Abstract
Sphingolipids are an important class of lipids present in all eukaryotic cells that regulate critical cellular processes. Disturbances in sphingolipid homeostasis have been linked to several diseases in humans. Ceramides are central in sphingolipid metabolism and are largely synthesized by six ceramide synthase (CerS) isoforms (CerS1–6), each with a preference for different fatty acyl chain lengths. Although the tissue distribution of CerS mRNA expression in humans and the roles of CerS isoforms in synthesizing ceramides with different acyl chain lengths are known, it is unknown how CerS expression dictates ceramides and downstream metabolites within tissues. In this study, we analyzed sphingolipid levels and CerS mRNA expression in 3-month-old C57BL/6J mouse brain, heart, kidney, liver, lung, and skeletal muscle. The results showed that CerS expression and sphingolipid species abundance varied by tissue and that CerS expression was a predictor of ceramide species within tissues. Interestingly, although CerS expression was not predictive of complex sphingolipid species within all tissues, composite scores for CerSs contributions to total sphingolipids measured in each tissue correlated to CerS expression. Lastly, we determined that the most abundant ceramide species in mouse tissues aligned with CerS mRNA expression in corresponding human tissues (based on chain length preference), suggesting that mice are relevant preclinical models for ceramide and sphingolipid research.
SIGNIFICANCE STATEMENT
The current study demonstrates that ceramide synthase (CerS) expression in specific tissues correlates not only with ceramide species but contributes to the generation of complex sphingolipids as well. As many of the CerSs and/or specific ceramide species have been implicated in disease, these studies suggest the potential for CerSs as therapeutic targets and the use of sphingolipid species as diagnostics in specific tissues.
Introduction
Sphingolipids, a major class of lipids present in all eukaryotic cells, are bioactive molecules that regulate critical cellular processes, such as cell cycle, apoptosis, differentiation, migration, proliferation, and senescence (Hannun and Obeid, 2018; Trayssac et al., 2018). Disturbances in sphingolipid homeostasis manifest in human pathologies, including type 2 diabetes–induced insulin resistance, cardiovascular disease, Alzheimer disease, and cancer. Previous studies have implicated sphingolipids as key regulators of cell biology. However, it is not fully understood how individual sphingolipid enzymes and their sphingolipid products impact physiology and disease.
Ceramides are central in sphingolipid metabolism and can be incorporated into various complex sphingolipids. Generation of ceramide occurs via one of six ceramide synthase (CerS) isoforms (CerS1–6). Each of the six CerSs preferentially incorporates specific fatty acyl-CoAs via N-acylation of sphingosine or dihydrosphingosine (Levy and Futerman, 2010). Moreover, these CerS are expressed in specific tissues, which can contribute to the unique roles of CerS isoforms in physiologic and pathophysiological states. CerS1 synthesizes C18- and C18:1-dihydroceramide [(dh)ceramide] and is mainly expressed in the brain and at low levels in skeletal muscle and testis (Levy and Futerman, 2010). CerS2, which is ubiquitously expressed, with highest expression in the kidney and liver (Cai et al., 2003), synthesizes long-chain and very-long-chain (VLC) (dh)ceramides (C20-C26). CerS3 synthesizes very-long-chain (dh)ceramides (C22–C26), primarily in skin and testis, with the highest expression in keratinocytes (Mizutani et al., 2006; Rabionet et al., 2008). CerS4, which exhibits the lowest expression of the CerSs, is expressed in skin, leukocytes, and liver (Laviad et al., 2008) and synthesizes C18- and C20-(dh)ceramide. CerS5 and C6 generate C14- and C16-(dh)ceramide (Levy and Futerman, 2010), with CerS5 being predominantly expressed in lung epithelial cells; however, the tissue distribution of CerS6 is less well studied. Ceramides with different N-acyl chains play distinct roles in biologic processes and may play different roles in disease states, therefore serving as prognostic markers (Zelnik et al., 2020).
Although the tissue distribution of CerS in humans and their roles in synthesizing specific ceramides are known, the mechanisms by which CerS expression regulates steady-state levels of tissue-specific ceramide species and other major sphingolipids are not completely understood. Therefore, in this study, we set out to determine expression levels of CerS, as well as other sphingolipid enzymes, in specific tissues in C57BL/6J mice. Expression of these enzymes was evaluated alongside lipidomics data from the same tissue types. This publication aims to serve as a resource to fill knowledge gaps in sphingolipid research and inspire future work in the field.
Materials and Methods
Animal Experiments and Lipidomics
Tissue samples were collected from 3-month-old male C57BL/6J mice purchased from Jackson Laboratory (Bar Harbor, ME). C57BL/6J mice were selected in part due to the fact that many sphingolipid enzyme-deficient mouse models were generated or backcrossed onto this strain. All mice were maintained on a 12:12-hour light/dark cycle and provided ad libitum access to standard chow food and water. All animal procedures were approved by the Institutional Animal Care and Use Committee of at the Medical University of South Carolina (and were conducted while L.J.S. and A.J.S. were employed at the Medical University of South Carolina) and followed the guidelines of the American Veterinary Medical Association. Tissue samples were collected immediately after euthanasia, snap frozen in liquid nitrogen, and maintained at −80 C.
Lipidomics were measured by liquid chromatograpy tandem mass spectrometry (LC-MS/MS) at the Medical University of South Carolina Lipidomics Facility in 1 mg of tissue and data were normalized to total protein (Bielawski et al., 2010). Briefly, previously snap-frozen tissues were isolated on ice and homogenized using a rotor homogenizer in buffer containing 20 mM Tris-HCl, pH 7.4, and complete protease inhibitors. After brief sonication and determination of protein concentration, 1 mg of protein homogenate was used, and lipids were extracted by the Lipidomics Core at the Medical University of South Carolina and analyzed on a Thermo Quantum Ultra triple quadrupole mass spectrometer tandem Thermo Accela HPLC (Thermo Finnegan, Waltham, MA). The analysis of all sphingolipids was performed in multiple reaction monitoring mode utilizing mass transitions from Bielawski et al. (2010) for all sphingolipids. The instrument was operated in positive ionization mode, with electrospray ionization (ESI) source operating at 400°C vaporizer temperature, heated capillary at 300°C, and spray voltage of 3500 V. Source gasses were maintained at 5, 10, and 40 for ion sweep, auxiliary, and Sheath gasses, respectively. Chromatographic separation was accomplished using a Thermo Accela HPLC system (Thermo Fisher Scientific, Waltham, MA). Chromatography employed a Peek Scientific C-8 column (4.6 mm × 150 mm × 3 μm). To enhance the reproducibility of analytes while maintaining a clear separation, the column temperature was set at 45°C. The mobile phases consisted of the following: Mobile phase A was composed of high-performance liquid chromatography–grade water containing 0.2% formic acid and 1 mM ammonium formate (pH 5.6), whereas mobile phase B (MPB) was composed of high-performance liquid chromatography–grade methanol with 0.2% formic acid and 1 mM ammonium formate (pH 5.6). The chromatographic conditions followed this gradient: Initially, upon sample injection, the mobile phase consisted of 82% MPB and remained constant for 2 minutes. It was then increased to 90% MPB by 4 minutes, further increased to 98% MPB by 10 minutes, and maintained at 98% MPB until 28 minutes. At this point, the mobile phase was reduced to 82% by 30 minutes and allowed to re-equilibrate for 5 minutes, resulting in a total gradient time of 35 minutes.
Prior to the analysis of sphingomyelin (SM), base hydrolysis was performed on an extraction aliquot. For the analysis of SM, a shorter gradient was employed. Briefly, after sample injection, the gradient increased from 90% MPB to 99% MPB within the first 7 minutes and was maintained at 99% MPB until 17 minutes into the gradient. Subsequently, the gradient was reverted to 90% MPB within 1 minute and allowed to equilibrate until 21 minutes.
A composite sphingolipid profile for each of the CerS was generated as follows. Due to the vastly different abundance of sphingolipid classes, sphingolipid data were first transformed to compositional data by summing all N-acyl chain lengths for a given species in a given sample. Then, each molecular species in that class and sample were divided by the sum to generate the percentage of total data. Next, the percentage totals of for potential downstream metabolic products of CerS activity (SMs and hexosylceramides) were averaged to generate a single representative value reflecting the CerS’ contribution to total sphingolipids measured in each tissue.
