Abstract
DNA topoisomerase II is a multidomain homodimeric enzyme that changes DNA topology by coupling ATP hydrolysis to the transport of one DNA helix through a transient double-stranded break in another. To investigate the biochemical properties of the individual domains of Leishmania donovani topoisomerase II, four truncation mutants were generated. Deletion of 178 aminoacids from the C-terminus (core and LdΔC1058) had no apparent effect on the DNA-binding or cleavage activities of the enzymes. However, when 429 aminoacids from the N-terminus and 451 aminoacids from the C-terminus were removed (LdΔNΔC), the enzyme was no longer active. Moreover, the removal of 429 aminoacids from the N-terminus (LdΔNΔC, core and LdΔN429) render the mutant proteins incapable of performing ATP hydrolysis. The mutant proteins show cleavage activities at wide range of KCl concentrations (25–350 mM). In addition, the mutant proteins, excepting LdΔNΔC, can also act on kDNA and linearize the minicircles. Surprisingly, the mutant proteins fail to show the formation of the enhanced cleavable complex in the presence of etoposide. Our findings suggest that the conformation required for interaction with the drug is absent in the mutant proteins. Here, we have also identified Tyr775 through direct sequencing of the DNA linked peptide as the catalytic residue implicated in DNA-breakage and rejoining. Taken together, our results demonstrate that topoisomerase II are functionally and mechanistically conserved enzymes and the variations in activity seem to reflect functional optimization for its physiological role during parasite genome replication.
INTRODUCTION
The participation of the DNA topoisomerases in a number of vital processes, including replication, transcription, genetic recombination, chromosomal decondensation and segregation is well documented (1). These enzymes catalyze the transient breakage of DNA strands to form enzyme-mediated gates through which an intact DNA strand or a pair of DNA strands of a double helix can pass. DNA topoisomerases can be divided into three sub-families, namely type IA, type IB and type II on the basis of sequence homology and mechanism of action (2). Type II DNA topoisomerases catalyze ATP-dependent passage of a DNA segment through a transient double-strand break in another segment. In such a reaction, the enzyme-mediated reversible cleavage is generated by the trans-esterification reactions between a pair of active site tyrosines and two DNA phosphodiester bonds staggered 4 bp apart. Eukaryotic topoisomerase II are dimeric, whereas its prokaryotic counterparts including gyrase and topo IV are A2B2 tetramers. Moreover, the N-terminal and the central parts of eukaryotic topoisomerase II are homologous to bacterial GyrB and GyrA subunits, respectively.
Even though cells cannot survive without the double-stranded DNA passage activity of topoisomerase II, the formation of a cleavage complex is potentially toxic. DNA polymerases or helicases can convert such transient topoII-DNA intermediates into permanent double-stranded DNA breaks that can eventually trigger cell death pathways (3). Besides its physiological activities, topoisomerase II is a target for a number of antineoplastic and antiparasitic drugs (4,5). Topoisomerase II inhibitors can be divided into topo II catalytic inhibitors and topo II poisons (6). Topoisomerase catalytic inhibitors like bis-dioxopiperizines do not stabilize DNA cleavage complexes whereas topo II poisons inhibit the enzyme by increasing the steady-state levels of DNA cleavage complexes (7,8).
Despite the importance of DNA cleavage/religation reaction to the function of topoisomerase, relatively little is known about the specific enzyme–DNA interaction that drives this reaction. Therefore, the molecular interaction of LdTOP2 and DNA remains an enigma. In conjunction with this fact, even though topoisomerase II from protozoan parasites have been cloned and sequenced (9), the mechanistic features of the enzyme have never been explored. In kinetoplastid parasites topoisomerase II undoubtedly function at several steps in the minicircle replication processes, decatenation of the parental circles from the network and recatenation of the daughter circles to the network (10). In Leishmania donovani, we had previously reported the existence of a type II topoisomerase containing 1236 aminoacids localized both in nucleus and kinetoplast (11). Given the importance of cleavage/religation reactions mediated by topoisomerase in the life cycle of the parasite, in the present study we report the properties of the DNA-binding domain of LdTOP2.
In human topoisomerase IIα a fragment from aminoacid 1–439 was shown to have an intrinsic ATPase activity (12), which could be stimulated by DNA. In another study, the core domain of drosophila topoisomerase II covering aminoacid 406–1207 was demonstrated to have wild-type levels of cleavage and religation activities (13). These results indicate that the individual domains in topoisomerase II preserve their intrinsic activities even when separated from the rest of the enzyme, demonstrating that they fold up as independent catalytic domains.
