Abstract
Previous studies have shown that protein-protein interactions among splicing factors may play an important role in pre-mRNA splicing. We report here identification and functional characterization of a new splicing factor, Sip1 (SC35-interacting protein 1). Sip1 was initially identified by virtue of its interaction with SC35, a splicing factor of the SR family. Sip1 interacts with not only several SR proteins but also with U1-70K and U2AF65, proteins associated with 5′ and 3′ splice sites, respectively. The predicted Sip1 sequence contains an arginine-serine-rich (RS) domain but does not have any known RNA-binding motifs, indicating that it is not a member of the SR family. Sip1 also contains a region with weak sequence similarity to the Drosophila splicing regulator suppressor of white apricot (SWAP). An essential role for Sip1 in pre-mRNA splicing was suggested by the observation that anti-Sip1 antibodies depleted splicing activity from HeLa nuclear extract. Purified recombinant Sip1 protein, but not other RS domain-containing proteins such as SC35, ASF/SF2, and U2AF65, restored the splicing activity of the Sip1-immunodepleted extract. Addition of U2AF65 protein further enhanced the splicing reconstitution by the Sip1 protein. Deficiency in the formation of both A and B splicing complexes in the Sip1-depleted nuclear extract indicates an important role of Sip1 in spliceosome assembly. Together, these results demonstrate that Sip1 is a novel RS domain-containing protein required for pre-mRNA splicing and that the functional role of Sip1 in splicing is distinct from those of known RS domain-containing splicing factors.
Pre-mRNA splicing takes place in spliceosomes, the large RNA-protein complexes containing pre-mRNA, U1, U2, U4/6, and U5 small nuclear ribonucleoprotein particles (snRNPs), and a large number of accessory protein factors (for reviews, see references 21, 22, 37, 44, and 48). It is increasingly clear that the protein factors are important for pre-mRNA splicing and that studies of these factors are essential for further understanding of molecular mechanisms of pre-mRNA splicing.
Most mammalian splicing factors have been identified by biochemical fractionation and purification (3, 15, 19, 31–36, 45, 69–71, 73), by using antibodies recognizing splicing factors (8, 9, 16, 17, 61, 66, 67, 74), and by sequence homology (25, 52, 74).
Splicing factors containing arginine-serine-rich (RS) domains have emerged as important players in pre-mRNA splicing. These include members of the SR family, both subunits of U2 auxiliary factor (U2AF), and the U1 snRNP protein U1-70K (for reviews, see references 18, 41, and 59). Drosophila alternative splicing regulators transformer (Tra), transformer 2 (Tra2), and suppressor of white apricot (SWAP) also contain RS domains (20, 40, 42). RS domains in these proteins play important roles in pre-mRNA splicing (7, 71, 75), in nuclear localization of these splicing proteins (23, 40), and in protein-RNA interactions (56, 60, 64). Previous studies by us and others have demonstrated that one mechanism whereby SR proteins function in splicing is to mediate specific protein-protein interactions among spliceosomal components and between general splicing factors and alternative splicing regulators (1, 1a, 6, 10, 27, 63, 74, 77). Such protein-protein interactions may play critical roles in splice site recognition and association (for reviews, see references 4, 18, 37, 41, 47 and 59). Specific interactions among the splicing factors also suggest that it is possible to identify new splicing factors by their interactions with known splicing factors.
Here we report identification of a new splicing factor, Sip1, by its interaction with the essential splicing factor SC35. The predicted Sip1 protein sequence contains an RS domain and a region with sequence similarity to the Drosophila splicing regulator, SWAP. We have expressed and purified recombinant Sip1 protein and raised polyclonal antibodies against the recombinant Sip1 protein. The anti-Sip1 antibodies specifically recognize a protein migrating at a molecular mass of approximately 210 kDa in HeLa nuclear extract. The anti-Sip1 antibodies sufficiently deplete Sip1 protein from the nuclear extract, and the Sip1-depleted extract is inactive in pre-mRNA splicing. Addition of recombinant Sip1 protein can partially restore splicing activity to the Sip1-depleted nuclear extract, indicating an essential role of Sip1 in pre-mRNA splicing. Other RS domain-containing proteins, including SC35, ASF/SF2, and U2AF65, cannot substitute for Sip1 in reconstituting splicing activity of the Sip1-depleted nuclear extract. However, addition of U2AF65 further increases splicing activity of Sip1-reconstituted nuclear extract, suggesting that there may be a functional interaction between Sip1 and U2AF65 in nuclear extract.
MATERIALS AND METHODS
Yeast two-hybrid interaction screening and protein-protein interaction assay.
The yeast two-hybrid interaction system including Saccharomyces cerevisiae EGY48, the yeast plasmids, and a HeLa cell cDNA library were kindly provided by R. Brent. SC35 was used as a bait to screen the HeLa cDNA library as described previously (63, 72).
To assay for pairwise interactions between Sip1 and other splicing proteins, yeast plasmids expressing individual splicing proteins as LexA fusion proteins were transformed into yeast strain EGY48 expressing Sip1 as a fusion protein containing the B42 activation domain (63, 72). The liquid assay for β-galactosidase activity was carried out with yeast extracts prepared from at least three independent colonies as described previously (63, 74). β-Galactosidase activities were normalized with protein concentrations of the corresponding yeast extracts. Background was defined as the amount of β-galactosidase activity detected in the yeast expressing the Sip1-activation domain fusion protein and the bait plasmid containing only the LexA without other cDNA sequences.
HeLa cell cDNA library screening and database search.
A cDNA fragment encoding Sip1 was isolated from the yeast two-hybrid library vector JG4-5 (72) by digestion with EcoRI and XhoI. This fragment was labeled by using Klenow enzyme in the presence of [32P]dCTP and used to screen a HeLa cell λZap library (Stratagene) according to standard procedures. Sequencing of cDNA clones was carried out by using a model 373A DNA sequencer with a PRISM Ready Reaction DyeDeoxy Terminator cycle sequencing kit (ABI). Database searches were carried out through the National Institutes of Health mail server, using BLASTN and BLASTP programs. Sequence comparison reveals that the carboxyl-terminal 4.3-kb sequence of Sip1 cDNA is almost identical to that of the human SR129 gene (accession no. Y11251), which was deposited as an unpublished sequence, and that the amino-terminal 1 kb of Sip1 cDNA is different from that of the SR129 gene. It is not clear yet whether Sip1 and SR129 represent different isoforms of the same gene or the Y11251 clone is a hybrid of two different genes. Sequence alignment was performed by using the ClustalW multiple sequence alignment program. Databank searches also revealed the presence of three murine EST cDNA clones which may represent the murine homolog of the SIP1 gene. No convincing yeast homolog has been identified.
Generation of Sip1 antisera.
cDNAs encoding different fragments of Sip1 protein were subcloned into the Escherichia coli expression vector pET28A (Novagen) to express His-tagged Sip1 fragments. One of the fragments (amino acid residues 227 to 782) was expressed at a relatively high level. This fragment contained mainly the central part of the Sip1 protein and only a small portion of the RS domain. This His-tagged Sip1 fragment was purified to near homogeneity by using nickel-agarose affinity chromatography in the presence of 6 M urea, and the Sip1 protein was further purified by preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The purified Sip1 protein isolated by SDS-PAGE was used to immunize two rabbits to generate polyclonal anti-Sip1 antisera.