Gene Expression
RNA was extracted and purified from eight tissue types, including brain, heart, kidney, liver, lung, and skeletal muscle. A total of 40 samples were used for RNA isolation, with five samples used per tissue type. RNA was isolated from all tissues using the E.Z.N.A. Total RNA Kit I from Omega Bio-tek, Inc. (Norcross, Georgia). RNA was isolated from blood using the SurePrep TrueTotal RNA Purification Kit from Thermo Fisher Scientific (Waltham, Massachusetts). The manufacturer’s protocol was followed for each. Total RNA was quantified using a NanoDrop spectrophotometer. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit from Thermo Fisher Scientific the per manufacturer’s protocol. TaqMan Real-Time PCR was then used to determine gene expression of the six isoforms of CerS in each tissue type against one control gene, β-2-microglobulin (B2M, Mm00437762_m1). The CerS experimental genes included CerS1 (Mm03024093_mH), Cers2 (Mm01258345_g1), CerS3 (Mm03990709_m1), CerS4 (Mm00482658_m1), CerS5 (Mm00510998_m1), and CerS6 (Mm00556165_m1). Four additional experimental genes were evaluated with TaqMan Real-Time PCR, including dihydroceramide desaturase (Degs)-1 (Mm00492146_m1), Degs2 (Mm00510313_m1), Sgms2 (Mm00512327_m1), and Ugcg (Mm00495925_m1). SYBR Real-Time PCR was used to determine the gene expression of 10 additional genes (reverse and forward primers listed in Table 1). Primers for acid ceramidase (gene) (Asah1), neutral ceramidase (Asah2), alkaline ceramidase (Acer)-1, Acer2, and Acer3 were obtained from Thermo Fisher Scientific, Invitrogen. Primers forβ-glucocerebrosidase (Gba), Gba2, sphingomyelin phosphodiesterase (Smpd)-2, Smpd3, and Smpd4 were obtained from Integrated DNA Technologies. Relative gene expression levels within each tissue were calculated using the –delta delta CT (2^-ΔΔCT). Relative expression levels for CerS1–6 and genes involved in ceramide synthesis (Degs1, Degs2, Gba, Gba2, Smpd2, Smpd3, and Smpd4) were normalized to CerS2 as CerS2 mRNA is known to be ubiquitously expressed and is highly abundant in many tissues (Levy and Futerman, 2010). Relative expression levels for genes involved in ceramide breakdown (Acer1, Acer2, Acer3, Asah1, Asah2, Ugcg, and Sgms2) were normalized to Acer3 due to its ubiquitous expression. Statistical analysis was performed using one-way or two-way ANOVA. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
TABLE 1.
Primer sequences for real-time primers
| Gene | Reverse Primer | Forward Primer |
|---|---|---|
| Asah1 | GGTAACATCCATCAAGGTTGTGAA | CTTTGCTACCATCTATGATGTCCTATCA |
| Asah2 | CCACGCTCACAAATGCCAT | GTGTCAGATATCAATTTGATGGGCTAT |
| Acer1 | TCTGGCATCTCATAC TTTGCATC | ATTCACTTCTACTACCTGCACAGCAT |
| Acer2 | ACGAAGCAAGGCAGATGAGAA | GCGACCAAGCCTTCTGTGA |
| Acer3 | GTATACAAATATGCAGCAGCTGTAAATCA | CACATGACTCTGAAATATGAAATGCA |
| Gba | ACACGGATGGAGAAGTCACAACT | ACCTACTCAGAAGCTGCTACTCAGATC |
| Gba2 | CTTCAGCTGTCCGGAAACCTT | GCAGCCACCATGATCCAAG |
| Smpd2 | TGATCAAATCGGACCTTTCCA | GCTGTGACCCTCACAGTGACAA |
| Smpd3 | TCTTTGGTCCTGAGGTGTGCTT | CCTCTAAGGGAGCTCTGTTTCTCA |
| Smpd4 | GTTGTGCAAAGGGCTTATTTATAGAGT | CTCACCTACAACAGCCTAGTTTCCTT |
Results
Brain CerSs Expression and Sphingolipidomics
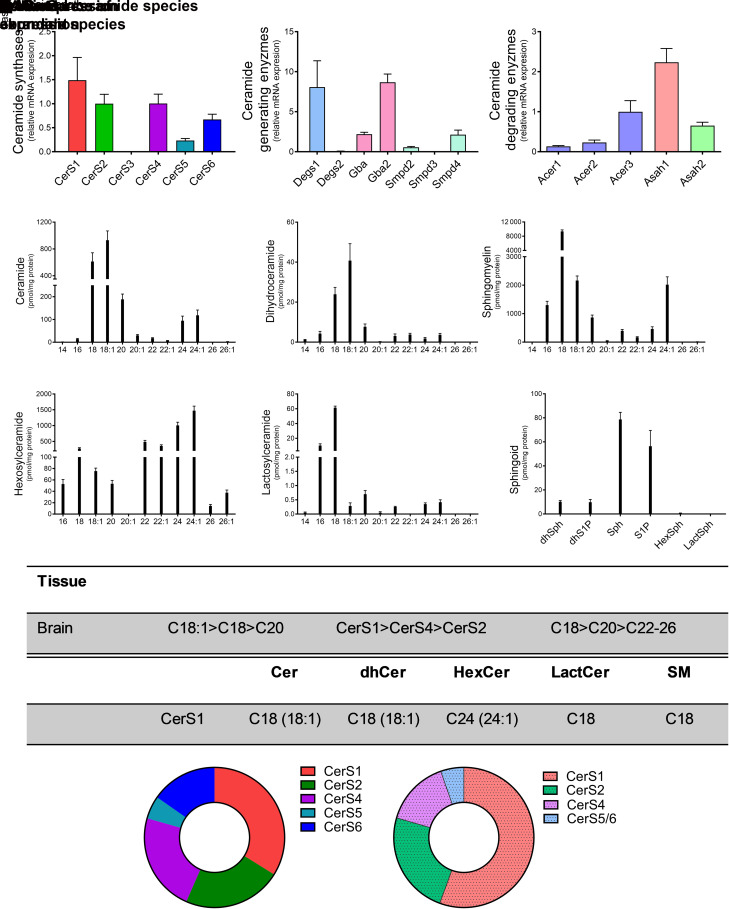
CerS1 has been shown to be highly expressed in CNS tissues (Ben-David and Futerman, 2010). Relative CerS expression levels in the brain demonstrated CerS1 to have the highest expression, with CerS2 and CerS4 closely following (Fig. 1A). Of the additional CerS enzymes, CerS5 and CerS6 displayed relatively low expression levels. CerS3 was undetectable in brain tissue. Several other enzymes also contribute to the generation and/or degradation of ceramide. Degs1 and β-glucocerebrosidase 2 (Gba2) were most abundant in the brain, with Degs2, GBA1, and Smpd 2, 3, and 4 (Smpd2, Smpd3, Smpd4) having markedly lower levels of relative expression (Fig. 1B). Of the ceramidases, Asah1 exhibited the highest expression of enzymes that deacylated ceramide, followed by Acer2 and Asah2 (Fig. 1C). Lipidomic analyses in the brain indicated that the two most abundant ceramide and dihydroceramide species contained 18-carbons—unsaturated C18:1-ceramide, closely followed by saturated C18-ceramide (Fig. 1, D and E). Similar to ceramides, C18- and C18:1 SMs were the most abundant SM species, but inversely to the ceramides, C18-SM was more abundant, with ∼4.3-fold increase over C18:1-SM (9356 ± 1042 pmol/mg protein versus 2164 ± 392.1 pmol/mg protein, respectively) (Fig. 1F). VLC hexosylceramides (C22-C24:1) were more abundant than either C18:1 or C18 hexosylceramides (Fig. 1G). In contrast to hexosylceramides, but consistent with CerS and SM, long-chain C16 and C18 lactosylceramides were the most abundant species. (Fig. 1H). Sphingosine and sphingosine-1-phosphate (S1P) levels were the most abundant sphingoid bases (Fig. 1I). In summary, CerS1 exhibited the highest expression in the brain, and C18 ceramides and SMs were the most abundant lipid species, demonstrating a correlation between CerS expression and acyl-chain length sphingolipids in the brain (Fig. 1, J and K). These data demonstrate the importance of CerS1 in the brain.
Fig. 1.
