Abstract
This study describes the localization and computational prediction of a binding site for the A3 adenosine receptor (A3AR) positive allosteric modulator 2-cyclohexyl-1H-imidazo[4,5-c]quinolin-4-(3,4-dichlorophenyl)amine (LUF6000). The work reveals an extrahelical lipid-facing binding pocket disparate from the orthosteric binding site that encompasses transmembrane domain (TMD) 1, TMD7, and Helix (H) 8, which was predicted by molecular modeling and validated by mutagenesis. According to the model, the nearly planar 1H-imidazo[4,5-c]quinolinamine ring system lies parallel to the transmembrane segments, inserted into an aromatic cage formed by π-π stacking interactions with the side chains of Y2847.55 in TMD7 and Y2938.54 in H8 and by π-NH bonding between Y2847.55 and the exocyclic amine. The 2-cyclohexyl group is positioned “upward” within a small hydrophobic subpocket created by residues in TMDs 1 and 7, while the 3,4-dichlorophenyl group extends toward the lipid interface. An H-bond between the N-1 amine of the heterocycle and the carbonyl of G291.49 further stabilizes the interaction. Molecular dynamics simulations predicted two metastable intermediates, one resembling a pose determined by molecular docking and a second involving transient interactions with Y2938.54; in simulations, each of these intermediates converges into the final bound state. Structure-activity-relationships for replacement of either of the identified exocyclic or endocyclic amines with heteroatoms lacking H-bond donating ability were consistent with the hypothetical pose. Thus, we characterized an allosteric pocket for 1H-imidazo[4,5-c]quinolin-4-amines that is consistent with data generated by orthogonal methods, which will aid in the rational design of improved A3AR positive allosteric modulators.
SIGNIFICANCE STATEMENT
Orthosteric A3AR agonists have advanced in clinical trials for inflammatory conditions, liver diseases, and cancer. Thus, the clinical appeal of selective receptor activation could extend to allosteric enhancers, which would induce site- and time-specific activation in the affected tissue. By identifying the allosteric site for known positive allosteric modulators, structure-based drug discovery modalities can be enabled to enhance the pharmacological properties of the 1H-imidazo[4,5-c]quinolin-4-amine class of A3AR positive allosteric modulators.
Introduction
Pharmaceutical targeting of the A3 adenosine receptor (A3AR) has emerged as a promising therapeutic approach for a broad spectrum of diseases that are driven by chronic inflammation. The A3AR is a Gi protein-coupled receptor that is abundantly expressed in various immune cell populations including granulocytic cells (neutrophils, eosinophils, basophils, mast cells), macrophages, and microglia, where it controls chemotaxis and cellular activation (Walker et al., 1997; Jordan et al., 1999; Hammarberg et al., 2003; Chen et al., 2006; Haskó et al., 2008; van der Hoeven et al., 2008; Ge et al., 2010; van der Hoeven et al., 2010; Antonioli et al., 2022; Gao et al., 2023). Moderately selective nucleoside agonists for the A3AR first developed nearly 30 years ago, including N6-(3-iodobenzyl)adenosine-5′-N-methylcarboxamide (IB-MECA) and its 2-chloro derivative Cl-IB-MECA, are currently in advanced clinical trials for the treatment of psoriasis, nonalcoholic steatohepatitis, and hepatocellular carcinoma (Fishman et al., 2023). Newer nucleoside agonists containing a rigid ribose substitution, some of which are greater than 10,000-fold selective verses the other adenosine receptor subtypes (A1, A2A, and A2BARs), are being developed for the treatment of neuropathic pain, stroke, and traumatic brain injury (Liston et al., 2020, 2022; Bozdemir et al., 2021).
As an alternative and more precise approach to target the A3AR, we have pursued the development of positive allosteric modulators (PAMs) for the clinically important A3AR. G protein-coupled receptor (GPCR) PAMs are ligands that act at an allosteric site outside the orthosteric binding site for the endogenous agonist, thereby magnifying signaling by increasing orthosteric agonist binding affinity resulting in an increase in potency and/or by increasing the orthosteric agonist signaling efficacy (Coughlin et al., 2019; Slosky et al., 2021; Wang et al., 2021). The advantages of allosteric modulators include the potential for greater specificity since allosteric sites are not subjected to evolutionary pressures to accommodate a shared ligand across receptor subtypes. The spatiotemporal specificity of PAMs, in principle, reduces the risk of unwanted, on-target side effects and the potential for efficacy loss due to receptor desensitization.
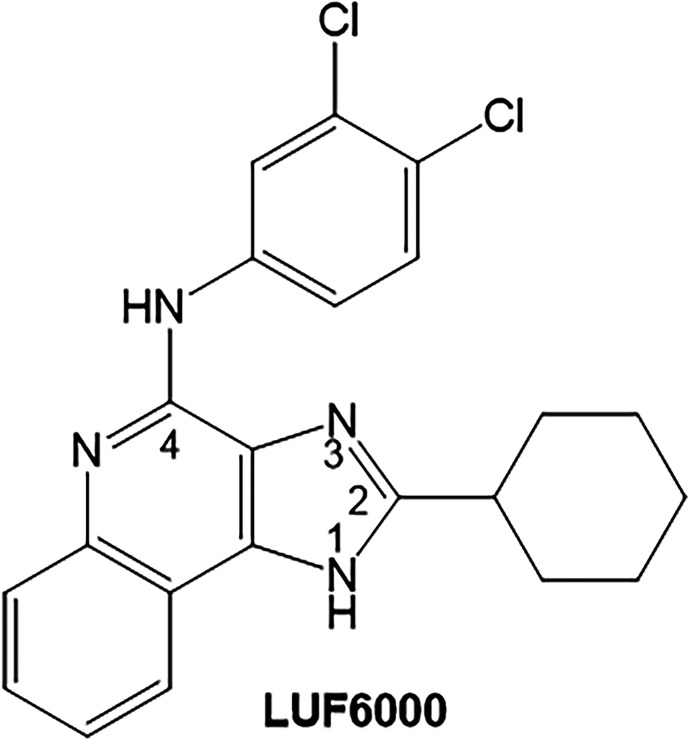
In prior work, we have developed an A3AR PAM, named LUF6000, and its congeners based on the 1H-imidazo[4,5-c]quinolin-4-amine chemical scaffold (Fig. 1) (Gao et al., 2002, 2008, 2011; Göblyös et al., 2006; Kim et al., 2009; Du et al., 2012, 2018; Fallot et al., 2022; Fisher et al., 2022). LUF6000 displays positive cooperativity with orthosteric agonists, enhancing their A3AR binding and potentiating agonist-induced downstream signaling. Uniquely, LUF6000 has been shown in transfected human embryonic kidney 293 (HEK293) cells to selectively enhance signaling by specific Gα protein isoforms (Gαi3 and Gα0A) without potentiating β-arrestin2 recruitment. This suggests that LUF6000 and its congeners may bias signaling in cells that coexpress the A3AR and specific Gα protein isoforms, such as spinal cord neurons involved in pain sensation and immune cells (Fisher et al., 2022). Unfortunately, at higher concentrations approaching 1 to 10 μM, essentially all of the 1H-imidazo[4,5-c]quinolin-4-amine derivatives thus far synthesized and characterized begin to reduce agonist potency due to a mechanism of action involving direct competition for orthosteric site binding. Another disadvantage of LUF6000 and all other similar 1H-imidazo[4,5-c]quinolin-4-amine derivatives we have investigated to date is that they are only weakly active at rodent A3ARs, thereby limiting the ability to test for efficacy in experimental animal models of disease (Du et al., 2018; Fallot et al., 2022; Fisher et al., 2022). Thus, we continue to search for improved derivatives with rodent activity and less propensity to reduce agonist potency; to date this endeavor is impeded by the lack of A3AR structural information to help guide drug design decisions.
Fig. 1.
Chemical structure of the 1H-imidazo[4,5-c]quinoline-4-amine A3AR positive allosteric modulator LUF6000.
Due to rapid advances in structural and computational biology, the diversity of allosteric binding sites for GPCRs is becoming increasingly appreciated. Locations of PAM binding sites that have been verified include the “extracellular vestibule” of muscarinic receptors comprising the extracellular face of the transmembrane domain (TMD) bundle that lines the path leading to the orthosteric site deeper within the transmembrane core (Burger et al., 2018; Nguyen et al., 2022; van der Westhuizen et al., 2021). However, a β2 adrenergic receptor PAM (Compound-6A) and a dopamine D1 receptor PAM (DETQ) each bind on their respective receptor’s inner surface in a pocket created by intracellular loop 2 and TMDs 3 and 4 (Wang et al., 2018; Liu et al., 2019). More recently, a cryogenic electron microscopic structure of the A1 adenosine receptor, complexed with adenosine, the αi2 G protein subunit, and the PAM MIPS521 was reported (Draper-Joyce et al., 2021). Here, MIPS521 was described to bind to an intramembrane extrahelical site that involves TMDs 1, 6, and 7. With each of these uniquely positioned, diverse sites, allosteric ligand binding is hypothesized to alter the receptor’s ability to transition between active and inactive states, providing positive cooperativity.
