347:52–64. doi: 10.1016/j.virol.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Herold S., Becker C., Ridge K.M., Budinger G.R. Influenza virus-induced lung injury: pathogenesis and implications for treatment. Eur. Respir. J. 2015;45:1463–1478. doi: 10.1183/09031936.00186214. [DOI] [PubMed] [Google Scholar]
- Hittinger M., Juntke J., Kletting S., Schneider-Daum N., de Souza Carvalho C., Lehr C.M. Preclinical safety and efficacy models for pulmonary drug delivery of antimicrobials with focus on in vitro models. Adv. Drug Deliv. Rev. 2015;85:44–56. doi: 10.1016/j.addr.2014.10.011. [DOI] [PubMed] [Google Scholar]
- Huo C., Tang Y., Li X., Han D., Gu Q., Su R., Liu Y., Reiter R.J., Liu G., Hu Y., Yang H. Melatonin alleviates lung injury in H1N1-infected mice by mast cell inactivation and cytokine storm suppression. PLoS Pathog. 2023;19 doi: 10.1371/journal.ppat.1011406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuliano A.D., Roguski K.M., Chang H.H., Muscatello D.J., Palekar R., Tempia S., Cohen C., Gran J.M., Schanzer D., Cowling B.J., Wu P., Kyncl J., Ang L.W., Park M., Redlberger-Fritz M., Yu H., Espenhain L., Krishnan A., Emukule G., van Asten L., Pereira da Silva S., Aungkulanon S., Buchholz U., Widdowson M.A., Bresee J.S., Global Seasonal Influenza-associated Mortality Collaborator N Estimates of global seasonal influenza-associated respiratory mortality: a modelling study. Lancet. 2018;391:1285–1300. doi: 10.1016/S0140-6736(17)33293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., Pillai P.S. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 2014;14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G.S., Jeon J.H., Choi Y.K., Jang S.Y., Park S.Y., Kim S.W., Byun J.K., Kim M.K., Lee S., Shin E.C., Lee I.K., Kang Y.N., Park K.G. Pyruvate dehydrogenase kinase regulates hepatitis C virus replication. Sci. Rep. 2016;6 doi: 10.1038/srep30846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyasu S., Kobayashi M., Goto Y., Hiraoka M., Harada H. Regulatory mechanisms of hypoxia-inducible factor 1 activity: two decades of knowledge. Cancer Sci. 2018;109:560–571. doi: 10.1111/cas.13483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R.M., Yuan W., Noah D.L., Latham A.G. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology. 2003;309:181–189. doi: 10.1016/s0042-6822(03)00119-3. [DOI] [PubMed] [Google Scholar]
- Li M., Chen Y., Chen T., Hu S., Chen L., Shen L., Li F., Yang J., Sun Y., Wang D., He L., Qin S., Shu Y. A host-based whole genome sequencing study reveals novel risk loci associated with severity of influenza a(H1N1)pdm09 infection. Emerg. Microbes Infect. 2021;10:123–131. doi: 10.1080/22221751.2020.1870412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.C., Chan M.C., Lee N. Clinical implications of antiviral resistance in influenza. Viruses. 2015;7:4929–4944. doi: 10.3390/v7092850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo F., Zou Z., Liu X., Ling M., Wang Q., Wang Q., Lu L., Shi L., Liu Y., Liu Q., Zhang A. Enhanced glycolysis, regulated by HIF-1α via MCT-4, promotes inflammation in arsenite-induced carcinogenesis. Carcinogenesis. 2017;38:615–626. doi: 10.1093/carcin/bgx034. [DOI] [PubMed] [Google Scholar]
- Mascanfroni I.D., Takenaka M.C., Yeste A., Patel B., Wu Y., Kenison J.E., Siddiqui S., Basso A.S., Otterbein L.E., Pardoll D.M., Pan F., Priel A., Clish C.B., Robson S.C., Quintana F.J. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat. Med. 2015;21:638–646. doi: 10.1038/nm.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matak P., Heinis M., Mathieu J.R., Corriden R., Cuvellier S., Delga S., Mounier R., Rouquette A., Raymond J., Lamarque D., Emile J.F., Nizet V., Touati E., Peyssonnaux C. Myeloid HIF-1 is protective in helicobacter pylori-mediated gastritis. J. Immunol. 2015;194:3259–3266. doi: 10.4049/jimmunol.1401260. [DOI] [PubMed] [Google Scholar]