In the present study, to further investigate the biochemical properties of the DNA-binding or core domain of LdTOP2, we have constructed four truncation mutants namely LdΔN429 (aminoacid residues from 430–1236), core (aminoacid residues 430–1058), LdΔC1058 (aminoacid residues 1–1058) and LdΔNΔC (aminoacid residues 430–785). The mutant proteins have been characterized with respect to ATPase, DNA-binding and cleavage activities. Our results indicate that the residues from 1–1058 which mimics the wild-type enzyme in complementing the temperature-sensitive topoisomerase II mutant yeast strain (14), retains its DNA-dependent ATPase activity and can also perform cleavage of supercoiled DNA. Moreover, the catalytic amino acid implicated in DNA breakage and reunion was mapped by sequencing the DNA bound protein. Taken together, this is the first report that demonstrates the domain demarcations of LdTOP2 and provides an insight into the mechanistic aspects of the core/DNA-binding domain of the protein and the catalytic residue implicated in DNA breakage and rejoining.
MATERIALS AND METHODS
Construction of truncation mutants
For construction of all truncation mutants, the regions corresponding to amino acids 430–785, 430–1058, 430–1236 and 1–1058 were amplified by PCR. For aminoacids 430–785 (1 kb DNA fragment), aminoacids 430–1058 (1.9 kb DNA fragment), aminoacids 430–1236 (2.4 kb DNA fragment), the sense primer used was 5′ GGGAATTCCATATGACAGAGGGTGACTCGGCG 3′ and the antisense primers were 5′ CGGGATCCCTTGTACAAGTCAAGGCG 3′, 5′CGGGATCCGATGTAGTCGAAGCTCTC3′ and 5′CGGGATCCCCCGCTTCACATGCTCCCCTTACA 3′, respectively. For the construction of 1–1058 (3.2 kb) mutant, the sense and the antisense primers used were 5′ GGGAATTCCATATGACAGACGCTTCCAAG- 3′ and 5′ CGGGATCCTCTAGAGAAGCTCTCGTCCACGCG- 3′ respectively. The amplified products were cloned in the NdeI and BamHI sites of bacterial expression vector pET16b (Novagen) and expressed in Escherichia coli BL21 (DE3) pLysS. The full length LdTOP2 was cloned in the BamHI and HindIII sites of expression vector pET28C, using sense primer 5′-CG GGATCCTCATGACAGACGCTTCCAAG-3′ and anti sense primer 5′-CCCAAGCTTCATCAAAACATGTGGCAG-3′. PCR was performed for 30 cycles of denaturation at 94°C for 45 s, annealing at 52.5°C and extension at 68°C for 1 kb/min using Pwo polymerase (Roche Applied Science) and 200 μM of each dNTP.
Expression and protein purification
Expression from constructs pET16b/1, pET16b/1.9, pET16b/2.4, pET16b/3.2 and pET28c/3.7 yielded 40, 70, 89, 116 and 137 kDa proteins, respectively. Purification of all the His-tagged proteins was done as follows. Briefly, the cells containing the recombinant plasmids were induced at 0.6 OD600 with 0.5 mM IPTG at 22°C for 10 h. The cells were harvested and resuspended in phosphate buffer (pH 7.8) containing 300 mM NaCl, 200 μg/ml lysozyme, 0.1% Triton X-100, 0.25% Sarkosyl and 1 mM PMSF. Final lysis was achieved by sonication on ice and the lysate was cleared by centrifugation at 12 000 rpm in a SS34 rotor for 20 min. The cleared lysate was passed through Ni- nitrilo triacetic acid (Ni-NTA) agarose column (Qiagen). After washing with phosphate buffer (pH 7.8) containing 300 mM NaCl and 30 mM imidazole, elution was done using 250 mM imidazole. For further purification, the fractions were pooled from Ni2+ column, dialyzed and loaded onto a 2 ml packed phosphocellulose column (P11 cellulose, Whatman). The column was washed with 10 ml of wash buffer (50 mM Tris–HCl, pH 7.5, 0.5 mM EDTA and 20% glycerol) containing 600 mM KCl and finally eluted with the same buffer containing 800 mM KCl. After elution the peak fractions were dialyzed overnight against storage buffer containing 10 mM Tris–HCl (pH 7.8), 100 mM NaCl, 5 mM MgCl2, 1 mM PMSF and 15% glycerol and stored at −70°C until further use.