Previously, we exploited species differences using a human (responding species)/mouse (nonresponding species) chimeric receptor approach to localize the LUF6000 binding pocket to the inner portions (with respect to the lipid bilayer) of the receptor (Fisher et al., 2022). In the current study, the binding region was narrowed further, informed by additional chimeric receptor studies, which was followed by induced-fit docking and molecular dynamics to pinpoint the binding site for LUF6000. This site is located in an extrahelical, lipid-facing pocket formed outside the receptor core similar to that described for the A1AR, except in this case binding interactions occur with TMD1, TMD7, and Helix (H)8.
Materials and Methods
Materials
Unless noted otherwise, all chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO). Mycoplasma-free HEK293 cells were purchased from the American Type Tissue Collection (#CRL-1573, Manassas, VA). The cells were further tested for possibility of mycoplasm contamination on a yearly basis. Cell culture media and additional components were from Thermo Fisher (Waltham, MA). LUF6000 was custom synthesized as previously described at a purity of > 95% (Göblyös et al., 2006).
Creation of HEK293 Cells Lines Expressing Adenosine Receptors
Full-length cDNAs encoding wild-type human (AY136749.1), wild-type mouse (NM_009631.4), human/mouse chimeric, and mutated A3ARs were synthesized commercially (Top Gene Technologies, St. Laurent, Quebec) and subcloned into pcDNA3.1 (#V79020, Invitrogen, Carlsbad, CA). Plasmids were transfected into HEK293 cells using TransIT-293 transfection reagent (#MIR 2704, Mirus Bio, Madison, WI) and selected using 2 mg/ml G418 (#G-418-5, Goldbio, St. Louis, MO) in cell culture media (DMEM with 10% fetal bovine serum, 100 units/ml penicillin, and 100 mg/ml streptomycin). Cell lines derived from individual clones were maintained in media containing 0.6 mg/ml G418. The expression level of each cell line expressing mutant receptors was similar (∼1,000–3,000 fmol/mg protein) based on saturation radioligand binding analysis (Supplemental Table 1).
Membrane Preparations
HEK293 cells were washed with PBS followed by homogenization in Buffer A containing 10 mM Na-HEPES (pH 7.4), 10 mM EDTA, and 1 mM benzamidine and centrifuged (27,000 × g) for 30 min at 4°C. Cell pellets were rehomogenized in Buffer A (except the EDTA concentration was reduced to 1 mM) and centrifuged. The supernatant was discarded, and cell pellets were resuspended in Buffer A (1 mM EDTA) containing 10% glucose and stored at -20°C.
Guanosine 5′-[γ-[35S]thio]triphosphate Binding Assays
HEK293 cell membranes (5–10 μg of protein) were pretreated with LUF6000 for 1 h in 100 μl of GTPγS binding buffer (50 mM Tris HCl at pH 7.4, 1 mM EGTA, 10 mM MgCl2, 100 mM NaCl, 0.004% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid, and 0.5% bovine serum albumin) in a 96-well, large-volume polypropylene plate (Du et al., 2018; Fisher et al., 2022). In all assays, ZM241385 and PSB-603 (#1026 and 3198, respectively, Tocris, Bristol, UK), each at a final concentration of 300 nM, were included in the assays to block A2BARs expressed endogenously in HEK293 cells. Adenosine deaminase (1 unit/ml; #10102105001, Roche Diagnostics, Mannheim, Germany) was also included to degrade adenosine present in the assay. The reactions were initiated by the addition of ∼0.2 nM guanosine 5′-[γ-[35S]thio]triphosphate ([35S]GTPγS) (#NEG030H250UC, Perkin-Elmer, Waltham, MA) and the selective A3AR agonist Cl-IB-MECA (#1104, Tocris, Bristol, UK) at the indicated concentrations, performed in triplicate, and incubated for 2 h at room temperature. At the end of the 2-h incubation period, the membranes were harvested by rapid filtration through Whatman GF/B filters (#FP-105, Brandel, Gaithersburg, MD) presoaked in GTPγS binding buffer containing 0.02% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid using a 96-well cell harvester (Brandel). Radioactivity trapped by the filter was measured by scintillation counting. Nonspecific binding was determined in the presence of 10 μM unlabeled GTPγS (#G8634, Sigma-Aldrich). Results with LUF6000 are normalized to the Emax value obtained in the presence of the vehicle.
[125I]I-AB-MECA Binding Assays
Cell membranes (50 μg) were incubated in 100 μl of binding buffer (50 mM Tris-HCl at pH 7.4, 10 mM MgCl2, 1 mM EDTA, and 1 unit/ml adenosine deaminase) containing ∼0.3 nM [125I]N6-(4-amino-3-iodobenzyl)-adenosine-5′-N-methylcarboxamide ([125I]I-AB-MECA) (Melman et al., 2008; Paoletta et al., 2013). The reactions, performed in triplicate, were incubated at room temperature for the times indicated, after which bound and free radioligand were separated by rapid filtration through GF/C glass fiber filters (#FP-205, Brandel). Radioactivity trapped by the filters was measured using a gamma counter. For dissociation studies, [125I]I-AB-MECA was incubated with membranes for 3 h to achieve equilibrium, after which the assays were initiated by the addition of adenosine-5′-N-ethylcarboxamide (100 μM; #1691, Tocris, Bristol, UK), along with LUF6000 (10 μM) or equivalent vehicle (DMSO). Specific [125I]I-AB-MECA binding was measured by rapid filtration at the indicated times. Nonspecific binding was measured by including 100 μM adenosine-5′-N-ethylcarboxamide for the duration of the assay. For equilibrium binding assays, membranes were incubated for 3 h with 6 to 8 concentrations of [125I]I-AB-MECA for 3 h before filtration; the specific activity of [125I]I-AB-MECA was reduced 10- to 20-fold with the nonradioactive compound. [125I]I-AB-MECA (specific activity ∼2,200 Ci/mmol) was prepared by radioiodination of AB-MECA (#28415, Cayman Chemical, Ann Arbor, MI) using the chloramine-T method and purified by high-pressure liquid chromatography (Auchampach et al., 1997).
Molecular Modeling
Detailed procedures for A3AR protein structure prediction, molecular docking, and molecular dynamics are described in detail in the Supplemental Material file.
Data Analysis
For [35S]GTPγS binding assays, EC50 and Emax values were calculated from data obtained from concentration-response curves according to: E = (EMax * x)/(EC50 + x), in which x is the concentration of Cl-IB-MECA. For [125I]I-AB-MECA dissociation binding assays, data were fit to a one-phase exponential decay model: Y = (Y0 – NS)e(-k * t), in which Y0 is specific binding at time 0, k is the dissociation rate constant, NS is nonspecific binding, and t is elapsed time. Data (Kd and Bmax values) from [125I]I-AB-MECA saturation binding assays were fit optimally to a one-site binding model described within the GraphPad (version 9.5.1) package that accounts for ligand depletion to determine Kd and Bmax values. All values are presented as the mean ± S.D. Data were compared using unpaired Student’s t tests or one-way ANOVA with Dunnett’s multiple comparison tests to identify concentrations of LUF6000 that produced changes in the EC50 and/or Emax of Cl-IB-MECA compared with vehicle (DMSO). A P value < 0.05 was considered statistically significant. Please note that this study was exploratory and not designed to test a prespecified null hypothesis. Therefore, calculated P values are descriptive only and must not be interpreted as hypothesis testing.