Expression and purification of recombinant Sip1 protein by using the baculovirus system.
Baculovirus expressing the full-length Sip1 protein as a His-tagged protein was prepared by using the pFastbac system (Gibco). The recombinant Sip1 protein was purified by ammonium sulfate precipitation (45 to 90% saturation) followed by nickel-agarose affinity chromatography according to the manufacturer’s instructions. The purified Sip1 protein was dialyzed against BC100 (20 mM HEPES [pH 7.6], 100 mM KCl) containing 10% glycerol and stored at −80°C in aliquots.
Western analyses.
Proteins were separated by SDS-PAGE (10% gel) and transferred to nitrocellulose filters. After blocking with 5% nonfat milk in Tris-buffered saline, primary antibodies (anti-Sip1 at 1:2,000 dilution and anti-U2AF65 at 1:1,500 dilution) were applied to the filters, and incubation was carried out at 4°C overnight. After four washes with Tris-buffered saline containing 0.05% Tween 20, horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G [F(ab′)2 fragment specific; Jackson Immunoresearch Laboratories] was added and incubated for 1 h at room temperature. After four washes, SuperSignal substrate solution (Pierce) was used to produce signals, and the filters were exposed to X-ray films.
In vitro splicing, splicing complex formation, immunodepletion, and complementation assays.
Splicing reactions were carried out with β-globin pre-mRNA prepared by in vitro transcription in the presence of [32P]UTP, using construct HβT7 as described previously (74). The labeled pre-mRNA was incubated with HeLa cell nuclear extract, HeLa nuclear extract depleted of Sip1, or the Sip1-depleted HeLa nuclear extract supplemented with recombinant proteins. The splicing products were analyzed on a 6% polyacrylamide gel containing 6 M urea and detected by autoradiography. For native gel analysis of splicing complexes, splicing reactions were performed with 32P-labeled pPIP85.A pre-mRNA (43) and separated on a nondenaturing 4% (80:1) polyacrylamide gel in 50 mM Tris-glycine. The gels were dried and exposed to X-ray films for autoradiography.
Anti-Sip1 antiserum or the preimmune serum was used for depletion experiments, performed by a protocol modified from that of Zuo and Maniatis (77). In this protocol, 0.5 ml (1:1 slurry) of protein A-Trisacryl (Pierce) was washed three times and blocked at 4°C for 1 h in immunodepletion (ID) wash buffer (20 mM HEPES [pH 7.8], 150 mM NaCl, 0.5% Nonidet P-40, 2 mM EDTA, 2 mg of bovine serum albumin per ml). The protein A-Trisacryl resin was then packed into a minicolumn and washed with 5 ml of ID wash buffer. Preimmune serum or anti-Sip1 antiserum was diluted with 2.5 ml of ID wash buffer and loaded onto the protein A-Trisacryl column three times. The columns were washed with 5 ml of ID wash buffer and 5 ml of BC100 (20 mM HEPES [pH 7.8], 100 mM KCl, 0.2 mM EDTA, 0.5 mM dithiothreitol). HeLa cell nuclear extract (0.5 ml at approximately 12 mg of protein per ml) was thawed, prewarmed to room temperature for 20 min, and passed five times at room temperature through an anti-Sip1 immunoaffinity column or a mock affinity column prepared with the preimmune serum. The extracts were then used or quick frozen and then stored as immunodepleted nuclear extracts. The individual batches of depleted extracts were examined by Western blotting using the anti-Sip1 and anti-U2AF65 antisera.
For the splicing complementation assay, recombinant SC35, ASF/SF2, and U2AF65 were purified. SC35 and ASF/SF2 were purified as described previously (63). Recombinant U2AF65 protein was purified from E. coli BL21(DE3)pLysS transformed with a plasmid expressing U2AF65 from a T7 vector (63). E. coli expressing the U2AF65 protein after isopropylthiogalactopyranoside induction was harvested and sonicated in a solution containing 20 mM HEPES (pH 8.0), 0.1M NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, and 5% glycerol. MgCl2 was added to a final concentration of 20 mM to precipitate the proteins. The recombinant U2AF65 protein was purified to near homogeneity on a MonoQ column with a 0.1 to 1 M NaCl gradient. The purified protein was dialyzed against BC100 containing 10% glycerol and stored in aliquots at −80°C.
Nucleotide sequence accession number.
The GenBank accession number for Sip1 cDNA is AF030234.
RESULTS
Identification of Sip1 by its interaction with the essential splicing factor SC35.
SC35 is an essential splicing factor of the SR family (16, 17; for a review, see reference 18). It interacts with several other known splicing factors (63, 74). Using SC35 as a bait in the yeast two-hybrid interaction cloning system (72), we have searched for proteins interacting with SC35. After screening approximately 2 × 106 independent colonies, we isolated several groups of cDNAs, including those encoding U1-70K, U2AF35, and two novel SC35-interacting proteins, named Sip1 and Sip2. In the previous study, we characterized specific interactions of SC35 with U1-70K and U2AF35, leading to the proposal of a model for SR protein function in spliceosome assembly (63). It was not clear whether the new proteins identified from the yeast two-hybrid screening were functionally important for splicing. We have now further characterized one of these new SC35-interacting proteins, Sip1.
Using the Sip1 cDNA fragment obtained from the yeast two-hybrid system, we screened a HeLa cell cDNA library and isolated 16 cDNA clones. After analyzing the sequences of these overlapping cDNAs, a probable full-length cDNA of 5,298 bp was obtained by ligation of two overlapping cDNAs. Based on a number of criteria, we conclude that this cDNA represents the full-length cDNA encoding Sip1 protein. First, the cDNA contains an open reading frame encoding 1,403 amino acids (Fig. 1), having a presumptive ATG initiation codon located 416 bp downstream from the 5′ end with surrounding sequence conforming to the consensus for mammalian translational initiation sequence (30). Second, there are stop codons present in all three reading frames upstream of the presumptive start codon. Third, the cDNA contains a polyadenylation signal in the 3′ untranslated region (62) with a poly(A) tail at the 3′ end. Fourth, a major band of approximately 5 kb, consistent with the cDNA size (5,298 bp), was detected in human tissues and cell lines by Northern blotting (Fig. 2). Finally, the protein products of the Sip1 cDNA obtained either from in vitro translation or from the baculovirus expression system migrate at the same size as the Sip1 protein present in HeLa cells as detected by Western blotting using anti-Sip1 antiserum (see Fig. 6; also data not shown). Taken together, these results indicate that the Sip1 protein sequence shown in Fig. 1 is very likely to be of full length.
FIG. 1.
Amino acid sequence of Sip1 predicted from the full-length cDNA sequence. SR or RS dipeptides are underlined. Eight imperfect repeats of RRSRSXSX are in boldface and underlined. The CTD-binding motif is doubly underlined.
FIG. 2.