CerS1 expression regulates sphingolipid acyl-chain length in the brain. Relative mRNA expression of (A) CerS isoforms, (B) ceramide-generating enzymes, and (C) ceramide-degrading enzymes were analyzed using real-time reverse-transcription polymerase chain reaction. (D–I) Sphingolipid levels were assessed using liquid chromatography tandem mass spectrometry (LC-MS/MS) and normalized to protein [(D) ceramides, (E) dihydroceramides, (F) SMs, (G) hexosylceramides (HexCer), (H) lactosylceramides (LactCer), and (I) sphingoid bases]. (J) Table detailing CerS expression and most abundant sphingolipid species in the brain. (K) Donut plots depicting CerS expression (left panel) and CerS contribution to ceramide species (right panel).
Heart CerSs Expression and Sphingolipidomics
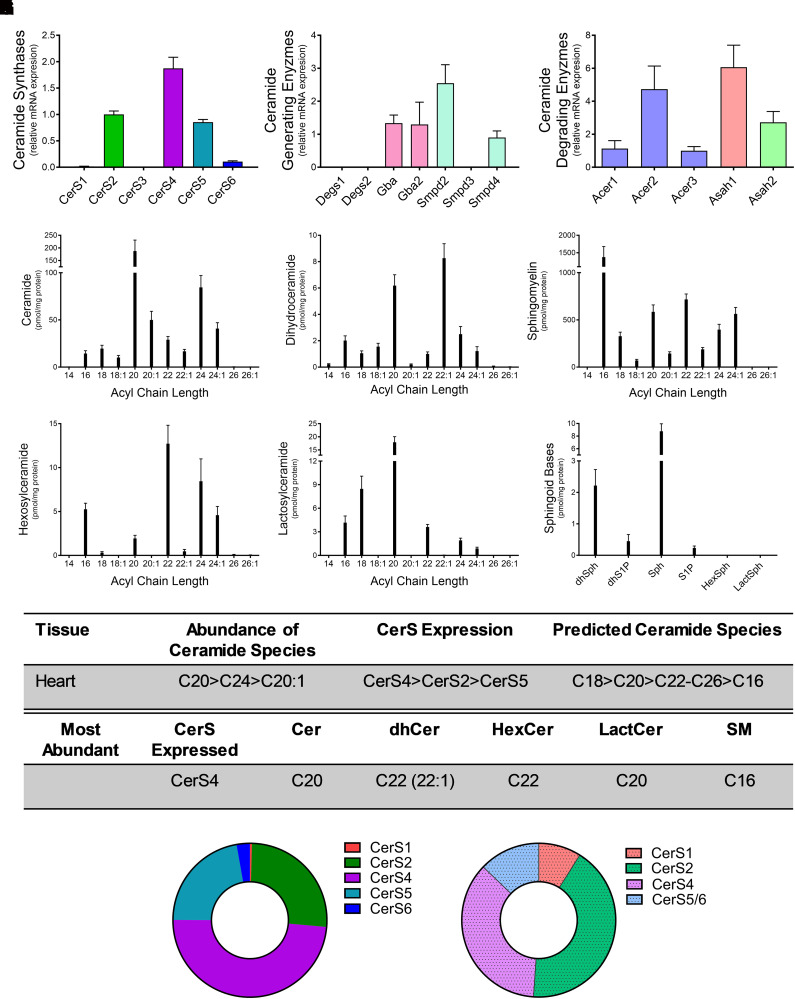
CerS expression and ceramide generation have been implicated in heart disease (Russo et al., 2012, 2013). Relative expression levels for CerS1-6 in the heart revealed CerS4 to have the highest expression (49%), followed by CerS2 (26%) and CerS5 (22%). The remaining isoforms were present at very small (3% or less) or undetectable levels (Fig. 2A). Smpd2 and Smpd4, as well as Gba and Gba2, were most abundant in the heart, with Degs1, Degs2, and Smpd3 having very low levels of expression (Fig. 2B). Of the enzymes that deacylated ceramides, Asah1, Acer3, and Asah2 exhibited the highest levels of relative expression in the heart, with Acer1 and Acer 2 expressed at lower levels (Fig. 2C). Lipidomic analyses in the heart demonstrated VLC C20 ceramide (187.6 ± 106.4 pmol/mg protein) and C22:1 dihydroceramide (8.27 ± 2.68 pmol/mg protein) to be the most abundant (Fig. 2, D and E). Contrary to the ceramides, long-chain C16 SM was the predominant SM species in the heart (Fig. 2F). Hexosyl- and lactosylceramides demonstrated similar patterns in acyl chain length as the ceramides, with VLC species having the highest abundance (Fig. 2, G and H). Sphingosine and dihydrosphingosine levels were also highest among the sphingoid bases (Fig. 2I). CerS4 exhibited the highest level of abundance in the heart, and, as predicted, this correlated with C20-ceramide and C20-lactosylceramide species (Fig. 2, J and K). However, unlike in the brain, C22 (C22:1)-dihydroceramides, C22-hexosylceramides, and C16-SM were more abundant. These deviations from predicted CerS4 chain lengths could be a result of the relatively high levels of expression in CerS2 and CerS5 in the heart.
Fig. 2.
CerS4 expression regulates sphingolipid acyl-chain length in the heart. Relative mRNA expression of (A) CerS isoforms, (B) ceramide-generating enzymes, and (C) ceramide-degrading enzymes were analyzed using real-time reverse-transcription polymerase chain reaction. (D–I) Sphingolipid levels were assessed using liquid chromatography tandem mass spectrometry (LC-MS/MS) and normalized to protein [(D) ceramides, (E) dihydroceramides, (F) SMs, (G) hexosylceramides (HexCer), (H) lactosylceramides (LactCer), and (I) sphingoid bases]. (J) Table detailing CerS expression and most abundant sphingolipid species in the heart. (K) Donut plots depicting CerS expression (left panel) and CerS contribution to ceramide species (right panel).
Kidney CerSs Expression and Sphingolipidomics
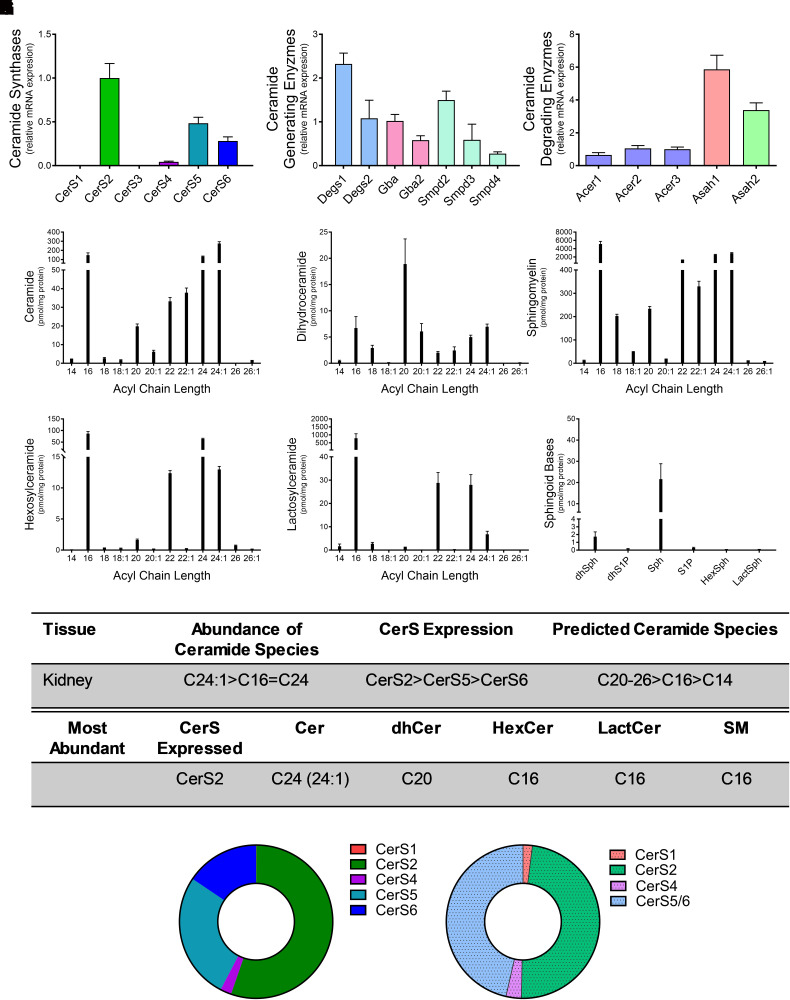
Generation of C16 ceramide by CerS5 and Cers6 has been implicated in kidney disease (Wang et al., 2023). Relative CerS expression levels in the kidney revealed CerS2 as the most highly expressed isoform (56%). CerS5 (27%) and CerS6 (16%) expression followed, with the remaining isoforms expressed at very low (≤2%) or undetectable levels (Fig. 3A). Analysis of additional enzymes that generate ceramide revealed the highest expression of Degs1, followed by Degs2, Gba, and Smpd2 (Fig. 3B). Asah1 displayed fivefold higher expression than Acer3, and Asah2 expression is also high, with a ∼3.5-fold higher expression than Acer3 (Fig. 3C). Similar to the heart, VLC ceramides and dihydroceramides were the most abundant in the kidney, with C24:1-ceramide (275.6 ± 46.5 pmol/mg protein; Fig. 3E) and C20 dihydroceramide (18.9 ± 11.7 pmol/mg protein; Fig. 3E) having the highest levels. C16 was the predominant species for all complex sphingolipid species analyzed (Figs. 3, F–H). Sphingosine levels were the highest of the sphingoid bases (21.6 ± 17.9 pmol/mg protein; Fig. 3I). Overall, CerS2 had the highest relative mRNA expression, and C24:1-ceramide had the highest total concentration among ceramide species (Fig. 3, J and K). CerS2 preferentially synthesizes long-chain and VLC ceramides, with chain lengths ranging from C20–C26. However, higher levels of C16 acyl-chains in complex sphingolipids are likely due to expression of CerS5 and CerS6 (Fig. 3K). These data support a correlation between CerS expression and ceramide species in the kidney.