Results
A3AR Human/Mouse Chimeras
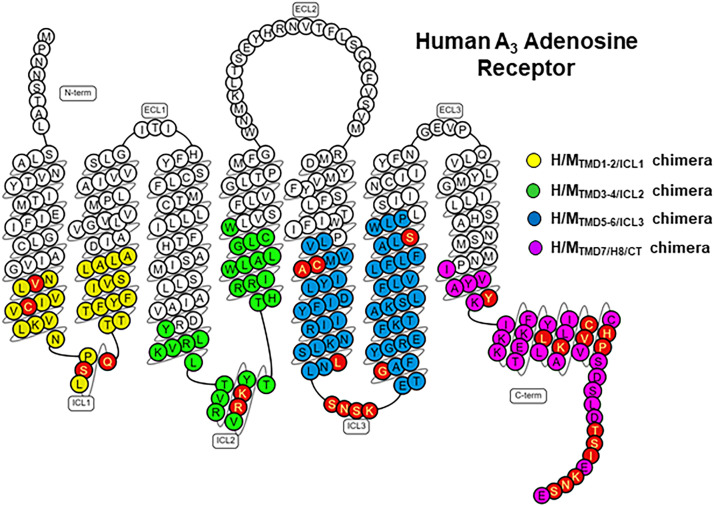
Our recently published work with human/mouse chimeras predicted the allosteric binding pocket for LUF6000 to be within the lower portions of the A3AR but did not delineate which of the TMDs and their corresponding intracellular loops is/are involved (Fisher et al., 2022). Therefore, we generated four new chimeric receptors depicted in Figs. 2 and 3A, in which we replaced the human A3AR sequence with the mouse sequence. The first three chimeras replaced each individual intracellular loop along with the lower half of each connecting TMD on either side with the mouse sequence, which were designated H/MTMD1-2/ICL1, H/MTMD3-4/ICL2, and H/MTMD5-6/ICL3. Separation between proximal and distal portions of the TMDs was ×.50 per the Ballesteros–Weinstein numbering system, where amino acid ×.50 is the most conserved amino acid within that TMD (Ballesteros and Weinstein, 1995). The fourth chimera (H/MTMD7/H8/CT) replaced the distal portion of TMD7, H8, and the C-terminus with the mouse sequence. Residue C3038.64 within H8 is a palmitoylation site and defines the border between H8 and the C-terminal tail. As shown in Fig. 2, the number of amino acid differences for each chimera were as follows: H/MTMD1-2/ICL1 = 4, H/MTMD3-4/ICL2 = 2, H/MTMD5-6/ICL3 = 9, and H/MTMD7/H8/CT = 1 in TMD7, 4 in H8, and 8 in the C-terminus (13 total).
Fig. 2.
Strategy for creating human/mouse chimeric receptors. (A) Snake diagram of the human A3AR amino acid sequence. Colored circles demarcate regions that were scanned for differences between the human and mouse sequences for the following chimeras: yellow = H/MTMD1-2/ICL1, green = H/MTMD3-4/ICL2, blue = H/MTMD5-6/ICL3, purple = H/MTMD7/H8/CT. Amino acids that differ between the two sequences and changed to the mouse sequence for the individual chimeras are highlighted in red. Figure prepared using the GPCRdb database (www.gpcrdb.org).
Fig. 3.
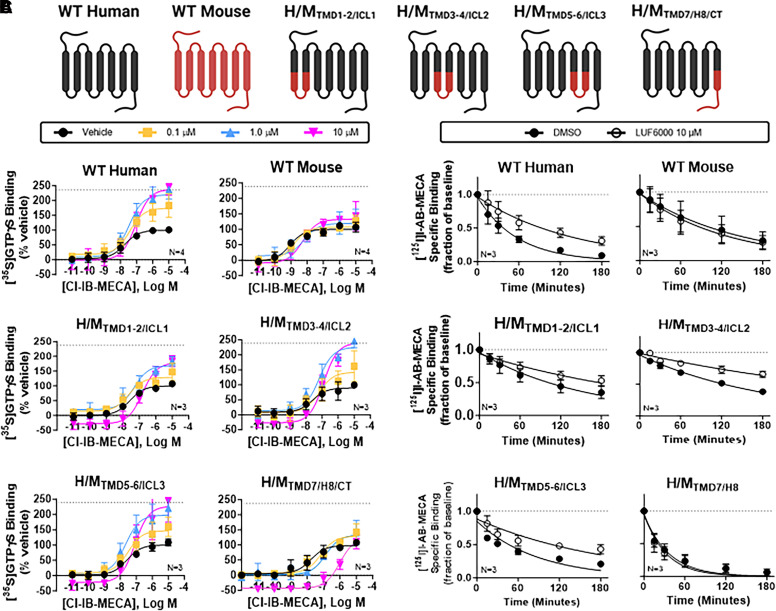
Characterization of human/mouse chimeric A3ARs. (A) Cartoon depicting the makeup of each chimeric receptor. (B) [35S]GTPγS binding assays. Concentration-response curves with Cl-IB-MECA were conducted in the presence of vehicle (DMSO, black) or 0.1 (yellow), 1 (blue), or 10 μM (magenta) LUF6000. Results were normalized to the Emax value of Cl-IB-MECA obtained in the presence of vehicle. The dotted line in each graph demarcates the Emax of Cl-IB-MECA in the presence of 10 μM LUF6000 with membranes from wild-type A3ARs (240 ± 11% of vehicle). (C) [125I]I-AB-MECA dissociation binding assays. The allosteric actions of LUF6000 were lost in assays with the H/MTMD7/H8/CT chimera and diminished in assays with the H/MTMD1-2/ICL1 chimera. EC50, Emax, and k values are reported in Supplemental Table 2. Data are presented as the mean ± S.D.
Each chimera was evaluated in [35S]GTPγS binding assays to assess G protein exchange activity. Concentration-response curves were performed with the A3AR-selective orthosteric agonist Cl-IB-MECA (10−11–10−5 M) in the presence of either vehicle (DMSO) or 0.1, 1, or 10 µM LUF6000. For each chimera, the potency of Cl-IB-MECA to stimulate [35S]GTPγS binding was similar (Supplemental Table 2). As shown in Fig. 3B and Supplemental Table 2, assays with the wild-type human A3AR LUF6000 increased the Emax of Cl-IB-MECA ∼2.4-fold (Supplemental Table 2), demonstrating efficacy enhancement while decreasing its EC50 ∼5-fold (from 19 to 90 nM). Our prior work indicates that the reduction in potency of Cl-IB-MECA produced by LUF6000 is due to competitive antagonism at the orthosteric binding site (Fisher et al., 2022). In contrast, LUF6000 failed to increase the Emax of Cl-IB-MECA in assays with the wild-type mouse A3AR, although it continued to compete for binding at the orthosteric site, resulting in a reduction in the potency of Cl-IB-MECA (from 1 to 5 nM).
Assays using isolated HEK293 cell membranes expressing H/MTMD3-4/ICL2 and H/MTMD5-6/ICL3 chimeras determined susceptibility to efficacy enhancement by LUF6000 similar to that observed with wild-type human A3ARs (Fig. 3B; Supplemental Table 2). Mild efficacy enhancement (∼79% increase in the Emax of Cl-IB-MECA at a concentration of 10 μM versus vehicle) was observed in assays using H/MTMD1-2/ICL1 membranes. Strikingly, however, when using H/MTMD7/H8/CT membranes LUF6000 failed to produce efficacy enhancement although it produced a substantial reduction in potency [EC50 of C-IB-MECA = 19 nM (vehicle) versus 966 nM (10 μM LUF6000)]. The loss in allosteric activity of LUF6000 unmasked its full antagonistic propensity (competition for orthosteric ligand binding), resulting in a dramatic rightward shift in the Cl-IB-MECA concentration-response curve. [125I]I-AB-MECA (agonist) radioligand dissociation binding assays, which detect the pure allosteric actions of LUF6000 uncomplicated by its orthosteric effects, corroborated findings from the G protein activation assays where LUF6000 at a concentration of 10 μM was found to slow the rate of [125I]I-AB-MECA dissociation in assays with the H/MTMD1-2/ICL1, H/MTMD3-4/ICL2, and H/MTMD5-6/ICL3 chimeras (Fig. 3C, Supplemental Table 2). However, with the H/MTMD7/H8/CT chimera, LUF6000 no longer slowed [125I]I-AB-MECA dissociation, indicating a loss of allosteric activity. Collectively, these results narrow the LUF6000 binding site to a region comprising the distal portions of TMD7 and H8. Results of the [35S]GTPγS binding assays also suggest potential interactions with TMD1 and/or intracellular loop 1. TMD2 was ruled out since there are no amino acid differences in the lower portion of TMD2 between the human and mouse.
Molecular Modeling
To map the LUF6000 binding site, we created a structural model of the A3AR using the AlphaFold multistate protocol (Heo and Feig, 2022). This is a modified version of AlphaFold (Jumper et al., 2021), which utilizes an annotated database of experimentally solved GPCR structures categorized according to their activation state. In this case, we modeled the human A3AR in its active conformation. Subsequently, docking was performed with LUF6000 using the standard precision protocol of the Glide program (Friesner et al., 2004). Considering the results of the chimeric receptor studies, the predicted topology of the receptor’s H8 region, and the hydrophobic properties of LUF6000, an extrahelical binding site was anticipated. Accordingly, the SiteMap tool (Halgren, 2007, 2009) was employed to search for hypothetical binding sites on the receptor surface (Supplemental Fig. 1), and the fourth-ranked site (site S4; SScore: 0.891, DScore 0.952) was found to be located at the interface among TMD1, TMD7, and H8, correlating precisely with the results of the human/mouse chimeric receptor studies. Thus, docking analysis with LUF6000 was concentrated on this region. Interestingly, four additional potential binding sites were identified from the SiteMap analysis in regions that are not consistent with the human/mouse chimeric receptor results (Supplemental Fig. 1). The top-ranked predicted binding site correlates with the putative orthosteric ligand binding site.