ATPase assay
ATPase measurements were carried out by the pyruvate kinase/lactate dehydrogenase assay described previously (12) with the following modifications. Reaction mixture (1 ml) contained: 0.1 mM NADH, 2 mM phosphoenolpyruvate, 3 units of pyruvate kinase and 4 units of lactate dehydrogenase. Reactions were initiated by mixing the reaction mixture and ATP, both pre-equilibrated to 30°C. Decrease in NADH concentration was monitored by measuring the absorbance at 340 nm in a UV-visible spectrophotometer (Shimadzu Corp.) All assays were performed in triplicate.
Electrophoretic mobility shift assay
The labeling of 36 mer oligonucleotide 1 (5′ATGAAATCTAACAATGCGCTCATCGTCATCCTCGGC3′) containing high affinity topoisomerase II binding site (15) was done using polynucleotide kinase (Invitrogen). The labeled oligonucleotide 1 was annealed to oligonucleotide 2 (5′ TACTTTAGATTGTTACGCGAGTAGCAGTAGGACCG 3′) in a buffer containing 40 mM Tris–HCl (pH 7.5), 20 mM MgCl2 and 50 mM NaCl at 70°C for 1 h and allowed to cool down slowly to room temperature. Briefly, the reaction was done in a 20 μl binding buffer containing 50 mM Tris–HCl (pH 7.5), 1 mM DTT, 4 mM MgCl2, 50 mM KCl and 15 μg/ml BSA and 5 pmol of 36 bp duplex oligonucleotide and varying concentrations of wild type and truncated LdTOP2 enzymes. Reaction mixtures were incubated at 4°C for 30 min and electrophoresed through 7% non-denaturing polyacrylamide gel (acrylamide:bis:: 29:1) in 0.25× TBE and autoradiographed. Quantitation of the unbound oligonucleotide was done by densitometry and Kd was estimated from protein concentration at which half of the duplex oligonucleotide was bound to the protein (16).
Cleavage reaction
Cleavage reaction was performed as described (17). A 20 μl reaction mixture contained 200 ng negatively supercoiled pRYG DNA, 10 mM Tris–HCl (pH 7.5), 100 mM KCl, 0.1 mM EDTA, 5 mM MgCl2, 0.5 mM DTT, 30 μg BSA and 10 pmol of enzymes. Reaction was started by incubating at 37°C for 30 min and terminated by the addition of 0.5% SDS and 10 mM EDTA. The mixture was further incubated with 100 μg/ml proteinase K at 37°C for 30 min and analyzed by electrophoresis on a 1% agarose gel. Ethidium bromide at a final concentration of 0.5 μg/ml was included in the gel to resolve the linear product (Form III) from the supercoiled molecule (Form I). Control assays always contained an amount of drug diluent (Dimethyl sulfoxide) equivalent to that present in drug containing reaction. Salt dependence of the mutant enzymes was determined by performing cleavage reaction as described above, using varying concentrations of salt. When KCl dependence was estimated, Mg2+ concentration was kept at 5 mM and for Ca2+ and Mg2+ requirement, KCl concentration was kept at 100 mM.
DNA relaxation
DNA relaxation reactions containing 0.5 μg of negatively supercoiled pRYG DNA were carried out in the absence or presence of etoposide in 25 mM Tris–HCl(pH 7.9), 10 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 2 mM ATP, 100 mM NaCl, 10% glycerol and 500 ng of recombinant L.donovani topoisomerase II. Following incubation at 37°C for 30 min, the reaction was terminated by the addition of loading buffer. The products were analyzed by gel electrophoresis in 1% agarose gel, stained with ethidium bromide and photographed under UV Transillumination.
Cleavage of kDNA
Plasmid pGEM4Z was used to clone the full-length L.donovani minicircles sequence resulting in the formation of pLURKE3 (18). The insert was excised with EcoRI and radiolabeled by random priming using [α32P]dCTP (Amersham pharmacia) and used as a probe. As described previously, (19) kinetoplast DNA (kDNA) was isolated from Leishmania strain UR6. About 200 ng kDNA was incubated with 50 pmol enzyme for 30 min at 37°C in the cleavage buffer mentioned above. DNA was transferred from agarose gel to nitro cellulose membrane. The membrane was hybridized with radiolabeled E3 probe.