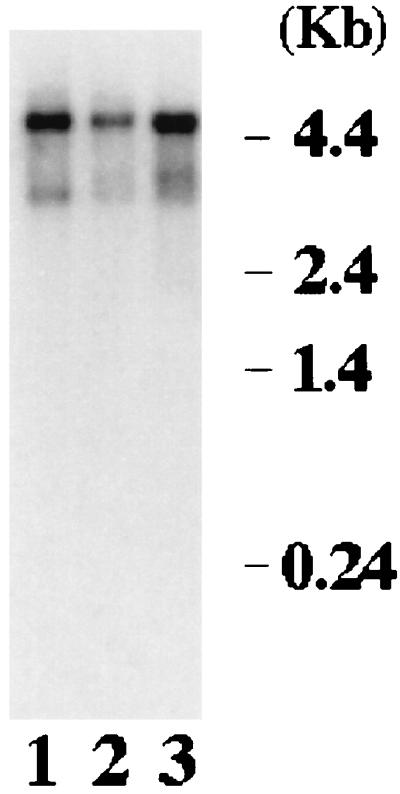
Northern analysis of Sip1 mRNA. Approximately 2 μg of poly(A)-selected RNA prepared from two human cell lines (HPB-ALL [lane 1] and Raji [lane 2]) or human thymus tissue (lane 3) was loaded in each lane. The RNA was transferred to a nitrocellulose filter, and the filter was probed with a 32P-labeled probe prepared from Sip1 cDNA. The autoradiograph of the filter is shown.
FIG. 6.
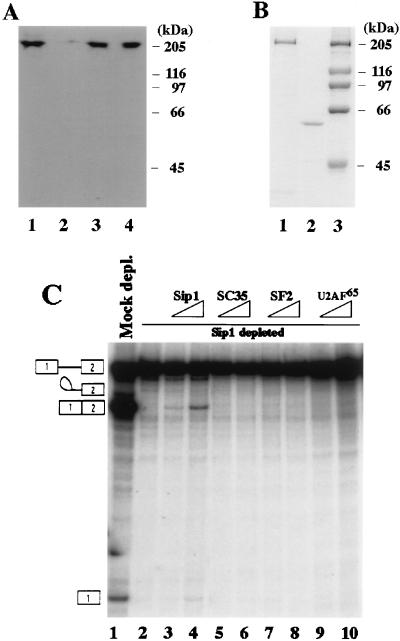
The essential role of Sip1 in pre-mRNA splicing is not redundant with that of SC35, ASF/SF2, or U2AF65. (A) Sip1 protein in HeLa cell nuclear extract could be depleted by anti-Sip1 but not preimmune serum. Western blotting analysis was performed with the anti-Sip1 antibody, using HeLa nuclear extract (lane 1), the extract after immunodepletion with anti-Sip1 (lane 2) or preimmune serum (lane 3), or the purified recombinant Sip1 protein from the baculovirus expression system (lane 4). (B) Coomassie blue staining of the purified recombinant Sip1 protein (lane 1), U2AF65 protein (lane 2), and protein molecular mass markers (lane 3). (C) The recombinant Sip1, but not SC35, ASF/SF2, or U2AF65, could partially restore splicing activity of the Sip1-depleted nuclear extract. In vitro splicing of human β-globin pre-mRNA was carried out for 90 min with HeLa nuclear extract after immunodepletion by the preimmune serum (Mock depl., lane 1) or anti-Sip1 (lanes 2 to 10). The splicing extract was supplemented with 100 and 300 ng of purified Sip1 (lanes 3 and 4), SC35 (lanes 5 and 6), ASF/SF2 (lanes 7 and 8), or U2AF65 (lanes 9 and 10). The splicing reaction products were analyzed on a 6% polyacrylamide gel.
Predicted sequence of Sip1 protein and its limited sequence similarities to known splicing factors.
The predicted Sip1 protein contains 1,403 amino acid residues (Fig. 1) with an estimated molecular mass of 157.9 kDa. It has a high percentage of serine (13.8%) and charged residues (8.3% lysine, 7.1% arginine, and 9.1% glutamine). Data bank searches reveal that Sip1 protein has certain sequence features of proteins involved in pre-mRNA splicing. First, the central portion of the Sip1 protein contains a domain rich in arginine-serine sequences. Eight imperfect repeats of RRSRSXSX are found in this RS domain, and this type of sequence motif is present in several SR proteins (8, 9, 13, 17, 19, 34, 52, 67), spliceosomal protein U1-70K (46, 54), and Drosophila splicing regulators Tra and SWAP (40, 42). In addition to this repeat motif, the arginine-serine-rich region of Sip1 shows primary sequence similarity to the RS domain of several SR proteins, with the highest percentage of amino acid identity to that of SRp75 (Fig. 3A). The sequence similarity in this region is not limited to SR dipeptides or the RRSRSXSX motif; there are 27% identity and 52% similarity over a stretch of 112 amino acid residues (Fig. 3A). Second, Sip1 shows 17% identity and 31% similarity to SWAP in a region of 158 residues from amino acids 273 to 431 (Fig. 3B). This region partially overlaps the first SURP module (12, 36, 53) of the Drosophila SWAP. The SURP motif has been identified in a number of splicing proteins, including SWAP, PRP21, SF3a120, and their homologs in different species (12, 36, 53). However, the highly conserved there is of the SURP motif are not present in Sip1, suggesting that Sip1 is not a member of this SURP family. Another feature of the Sip1 protein is that at the very carboxyl terminus there is an 80-amino-acid motif (Fig. 1) which has been proposed to be the domain on two rat proteins which interacts with the carboxyl-terminal domain (CTD) of RNA polymerase II (65). The presence of this putative CTD-binding domain in Sip1 protein raises the possibility that Sip1 is involved in communication between the splicing machinery and the transcription machinery.
FIG. 3.
Sequence similarities between Sip1 and SRp75 (A) and between Sip1 and Drosophila SWAP (B). Identical amino acid residues are in boldface and underlined. Conserved (I-L-V, D-E, K-R, S-T) amino acids are underlined.
Interaction of Sip1 with U2AF65, U1-70K, and SR proteins.
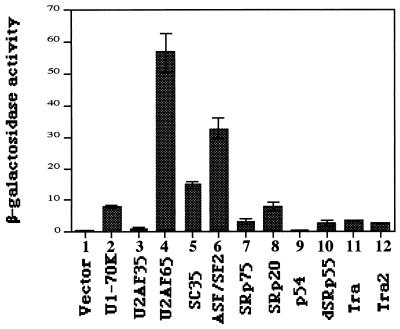
Since splicing factors containing RS domains have been shown to mediate protein-protein interactions important for splicing, we examined protein-protein interactions of Sip1 with other splicing factors, especially the proteins containing RS domains. In the yeast two-hybrid interaction assays, Sip1 interacts not only with SR proteins SC35, ASF/SF2, SRp75, and SRp20 but also with U1-70K and U2AF65 (Fig. 4). The Drosophila splicing regulators Tra and Tra2 as well as the Drosophila SR protein dSRp55 also interact with Sip1 in this assay. These interactions are not due to nonspecific association between the arginine-serine-rich sequences because no significant interactions were detected between Sip1 and U2AF35 or p54, although both of these proteins contain RS domains. Therefore, the profile of protein-protein interactions of Sip1 is different from that observed with SR proteins such as SC35 and ASF/SF2, consistent with the structural differences between Sip1 and SR proteins.
FIG. 4.