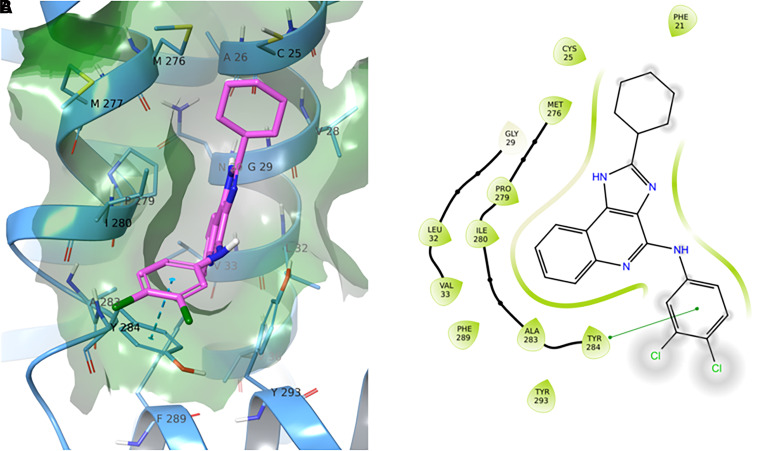
To generate possible LUF6000 binding poses within the S4 allosteric pocket, we carried out molecular docking analyses with the Glide program (Friesner et al., 2004), and calculations of the top five-ranked poses are provided in Supplemental Fig. 2. Because four of the poses (P1, P2, P3, and P5) were degenerate, the top-ranked P1 pose was selected as the putative LUF6000 binding mode, as depicted in Fig. 4. According to the model, the nearly planar 1H-imidazo[4,5-c]quinolin-4-amine ring system of LUF6000 lies parallel to the transmembrane segments, inserted into an aromatic pocket formed by Y2847.55, F2898.50, and Y2938.54. The 2-cyclohexyl group is positioned “upward” within a small hydrophobic subpocket (S4’) created by residues M2767.47, M2777.48, P2797.50, C251.45, V1.48, and G291.49, while the 3,4-dichlorophenyl group extends toward the lipid interface via a π-π stacking interaction with the aromatic sidechain of Y2847.55. Notably, no stabilizing H-bond interactions between LUF6000 and residues within the binding pocket were predicted. This was surprising considering the proximity of several highly interactive residues, notably Y2847.55 and Y2938.54. Thus, the pose suggests weak binding interactions. Because of the lack of H-bonding with the representative P1 pose, we investigated the merits of pose P4 (Supplemental Fig. 3), which is similar except the orientation of the heterocyclic scaffold is flipped such that the 3,4-dichlorophenyl group associates with the hydrophobic subpocket and the 2-cyclohexyl group extends toward the lipid interface without a π-π stacking interaction. Similarly, with this pose no H-bonding was predicted.
Fig. 4.
Docking of LUF6000 with the AlphaFold model of the A3AR. The top ranked pose (P1) for LUF6000 within the S4 allosteric site is shown. Panel A reports a three-dimensional representation of the pose, where LUF6000 is depicted in green and surrounding receptor residues within a 5 Å radius are shown in cornflower blue. Connolly surface is projected and colored according to the residue type coloring scheme of Maestro. Panel B illustrates the bidimensional interaction scheme for LUF6000 within the S4 pocket as provided by Maestro. A π-π stacking interaction is shown between Y2847.55 and the 3,4-dichlorophenyl group of LUF6000. The 2-cyclohexyl group is positioned within a hydrophobic subpocket formed by residues in TMDs 1 and 7.
To further refine pose P1, multiple independent MD simulations were performed to reveal whether additional stabilizing interactions may form in a dynamic environment. Since the active-state conformation of GPCRs can rapidly devolve if left unrestrained (Latorraca et al., 2017), simulations were performed with the AlphaFold multistate A3AR model complexed with a homology model of the Gαi subunit, based on the active-state cryo-electron microscopy structure of the A1AR bound to adenosine and complexed with the Gαi2 subunit as a template (PDB ID: 7LD4; Draper-Joyce et al., 2021). In addition, C3038.64 in H8 was palmitoylated due to its proximity to the putative LUF6000 binding site. Simulations were conducted identically either without (apo) or with (holo) LUF6000 bound to the S4 allosteric site, where the systems were 1) embedded within a phosphatidylcholine bilayer solvated with explicit water molecules containing a physiologic concentration of sodium chloride, 2) equilibrated for 40 ns, and 3) subjected to three independent MD replicates of 400 ns duration. Trajectories from the independent simulations were clustered based on the pairwise root mean square deviation (RMSD) matrix of the receptor backbone (apo system) or LUF6000 coordinates (holo system) with the TTCLUST Python package (Tubiana et al., 2018). For each cluster, the population size, expressed as a percentage of trajectory frames within the cluster, is reported.
MD Simulations with the Apo System
Cluster analyses of the apo system revealed an equilibrium between an open conformation of the binding site and a closed conformation, depending on the relative orientation of Y2847.55 (Supplemental Fig. 4). In the closed conformation, Y2847.55 stacks against Y2938.54 and H-bonds with the backbone carbonyl of G291.49, completely closing the pocket and transforming it into a flat hydrophobic surface. In contrast, Y2847.55 projects away from Y2938.54 in the open conformation, allowing access to the pocket. The presence of an equilibrium between these two alternate conformations lends support to the proposed P1 docking pose, whereby components of LUF6000 may interact with Y2847.55 and Y2938.54 in the open conformation. Switching between the open and closed conformations can be observed in Supplemental Video 1, which reports a superposition between the three apo MD trajectories.
MD Simulations with the Holo System
Simulations performed on the holo system revealed instability of the P1 pose over long simulation times (see Supplemental Video 2 showing superposition of the three trajectories), as illustrated by both the ligand RMSD and interaction fingerprint similarity (Pavan et al., 2022) plots reported in Supplemental Fig. 5.
Specifically, in the first replicate (MD1) LUF6000 escapes the aromatic cage formed by Y2847.55, F2898.50, and Y2938.54 to rearrange itself into a more membrane-exposed conformation stabilized by a direct H-bond between the exocyclic nitrogen and Y2938.54, π-π stacking with Y2938.54, and a water-bridged H-bond with the unpaired carbonyl of M2767.47 and the N-1 nitrogen of the 1H-imidazo[4,5-c]quinolin-4-amine heterocycle (Supplemental Fig. 6, Supplemental Video 3). Notably, the presence of a water molecule in the small hydrophobic cleft between M2767.47 and G251.45 was also observed in two high-resolution GPCR structures (5IU4 of an inactive-state A2AAR in complex with ZM241385 and 5WIU of an inactive-state D4 dopamine receptor in complex with nemonapride) used in the work of Venkatakrishnan and colleagues (Venkatakrishnan et al., 2019) to map conserved water molecules in GPCRs.
In the second replicate (MD2), which demonstrated the most stability based on its RMSD profile (Supplemental Fig. 5), LUF6000 maintains a conformation similar to the docking pose for most of the simulation but then eventually loses contact with Y2938.54 (Supplemental Fig. 7, Supplemental Video 4). Although LUF6000 does not deviate from the starting position, the pocket does not stabilize, suggesting that the final position observed in this simulation represents a metastable intermediate rather than the bound state.
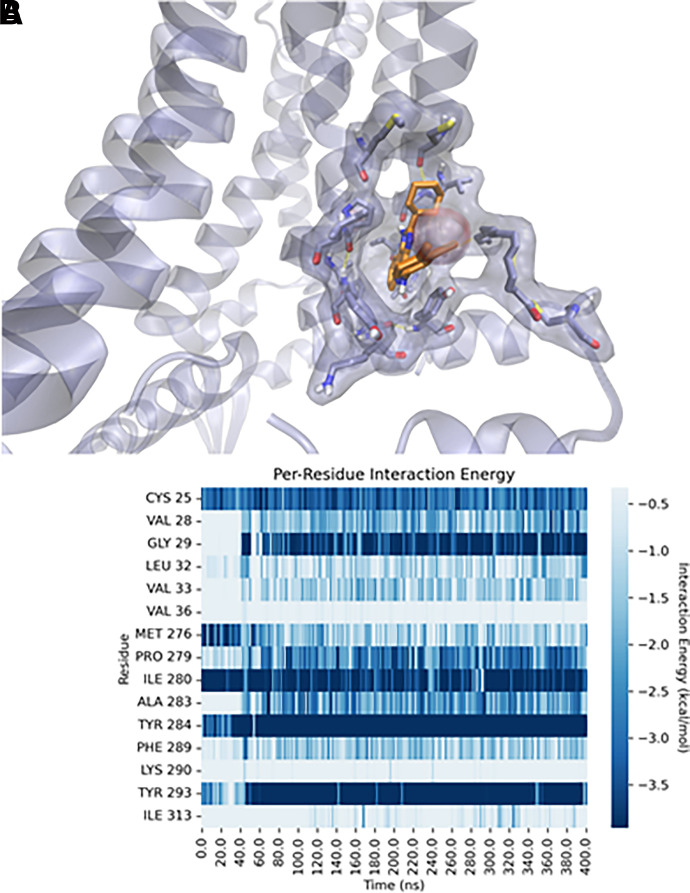
Finally, in the third simulation (MD3), LUF6000 transiently occupies the same metastable state observed at the end of the MD2 trial before rapidly converting to a different conformation that is maintained for the remainder of the trajectory (Fig. 5; Supplemental Video 5). Specifically, after ∼50 ns, F2898.50 flips away from its resting position and participates in π-π stacking with F482.40. This movement allows LUF6000 to insert deeper into the pocket, forming a stable H-bond (persistence = 21%) between the N-1 nitrogen of the 1H-imidazo[4,5-c]quinolin-4-amine heterocycle and the backbone carbonyl of G291.49. As a result, the heterocycle of LUF6000 forms a stabilizing π-π stacking interaction with Y2938.54, whereas the exocyclic amine and the 3,4-dichlorophenyl group, respectively, form transient NH-π bonding and π-π stacking with Y2847.55. In this position, the 2-cyclohexyl substituent is better accommodated into the hydrophobic S4' subpocket, forcing expulsion of the previously accommodated water molecule (Fig. 5; Supplemental Video 5).