Covalent complex between truncated LdTOP2 enzymes and radiolabeled DNA
Covalent complex between protein and DNA was formed as described (20). Reactions were initiated with 50 pmol truncated LdTOP2 enzymes (70, 89 and 116 kDa) in the cleavage buffer mentioned above, containing 4ng 32P-labeled negatively supercoiled pRYG plasmid DNA. Reactions were quenched with NaOH and neutralized with HCl. DNase (FPLC pure, Amersham pharmacia) treatment was done for 1 h at 37°C. Reactions were terminated by the addition of 0.5% SDS and 10 mM EDTA before the complexes were processed through two cycles of a Sephadex G-25 spin column and proteins were precipitated and separated by SDS–PAGE. The gel was dried and autoradiographed to expose the protein bands.
For thin layer chromatography, the protein-DNA complexes purified through G-25 column were treated with proteinase K at 37°C for 30 min the digested products were vaccum dried and re-suspended in 30 μl of constant boiling with 6N HCl. After removal of HCl, the hydrolysates were dried under vaccum and redissolved in H2O containing unlabeled phosphoserine, phosphotyrosine and phosphothreonine. The hydrolysates were spotted on silica gel plate and chromatographed in a solvent containing ethanol/n-butyl alcohol/ammonia/water (volume ratios 4:1/:2:3). After drying, the plates were sprayed with 0.25% ninhydrin in acetone and heated to 65°C to visualize the phospho aminoacid standards. The radioactive signals of the phospho aminoacid were detected by autoradiography (21).
Preparation of LdTOP2-derived tryptic peptide covalently linked to DNA
A 100 μl mixture containing the protein and 32P labeled pRYG DNA was incubated in the above-mentioned cleavage buffer at 37°C for 10 min. SDS was added to a final concentration of 0.5% to form a covalent complex between DNA and protein. The protein DNA complex was subjected to SDS–PAGE electrophoresis and stained with Coomassie brilliant blue. The protein band was excised and in gel trypsin digestion was performed. The trypsin-digested peptides were fractionated by reverse phase HPLC. Aliquots of the fractions were assayed for 32P. The positive fraction was subjected to Edman sequencing.
RESULTS
Expression of N-terminal deletion mutants and core domain of LdTOP2
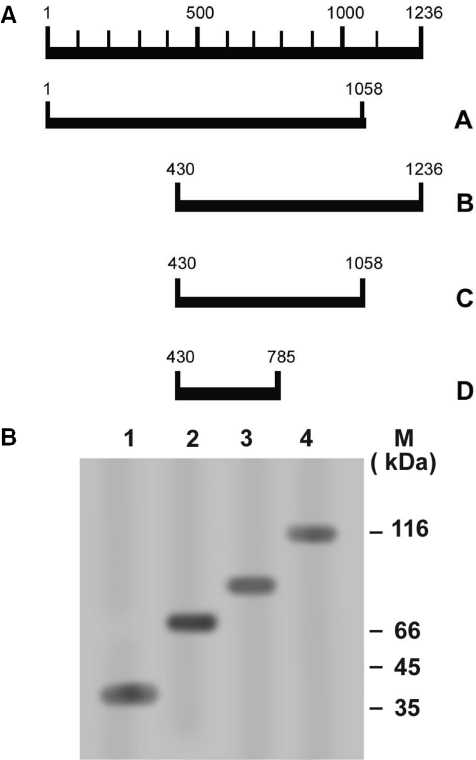
In order to study the function of core domain of kinetoplastid toposiomerase II, we constructed four truncation mutants of L.donovani topoisomerase II (1236 amino acids). The truncation start points and end points were determined on the basis of our previous analysis, where the amino terminal 1–385 residues have an intrinsic ATPase activity (T. Sengupta, M. Mukherjee, A. Das, C. Mandal, R. Das, T. Mukherjee and H. K. Majumder, unpublished data) and 1–1058 amino acids can decatenate kDNA and complement temperature-sensitive mutant yeast strain (14). So, to further investigate the interaction of DNA with the core domain (homologous to the core/DNA-binding domain of other eukaryotic topoisomerase II), we constructed four truncation mutants of LdTOP2 containing amino acids 430–785 (LdΔNΔC), 430–1058(core), 430–1236 (LdΔN429) and 1–1058 (LdΔC1058) (Figure 1A). All the mutants were generated by PCR amplification of the full-length LdTOP2 gene using site-specific primers. The PCR products were cloned in pET16b and transformed in E.coli BL21 (DE3) plysS. The bacterial cells harboring the truncation constructs were induced with 0.5 mM IPTG and the resultant overexpressed proteins were purified by Ni-NTA agarose, as the mutant enzymes had six histidine residues at their N-terminus, and the wild-type LdTOP2 had histidine tagged both at the N-terminus and the C-terminus. Further purification was achieved by passing the Ni-NTA eluate through the phosphocellulose column as described in Materials and methods (Figure 1B).