Interactions between Sip1 and different RS domain-containing splicing factors (as indicated under the x axis) detected by the yeast two-hybrid interaction assay. Quantitative liquid β-galactosidase assays were performed with yeast extracts from at least three independent yeast isolates for each combination. Relative β-galactosidase activities shown represent fold activation above background (see Materials and Methods).
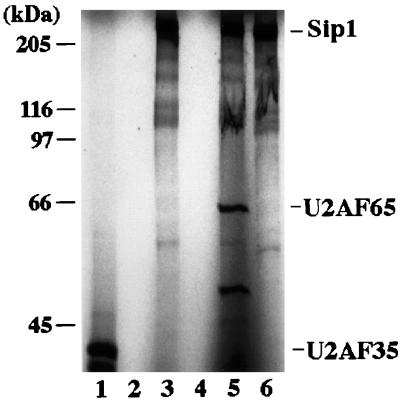
To confirm the results from the yeast two-hybrid system, we examined protein-protein interactions in vitro. Sip1, U2AF65, and U2AF35 proteins were labeled by in vitro translation in the presence of [35S]methionine. Coimmunoprecipitation experiments were carried out to examine possible protein-protein interactions between Sip1 and U2AF65 or U2AF35 (Fig. 5). While the anti-Sip1 antibody (see below and Materials and Methods) immunoprecipitated the in vitro-translated Sip1 protein (lane 6), it did not precipitate either U2AF35 (lane 2) or U2AF65 (lane 4). When Sip1 was coincubated with U2AF65, the anti-Sip1 antibody precipitated both U2AF65 and Sip1 (lane 5). However, when Sip1 was coincubated with U2AF35, the anti-Sip1 antibody precipitated Sip1 but not U2AF35 (lane 3). These results demonstrate that Sip1 interacts with U2AF65 but not U2AF35. The coprecipitation of Sip1 and U2AF65 was not dependent on the presence of RNA, because RNase treatment did not affect the coprecipitation (data not shown). Similar immunoprecipitation experiments indicate that Sip1 also interacts with U1-70K and SC35 in vitro. The results from the yeast two-hybrid system and coimmunoprecipitation experiments were thus consistent in revealing specific protein-protein interactions between Sip1 and these known splicing factors. The observation that Sip1 interacts with a number of essential splicing factors strongly suggests that Sip1 may be involved in splicing.
FIG. 5.
Interaction of Sip1 with U2AF65 but not U2AF35 as assayed by coimmunoprecipitation. Sip1, U2AF65, and U2AF35 were labeled with [35S]methionine by in vitro translation reactions. Individual proteins alone or proteins after coincubation were immunoprecipitated with anti-Sip1 antiserum (lanes 2 to 6). Lane 1 contains U2AF35 immunoprecipitated by anti-FLAG antibody, which recognizes the epitope tag on the in vitro-translated U2AF35. Lanes 2 and 4 contain immunoprecipitates from U2AF35 (lane 2) and U2AF65 (lane 4) in vitro translation products precipitated by anti-Sip1 antibody, showing that this antibody does not cross-react with either U2AF35 or U2AF65. Lanes 3 and 5 contain immunoprecipitates formed after coincubation of Sip1 with U2AF35 (lane 3) or with U2AF65 (lane 5), using anti-Sip1 antibody. Lane 6 contains Sip1 in vitro translation products precipitated by anti-Sip1.
Expression of recombinant Sip1 protein and generation of anti-Sip1 antiserum.
To investigate the biochemical function of Sip1, we decided to express recombinant Sip1 protein and generate specific antibodies recognizing Sip1. After several attempts, we were unable to express the full-length Sip1 in E. coli. However, a fragment of Sip1 containing amino acid residues 227 to 782 was expressed as a His-tagged protein in E. coli. The His-tagged Sip1 protein fragment was purified to apparent homogeneity and was used to raise polyclonal antibodies in rabbits.
The anti-Sip1 antibodies specifically recognize a protein in HeLa cell nuclear extract of approximately 210 kDa (Fig. 6A), larger than the molecular mass predicted from the full-length cDNA (158 kDa). The 210-kDa protein was confirmed to be Sip1 by the observation that both the in vitro-translated product prepared from the full-length Sip1 cDNA and recombinant Sip1 protein purified from the baculovirus system migrated at a molecular mass of approximately 210 kDa (Fig. 5, 6A, and 6B). The specificity of the anti-Sip1 antibodies was also shown by the findings that the antibodies precipitated the 210-kDa protein prepared by in vitro translation of the mRNA made from the full-length Sip1 cDNA but did not precipitate other RS domain-containing proteins or unrelated proteins and that Western blotting using the antibodies detected only one protein band of 210 kDa in the nuclear extracts prepared from HeLa cells and did not cross-react with other proteins (Fig. 6A and data not shown). These results suggest that the difference between the predicted molecular mass and the apparent size determined by SDS-PAGE may be due to posttranslational modification such as phosphorylation, which is known to cause aberrant migration of several RS domain-containing proteins in SDS-PAGE (for a review, see reference 18).
For functional studies, we expressed a full-length recombinant Sip1 protein in the baculovirus system. The recombinant Sip1 protein from the baculovirus system was also recognized by the anti-Sip1 antiserum (Fig. 6A, lane 4) and migrated similarly to the Sip1 protein in HeLa nuclear extract (Fig. 6A, lanes 1 and 3), suggesting that the baculovirus-expressed recombinant Sip1 protein underwent posttranslational modification(s) similar to that in HeLa cells.
Sip1 protein as an essential component in HeLa nuclear extract for its pre-mRNA splicing activity.
The observation that Sip1 not only has sequence features of proteins involved in pre-mRNA splicing but also interacts with several proteins important for splicing, including SR proteins and U2AF65, suggests that Sip1 plays a role in splicing. To test this, we first investigated whether depletion of Sip1 from HeLa cell nuclear extract could affect the splicing activity. Under appropriate conditions, 80 to 90% of Sip1 protein from the nuclear extract could be removed by immunodepletion using the anti-Sip1 antiserum (Fig. 6A; compare lane 2 with lane 1), whereas the level of Sip1 was not reduced in the mock-depleted extract with the preimmune serum (Fig. 6A; compare lane 3 with lane 1). We examined the effect of immunodepletion on the splicing activity of HeLa nuclear extract. Splicing reactions were carried out with β-globin pre-mRNA prepared by in vitro transcription in the presence of [32P]UTP with the HβT7 construct as described previously (74). When the labeled pre-mRNA was incubated with HeLa nuclear extract, the β-globin pre-mRNA was efficiently spliced (Fig. 7B, lanes 8 and 16). The splicing activity was not affected in the nuclear extract treated with the preimmune serum (Fig. 6C, lane 1; Fig. 7B, lanes 1 and 9). By contrast, β-globin pre-mRNA was not spliced by the nuclear extract immunodepleted with the anti-Sip1 antiserum (Fig. 6C, lane 2; Fig. 7B, lanes 2 and 10).
FIG. 7.