Fig. 5.
Final state of the third molecular dynamics (MD3) refinement performed on the P1 docking pose proposed to represent the bound state of LUF6000 with the S4 site. Panel A shows a three-dimensional representation of the final state of the simulation. Panel B shows per-residue decomposition of the receptor-ligand interaction energy throughout the 400 ns simulation.
Clustering analyses of the holo trials support claims derived from visual inspection of the individual trajectories (Supplemental Fig. 8). Specifically, clusters C1, C3, and C4 represent degenerate solutions loosely resembling the docking pose, with LUF6000 π-π stacking and forming a series of close-range hydrophobic interactions with residues surrounding the pocket. With each of these clusters, contact with Y2938.54 is absent, resulting in positioning of the distal portion of H8 further away from the cytosolic ends of TMDs 7 and 1. Cluster C2, which is the most highly populated cluster and is predicted to represent the final bound state, closely resembles the final state of MD3, with LUF6000 deeply buried within the site S4 pocket, whereas cluster C5 resembles the final state observed in MD1, where LUF6000 becomes more membrane exposed. Interestingly, in both of these clusters, H8 is tilted “upwards” closer in space to TM7 and TM1, presumably as a result of LUF6000 functioning as a molecular glue upon establishment of additional interactions with Y2938.54. Considering that Lu and colleagues (Lu et al., 2021) recently proposed that tilting of H8 is a requirement for the angiotensin-II receptor to become activated, cluster C2 provides an attractive explanation for LUF6000's pharmacological actions as a positive allosteric modulator. Moreover, it is reasonable to assume that the remaining clusters (C1/C3/C4 defined as metastate 1 and C5 defined as metastate 2) represent intermediate binding states that precede the bound state (cluster 2). To directly test this hypothesis, we extended the MD1 simulation for an additional 100 ns to determine whether the C5 conformation converts to a bound state. Strikingly, LUF6000 rapidly evolved toward a metastable state 1-like state before eventually converging to a C2-like bound conformation (Supplemental Video 6). This finding supports the idea that H-bonding and π-π stacking interactions with both Y2847.55 and Y2938.54 are pivotal in attracting and maneuvering LUF6000 into the S4 binding pocket.
Additional Mutagenesis
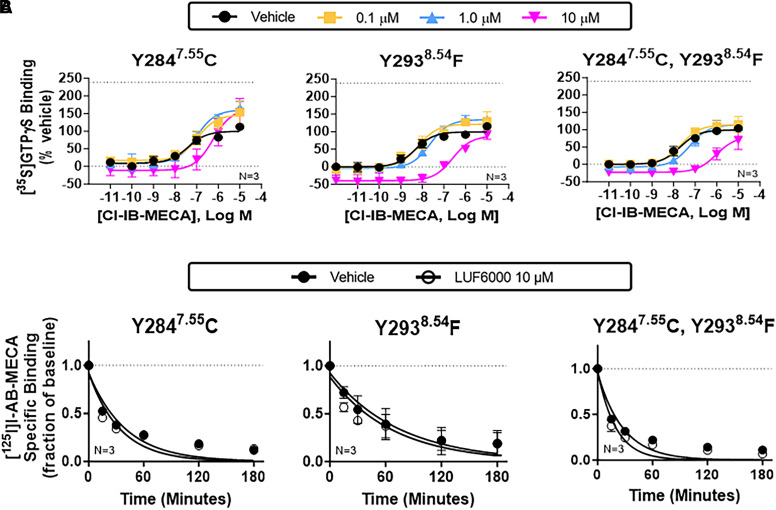
The predicted binding mode was validated by mutagenesis experiments, wherein allosteric enhancing activity of LUF6000 was greatly reduced in [35S]GTPγS exchange and [125I]I-AB-MECA dissociation binding assays when Y2847.55 was mutated to a cysteine correlating with the mouse sequence (Fig. 6; Supplemental Table 2). In addition, PAM activity of LUF6000 was lost entirely when Y2938.54 was changed to phenylalanine (Fig. 6; Supplemental Table 2). However, with both tyrosine mutants, the orthosteric competitive nature of LUF6000 persisted. According to the model, the dramatic effect of the loss of a single oxygen atom in the Y2938.54 mutation supports the hypothesis that the Y2938.54 hydroxyl group participates in H-bonding with the exocyclic nitrogen of LUF6000 during transition to the final bound state.
Fig. 6.
Characterization of Y2847.55C and Y2938.54F mutant receptors. [35S]GTPγS (A) and [125I]I-AB-MECA dissociation (B) binding assays with HEK293 cell membranes expressing Y2847.55C or Y2938.54F mutant receptors alone or in combination. The allosteric actions of LUF6000 were lost with either mutation, supporting the participation of π-π stacking with Y2847.55 and Y2938.54 and H-bonding between Y2938.54 and the exocyclic nitrogen of LUF6000 as critical interactions for LUF6000 binding to the proposed S4 allosteric site. For the [35S]GTPγS binding assays, concentration-response curves with Cl-IB-MECA were conducted in the presence of vehicle (DMSO, black) or 0.1 (yellow), 1 (blue), or 10 μM (magenta) LUF6000. Results were normalized to the Emax value of Cl-IB-MECA obtained in the presence of vehicle. The dotted line in each graph demarcates the Emax of Cl-IB-MECA in the presence of 10 μM LUF6000 with membranes from wild-type A3ARs (240 ± 11% of vehicle). EC50, Emax, and k values are reported in Supplemental Table 2. Data are presented as the mean ± S.D.