Figure 1.
Schematic representation of topoisomerase II of L.donovani and purification of the truncated proteins. (A) A coordinate of Leishmania TOP2 peptide. (A) LdΔC1058, (B) LdΔN429, (C) Core, (D) LdΔNΔC protein. (B) 10% SDS-polyacrylamide gel showing purification of the truncation proteins. Lanes 1–4, 2 μg of 40, 70, 89 and 116 kDa, respectively, purified through Ni-NTA and phosphocellulose columns. The gel was stained by silver stain. Sizes of molecular mass markers (MBI Fermentas) in kDa are shown.
ATPase activity
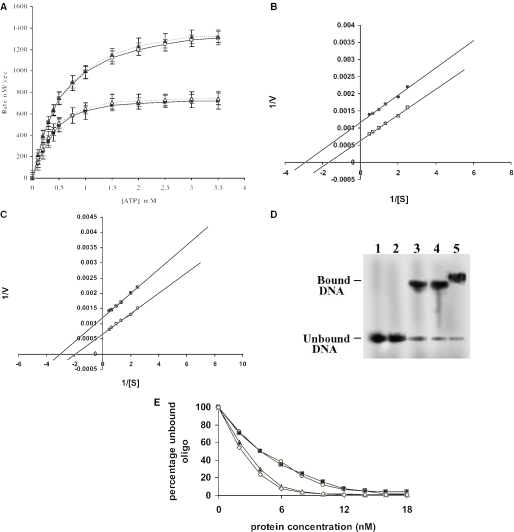
It is known that upon binding to ATP, topoisomerase II undergoes a conformational change that triggers double-stranded DNA passage (20). We checked if any of the mutant proteins retain their ATPase activities. Apart from the wild-type enzyme, only the 116 kDa enzyme (1–1058 aminoacids residues) had an ATPase activity. Moreover, the ATPase activity of this protein was found to be stimulated by DNA (Figure 2A). The KM of the 116 kDa protein was found to be 0.33 mM and 0.55 mM in the presence and absence of DNA, respectively (Figure 2B). DNA was found to stimulate its activity 2-fold. These values are comparable with those of the wild-type enzyme having KM of 0.32 mM and 0.50 mM in the presence and absence of DNA, respectively (Figure 2C).
Figure 2.
Properties of the mutant proteins. (A) Dependence of ATPase rate of the 116 and 137 kDa protein on ATP concentration at constant enzyme concentration. 116 kDa (open square), 137 kDa (black triangle) in presence of DNA, 116 kDa (black square), 137 kDa (open triangle) in absence of DNA. The data shown are mean ± SD of three independent experiments. (B) The KM was calculated from the Lineweaver–Burk plot, 116 kDa in absence (open square), and in presence (black circle) of DNA respectively. (C) 137 kDa in absence (open circle) and in presence (black square) of DNA respectively. (D) DNA-binding activity of the truncated LdTOP2 proteins. Lane 1, labeled 36mer duplex oligonucleotide; lanes 2–5, 36mer oligonucleotide incubated with 40, 70, 89 and 116 kDa. (E) Graphical representation of the native gel shift assay. The percentage of unbound oligonucleotide present in gel was quantified by film densitometry and plotted against protein concentration. The kd value was calculated from this graph, core (open circle), LdΔC1058 (black triangle), LdΔN429 (black square) and 137 kDa (open diamond) respectively.