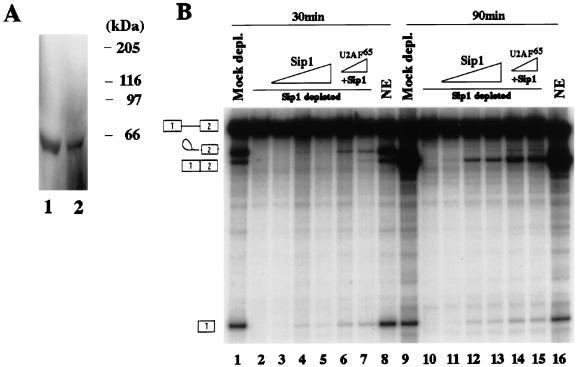
Sip1 interacts with U2AF65 in HeLa cell nuclear extract, and U2AF65 enhances splicing activity of the Sip1-reconstituted nuclear extract. (A) The level of U2AF65 in the Sip1-immunodepleted nuclear extracts was less than in the mock-depleted nuclear extracts as detected by Western blotting. Approximately 150 μg of nuclear extracts which had been immunodepleted by using either preimmune (lane 1) or anti-Sip1 (lane 2) serum was fractionated by SDS-PAGE (10% gel), transferred to a nitrocellulose membrane, and probed with anti-U2AF65 antibody (77). (B) U2AF65 further enhanced the splicing reconstitution by Sip1 in the Sip1-depleted nuclear extract. Lanes 1 to 9 contain the products of in vitro splicing of human β-globin pre-mRNA after a 30-min incubation; lanes 9 to 16 are the corresponding reaction products after a 90-min incubation. The splicing reaction products obtained with untreated HeLa nuclear extract (NE) are shown in lanes 8 and 16. The splicing products obtained with HeLa nuclear extract after immunodepletion by the preimmune serum (Mock depl.) are shown in lanes 1 and 9. Lanes 2 to 5 and 10 to 13 contain the splicing reaction products obtained with Sip1-immunodepleted HeLa nuclear extract supplemented with 0, 100, 200, and 300 ng of purified recombinant Sip1 protein after 30- or 90-min incubation. Lanes 6 and 7 and lanes 14 and 15 show the splicing products obtained with the Sip1-depleted extract supplemented with 200 ng of purified Sip1 and 100 or 200 ng of purified U2AF65 protein, respectively.
To test whether Sip1 was essential for splicing, the recombinant Sip1 protein purified from the baculovirus system was added back to the immunodepleted nuclear extract. Addition of other RS domain-containing proteins, including SC35, ASF/SF2, or U2AF65, had no effect on the splicing activity of the Sip1-depleted extract (Fig. 6C, lanes 5 to 10). The recombinant Sip1 protein which had been purified to apparent homogeneity (Fig. 6B, lane 1), on the other hand, could restore the splicing activity of the Sip1-depleted extract (Fig. 6C, lanes 3 and 4; Fig. 7B). In the range of 100 to 300 ng of Sip1 protein, the restoration of splicing activity was dependent on the amount of Sip1 protein added (Fig. 6C, lanes 3 and 4; Fig. 7B, lanes 3 to 5 and 11 to 13). Further increases in the amount of Sip1 protein did not increase the splicing activity further (data not shown). The restoration of the splicing activity is due to the activity of the purified recombinant Sip1 protein but not due to contaminating proteins from Sf9 cells because mock protein preparations and other RS domain-containing proteins purified from Sf9 cells did not show any activity, and pretreatment of the purified recombinant Sip1 protein with the anti-Sip1 antibody completely removed the splicing reconstitution activity (data not shown). The observation that addition of purified recombinant Sip1 protein but not SC35, ASF/SF2, or U2AF65 protein to the Sip1-depleted extract could restore the splicing activity indicates that the lack of splicing activity in the Sip1-depleted extract was due to Sip1 deficiency in the extract. These results demonstrate that Sip1 protein is essential for splicing and that the functional role of Sip1 in splicing is not redundant with that of known RS domain-containing proteins such as SC35, ASF/SF2 or U2AF65.
U2AF65 further enhances splicing activity in the Sip1-reconstituted nuclear extract.
We noticed that the addition of Sip1 could only partially restore the splicing activity to the nuclear extract immunodepleted by the anti-Sip1 antiserum. To determine whether this was due to the absence or reduction of other splicing factors which interact with Sip1, we examined the protein level of U2AF65 in the mock- and Sip1-immunodepleted nuclear extracts because U2AF65 interacts strongly with Sip1 in both the yeast two-hybrid assay and in vitro biochemical experiments (Fig. 4 and 5). Western blotting experiments using anti-U2AF65 (77) showed that the level of U2AF65 was consistently lower in Sip1-immunodepleted extracts than in the mock-immunodepleted extracts with the preimmune serum (Fig. 7A). Consistent with this, U2AF65 protein was detectable in the beads by Western blotting with anti-U2AF65 after immunoprecipitation of the nuclear extract by anti-Sip1 antibodies (data not shown). Because the anti-Sip1 antibody did not cross-react with U2AF65 either by Western blotting or in the very sensitive immunoprecipitation experiments with 35S-labeled U2AF65 protein (Fig. 6 and 5), U2AF65 protein itself was not directly precipitated by the anti-Sip1 antibody in the absence of Sip1 (Fig. 5 and data not shown). The most likely explanation is that the level of U2AF65 was reduced in the Sip1-immunodepleted extract because of protein-protein interaction between U2AF65 and Sip1. We then tested whether combination of Sip1 and U2AF65 could further increase the splicing activity of the Sip1-immunodepleted extracts. While the addition of U2AF65 alone to the Sip1-depleted extract did not lead to any detectable splicing activity (Fig. 6C, lanes 9 and 10), addition of 100 to 200 ng of U2AF65 protein together with 200 ng of Sip1 protein further enhanced splicing activity (Fig. 7B; compare lanes 6 and 7 with lane 5 and lanes 14 and 15 with lane 13). Together, these results suggest that U2AF65 may interact with Sip1 in the spliceosome and functionally cooperate with Sip1 in the splicing reaction.
That Sip1 may functionally interact with other splicing factors was suggested by the fact that the level of splicing activity after both Sip1 and U2AF65 addition was only about 20 to 30% of that detected with the untreated nuclear extract (Fig. 7B, lanes 8 and 16) or with the mock-depleted nuclear extract with the preimmune serum (Fig. 7B, lanes 1 and 9). The possibility of functional interactions between Sip1 and other splicing factors in the splicing reaction is consistent with the protein-protein interaction profile revealed by the yeast two-hybrid assay and immunoprecipitation studies.
Sip1 is involved in spliceosome assembly.
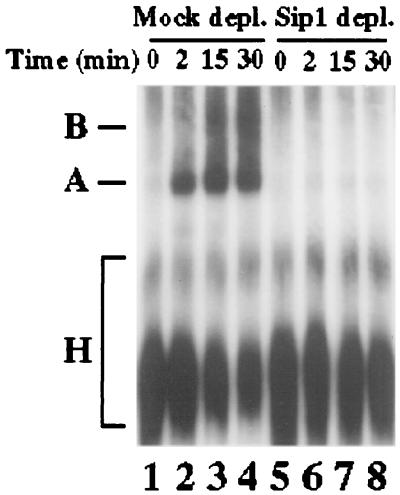
To dissect the functional mechanism of Sip1 in splicing, we examined protein-protein interactions between Sip1 and other splicing factors, especially the proteins involved in the early steps of spliceosome assembly such as U1-70K, SR proteins, and U2AF65, as described above (Fig. 4 and 5). Interaction of Sip1 with several proteins essential for early steps of spliceosome assembly strongly suggests that Sip1 may be involved in this process. We therefore investigated the role of Sip1 in spliceosome assembly. Nonspecific H complex, prespliceosomal complex A, and spliceosomal complex B can be separated by native gel electrophoresis (28, 29). Using 32P-labeled pPIP85.A pre-mRNA (43), we demonstrated that in the Sip1-depleted nuclear extract, the formation of both A and B complexes was deficient whereas the formation of these splicing complexes in the mock-depleted nuclear extract was not affected (Fig. 8), indicating that Sip1 plays an important role in spliceosome assembly.