Fig. 3.
CerS2 expression regulates sphingolipid acyl-chain length in the kidney. Relative mRNA expression of (A) CerS isoforms, (B) ceramide-generating enzymes, and (C) ceramide-degrading enzymes were analyzed using real-time reverse-transcription polymerase chain reaction. (D–I) Sphingolipid levels were assessed using liquid chromatography tandem mass spectrometry (LC-MS/MS) and normalized to protein [(D) ceramides, (E) dihydroceramides, (F) SMs, (G) hexosylceramides (HexCer), (H) lactosylceramides (LactCer), and (I) sphingoid bases]. (J) Table detailing CerS expression and most abundant sphingolipid species in the kidney. (K) Donut plots depicting CerS expression (left panel) and CerS contribution to ceramide species (right panel).
Liver CerSs Expression and Sphingolipidomics
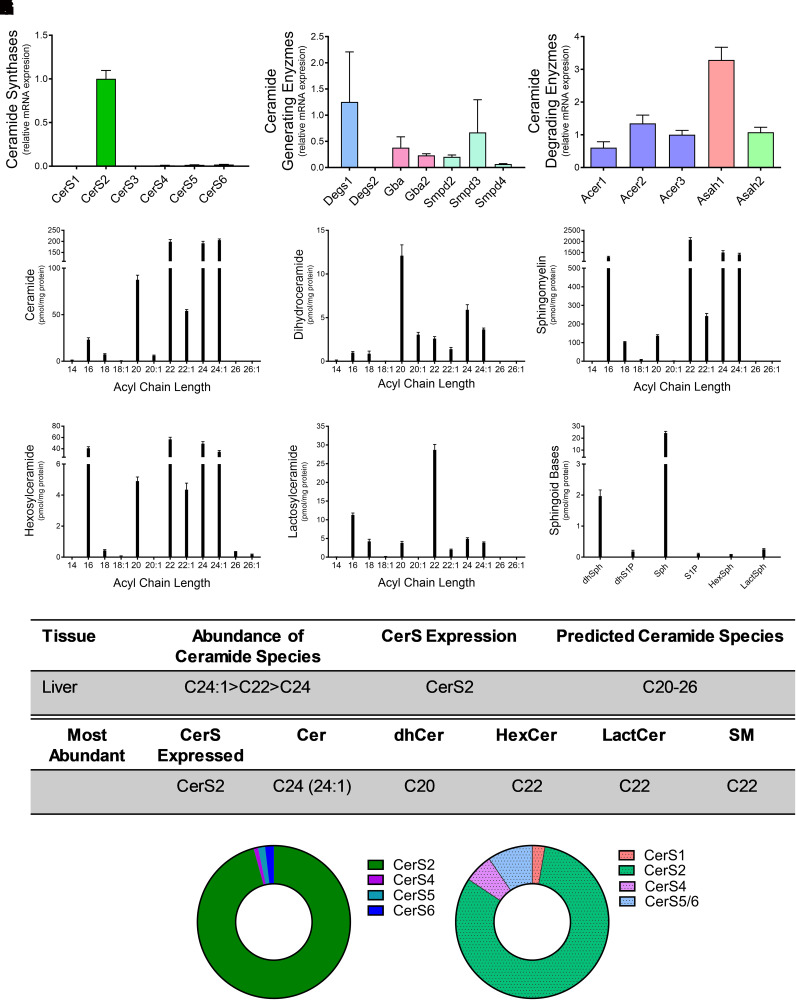
Loss of CerS2 in a murine model resulted in significant pathology with age (Pewzner-Jung et al., 2010). Relative CerS expression levels in the liver demonstrated that ∼96% of CerS in the liver was CerS2, with all other isoforms having negligible expression (Fig. 4A). Among genes involved in ceramide synthesis, Degs1 demonstrated similar levels of expression to that of CerS2 (Fig. 4B). Among genes involved in ceramide breakdown, Asah1 expression was highest (Fig. 4C). VLC ceramides and dihydroceramides, C20, 22, C24, and C24:1, were the most abundant species measured in the liver (Fig. 4, D and E). Similar trends in VLC SM, hexosylceramide, and lactosylceramide were determined, with C22 being the predominant species in all complex sphingolipids (Figs. 4, F–H). Similar to measurements in other organ systems, sphingosine was the most abundant sphingoid base measured (Fig. 4I). Overall, CerS2 had the highest relative mRNA expression, and C24:1-ceramide was the most abundant ceramide species (Fig. 4, J and K). Similar to the kidney, complex sphingolipids all exhibited the same acyl-chain (C22) as the predominant species. These data demonstrate the significance of CerS2 expression and the coupling of VLC sphingolipids in the liver.
Fig. 4.
CerS2 expression regulates sphingolipid acyl-chain length in the liver. Relative mRNA expression of (A) CerS isoforms, (B) ceramide-generating enzymes, and (C) ceramide-degrading enzymes were analyzed using real-time reverse-transcription polymerase chain reaction. (D–I) Sphingolipid levels were assessed using liquid chromatography tandem mass spectrometry (LC-MS/MS) and normalized to protein [(D) ceramides, (E) dihydroceramides, (F) SMs, (G) hexosylceramides (HexCer), (H) lactosylceramides (LactCer), and (I) sphingoid bases]. (J) Table detailing CerS expression and most abundant sphingolipid species in the liver. (K) Donut plots depicting CerS expression (left panel) and CerS contribution to ceramide species (right panel).