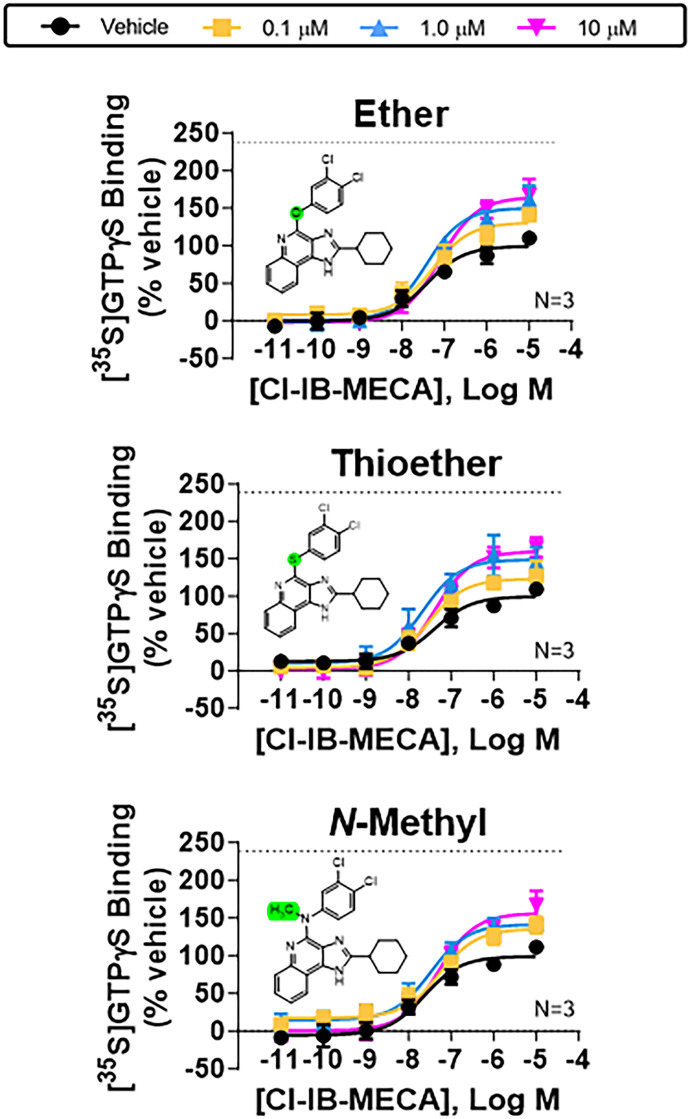
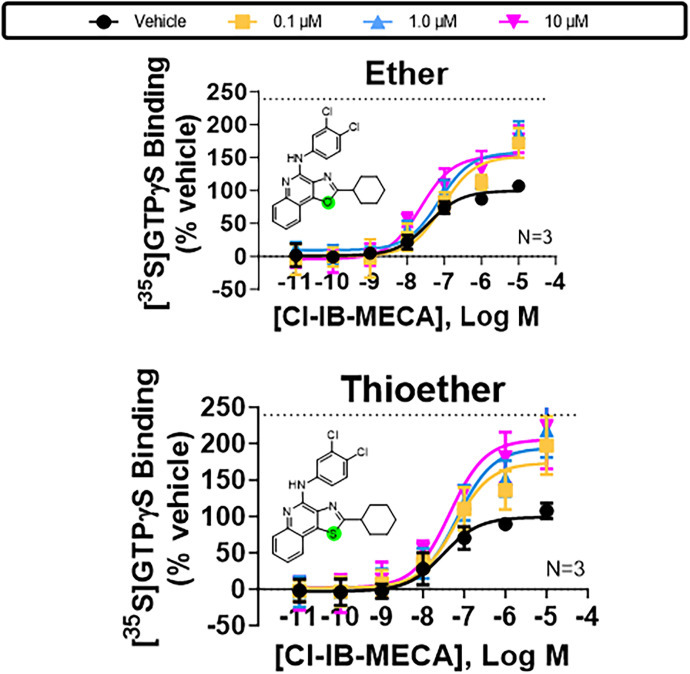
The possibility of a specific H-bond between the exocyclic NH, acting as a donor, and Y2938.54 was further strengthened by the observation that enhancing activity was reduced with LUF6000 derivatives in which the exocyclic nitrogen was replaced with other heteroatoms or methylated (Fig. 7; Supplemental Table 3). We synthesized analogs with ether, thioether, or methylamino substitutions in place of the exocyclic NH as described in the Supplemental Chemical Synthesis section in the Supplemental Material file; each of these analogs cannot donate a H-bond to the receptor protein. Correspondingly, we also synthesized ether and thioether derivatives of LUF6000 at position 1 of the heterocycle that lack the ability to donate an H-bond, which were also found to exhibit submaximal PAM activity (Fig. 8; Supplemental Table 3).
Fig. 7.
Characterization of exocyclic ether, thioether, and N-methyl derivatives of LUF6000 with wild-type A3ARs. [35S]GTPγS binding assays assessing effects of increasing concentrations of the indicated exocyclic ether, thioether, or N-methyl derivatives of LUF6000 with HEK293 cell membranes expressing wild-type A3ARs. Allosteric actions of all the derivatives were diminished compared with LUF6000, supporting the hypothesis for H-bonding between the exocyclic nitrogen of LUF6000 and the hydroxyl of Y2938.54. Concentration-response curves with Cl-IB-MECA were conducted in the presence of vehicle (DMSO, black) or 0.1 (yellow), 1 (blue), or 10 μM (magenta) of the indicated derivatives. The dotted line in each graph demarcates the Emax of Cl-IB-MECA in the presence of 10 μM LUF6000 with membranes from wild-type A3ARs (240 ± 11% of vehicle). EC50 and Emax values are reported in Supplemental Table 7. Data are presented as the mean ± S.D.
Fig. 8.
Characterization of ether and thioether derivatives of LUF6000 at the N-1 position of the imidazole ring with wild-type A3ARs. [35S]GTPγS binding assays assessing effects of increasing concentrations of the indicated ether and thioether derivatives of LUF6000 with HEK293 cell membranes expressing wild-type A3ARs. Allosteric actions of both derivatives were diminished as compared with LUF6000, supporting the hypothesis for H-bonding between the N-1 nitrogen of the heterocycle of LUF6000 and the hydroxyl of Y2938.54. Concentration-response curves with Cl-IB-MECA were conducted in the presence of vehicle (DMSO, black) or 0.1 (yellow), 1 (blue), or 10 μM (magenta) of the derivatives. The dotted line in each graph demarcates the Emax of Cl-IB-MECA in the presence of 10 μM LUF6000 with membranes from wild-type A3ARs (240 ± 11% of vehicle). EC50 and Emax values are reported in Supplemental Table 7. Data are presented as mean ± S.D.
Discussion
This study describes a putative binding region and molecular interactions for the prototypical 1H-imidazo[4,5-c]quinolin-4-amine allosteric modulator LUF6000. Molecular modeling, guided and further supported by mutagenesis and pharmacological studies, predicts that LUF6000 occupies an extrahelical, lipid-facing allosteric pocket formed by TMD1, TMD7, and H8. The putative binding region does not overlap with the orthosteric binding site, and no amino acid residues are shared between the two sites.
This putative binding mode is contrary to prior computational modeling predictions with the A3AR (Gao et al., 2003; Deganutti et al., 2015) but is highly consistent with structure-activity-relationship information accumulated by our group with 1H-imidazo[4,5-c]quinolin-4-amine PAMs. For full PAM activity, hydrophobic substitutions are required at the 2- and 4-amine positions of the 1H-imidazo[4,5-c]quinolin-4-amine heterocycle, whereas polar substitutions are not tolerated (Gao et al., 2002; Göblyös et al., 2006; Kim et al., 2009; Fallot et al., 2022; Fisher et al., 2022). Concerning the 2-position, this requirement was demonstrated in comprehensive evaluation of 1H-imidazo[4,5-c]quinolin-4-amine derivatives with 2-cycloalkyl substitutions ranging from 3 to 12 carbons, wherein full PAM activity was achieved with 2-cyclohexyl or 2-cycloheptyl substitutions; activity progressively diminished with the addition of larger or smaller substitutions (Fisher et al., 2022). Additional work established that PAM activity is lost upon introducing nitrogen or oxygen to comparable ring systems at the 2-position to increase polarity (Fallot et al., 2022). However, less structure-activity-relationship work has focused on the 4-position. Nevertheless, we previously showed that an aryl substituent is greatly favored over heterocyclic ring systems with greater polarity (Göblyös et al., 2006; Kim et al., 2009).
Considering the extensive prior structure-activity-relationship work and the proposed binding pose of LUF6000 described in this study, we present the hypothesis that the 1H-imidazo[4,5-c]quinolin-4-amine ring system inserts into an aromatic cage formed by Y2847.55 in TMD7 and Y2938.54 in H8 (Figs. 4 and 5), which becomes accessible when Y2847.55 positions to an open conformation. The 2-cyclohexyl group is situated within a hydrophobic subpocket created by residues in TMDs 1 and 7, while the 3,4-dichlorohexyl group extends toward the lipid bilayer, which due to its hydrophobic nature may provide additional stabilization. A3AR-specificity in relation to the other adenosine receptor subtypes, demonstrated in prior published work (Fallot et al., 2022), is explained by replacing the critical Y2938.54 with phenylalanine, whereas nonresponsiveness of the mouse A3AR to LUF6000 is explained by replacing the critical Y2847.55 with a cysteine. Based on the chimeric receptor studies, we exchanged the entire distal portions of the mouse A3AR sequence with the human sequence, which included the distal half of TMD7, H8, and the C-terminus. However, responsiveness to LUF6000 was not conferred with the newly created chimeric receptors in [35S]GTPγS exchange assays (Supplemental Fig. 9, Supplemental Table 2). Thus, amino acid differences in other regions of the receptor, presumably TMD1 based on the previous chimeric receptor and modeling results, also appear to contribute to species differences.
In a prior mutagenesis study conducted by our group 20 years ago when the structural assessment of GPCRs was in its infancy (Gao et al., 2003), we explored the participation of several amino acid residues in mediating the allosteric actions of three structurally dissimilar PAMs. DU124183, a 1H-imidazo[4,5-c]quinolin-4-amine derivative that contains 2-cyclopentyl and 4-aminophenyl substituents (Supplemental Fig. 10), was included in this investigation. We reported that the allosteric actions of DU124183, detected by slowed [125I]I-AB-MECA dissociation, were reduced with each of the following mutants: N30A1.50, D58N2.50, D107N3.49, F182A5.43, and N274A7.45 (Gao et al., 2003). Considering that within the family of adenosine receptors it is now known that each of these residues are required for receptor rearrangement, sodium binding, or orthosteric ligand binding, loss of activity of DU124183 with each of these mutations is predicted to affect the receptor’s ability to transition between active and inactive states or affect cooperative interactions between the orthosteric and allosteric sites rather than the ability of DU124183 to bind to its allosteric site. Notably, the mutant receptors in this original study were not investigated in assays to assess G protein activation, and each of the amino acids is conserved among the human and mouse sequences (conserved in all species where the A3AR sequence has been reported).