The truncation mutants bind to DNA in gel retardation assay
A polyacrylamide gel retardation assay was carried out to study the DNA binding characteristics of LdTOP2 truncation mutants. The linear DNA substrate was a 36 bp duplex containing high affinity topoisomerase II cleavage site (15). The ability of LdTOP2 truncation mutants to bind the 36 bp duplex was tested under standard gel retardation assay condition, as described in Materials and Methods. The constructs containing amino acid 1–1058 (116 kDa), 430–1058 (70 kDa) and 430–1236 (89 kDa) were able to bind to the oligonucleotide but the construct containing amino acids 430–785 (40 kDa) failed to bind to the duplex DNA. The electrophoretic patterns were typical of the binding studies (Figure 2D). The bound DNA migrated as a high molecular weight species as compared to the unbound DNA. Since the electrophoretic mobility of the complexed DNA was greatly retarded as compared to that of the free substrate, the amount of unbound DNA could be quantitated directly by densitometric scanning. This quantitation, in turn, allowed the straightforward calculation of bound plasmid molecules by subtraction from the total DNA employed (Figure 2E). The Kd value of LdΔC1058, LdΔN429 and core were found to be 3.3, 4.09 and 4.2 nM, respectively, while that of the wild-type enzyme was 3.0 nM.
Analysis of the cleavage activity of the mutant proteins
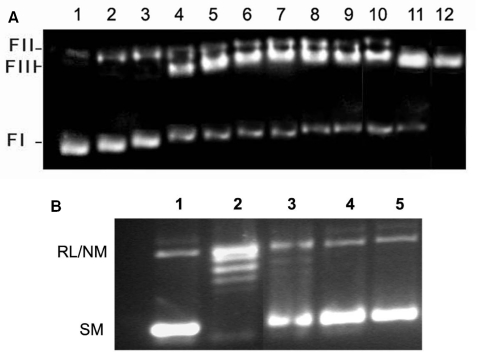
An essential step in the relaxation of supercoiled DNA is the cleavage of DNA in both strands by topoisomerase II. To test the possibility that a mutant enzyme can nevertheless cleave DNA via trans-esterification and may possess a normal level of cleavage activity, the purified mutant enzymes were assayed for their cleavage of plasmid DNA. The 40 kDa protein (430–785 amino acids residues) which does not bind to DNA also fails to show any cleavage of supercoiled DNA (Figure 3A), whereas all other purified proteins (70, 89 and 116 kDa and and the wild-type LdTOP2) cleave supercoiled DNA. However, etoposide enhanced the formation of cleavable complex only by the wild-type LdTOP2 protein and did not have any effect on the activities of the mutant proteins (Figure 3A). Furthermore, to show that the mutant enzymes generated linear DNA (Form III) and not the nicked form (Form II), pRYG DNA treated with restriction endonuclease HindIII was used as a marker (Figure 3A). The wild-type LdTOP2 was sensitive to etoposide (Figure 3B). LdTOP2 relaxed supercoiled pRYG DNA (Lane 1) and the formation of topological isomers (lane 2) was inhibited by etoposide (lanes 3–5).
Figure 3.
Comparison of cleavage activity of the truncated proteins. (A) Lane 1: negatively supercoiled pRYG DNA; lanes 2, 4, 6, 8 and 10, cleavage by 40, 70, 89 and 116 kDa and the full length LdTOP2 protein respectively. Lanes 3, 5, 7, 9 and 11, same as lanes 2, 4, 6, 8 and 10 but in the presence of 20 μg/ml etoposide respectively. Lane 12, pRYG DNA digested with Hind III restriction enzyme. (B) Lane 1: negatively supercoiled pRYG DNA only, Lane 2: same as lane 1 but with 500 ng topoisomerase II, Lane 3, 4 and 5 same as above but with 20, 50 and 100 μg/ml etoposide respectively. Positions of supercoiled monomer (SM) and relaxed and nicked monomer are indicated (RL/NM).