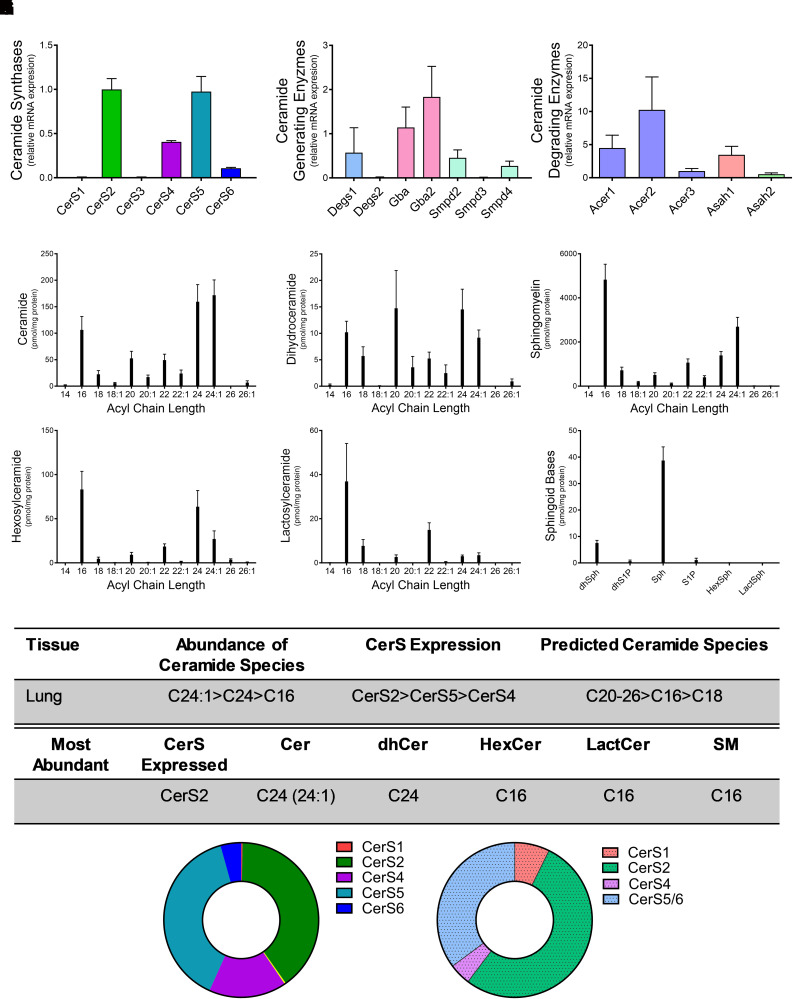
Lung CerSs Expression and Sphingolipidomics
Ceramides in the lung have been implicated in several diseases, including asthma (Choi et al., 2020). Examination of CerS expression in lung tissues demonstrated that CerS2 (40%) and CerS5 (39%) were highly expressed, followed by CerS4 (16%) and CerS6 (6%) (Fig. 5A). Gba and Gba2, which also generate ceramide, also exhibited relatively high expression levels in the lung (Fig. 5B). Acer2 and Acer1 had high relative expression in the lung, with levels ninefold and fourfold, respectively, higher than CerS2 (Fig. 5C). As anticipated based on CerS expression, C24:1 (171.8 ± 18.9 pmol/mg protein), C24 (159.5 ± 32.3 pmol/mg protein), and C16 (106.3 ± 25.2 pmol/mg protein) ceramides were the most abundant in the lungs (Fig. 5D). Similar to ceramides, C24-dihydroceramide had the highest concentration for dihydroceramide species. Unlike the ceramides, dihydroceramides with C20 chain length were also a major species and were almost as abundant as the C24-dihydroceramides (Fig. 5E). Similar to the kidney, C16 was the most abundant species for complex sphingolipids, including SM, hexosylceramide, and lactosylceramide (Fig. 5, F–H), and sphingosine levels were highest of the sphingoid bases (Fig. 5I). CerS2 had the highest relative mRNA expression, and C24:1-ceramide had the highest total concentration among ceramide species (Fig. 5K). However, CerS5 also exhibited high expression, and C16 species were the predominant acyl-chain for complex sphingolipids. These data suggest that CerS2 may generate the N-acyl profiles for ceramides and dihydroceramides, whereas CerS5-generated ceramides are preferentially used for complex sphingolipids.
Fig. 5.
CerS2 and CerS5 expression regulate sphingolipid acyl-chain length in the lung. Relative mRNA expression of (A) CerS isoforms, (B) ceramide-generating enzymes, and (C) ceramide-degrading enzymes were analyzed using real-time reverse-transcription polymerase chain reaction. (D–I) Sphingolipid levels were assessed using liquid chromatography tandem mass spectrometry (LC-MS/MS) and normalized to protein [(D) ceramides, (E) dihydroceramides, (F) SMs, G) hexosylceramides (HexCer), (H) lactosylceramides (LactCer), and (I) sphingoid bases]. (J) Table detailing CerS expression and most abundant sphingolipid species in the lung. (K) Donut plots depicting CerS expression (left panel) and CerS contribution to ceramide species (right panel).
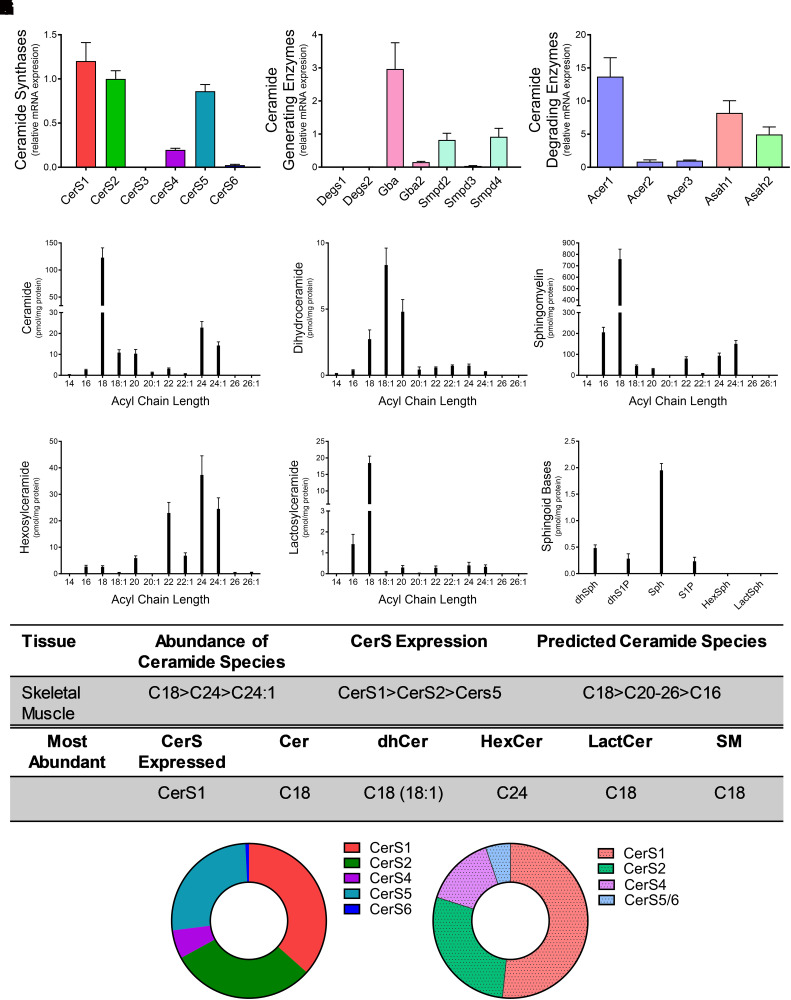
Skeletal Muscle CerSs Expression and Sphingolipidomics
CerS and specific ceramide species have been implicated in metabolic disease in skeletal muscle and in sarcopenia (Gosejacob et al., 2016; Turpin-Nolan et al., 2019; Tosetti et al., 2020). Interestingly CerS1 (37%) was the most highly expressed CerS, with CerS2 (30%) and CerS5 (26%) also expressed at relatively high levels (Fig. 6A). Gba was also highly expressed in skeletal muscle (Fig. 6B). Acer1, Asah1, and Asah2 were highly expressed relative to expression of both Acer2 and Acer3 (Fig. 6C). Lipidomics revealed C18 as the most abundant acyl chain length for all sphingolipids except hexosylceramide (Fig. 6, D–H; C18 ceramide 122.9 ± 44.3 pmol/mg protein, C18:1 dihydroceramide 2.73 ± 1.7 pmol/mg protein, C18-SM 758.1 ± 214.4 pmol/mg protein, and C18 lactosylceramide 18.45 ± 5.1 pmol/mg protein). Interestingly, C24 was the most abundant species for hexosylceramide (Fig. 6G). Sphingosine displayed the greatest concentration among the sphingoid bases (Fig. 6I). CerS1 exhibited the highest relative mRNA expression, and C18 was the predominant species for all sphingolipids measured (Fig. 6, J and K). These data demonstrate a significant contribution of CerS1 in skeletal muscle for the generation of sphingolipids C18.
Fig. 6.
CerS1 and CerS2 expression regulate sphingolipid acyl-chain length in the skeletal muscle. Relative mRNA expression of (A) CerS isoforms, (B) ceramide-generating enzymes, and (C) ceramide-degrading enzymes were analyzed using real-time reverse-transcription polymerase chain reaction. (D–I) Sphingolipid levels were assessed using liquid chromatography tandem mass spectrometry (LC-MS/MS) and normalized to protein [(D) ceramides, (E) dihydroceramides, (F) SMs, (G) hexosylceramides (HexCer), (H) lactosylceramides (LactCer), and (I) sphingoid bases]. (J) Table detailing CerS expression and most abundant sphingolipid species in the skeletal muscle. (K) Donut plots depicting CerS expression (left panel) and CerS contribution to ceramide species (right panel).