At this time, we can only speculate on the molecular mechanism of LUF6000s allosteric effects. As described with other PAMs (Burger et al., 2018; Wang et al., 2018; Liu et al., 2019; van der Westhuizen et al., 2021; Nguyen et al., 2022), LUF6000 may favorably alter the conformational equilibrium of the receptor toward an active compared with the inactive state. Regarding the antagonistic nature of LUF6000 that occurs at concentrations approaching 10 μM, we have previously found this to be consistent with competitive inhibition at the orthosteric binding site rather than negative allosterism (Fallot et al., 2022; Fisher et al., 2022). This was confirmed using “inner” and “outer” (with respect to the plasma membrane) human/mouse chimeric receptors, where the positive allosteric activity of LUF6000 was lost when the inner portions of the receptor were comprised of the mouse sequence, yet its competitive antagonistic actions persisted (Fisher et al., 2022). This conclusion was further supported by studies with the antagonist radioligand 125I-ABOPX, where LUF6000 was found to reduce specific binding to zero in equilibrium binding assays, and by docking studies with a homology model of the A3AR where LUF6000 could be accommodated into the canonical AR orthosteric binding site as we described (Fallot et al., 2022; Fisher et al., 2022).
The allosteric site described herein correlates closely with orphan site 9 (OS9) predicted through a computational study aimed to define the GPCR “pocketome” (Hedderich et al., 2022). Through exhaustive docking analysis of a library of small molecule probes with structures from 113 unique receptors (557 unique structures), this study confirmed all previously identified allosteric pockets and revealed 9 untargeted sites termed orphan sites. Like the proposed LUF6000 binding site we have described, OS9 is an exofacial site that lies between TMDs 1 and 7 above H8. In a computational study aimed to describe activation mechanisms of the angiotensin type II receptor, Lu and colleagues (Lu et al., 2021) identified a “cryptic” binding pocket similar to OS9, which was termed pocket P6. Via mutational analysis of representative receptors (angiotensin II, M3 muscarinic, and β2 adrenergic), both of these studies provided evidence that amino acids surrounding this site have a robust effect on both G protein activation and β-arrestin recruitment, suggesting the modulatory potential of ligands that occupy this site (Lu et al., 2021; Hedderich et al., 2022). The position and shape of the OS9 pocket was predicted to be highly conserved and was proposed to be a pan-class A GPCR pocket.
In conclusion, here we have provided computational, mutagenesis, and pharmacological evidence supporting an extrahelical binding site for the 1H-imidazo[4,5-c]quinolin-4-amine A3AR PAM LUF6000. This site is likely shared among other GPCRs based on past computational predictions of potential allosteric sites for the broad family of GPCRs. This study has provided new structural insights for the A3AR and will aid rational approaches to design improved allosteric ligands.
Data Availability
The authors declare that all the data supporting the findings of this study are available within the paper and Supplemental Material. A PDB structure reporting the heavy atom coordinates of the human A3AR model bound to LUF6000 (obtained through InducedFit Docking) is in a separate Supplemental Material file.
Abbreviations
- A3AR
A3 adenosine receptor
- AB-MECA
N6-4-aminobenzyl)adenosine-5′-N-methylcarboxamide
- Cl-IB-MECA
2-chloro-N6-(3-iodobenzyl)adenosine-5′-N-methylcarboxamide
- GPCR
G protein-coupled receptor
- H
Helix
- HEK293
human embryonic kidney 293
- [125I]I-AB-MECA
N6-(4-amino-3-[125I]iodobenzyl)adenosine-5′-N-methylcarboxamide
- IB-MECA
N6-(3-iodobenzyl)adenosine-5′-N-methylcarboxamide
- LUF6000
2-cyclohexyl-1H-imidazo[4,5-c]quinolin-4-(3,4-dichlorophenyl)amine
- OS9
orphan site 9
- PAM
positive allosteric modulator
- RMSD
root mean square deviation
- [35S]GTPγS
guanosine 5′-[γ-[35S]thio]triphosphate
- TMD
transmembrane domain
Authorship Contributions
Participated in research design: Fisher, Pavan, Salmaso, Keyes, Wan, Pradhan, Gao, Smith, Jacobson, Auchampach.
Conducted experiments: Fisher, Pavan, Salmaso, Keyes, Wan, Auchampach.
Contributed new reagents: Keyes, Pradhan, Smith, Jacobson.
Performed data analysis: Fisher, Pavan, Salmaso, Keyes, Wan, Auchampach.
Wrote or contributed to the writing of the manuscript: Fisher, Pavan, Jacobson, Auchampach.
Footnotes
This research was supported in part by National Institutes of Health National Heart, Lung, and Blood Institute [Grant R01 HL133589] (to J.A.A.), National Institutes of General Medicine Sciences [Grant R35 GM128840] (to B.C.S.), National Heart, Lung, and Blood Institute [Grant F31 HL160193] (to C.L.F.), the National Institute of Diabetes and Digestive and Kidney Diseases Intramural Research Program [Grant ZIADK031117] (to K.A.J.), the American Heart Association (Predoctoral Fellowship) [Grant 898217] (to C.L.F.), and the Medical College of Wisconsin Therapeutic Accelerator Program (to J.A.A.).
No author has an actual or perceived conflict of interest with the contents of this article.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Antonioli L, Pacher P, Haskó G (2022) Adenosine and inflammation: it’s time to (re)solve the problem. Trends Pharmacol Sci 43:43–55. [DOI] [PubMed] [Google Scholar]
- Auchampach JA, Jin X, Wan TC, Caughey GH, Linden J (1997) Canine mast cell adenosine receptors: cloning and expression of the A3 receptor and evidence that degranulation is mediated by the A2B receptor. Mol Pharmacol 52:846–860. [DOI] [PubMed] [Google Scholar]
- Ballesteros JA, Weinstein H (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods in Neirosciences 25:366–428. [Google Scholar]
- Bozdemir E, Vigil FA, Chun SH, Espinoza L, Bugay V, Khoury SM, Holstein DM, Stoja A, Lozano D, Tunca C, et al. (2021) Neuroprotective roles of the adenosine A3 receptor agonist AST-004 in mouse model of traumatic brain injury. Neurotherapeutics 18:2707–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger WAC, Sexton PM, Christopoulos A, Thal DM (2018) Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors. J Gen Physiol 150:1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Corriden R, Inoue Y, Yip L, Hashiguchi N, Zinkernagel A, Nizet V, Insel PA, Junger WG (2006) ATP release guides neutrophil chemotaxis via P2Y2 and A3 receptors. Science 314:1792–1795. [DOI] [PubMed] [Google Scholar]
- Coughlin Q, Hopper AT, Blanco MJ, Tirunagaru V, Robichaud AJ, Doller D (2019) Allosteric modalities for membrane-bound receptors: insights from drug hunting for brain diseases. J Med Chem 62:5979–6002. [DOI] [PubMed] [Google Scholar]
- Deganutti G, Cuzzolin A, Ciancetta A, Moro S (2015) Understanding allosteric interactions in G protein-coupled receptors using supervised molecular dynamics: a prototype study analysing the human A3 adenosine receptor positive allosteric modulator LUF6000. Bioorg Med Chem 23:4065–4071. [DOI] [PubMed] [Google Scholar]
- Draper-Joyce CJ, Bhola R, Wang J, Bhattarai A, Nguyen ATN, Cowie-Kent I, O’Sullivan K, Chia LY, Venugopal H, Valant C, et al. (2021) Positive allosteric mechanisms of adenosine A1 receptor-mediated analgesia. Nature 597:571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Gao ZG, Nithipatikom K, IJzerman AP, Veldhoven JP, Jacobson KA, Gross GJ, Auchampach JA (2012) Protection from myocardial ischemia/reperfusion injury by a positive allosteric modulator of the A3 adenosine receptor. J Pharmacol Exp Ther 340:210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Gao ZG, Paoletta S, Wan TC, Gizewski ET, Barbour S, van Veldhoven JPD, IJzerman AP, Jacobson KA, Auchampach JA (2018) Species differences and mechanism of action of A3 adenosine receptor allosteric modulators. Purinergic Signal 14:59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallot LB, Suresh RR, Fisher CL, Salmaso V, O’Connor RD, Kaufman N, Gao ZG, Auchampach JA, Jacobson KA (2022) Structure-activity studies of 1H-imidazo[4,5-c]quinolin-4-amine derivatives as A3 adenosine receptor positive allosteric modulators. J Med Chem 65:15238–15262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CL, Fallot LB, Wan TC, Keyes RF, Suresh RR, Rothwell AC, Gao ZG, McCorvy JD, Smith BC, Jacobson KA, et al. (2022) Characterization of dual-acting A3 adenosine receptor positive allosteric modulators that preferentially enhance adenosine-induced Gαi3 and GαoA isoprotein activation. ACS Pharmacol Transl Sci 5:625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman P, Stemmer SM, Bareket-Samish A, Silverman MH, Kerns WD (2023) Targeting the A3 adenosine receptor to treat hepatocellular carcinoma: anti-cancer and hepatoprotective effects. Purinergic Signal 19:513–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, et al. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47:1739–1749. [DOI] [PubMed] [Google Scholar]