FIG. 8.
Involvement of Sip1 in spliceosome assembly. The splicing complexes were formed with 32P-labeled pPIP85.A pre-mRNA at different time points in the presence of ATP, using nuclear extracts which were immunodepleted (depl.) by using the preimmune or anti-Sip1 serum (lanes 1 to 4 and 5 to 8, respectively). The splicing complexes were separated on a native 4% polyacrylamide gel by electrophoresis. The autoradiograph of the gel is shown. Positions of A and B complexes and nonspecific H complex are indicated.
Our results demonstrate that depletion of Sip1 leads to blockage of splicing prior to the first step without any detectable cleaved first exon or lariat intermediate (Fig. 6 and 7) and that assembly of both A and B splicing complexes is defective in the Sip1-depleted nuclear extracts (Fig. 8). Because Sip1 interacts with several proteins essential for the earliest steps of spliceosome assembly such as U2AF65 and SR proteins, it is very likely that Sip1 is involved in these earliest steps of spliceosome assembly.
DISCUSSION
We have demonstrated that it is possible to identify new proteins involved in pre-mRNA splicing through their specific interactions with other splicing factors. A protein identified by this approach, Sip1, has been shown to be a new factor essential for mammalian pre-mRNA splicing.
Structural features of Sip1 protein.
Sip1 is a novel protein with sequence similarity to previously identified splicing factors. It contains eight imperfect RRSRSXSX repeats, a motif present in the RS domain of several proteins involved in constitutive splicing and alternative splicing regulation, such as SR proteins and the Drosophila splicing regulators Tra and SWAP. RS domains have been implicated in both protein-protein and protein-RNA interactions (1, 23, 27, 40, 56, 60, 63, 77). We are currently investigating whether the RS domain of Sip1 plays a role in mediating protein-RNA and protein-protein interactions. Several proteins such as U2AF35 and Tra do not contain an RNA-binding motif (42, 73), yet they play important roles in splicing and alternative splicing regulation (42, 77). Sip1 has additional sequence similarity outside of the RS domain with the Drosophila splicing regulator SWAP. We will further investigate the functional significance of this sequence similarity.
Sip1 protein migrates at a molecular mass of approximately 210 kDa, similar to the apparent molecular masses of the U5 snRNP-specific 200- and 220-kDa proteins. Recent information on the partial peptide sequences of these two proteins shows that the 200-kDa protein is a member of DEXH-box family of putative RNA helicases and the 220-kDa protein is a homolog of yeast PRP8 (38). Thus, Sip1 is distinct from these proteins. In affinity-purified spliceosomes assembled on adenovirus major late promoter pre-mRNA or tropomysin pre-mRNA and spliceosomes purified by gel filtration, in addition to the doublet corresponding to the U5-specific proteins 200 and 220 kDa, there appear to be a few protein spots in the 200-kDa range as detected by silver staining of two-dimensional gels (2). In addition, a group of SR protein-related polypeptides have been identified (5, 5a). It remains to be determined whether Sip1 is one of these spliceosome-associated proteins or SR protein-related polypeptides.
The presence of the 80-amino-acid RNA polymerase II CTD-binding motif in the Sip1 protein suggests that Sip1 may be able to interact with the RNA polymerase II CTD, which has been implicated in pre-mRNA processing (65), and play a role in linking the processes of transcription and pre-mRNA splicing. This possibility will be further investigated.
Interactions between Sip1 and spliceosomal components.
We have tested whether Sip1 is a spliceosomal snRNP by examining the presence of snRNAs including U1, U2, U4, U5, and U6 in the immunoprecipitates from HeLa nuclear extracts formed by anti-Sip1 antibodies. We found that no significant amount of spliceosomal snRNPs could be detected in the Sip1 immunoprecipitates (74a), suggesting that Sip1 is not an snRNP protein.
Systematic examination of protein-protein interactions reveals that Sip1 can directly and specifically interact with several known splicing factors. This protein-protein interaction profile is distinct from that of SR proteins such as SC35 and ASF/SF2. Previous studies show that SC35 and ASF/SF2 interact with U1-70K, U2AF35, and several SR proteins, including p54 (63, 74). Similar to SC35 and ASF/SF2, Sip1 interacts with U1-70K, a component of U1 snRNP associated with the 5′ splice site. However, of the proteins associated with the 3′ splice site, Sip1 can interact with U2AF65 but not with U2AF35. Among the SR proteins, Sip1 can interact with SC35 and ASF/SF2 but not with p54. Sip1 is also different from U2AF35 in protein-protein interactions. Both Sip1 and U2AF35 interact with U2AF65 and with several SR proteins; however, Sip1 interacts with U1-70K, whereas U2AF35 does not directly interact with U1-70K (63). Consistent with the finding that Sip1 interacts with multiple spliceosomal proteins, we have observed that anti-Sip1 antibody but not the preimmune serum can efficiently precipitate splicing complexes containing pre-mRNA and splicing products (74a), indicating that Sip1 protein is associated with the spliceosome.
Although U2AF65 could not be directly immunoprecipitated by the anti-Sip1 antibody, the level of U2AF65 was reduced in the Sip1-immunodepleted nuclear extracts. Addition of U2AF65 alone did not have any detectable effect on splicing activity of the Sip1-depleted extract; however, addition of both Sip1 and U2AF65 proteins to the Sip1-depleted extract could restore the splicing activity to a level higher than that achieved by addition of Sip1 protein alone. The most likely explanation for this result is that Sip1 and U2AF65 are associated with each other in HeLa nuclear extract. Addition of both proteins, and perhaps other splicing factors, is required to fully restore the splicing activity of the depleted nuclear extract.
An essential role for Sip1 in pre-mRNA splicing.
A role for Sip1 in splicing was first suggested by the observation that the anti-Sip1 antiserum could deplete splicing activity of HeLa nuclear extract. Purified recombinant Sip1 protein could restore the splicing activity to the Sip1-immunodepleted nuclear extract, demonstrating that Sip1 is indeed required for splicing. That Sip1 is a general splicing factor is supported by the observation that Sip1 is required for splicing of not only human β-globin pre-mRNA substrate but also other model substrates tested, including pPIP85.A (43, 74a). The exact role of Sip1 is not clear, but it appears to be distinct from that of other RS domain-containing splicing factors. This conclusion was based on our findings that the protein-protein interaction profile of Sip1 is different from those of other splicing factors and that other RS domain-containing proteins, including SC35, ASF/SF2, and U2AF65, could not restore the splicing activity of the Sip1-immunodepleted extract.