Discussion
In this study, we analyzed relative mRNA expression levels of CerS1-6 enzymes and their N-acyl chain ceramide products in six major tissues of C57BL6 mice. We determined correlations between expression of CerS isoforms and the acyl-chain composition for ceramides and dihydroceramides in the tissues analyzed. However, CerS expression and N-acyl chain distribution across complex sphingolipids did not always correlate. This incongruence suggests that availability of specific ceramide species does not singularly determine species generation of complex sphingolipids and that the mechanism of complex sphingolipid species formation is not purely substrate driven. This could result from either substrate preference of SM synthases and glucosylceramide synthase and/or access of these enzymes to specific substrates based on fine tuning of subcellular location of specific ceramides.
In kidney tissue, CerS2 displayed the highest relative mRNA expression. Given that CerS2 preferentially synthesizes ceramides with carbon chain lengths ranging from C20–C26, we expected to see higher amounts of these chain lengths in other complex sphingolipids within the kidney as well, but we only found this to be true for ceramide (C24:1) and dihydroceramide (C20). These data may indicate that lipids produced in the kidney via CerS2 are responsible for the lower levels of circulating VLC ceramide previous observed in diabetic kidney disease by Klein et al. (2014). Though, C16 was found to be the most abundant species for hexosylceramide, lactosylceramide, and SM in the kidney. Interestingly, another report found correlation between chronic kidney disease and C16 SM in circulation (Liu et al., 2016), further indicating that the sphingolipid N-acyl profile of the kidneys may play a larger role in remodeling circulation bioactive lipids in kidney disease states. We suspect that C16-ceramide is being preferentially used to synthesize complex sphingolipids in the kidney rather than the predominant ceramide species with 24-carbon chains. C20-dihydroceramide is also the most abundant of the dihydroceramide species but not for the ceramides or complex sphingolipid species; as dihydroceramides are generated by CerS in the de novo synthesis pathway, this suggests that Degs could have an acyl chain length specificity and that the C20-dihydroceramides are not being metabolized to the corresponding ceramides and complex sphingolipids. The lung displayed similar patterns to the kidney. Functions of the lung and kidney are known to be intimately related in health and disease (Visconti et al., 2016). The similarities in lung and kidney sphingolipid profiles in our study suggests the potential for CerS2 as a potential enzyme of interest in pulmonary-renal crosstalk in health and disease.
Notably, the liver demonstrated congruence of ceramide N-acyl chain distribution and CerS mRNA expression. The most abundant species for each complex sphingolipid in liver aligned with the chain lengths synthesized by CerS2, with VLC sphingolipids predominant in liver. Unlike other tissues, the predominant CerS isoform in liver constituted 96% of CerS expression. The kidney, the tissue with the next highest single isoform expression, only demonstrated CerS2 constituting 55% of CerS expression. This could suggest a positive correlation between CerS expression and relative abundance of individual species across sphingolipid classes. Despite this consistency in liver tissue, the most abundant ceramide chain length (C24) was not the most abundant across other classes of sphingolipids. Instead, complex sphingolipids with 22-carbon chains were highest, further supporting the idea that species other than the most abundant are preferentially used to synthesize complex sphingolipids.
Other enzymes included in our study are also known to demonstrate higher affinity for sphingolipid species of distinct chain lengths. Acid ceramidase (protein) (aCDase), the enzyme responsible for the hydrolysis of ceramide to sphingosine at acidic pH, preferentially binds unsaturated ceramides with N-acyl chain lengths ranging from C6 to C16 (Parveen et al., 2019). Notably, Asah1, the gene encoding aCDase, was consistently highly expressed in all tissues. High expression of Asah1 suggests that high levels of aCDase likely contribute to the consistently high levels of sphingosine within tissues. C14- and C16-ceramide displayed low concentrations throughout most tissues (<20 pmol/mg protein), apart from the lung and kidney (>100 pmol/mg protein). This could be due to specific ceramide species’ consumption by aCDase in specific tissues to replenish sphingosine. Additional enzymes involved in sphingosine generation, including Acer1 and Acer2, were also found to be highly expressed in multiple tissues. Acer1 encodes alkaline ceramidase 1, which preferentially utilizes ceramides with acyl chain lengths of C24 (Sun et al., 2008). Acer2 encodes alkaline ceramidase 2 and demonstrated higher affinity for C16-C24:1 ceramides (Sun et al., 2010). Regeneration of sphingosine from ceramide may be critical within organs, especially in skeletal muscle and kidney. Notably, sphingosine is subsequently phosphorylated by sphingosine kinases to S1P. S1P plays a role in a variety of diseases. One limitation of the current study is that blood levels of sphingolipids were not examined. This is particularly relevant to S1P as it is secreted and has extracellular roles.
The influence of CerS expression on N-acyl chain sphingolipids has been previously examined in the brain, kidney, liver, skeletal muscle, and testis (Laviad et al., 2008). This work demonstrated the importance of CerS2 in several tissues, including kidney and liver. Similar to our studies, CerS2 was the most abundant CerS in both the kidney and liver. There were differences between N-acyl chain abundance across sphingolipid classes in these studies; however, both studies demonstrated that high levels of CerS2 and CerS5 generated sphingolipids. CerS1 expression in our studies and by Laviad et al. (2008) was the most abundant isoform in skeletal muscle tissues, with high levels of C18 ceramide and SM. Interestingly, both studies also showed that VLC (C24 or C24:1) hexosylceramides were the most abundant. CerS1 was also found to be the most abundant isoform in the brain in both studies, with very similar trends in N-acyl chain lengths of sphingolipids. Studies by Laviad et al. (2008) were conducted in 6–8-week-old female mice, whereas our studies were conducted in 12-week-old (3 month) male mice. Together, these studies highlight the importance of CerS in specific tissues, with some of the differences noted between the studies as potentially being sex based.
CerS expression and ceramide generation were not uniform across tissues, suggesting physiologic importance for specific CerS in specific tissues. Petrache et al. (2013) reported high expression of CerS2 and CerS5, and the respective C24 and C16-ceramides, in the lungs of both mice and humans, similar to our measurements of CerS2 and VLC ceramides in lungs. Global knockout of CerS2 not only resulted in a concomitant decrease in C24-ceramide but increased inflammation and reduced lung function, suggesting a crucial role for CerS2 in lung health (Petrache et al., 2013). CerS2 expression and VLC sphingolipid levels are also high in liver and kidney tissues [Figs. 3 and 4; (Pewzner-Jung et al., 2010)]. Indeed, mice lacking CerS2 exhibited significant decreases in VLC ceramides, hexosylceramides, and SMs. In addition, these mice developed hepatocarcinomas beginning at 7 months of age (Imgrund et al., 2009). Similar to the liver, kidney tissues in these mice also demonstrated significantly decreased VLC sphingolipids and a concomitant increased C16 sphingolipids, demonstrating increased CerS5(6) activity in these tissues in the absence of CerS2. Alterations in these lipids in the murine kidney were also suggested to alter kidney architecture with age. In humans, a genetic variant in CerS2 has been linked to increased albuminuria in diabetic patients (Shiffman et al., 2014). These studies further suggest that CerS expression and sphingolipid levels in specific tissues play a significant role in health and disease.
CerS1 and C18-sphignolipids (ceramide and SM) have been shown to be important in brain tissue. Studies in humans have implicated that deficiency in CerS1 activity and C18 sphingolipids resulted in epilepsy and dementia (Vanni et al., 2014). Loss of CerS1 in mice also resulted in neurodegeneration due to Purkinje cell loss (Zhao et al., 2011; Ginkel et al., 2012). Interestingly, C22-C24:1 hexosylceramides were more abundant than either C18:1 or C18 in the brain in both our studies and those conducted by Laviad et al. (2008), suggesting a significant role for CerS2 in CNS. Indeed, mice with global CerS2 deficiency developed encephalopathy with a reduction in VLC hexosylceramides in both total brain lysates and purified myelin (Ben-David et al., 2011). These mice also demonstrate progressive loss of myelin beginning in early adulthood (Imgrund et al., 2009). Together, these studies and others suggest that CerS2 and their associated complex sphingolipids may be important in maintaining brain longevity. CerS1 is also highly expressed in skeletal muscle (Fig. 6). Mice with global or muscle-specific knockout of CerS1 exhibited decreased levels of C18-ceramide, muscle atrophy, and impaired strength (Tosetti et al., 2020). Further, CerS1 expression declined during aging in both mouse and human skeletal muscle, which may have important implications in age-related muscle wasting and sarcopenia (Tosetti et al., 2020). Together, these studies reinforce the importance of specific CerS and specific N-acyl chain sphingolipids in regulating homeostasis and function in brain and skeletal muscle.