- Gao ZG, Auchampach JA, Jacobson KA (2023) Species dependence of A(3) adenosine receptor pharmacology and function. Purinergic Signal 19:523–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZG, Kim SG, Soltysiak KA, Melman N, IJzerman AP, Jacobson KA (2002) Selective allosteric enhancement of agonist binding and function at human A3 adenosine receptors by a series of imidazoquinoline derivatives. Mol Pharmacol 62:81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZG, Kim SK, Gross AS, Chen A, Blaustein JB, Jacobson KA (2003) Identification of essential residues involved in the allosteric modulation of the human A(3) adenosine receptor. Mol Pharmacol 63:1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZG, Verzijl D, Zweemer A, Ye K, Göblyös A, IJzerman AP, Jacobson KA (2011) Functionally biased modulation of A(3) adenosine receptor agonist efficacy and potency by imidazoquinolinamine allosteric enhancers. Biochem Pharmacol 82:658–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZG, Ye K, Göblyös A, IJzerman AP, Jacobson KA (2008) Flexible modulation of agonist efficacy at the human A3 adenosine receptor by the imidazoquinoline allosteric enhancer LUF6000. BMC Pharmacol 8:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge ZD, van der Hoeven D, Maas JE, Wan TC, Auchampach JA (2010) A(3) adenosine receptor activation during reperfusion reduces infarct size through actions on bone marrow-derived cells. J Mol Cell Cardiol 49:280–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göblyös A, Gao ZG, Brussee J, Connestari R, Santiago SN, Ye K, IJzerman AP, Jacobson KA (2006) Structure-activity relationships of new 1H-imidazo[4,5-c]quinolin-4-amine derivatives as allosteric enhancers of the A3 adenosine receptor. J Med Chem 49:3354–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halgren T (2007) New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des 69:146–148. [DOI] [PubMed] [Google Scholar]
- Halgren TA (2009) Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 49:377–389. [DOI] [PubMed] [Google Scholar]
- Hammarberg C, Schulte G, Fredholm BB (2003) Evidence for functional adenosine A3 receptors in microglia cells. J Neurochem 86:1051–1054. [DOI] [PubMed] [Google Scholar]
- Haskó G, Linden J, Cronstein B, Pacher P (2008) Adenosine receptors: therapeutic aspects for inflammatory and immune diseases. Nat Rev Drug Discov 7:759–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedderich JB, Persechino M, Becker K, Heydenreich FM, Gutermuth T, Bouvier M, Bünemann M, Kolb P (2022) The pocketome of G-protein-coupled receptors reveals previously untargeted allosteric sites. Nat Commun 13:2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo L, Feig M (2022) Multi-state modeling of G-protein coupled receptors at experimental accuracy. Proteins 90:1873–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan JE, Thourani VH, Auchampach JA, Robinson JA, Wang NP, Vinten-Johansen J (1999) A(3) adenosine receptor activation attenuates neutrophil function and neutrophil-mediated reperfusion injury. Am J Physiol 277:H1895–H1905. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, de Castro S, Gao ZG, IJzerman AP, Jacobson KA (2009) Novel 2- and 4-substituted 1H-imidazo[4,5-c]quinolin-4-amine derivatives as allosteric modulators of the A3 adenosine receptor. J Med Chem 52:2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorraca NR, Venkatakrishnan AJ, Dror RO (2017) GPCR dynamics: structures in motion. Chem Rev 117:139–155. [DOI] [PubMed] [Google Scholar]
- Liston TE, Hama A, Boltze J, Poe RB, Natsume T, Hayashi I, Takamatsu H, Korinek WS, Lechleiter JD (2022) Adenosine A1R/A3R (adenosine A1 and A3 receptor) agonist AST-004 reduces brain infarction in a nonhuman primate model of stroke. Stroke 53:238–248. [DOI] [PubMed] [Google Scholar]
- Liston TE, Hinz S, Müller CE, Holstein DM, Wendling J, Melton RJ, Campbell M, Korinek WS, Suresh RR, Sethre-Hofstad DA, et al. (2020) Nucleotide P2Y1 receptor agonists are in vitro and in vivo prodrugs of A1/A3 adenosine receptor agonists: implications for roles of P2Y1 and A1/A3 receptors in physiology and pathology. Purinergic Signal 16:543–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Masoudi A, Kahsai AW, Huang LY, Pani B, Staus DP, Shim PJ, Hirata K, Simhal RK, Schwalb AM, et al. (2019) Mechanism of β2AR regulation by an intracellular positive allosteric modulator. Science 364:1283–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, He X, Yang Z, Chai Z, Zhou S, Wang J, Rehman AU, Ni D, Pu J, Sun J, et al. (2021) Activation pathway of a G protein-coupled receptor uncovers conformational intermediates as targets for allosteric drug design. Nat Commun 12:4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman A, Gao ZG, Kumar D, Wan TC, Gizewski E, Auchampach JA, Jacobson KA (2008) Design of (N)-methanocarba adenosine 5′-uronamides as species-independent A3 receptor-selective agonists. Bioorg Med Chem Lett 18:2813–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HTM, van der Westhuizen ET, Langmead CJ, Tobin AB, Sexton PM, Christopoulos A, Valant C (2022) Opportunities and challenges for the development of M(1) muscarinic receptor positive allosteric modulators in the treatment for neurocognitive deficits. Br J Pharmacol, in press. [DOI] [PubMed]
- Paoletta S, Tosh DK, Finley A, Gizewski ET, Moss SM, Gao ZG, Auchampach JA, Salvemini D, Jacobson KA (2013) Rational design of sulfonated A3 adenosine receptor-selective nucleosides as pharmacological tools to study chronic neuropathic pain. J Med Chem 56:5949–5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavan M, Menin S, Bassani D, Sturlese M, Moro S (2022) Implementing a scoring function based on interaction fingerprint for Autogrow4: protein kinase CK1δ as a case study. Front Mol Biosci 9:909499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slosky LM, Caron MG, Barak LS (2021) Biased allosteric modulators: new frontiers in GPCR drug discovery. Trends Pharmacol Sci 42:283–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubiana T, Carvaillo JC, Boulard Y, Bressanelli S (2018) TTClust: a versatile molecular simulation trajectory clustering program with graphical summaries. J Chem Inf Model 58:2178–2182. [DOI] [PubMed] [Google Scholar]
- van der Hoeven D, Gizewski ET, Auchampach JA (2010) Activation of the A(3) adenosine receptor inhibits fMLP-induced Rac activation in mouse bone marrow neutrophils. Biochem Pharmacol 79:1667–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoeven D, Wan TC, Auchampach JA (2008) Activation of the A(3) adenosine receptor suppresses superoxide production and chemotaxis of mouse bone marrow neutrophils. Mol Pharmacol 74:685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Westhuizen ET, Choy KHC, Valant C, McKenzie-Nickson S, Bradley SJ, Tobin AB, Sexton PM, Christopoulos A (2021) Fine tuning muscarinic acetylcholine receptor signaling through allostery and bias. Front Pharmacol 11:606656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Ma AK, Fonseca R, Latorraca NR, Kelly B, Betz RM, Asawa C, Kobilka BK, Dror RO (2019) Diverse GPCRs exhibit conserved water networks for stabilization and activation. Proc Natl Acad Sci USA 116:3288–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BA, Jacobson MA, Knight DA, Salvatore CA, Weir T, Zhou D, Bai TR (1997) Adenosine A3 receptor expression and function in eosinophils. Am J Respir Cell Mol Biol 16:531–537. [DOI] [PubMed] [Google Scholar]
- Wang X, Heinz BA, Qian YW, Carter JH, Gadski RA, Beavers LS, Little SP, Yang CR, Beck JP, Hao J, et al. (2018) Intracellular binding site for a positive allosteric modulator of the dopamine D1 receptor. Mol Pharmacol 94:1232–1245. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yu Z, Xiao W, Lu S, Zhang J (2021) Allosteric binding sites at the receptor-lipid bilayer interface: novel targets for GPCR drug discovery. Drug Discov Today 26:690–703. [DOI] [PubMed] [Google Scholar]