The exact step at which Sip1 functions in the splicing reaction remains to be investigated. In the Sip1-immunodepleted extract, both the first step and the second step of the splicing reaction were blocked. It is possible that blockage of the second step is the consequence of the defect in the first step of the splicing reaction. Alternatively, Sip1 may be directly involved in both steps of the splicing reaction.
The ability of Sip1 to interact with U2AF65, U1-70K, and several SR proteins suggests a possible role of Sip1 in the earliest steps of spliceosome assembly. Indeed, in the Sip1-depleted nuclear extract, the formation of both A and B complexes is deficient. U2AF has been shown to be required for A-complex formation (51). Because Sip1 interacts with U2AF65, it is possible that the role of Sip1 in A-complex formation is due to its interaction with U2AF65. However, in the Sip1-depleted nuclear extract, a significant amount of U2AF65 is detected, although less than in the mock-depleted extract. This finding suggests that Sip1 may have a direct role in A-complex formation in addition to its interaction with U2AF. The finding that Sip1 interacts with both U1-70K and U2AF65 suggests that Sip1 may be involved in establishing interactions between the 5′ and 3′ splice sites during spliceosome assembly. It is also conceivable that by interacting with both U2AF65 and SR proteins, Sip1 may be able to form a bridge between U2AF65 and SR proteins associated with exon elements, thus mediating interactions across the exon in the exon definition model (4). Previous work has shown that U1 snRNP targets U2AF65 to the 3′ splice site by interactions spanning the exon (24). Sip1 can interact with both U1-70K and U2AF65 and therefore may play a role in the network of interactions spanning the exon. Similarly, by interacting with U1-70K, SR proteins, and U2AF65, Sip1 may participate in the multiple protein-protein interactions across the intron (63). Interaction with U1-70K would allow Sip1 to influence recognition of the 5′ splice site by the U1 snRNP, while interaction between Sip1 and U2AF65 may influence the recruitment of U2 snRNP to the branch site.
SR proteins are critical for pre-mRNA splicing by acting at multiple steps of spliceosome assembly. Previous studies have demonstrated that SR proteins can enhance interaction of U1 snRNP with the 5′ splice site (14, 26, 27, 55, 68) and that at high concentrations, SR proteins can abrogate the requirement for U1 snRNP in pre-mRNA splicing (11, 57, 58). In addition, SR proteins can recruit U2 snRNP to the branch site of a pre-mRNA which lacks a 5′ splice site (56) and escort U4/6.U5 tri-snRNP to the spliceosome (49). It is therefore possible that by interacting with the SR proteins, Sip1 can also function at multiple steps of spliceosome assembly.
ACKNOWLEDGMENTS
We thank R. Brent for providing the yeast two-hybrid system, T. Maniatis and P. Zuo for providing anti-U2AF65 antiserum, R.-M. Xu for helpful suggestions about protein purification, S. Noblitt for technical assistance, and D. Black, J. Bruzik, A. Goate, and Y. Rao for critical reading of the manuscript.
This work was supported by a grant from NIH (RO1 GM53945), an institutional grant through Washington University from Howard Hughes Medical Institute (76296-538202), and a special fellowship from the Leukemia Society of America.
REFERENCES
- 1.Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by Tra2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 1a.Beggs J D, Hodges P E. U2 fulfills a commitment. Curr Biol. 1994;4:264–267. doi: 10.1016/s0960-9822(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 2.Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 3.Bennett M, Reed R. Correspondence between a mammalian spliceosome component and an essential yeast splicing factor. Science. 1992;262:105–108. doi: 10.1126/science.8211113. [DOI] [PubMed] [Google Scholar]
- 4.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;2:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 5.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Blencowe B J, Issner R, Kim J, McCaw P, Sharp P A. New proteins related to the Ser-Arg family of splicing factors. RNA. 1995;1:852–865. [PMC free article] [PubMed] [Google Scholar]
- 6.Bruzik J P, Maniatis T. Enhancer-dependent interaction between 5′ and 3′ splice sites in trans. Proc Natl Acad Sci USA. 1995;92:7056–7059. doi: 10.1073/pnas.92.15.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cáceres J F, Krainer A R. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J. 1993;12:4715–4726. doi: 10.1002/j.1460-2075.1993.tb06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavaloc Y, Popielarz M, Fuchs J P, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaudhary N, McMahon C, Blobel G. Primary structure of a human arginine-rich nuclear protein that colocalizes with spliceosome components. Proc Natl Acad Sci USA. 1991;88:8189–8193. doi: 10.1073/pnas.88.18.8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiara M D, Reed R. A two-step mechanism for 5′ and 3′ splice-site pairing. Nature. 1995;375:510–513. doi: 10.1038/375510a0. [DOI] [PubMed] [Google Scholar]
- 11.Crispino J D, Blencowe B J, Sharp P A. Complementation by SR proteins of pre-mRNA splicing reactions depleted of U1 snRNP. Science. 1994;265:1866–1869. doi: 10.1126/science.8091213. [DOI] [PubMed] [Google Scholar]
- 12.Denhez F, Lafyatis R. Conservation of regulated alternative splicing and identification of functional domains in vertebrate homologs to Drosophila splicing regulator, suppressor of white apricot. J Biol Chem. 1994;269:16170–16179. [PubMed] [Google Scholar]
- 13.Diamond R H, Du K, Lee V M, Mohn K L, Haber B A, Tewari D S, Taub R. Novel delayed-early and highly insulin-induced growth response genes: identification of HRS, a potential regulator of alternative pre-mRNA splicing. J Biol Chem. 1993;268:15185–15192. [PubMed] [Google Scholar]
- 14.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fetzer S, Jurgen L, Will C L, Lührmann R. The U4/U6.U5 tri-snRNP-specific 27K protein is a novel SR protein that can be phosphorylated by the snRNP-associated protein kinase. RNA. 1997;3:344–355. [PMC free article] [PubMed] [Google Scholar]
- 16.Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 17.Fu X-D, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 18.Fu X-D. The superfamily of arginine/serine rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 19.Ge H, Zuo P, Manley J L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 20.Goralski T J, Edström J-E, Baker B S. The sex determination locus transformer-2 of Drosophila encodes a polypeptide with similarity to RNA binding proteins. Cell. 1989;56:1011–1018. doi: 10.1016/0092-8674(89)90634-x. [DOI] [PubMed] [Google Scholar]
- 21.Green M R. Biochemical mechanisms of constitutive and regulated pre-mRNA splicing. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- 22.Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 23.Hedley M-L, Amrein H, Maniatis T. An amino acid sequence motif sufficient for subnuclear localization of an arginine/serine-rich splicing factor. Proc Natl Acad Sci USA. 1995;92:11524–11528. doi: 10.1073/pnas.92.25.11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman B E, Grabowski P J. U1 snRNP targets an essential splicing factor, U2AF65, to the 3′ splice site by a network of interactions spanning the exon. Genes Dev. 1992;6:2554–2568. doi: 10.1101/gad.6.12b.2554. [DOI] [PubMed] [Google Scholar]
- 25.Horowitz D S, Krainer A R. A human protein required for the second step of pre-mRNA splicing is functionally related to a yeast splicing factor. Genes Dev. 1997;11:139–151. doi: 10.1101/gad.11.1.139. [DOI] [PubMed] [Google Scholar]