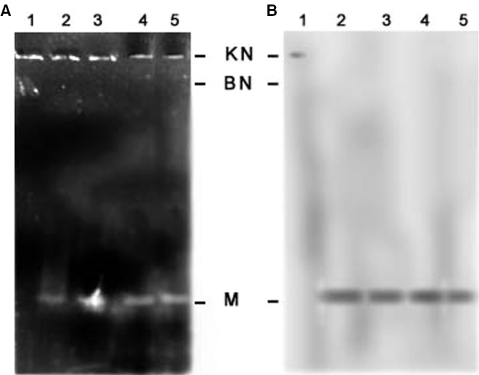
Topoisomerase II is the key enzyme involved in DNA decatenation (22). Virtually every step in the replication of the massive kDNA network of kinetoplastid parasites entails alteration in DNA topology. Such reactions are catalyzed by DNA topoisomerases. Many compounds active against eukaryotic topoisomerase II have been reported to cause linearization of the minicircles (10). To explore whether the mutant enzymes having DNA-binding and cleavage activities could interact with kinetoplast DNA of Leishmania, we investigated if the mutant enzymes could linearize kDNA. As described earlier kDNA isolated from L.donovani was incubated with the mutant enzymes in the cleavage buffer. After a 30-min incubation at 37°C, the reaction was terminated by the addition of SDS and EDTA and the cleavage products were subjected to proteinase K digestion. The DNA was resolved by agarose gel electrophoresis and transferred to a nitrocellulose membrane. To examine the minicircle DNA, the blot was probed with 32P-labeled E3. The predominant forms were found to be linearized (Form III) minicircles (Figure 4A and B). To show that the mutant enzymes generated linearized minicircles, kDNA treated with restriction endonuclease PstI was used as control. A comparison of the activities of all mutants have been summarized in Table 1.
Figure 4.
Topoisomerase II induced linearization of minicircles. (A) Lane 1, kDNA isolated from Leishmania strain UR6. Lanes 2–4 same as lane 1 but incubated with core, LdΔN429, LdΔC1058. Lane 5, kDNA isolated from Leishmania strain UR6 digested with restriction endonuclease PstI. (B) The agarose gel in panel (A) was transferred to nitrocellulose and probed with linearized E3 as described in Materials and Methods to detect the free minicircles.
Table 1.
Comparison of the activities of wild type and mutant LdTop2 proteins
| Mutants | Aminoacids deleted | ATPase Activity | DNA-binding | Cleavage activity |
|---|---|---|---|---|
| LdΔNΔC (40 kDa) | 429 from N-terminus 451 from C-terminus (residues 430–785) | − | − | − |
| Core (70 kDa) | 429 from N-terminus 178 from C-terminus (residues 430–1058) | − | + | + |
| LdΔN429 (89 kDa) | 429 from N-terminus (residues 430–1236) | − | + | + |
| LdΔC1058 (116 kDa) | 178 from C-terminus (residues 1–1058) | + | + | + |
| LdTOP2 (wild type) | 1236 amino acid | + | + | + |
Salt dependence of cleavage reaction
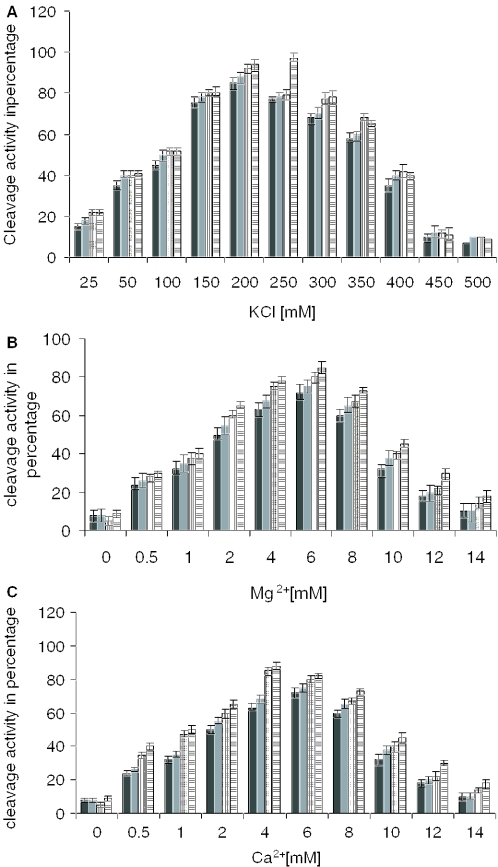
To investigate the characteristics of the truncated proteins in the cleavage of DNA, assays were performed under various KCl concentrations keeping the wild-type enzyme as control. The analysis revealed that the mutant enzymes were active over a wide range of salt concentrations from 25–300 mM, while the activity is inhibited at 400 mM KCl concentration (Figure 5A).
Figure 5.