Untargeted metabolomic and lipidomic analyses have significantly increased potential implications for many lipid species, especially sphingolipids, in disease. A sphingolipid map qualitatively and quantifying analyzing 114 sphingolipids in 21 different tissues in C57BL/6 mice was recently reported (Muralidharan et al., 2021). Though this study did not examine enzyme expression, tissue lipid compositions were similar in both studies. Similar to the ceramide measurements reported here and by Laviad et al. (2008), Muralidharan et al. reported C18 and C18:1 acyl-chain ceramides to be most abundant in the brain and skeletal muscle as well as high levels of VLC ceramides in liver and kidney tissues. Importantly, Muralidharan et al., examined sex-specific differences and determined significant sex-based differences in several species of hexosylceramides (as well as other sphingolipids) in kidney, skin, liver, lung, and brown adipose tissues as well as plasma. These studies also demonstrated significant increases in many species of ceramides, SMs, and hexosylceramides upon high-fat diet feeding. Though our studies only used male mice, and Laviad et al. (2008) only used female mice, future studies could be geared at defining sex-based differences in basal sphingolipid enzymes and sphingolipid levels.
There are several important limitations of the current study that point to the need for more in-depth future studies. Several organs, including spleen, intestine, skin, and fat, were not analyzed. As sphingolipids play an important role in these organs, future studies should expand on the current analysis to determine CerS-sphingolipid chain length correlation across sphingolipid classes. Another limitation of the current study is that total tissue homogenate was analyzed, and thus the level of sphingolipids in specific cell types was not determined. There is a paucity of cell type–specific CerS expression and sphingolipid data in the literature, and this will be important to focus on to gain mechanistic insight into the role of specific CerS isoforms in development and disease. Third, hexosylceramides were analyzed, which are a total of glucosyl- and galactosylceramides. Future studies will need to independently quantify glucosylceramides and galactosylceramides as well as their downstream complex metabolites as they are important contributors to disease, and very little is known with regard to the contribution of the various CerS to complex glycosphingolipid chain lengths. In addition, our study does not include data for chain lengths less than C14, data for C14- and C16-unsaturated species, or data for incorporation of additional N-acyl chain lengths into the sphingoid backbone or bases. CerS activity and acyl-chain composition of ceramides have also been shown to reflect activity of fatty acyl-CoA elongases. In addition, intracellular trafficking of ceramide also contributes to the generation of SM and glycosphingolipids. Though not measured in our studies, ceramide transport protein has higher affinity for long-chain ceramides (Kumagai et al., 2005) and could also contribute to species of SM and glycosphingolipids in specific tissues. Comprehensive lipidomics, expression of additional lipid-generating enzymes, and their activities would add significant knowledge to composition of lipid species in future studies. Another limitation is that our analyses were restricted to gene expression, which does not always accurately reflect protein levels or post-translation modifications that impact enzyme activity. This is particularly relevant to the CerS that are known to form heterodimers, which is important for their activity (Laviad et al., 2012). Finally, the lipidomics herein are steady-state measurements. Future studies are needed that perform in vivo pulse-chase labeling and perhaps the contribution of various metabolites or dietary fatty acids to sphingolipid metabolism in particular tissues.
The current study is meant to serve as a reference for the sphingolipid community. These data clearly demonstrate significant correlations between CerS expression, even at the mRNA level, and ceramide generation in specific tissues. As detailed above, many of these CerS and ceramide species have also been implicated in disease states in specific tissues. CerS1 knockout mice exhibit significant Purkinje cell loss and cerebellar ataxia (Zhao et al., 2011; Ginkel et al., 2012). CerS2-deficient mice exhibit disease states in multiple organ systems, including the brain (Imgrund et al., 2009; Ben-David et al., 2011), liver (Pewzner-Jung et al., 2010), and lungs (Petrache et al., 2013). CerS3, though not detected in the tissues examined in these studies, has critical implications in skin barrier function (Jennemann et al., 2012). CerS5 and CerS6 knockout mice are protected from diet-induced obesity and metabolic syndrome (Turpin et al., 2014; Gosejacob et al., 2016). These studies demonstrate the potential for modulation of specific CerSs as therapeutic interventions.
Data Availability
The authors declare that all the data supporting the findings of this study are contained within the paper.
Abbreviations
- (dh)ceramide
dihydroceramide
- Acer
alkaline ceramidase
- aCDase
acid ceramidase (protein)
- Asah1
acid ceramidase (gene)
- Asah2
neutral ceramidase
- CerS
ceramide synthase
- Degs
dihydroceramide desaturase
- Gba
β-glucocerebrosidase 2
- MPB
mobile phase B
- S1P
sphingosine-1-phosphate
- SM
sphingomyelin
- SMPD
sphingomyelin phosphodiesterase
- VLC
very long chain
Authorship Contributions
Participated in research design: Richardson, Humphrey, Sears, Hoffman, Orwick, M.A. Doll, Hernandez-Corbacho, Obeid, Hannun, A.J. Snider, Siskind.
Conducted experiments: Richardson, Humphrey, Hoffman, Orwick, M.A. Doll, Hernandez-Corbacho, A.J. Snider, Siskind.
Contributed new reagents or analytic tools: Xia, J.M. Snider.
Performed data analysis: Richardson, Humphrey, Sears, Hoffman, Orwick, M.A. Doll, Xia, Hernandez-Corbacho, J.M. Snider, A.J. Snider, Siskind.
Wrote or contributed to the writing of the manuscript: Richardson, Humphrey, Sears, Hoffman, Orwick, M.A. Doll, C.L. Doll, Xia, J.M. Snider, Hannun, A.J. Snider, Siskind.
Footnotes
Lipidomic analyses in this publication were provided by the Medical University of South Carolina Lipidomics Shared Resource, Hollings Cancer Center, Medical University of South Carolina and were supported in part by National Institutes of Health National Cancer Institute [Grants P30 CA138313 and P30 GM103339]. Composite scoring analyses were provided by the University of Arizona Cancer Center Analytical Chemistry Shared Resource Core and were supported in part by National Institutes of Health National Cancer Institute [Grant P30 CA023074]. Research reported in this publication was supported in part by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases [Grant T35 DK072923] (to W.J.R.), [Grant R01 DK093462] (to L.J.S.), and [Grant R01 DK130971] (to A.J.S.) and the Jewish Heritage Foundation for Excellence Faculty Recruitment and Retention Program (to L.J.S.). The contents of this manuscript do not represent the views of the US Department of Veterans Affairs or the US Government.
No author has an actual or perceived conflict of interest with the contents of this article.
1W.J.R. and S.B.H. contributed equally to this work.
Lina M. Obeid died during the preparation of this article.