- 26.Jamison S F, Pasman Z, Wang J, Will C, Lührmann R, Manley J L, Garcia-Blanco M A. U1 snRNP-ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res. 1995;23:3260–3267. doi: 10.1093/nar/23.16.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′ splice site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 28.Konarska M M, Sharp P A. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 29.Konarska M M, Sharp P A. Interactions between snRNPs in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 30.Kozak M. The scanning model for translation. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krainer A R, Maniatis T. Multiple factors including the small nuclear ribonucleoproteins U1 and U2 are necessary for pre-mRNA splicing in vitro. Cell. 1985;42:725–736. doi: 10.1016/0092-8674(85)90269-7. [DOI] [PubMed] [Google Scholar]
- 32.Krainer A R, Conway G C, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- 33.Krainer A R, Conway G C, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 34.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 35.Krämer A, Legrain P, Mulhauser F, Groning K, Brosi R, Bilbe G. Splicing factor SF3a60 is the mammalian homologue of PRP9 of S. cerevisiae: the conserved zinc finger-like motif is functionally exchangeable in vivo. Nucleic Acids Res. 1994;22:5223–5228. doi: 10.1093/nar/22.24.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krämer A, Mulhauser F, Wersig C, Gröning K, Bilbe G. Mammalian splicing factor SF3a represents a new member of the surp family of proteins and is homologous to the essential splicing factor PRP21p of S. cerevisiae. RNA. 1995;1:260–272. [PMC free article] [PubMed] [Google Scholar]
- 37.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 38.Lauber J, Fabrizio P, Teigelkamp S, Lane W S, Hartmann E, Lührmann R. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in S. cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- 39.Lerner E A, Lerner M R, Janeway C A, Steitz J A. Monoclonal antibodies to nucleic acid-containing cellular constituents; probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Bingham P M. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 41.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 42.McKeown M, Belote J M, Baker B. A molecular analysis of tra, a gene in D. melanogaster that controls female sexual differentiation. Cell. 1987;48:489–499. doi: 10.1016/0092-8674(87)90199-1. [DOI] [PubMed] [Google Scholar]
- 43.Moore J M, Sharp P A. Site-specific modification of pre-mRNA: the 2′ hydroxyl groups at the splice sites. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 44.Moore J M, Query C C, Sharp P A. Splicing of precursor to messenger RNAs by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–358. [Google Scholar]
- 45.Patton J G, Mayer S A, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of a polypyrimidine-tract binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 46.Query C C, Bentley R C, Keene J D. A common RNA recognition motif identified within a defined U1 RNA binding domain of the 70K U1 snRNP protein. Cell. 1989;57:89–101. doi: 10.1016/0092-8674(89)90175-x. [DOI] [PubMed] [Google Scholar]
- 47.Reed R. Initial splice site recognition and base pairing during pre-mRNA splicing. Curr Opin Genet Dev. 1996;6:215–220. doi: 10.1016/s0959-437x(96)80053-0. [DOI] [PubMed] [Google Scholar]
- 48.Rio D C. Splicing of pre-mRNA: mechanism, regulation and role in development. Curr Opin Genet Dev. 1993;3:574–584. doi: 10.1016/0959-437x(93)90093-5. [DOI] [PubMed] [Google Scholar]
- 49.Roscigno R F, Garcia-Blanco M A. SR proteins escort U4/6.U5 tri-snRNP to the spliceosome. RNA. 1995;1:692–706. [PMC free article] [PubMed] [Google Scholar]
- 50.Roth M B, Murphy C, Gall J G. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ruskin B, Zamore P D, Green M R. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- 52.Screaton G R, Caceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spikes D A, Kramer J, Bingham P, Van Doren K. SWAP family pre-mRNA splicing regulators are a novel, ancient protein family sharing a highly conserved sequence motif with prp21 family of constitutive splicing factors. Nucleic Acids Res. 1994;22:4510–4519. doi: 10.1093/nar/22.21.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spritz R A, Strucnk K, Surowy C S, Hoch S O, Barton D E, Francke U. The human U1-70k snRNP protein: cDNA cloning, chromosomal localization, expression, alternative splicing, and RNA-binding. Nucleic Acids Res. 1987;15:10373–10391. doi: 10.1093/nar/15.24.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tacke R, Chen Y, Manley J L. Sequence specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94:1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarn W Y, Steitz J A. SR proteins can compensate for the loss of U1 snRNP functions in vitro. Genes Dev. 1994;8:2704–2717. doi: 10.1101/gad.8.22.2704. [DOI] [PubMed] [Google Scholar]
- 58.Tarn W Y, Steitz J A. Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc Natl Acad Sci USA. 1995;92:2504–2508. doi: 10.1073/pnas.92.7.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Valcarcel J, Green M R. The SR protein family: pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- 60.Valcarcel J, Gaur R K, Singh R, Green M R. Interaction of U2AF65 RS region with pre-mRNA of branch point and promotion of base pairing with U2 snRNA. Science. 1996;273:1706–1709. doi: 10.1126/science.273.5282.1706. [DOI] [PubMed] [Google Scholar]
- 61.Vellard M, Sureau A, Soret J, Martinerie C, Perbal B. A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc Natl Acad Sci USA. 1992;89:2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wahle E, Keller W. The biochemistry of 3′ end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 63.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 64.Xiao S H, Manley J L. Phosphorylation of the ASF/SF2 RS domain affects both protein-protein and protein-RNA interactions and is necessary for splicing. Genes Dev. 1997;11:334–344. doi: 10.1101/gad.11.3.334. [DOI] [PubMed] [Google Scholar]
- 65.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zahler A M, Lane W S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 67.Zahler A M, Neugebauer K M, Stolk J A, Roth M B. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zahler A M, Roth M B. Distinct functions of SR proteins in recruitment of U1 small nuclear ribonucleoprotein to alternative 5′ splice sites. Proc Natl Acad Sci USA. 1995;92:2642–2646. doi: 10.1073/pnas.92.7.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zamore P D, Green M R. Identification, purification and characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci USA. 1989;86:9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zamore P D, Green M R. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intracellular distribution. EMBO J. 1991;10:207–214. doi: 10.1002/j.1460-2075.1991.tb07937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zamore P D, Patton J G, Green M R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 72.Zervos A S, Gyuris J, Brent R. Mix1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 73.Zhang M, Zamore P D, Carmo-Fonseca M, Lamond A I, Green M R. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89:8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang W-J, Wu J Y. Functional properties of p54, a novel SR protein active in constitutive and alternative splicing. Mol Cell Biol. 1996;16:5400–5408. doi: 10.1128/mcb.16.10.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74a.Zhang, W.-J., and J. Y. Wu. Unpublished result.
- 75.Zuo P, Manley J L. Functional domains of the human splicing factor ASF/SF2. EMBO J. 1993;12:4727–4737. doi: 10.1002/j.1460-2075.1993.tb06161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zuo P, Manley J L. The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5′ splice sites. Proc Natl Acad Sci USA. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev. 1996;10:1356–1368. doi: 10.1101/gad.10.11.1356. [DOI] [PubMed] [Google Scholar]