- Matsuoka T., Sato T., Akita T., Yanagida J., Ohge H., Kuwabara M., Tanaka J. High vaccination coverage among children during influenza a(H1N1)pdm09 as a potential factor of herd immunity. Int. J. Environ. Res. Public Health. 2016;13:1017. doi: 10.3390/ijerph13101017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. Cardif is an adaptor protein in the rig-i antiviral pathway and is targeted by hepatitis c virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Miyasaka A., Oda K., Ikeda Y., Sone K., Fukuda T., Inaba K., Makii C., Enomoto A., Hosoya N., Tanikawa M., Uehara Y., Arimoto T., Kuramoto H., Wada-Hiraike O., Miyagawa K., Yano T., Kawana K., Osuga Y., Fujii T. Pi3k/mtor pathway inhibition overcomes radioresistance via suppression of the hif1-alpha/vegf pathway in endometrial cancer. Gynecol. Oncol. 2015;138:174–180. doi: 10.1016/j.ygyno.2015.04.015. [DOI] [PubMed] [Google Scholar]
- Morris D.R., Qu Y., Agrawal A., Garofalo R.P., Casola A. HIF-1α modulates core metabolism and virus replication in primary airway epithelial cells infected with respiratory syncytial virus. Viruses. 2020;12:1088. doi: 10.3390/v12101088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdaca G., Paladin F., Tonacci A., Isola S., Allegra A., Gangemi S. The potential role of cytokine storm pathway in the clinical course of viral respiratory pandemic. Biomedicines. 2021;9:1688. doi: 10.3390/biomedicines9111688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papandreou I., Cairns R.A., Fontana L., Lim A.L., Denko N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006;3:187–197. doi: 10.1016/j.cmet.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Passalacqua K.D., Lu J., Goodfellow I., Kolawole A.O., Arche J.R., Maddox R.J., Carnahan K.E., O'Riordan M.X.D., Wobus C.E. Glycolysis is an intrinsic factor for optimal replication of a norovirus. mBio. 2019;10 doi: 10.1128/mBio.02175-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleschka S. Overview of influenza viruses. Curr. Top. Microbiol. Immunol. 2013;370:1–20. doi: 10.1007/82_2012_272. [DOI] [PubMed] [Google Scholar]
- Ren L., Zhang W., Zhang J., Zhang J., Zhang H., Zhu Y., Meng X., Yi Z., Wang R. Influenza a virus (H1N1) infection induces glycolysis to facilitate viral replication. Virol. Sin. 2021;36:1532–1542. doi: 10.1007/s12250-021-00433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Zhang W., Han P., Zhang J., Zhu Y., Meng X., Zhang J., Hu Y., Yi Z., Wang R. Influenza a virus (H1N1) triggers a hypoxic response by stabilizing hypoxia-inducible factor-1alpha via inhibition of proteasome. Virology. 2019;530:51–58. doi: 10.1016/j.virol.2019.02.010. [DOI] [PubMed] [Google Scholar]
- Rondina M.T., Tatsumi K., Bastarache J.A., Mackman N. Microvesicle tissue factor activity and interleukin-8 levels are associated with mortality in patients with influenza a/H1N1 infection. Crit. Care Med. 2016;44:e574–e578. doi: 10.1097/CCM.0000000000001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarda C., Palma P., Rello J. Severe influenza: overview in critically ill patients. Curr. Opin. Crit. Care. 2019;25:449–457. doi: 10.1097/MCC.0000000000000638. [DOI] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., Chen Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Shapira S.D., Gat-Viks I., Shum B.O., Dricot A., de Grace M.M., Wu L., Gupta P.B., Hao T., Silver S.J., Root D.E., Hill D.E., Regev A., Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A.L., Wang H., Zhang Z., Millien G., Tyagi M., Hongpaisan J. HIV promotes neurocognitive impairment by damaging the hippocampal microvessels. Mol. Neurobiol. 2022;59:4966–4986. doi: 10.1007/s12035-022-02890-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K.R., Kasper J., van der Aa S., Andeweg A.C., Zaaraoui-Boutahar F., Goeijenbier M., Richard M., Herold S., Becker C., Scott D.P., Limpens R.W., Koster A.J., Barcena M., Fouchier R.A., Kirkpatrick C.J., Kuiken T. Influenza virus damages the alveolar barrier by disrupting epithelial cell tight junctions. Eur. Respir. J. 2016;47:954–966. doi: 10.1183/13993003.01282-2015. [DOI] [PubMed] [Google Scholar]
- Song J., Li M., Li C., Liu K., Zhu Y., Zhang H. Friend or foe: RIG- I like receptors and diseases. Autoimmun. Rev. 2022;21 doi: 10.1016/j.autrev.2022.103161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper A., Kimura I., Nagaoka S., Konno Y., Yamamoto K., Koyanagi Y., Sato K. Type I interferon responses by HIV-1 infection: association with disease progression and control. Front. Immunol. 2017;8:1823. doi: 10.3389/fimmu.2017.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Meng X., Pu Y., Sun F., Man Z., Zhang J., Yin L., Pu Y. Overexpression of HIF-1a could partially protect K562 cells from 1,4-benzoquinone induced toxicity by inhibiting ROS, apoptosis and enhancing glycolysis. Toxicol. In Vitro. 2019;55:18–23. doi: 10.1016/j.tiv.2018.11.005. [DOI] [PubMed] [Google Scholar]