References
- Ben-David O, Futerman AH (2010) The role of the ceramide acyl chain length in neurodegeneration: involvement of ceramide synthases. Neuromolecular Med 12:341–350. [DOI] [PubMed] [Google Scholar]
- Ben-David OPewzner-Jung YBrenner OLaviad ELKogot-Levin AWeissberg IBiton IEPienik RWang EKelly S, et al. (2011) Encephalopathy caused by ablation of very long acyl chain ceramide synthesis may be largely due to reduced galactosylceramide levels. J Biol Chem 286:30022–30033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski J, Pierce JS, Snider J, Rembiesa B, Szulc ZM, Bielawska A (2010) Sphingolipid analysis by high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS). Adv Exp Med Biol 688:46–59. [DOI] [PubMed] [Google Scholar]
- Cai XF, Tao Z, Yan ZQ, Yang SL, Gong Y (2003) Molecular cloning, characterisation and tissue-specific expression of human LAG3, a member of the novel Lag1 protein family. DNA Seq 14:79–86. [DOI] [PubMed] [Google Scholar]
- Choi Y, Kim M, Kim SJ, Yoo HJ, Kim SH, Park HS (2020) Metabolic shift favoring C18:0 ceramide accumulation in obese asthma. Allergy 75:2858–2866. [DOI] [PubMed] [Google Scholar]
- Ginkel CHartmann Dvom Dorp KZlomuzica AFarwanah HEckhardt MSandhoff RDegen JRabionet MDere E, et al. (2012) Ablation of neuronal ceramide synthase 1 in mice decreases ganglioside levels and expression of myelin-associated glycoprotein in oligodendrocytes. J Biol Chem 287:41888–41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosejacob D, Jäger PS, Vom Dorp K, Frejno M, Carstensen AC, Köhnke M, Degen J, Dörmann P, Hoch M (2016) Ceramide Synthase 5 Is Essential to Maintain C16:0-Ceramide Pools and Contributes to the Development of Diet-induced Obesity. J Biol Chem 291:6989–7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM (2018) Sphingolipids and their metabolism in physiology and disease. Nat Rev Mol Cell Biol 19:175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imgrund S, Hartmann D, Farwanah H, Eckhardt M, Sandhoff R, Degen J, Gieselmann V, Sandhoff K, Willecke K (2009) Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J Biol Chem 284:33549–33560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennemann RRabionet MGorgas KEpstein SDalpke ARothermel UBayerle Avan der Hoeven FImgrund SKirsch J, et al. (2012) Loss of ceramide synthase 3 causes lethal skin barrier disruption. Hum Mol Genet 21:586–608. [DOI] [PubMed] [Google Scholar]
- Klein RL, Hammad SM, Baker NL, Hunt KJ, Al Gadban MM, Cleary PA, Virella G, Lopes-Virella MF; DCCT/EDIC Research Group (2014) Decreased plasma levels of select very long chain ceramide species are associated with the development of nephropathy in type 1 diabetes. Metabolism 63:1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai K, Yasuda S, Okemoto K, Nishijima M, Kobayashi S, Hanada K (2005) CERT mediates intermembrane transfer of various molecular species of ceramides. J Biol Chem 280:6488–6495. [DOI] [PubMed] [Google Scholar]
- Laviad EL, Albee L, Pankova-Kholmyansky I, Epstein S, Park H, Merrill AH Jr, Futerman AH (2008) Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J Biol Chem 283:5677–5684. [DOI] [PubMed] [Google Scholar]
- Laviad EL, Kelly S, Merrill AH Jr, Futerman AH (2012) Modulation of ceramide synthase activity via dimerization. J Biol Chem 287:21025–21033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Futerman AH (2010) Mammalian ceramide synthases. IUBMB Life 62:347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JJGhosh SKovalik JPChing JChoi HWTavintharan SOng CNSum CFSummers SATai ES, et al. (2016) Profiling of Plasma Metabolites Suggests Altered Mitochondrial Fuel Usage and Remodeling of Sphingolipid Metabolism in Individuals With Type 2 Diabetes and Kidney Disease. Kidney Int Rep 2:470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani Y, Kihara A, Igarashi Y (2006) LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem J 398:531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan S, Shimobayashi M, Ji S, Burla B, Hall MN, Wenk MR, Torta F (2021) A reference map of sphingolipids in murine tissues. Cell Rep 35:109250. [DOI] [PubMed] [Google Scholar]
- Parveen F, Bender D, Law SH, Mishra VK, Chen CC, Ke LY (2019) Role of Ceramidases in Sphingolipid Metabolism and Human Diseases. Cells 8:1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrache I, Kamocki K, Poirier C, Pewzner-Jung Y, Laviad EL, Schweitzer KS, Van Demark M, Justice MJ, Hubbard WC, Futerman AH (2013) Ceramide synthases expression and role of ceramide synthase-2 in the lung: insight from human lung cells and mouse models. PLoS One 8:e62968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pewzner-Jung YBrenner OBraun SLaviad ELBen-Dor SFeldmesser EHorn-Saban SAmann-Zalcenstein DRaanan CBerkutzki T, et al. (2010) A critical role for ceramide synthase 2 in liver homeostasis: II. insights into molecular changes leading to hepatopathy. J Biol Chem 285:10911–10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabionet Mvan der Spoel ACChuang CCvon Tümpling-Radosta BLitjens MBouwmeester DHellbusch CCKörner CWiegandt HGorgas K, et al. (2008) Male germ cells require polyenoic sphingolipids with complex glycosylation for completion of meiosis: a link to ceramide synthase-3. J Biol Chem 283:13357–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SB, Baicu CF, Van Laer A, Geng T, Kasiganesan H, Zile MR, Cowart LA (2012) Ceramide synthase 5 mediates lipid-induced autophagy and hypertrophy in cardiomyocytes. J Clin Invest 122:3919–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SB, Tidhar R, Futerman AH, Cowart LA (2013) Myristate-derived d16:0 sphingolipids constitute a cardiac sphingolipid pool with distinct synthetic routes and functional properties. J Biol Chem 288:13397–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffman D, Pare G, Oberbauer R, Louie JZ, Rowland CM, Devlin JJ, Mann JF, McQueen MJ (2014) A gene variant in CERS2 is associated with rate of increase in albuminuria in patients with diabetes from ONTARGET and TRANSCEND. PLoS One 9:e106631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Jin J, Xu R, Hu W, Szulc ZM, Bielawski J, Obeid LM, Mao C (2010) Substrate specificity, membrane topology, and activity regulation of human alkaline ceramidase 2 (ACER2). J Biol Chem 285:8995–9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Xu R, Hu W, Jin J, Crellin HA, Bielawski J, Szulc ZM, Thiers BH, Obeid LM, Mao C (2008) Upregulation of the human alkaline ceramidase 1 and acid ceramidase mediates calcium-induced differentiation of epidermal keratinocytes. J Invest Dermatol 128:389–397. [DOI] [PubMed] [Google Scholar]
- Tosetti BBrodesser SBrunn ADeckert MBlüher MDoehner WAnker SDWenzel DFleischmann BPongratz C, et al. (2020) A tissue-specific screen of ceramide expression in aged mice identifies ceramide synthase-1 and ceramide synthase-5 as potential regulators of fiber size and strength in skeletal muscle. Aging Cell 19:e13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trayssac M, Hannun YA, Obeid LM (2018) Role of sphingolipids in senescence: implication in aging and age-related diseases. J Clin Invest 128:2702–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin-Nolan SM, Hammerschmidt P, Chen W, Jais A, Timper K, Awazawa M, Brodesser S, Brüning JC (2019) CerS1-Derived C18:0 Ceramide in Skeletal Muscle Promotes Obesity-Induced Insulin Resistance. Cell Rep 26:1–10.e7. [DOI] [PubMed] [Google Scholar]
- Turpin SMNicholls HTWillmes DMMourier ABrodesser SWunderlich CMMauer JXu EHammerschmidt PBrönneke HS, et al. (2014) Obesity-induced CerS6-dependent C16:0 ceramide production promotes weight gain and glucose intolerance. Cell Metab 20:678–686. [DOI] [PubMed] [Google Scholar]
- Vanni NFruscione FFerlazzo EStriano PRobbiano ATraverso MSander TFalace AGazzerro EBramanti P, et al. (2014) Impairment of ceramide synthesis causes a novel progressive myoclonus epilepsy. Ann Neurol 76:206–212. [DOI] [PubMed] [Google Scholar]
- Visconti L, Santoro D, Cernaro V, Buemi M, Lacquaniti A (2016) Kidney-lung connections in acute and chronic diseases: current perspectives. J Nephrol 29:341–348. [DOI] [PubMed] [Google Scholar]
- Wang XSong MLi XSu CYang YWang KLiu CZheng ZJia YRen S, et al. (2023) CERS6-derived ceramides aggravate kidney fibrosis by inhibiting PINK1-mediated mitophagy in diabetic kidney disease. Am J Physiol Cell Physiol 325:C538–C549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelnik ID, Volpert G, Viiri LE, Kauhanen D, Arazi T, Aalto-Setälä K, Laaksonen R, Futerman AH (2020) Different rates of flux through the biosynthetic pathway for long-chain versus very-long-chain sphingolipids. J Lipid Res 61:1341–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Spassieva SD, Jucius TJ, Shultz LD, Shick HE, Macklin WB, Hannun YA, Obeid LM, Ackerman SL (2011) A deficiency of ceramide biosynthesis causes cerebellar purkinje cell neurodegeneration and lipofuscin accumulation. PLoS Genet 7:e1002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors declare that all the data supporting the findings of this study are contained within the paper.