- Tai T.C., Wong-Faull D.C., Claycomb R., Wong D.L. Hypoxic stress-induced changes in adrenergic function: role of HIF1 alpha. J. Neurochem. 2009;109:513–524. doi: 10.1111/j.1471-4159.2009.05978.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Talon J., Horvath C.M., Polley R., Basler C.F., Muster T., Palese P., Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza a virus ns1 protein. J. Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyrsted J., Storgaard J., Blay-Cadanet J., Heinz A., Thielke A.L., Crotta S., de Paoli F., Olagnier D., Wack A., Hiller K., Hansen A.L., Holm C.K. Influenza a induces lactate formation to inhibit type I IFN in primary human airway epithelium. iScience. 2021;24 doi: 10.1016/j.isci.2021.103300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triner D., Shah Y.M. Hypoxia-inducible factors: a central link between inflammation and cancer. J. Clin. Invest. 2016;126:3689–3698. doi: 10.1172/JCI84430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeki T. Antiviral treatment for patients hospitalized with 2009 pandemic influenza a (H1N1) N. Engl. J. Med. 2009;361:e110. doi: 10.1056/NEJMopv0910738. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Watanabe S., Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y., Kim T., Brumwell A.N., Driver I.H., Wei Y., Tan V., Jackson J.R., Xu J., Lee D.K., Gotts J.E., Matthay M.A., Shannon J.M., Chapman H.A., Vaughan A.E. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat. Cell Biol. 2017;19:904–914. doi: 10.1038/ncb3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Huan L., Guo T., Wu Y., Liu Y., Wang Q., Huang S., Xu Y., Liang L., He X. LncRNA SNHG11 facilitates tumor metastasis by interacting with and stabilizing HIF-1α. Oncogene. 2020;39:7005–7018. doi: 10.1038/s41388-020-01512-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalpoor H., Akbari A., Nabi-Afjadi M., Forghaniesfidvajani R., Tavakol C., Barzegar Z., Iravanpour F., Hosseini M., Mousavi S.R., Farrokhi M.R. Hypoxia-inducible factor 1 alpha (HIF-1α) stimulated and P2X7 receptor activated by COVID-19, as a potential therapeutic target and risk factor for epilepsy. Hum. Cell. 2022;35:1338–1345. doi: 10.1007/s13577-022-00747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.L., Hu X., Zhang W., Tam K.Y. Unexpected discovery of dichloroacetate derived adenosine triphosphate competitors targeting pyruvate dehydrogenase kinase to inhibit cancer proliferation. J. Med. Chem. 2016;59:3562–3568. doi: 10.1021/acs.jmedchem.5b01828. [DOI] [PubMed] [Google Scholar]
- Zhang W., Wang G., Xu Z.G., Tu H., Hu F., Dai J., Chang Y., Chen Y., Lu Y., Zeng H., Cai Z., Han F., Xu C., Jin G., Sun L., Pan B.S., Lai S.W., Hsu C.C., Xu J., Chen Z.Z., Li H.Y., Seth P., Hu J., Zhang X., Li H., Lin H.K. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell. 2019;178:176–189 e115. doi: 10.1016/j.cell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Chen J., Cheng L., Xu K., Yang Y., Su X. Deficiency of HIF-1α enhances influenza A virus replication by promoting autophagy in alveolar type II epithelial cells. Emerg. Microbes Infect. 2020;9:691–706. doi: 10.1080/22221751.2020.1742585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li Y.J., Gao S.Y., Wang X.Z., Wang P.Y., Yan Y.F., Xie S.Y., Lv C.J. Sulindac has strong antifibrotic effects by suppressing STAT3-related miR-21. J. Cell. Mol. Med. 2015;19:1103–1113. doi: 10.1111/jcmm.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Rao M., Wallis R.S., Kaufmann S.H., Rustomjee R., Mwaba P., Vilaplana C., Yeboah-Manu D., Chakaya J., Ippolito G., Azhar E., Hoelscher M., Maeurer M., Host-Directed Therapies Network Consortium Host-directed therapies for infectious diseases: current status, recent progress, and future prospects. Lancet Infect. Dis. 2016;16:e47–e63. doi: 10.1016/S1473-3099(16)00078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this research are reported in the main text and figures and in the Supplementary Data. All reagents and materials generated in this study are available from the corresponding author.