Salt dependence of cleavage activity of the mutant proteins. (A) Graphical representation of the extent of covalent complex formation plotted as a function of KCl concentration. (B) Graphical representation of the extent of covalent complex formation plotted as a function of Mg2+ concentration. (C) Graphical representation of the extent of covalent complex formation plotted as a function of Ca2+ concentration where vertical shaded bar is LdΔC1058, gray bar is LdNΔ429, black bar is core protein and horizontal shaded bar is 137 kDa protein in all cases. Percentage of cleavage was calculated as described above.
Topoisomerase II requires divalent cations at two separate catalytic centers, the DNA cleavage/religation center and the ATP binding/hydrolysis center. In our previous study (T. Sengupta, M. Mukherjee, A. Das, C. Mandal, R. Das, T. Mukherjee and H. K. Majumder, unpublished data), we have observed that Mg2+ is required for enzyme-mediated ATP hydrolysis by the N-terminal domain of LdTOP2. In the present study, we show the effect of Mg2+ and Ca2+ on the cleavage activity mediated by the core domain of the enzyme. Our results show that Mg2+ as well as Ca2+ can support cleavage activity mediated by LdTOP2 (Figure 5B and C), the optimum conditions for cleavage being 4 mM Ca2+ and 6 mM Mg2+, respectively.
Active site mapping of L.donovani topoisomerase II
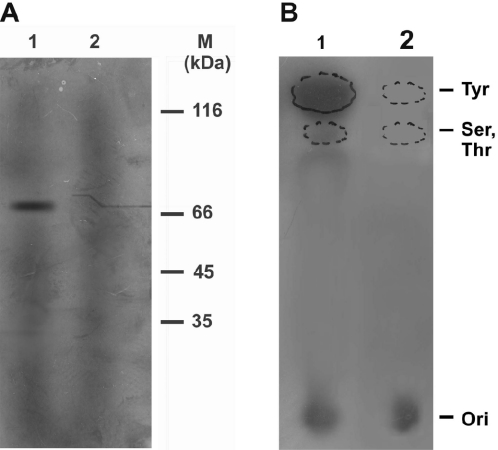
Since all the proteins were observed to have similar binding and cleavage patterns, the 70 kDa core protein was selected for active site mapping and was incubated with 32P-labeled pRYG DNA in cleavage buffer. After incubation at 37°C for 30 min, the mixture was treated with KOH to capture the covalent protein-DNA complex, then digested with protease-free DNase I and separated by SDS–PAGE. This procedure yields a single broad band migrating at 70 kDa (Figure 6A), thus localizing the catalytic residue in the 70 kDa core domain.
Figure 6.
Radiolabeling of the core domain. (A) 70 kDa (core) enzyme was incubated with 32P-labeled pRYG DNA, denatured, digested with DNaseI, separated by SDS–PAGE and visualized by autoradiography (lane 1). Lane 2 same as lane 1 but subjected to SDS–PAGE after protease treatment. (B) Identification of phospho aminoacid linkage in the hydrolysate of core LdTOP2-DNA covalent complexes. Lane 1 represents the migration of 32P-labeled acid hydrolysates. Lane 2 shows migration profile of standard cold phosphoaminoacids. The positions of the marker compounds (seen by ninhydrin staining) are indicated by dotted lines. The point of application of samples is marked as Ori.
In order to investigate the chemical nature of the covalent linkage formed between the substrate DNA and LdTOP2, biochemical mapping studies were carried out by labeling the purified enzyme as described above. Acid hydrolysis of the labeled oligonucleotide-protein complex yielded product that was found to co-chromatograph with chemically synthesized phosphotyrosine on thin layer silica gel in the solvent system mentioned in Materials and Methods (Figure 6B). Thus the covalent linkage formed between the L.donovani enzyme and DNA was Tyr-P, as observed, for other eukaryotic topoisomerase II.
An unambiguous way of identifying the active site of topoisomerase II for the breakage and rejoining of its substrate is to form the covalent enzyme-DNA complex, digest the complex with an endopeptidase and sequence the DNA-linked peptide. The peptide sequence directly reveals the position of the PO4-linked amino acid in the topoisomerase polypeptide chain. The sequencing of the active site residue of LdTOP2 was done as described in Materials and Methods. To minimize the interference of DNA with the aminoacid sequencing, the protein was kept in molar excess. The protein-DNA complex was subjected to SDS–PAGE and stained with Coomassie brilliant blue. The protein band was subsequently excised and subjected to ingel trypsin digestion. The resultant peptides were analyzed for the presence of 32P and the positive fractions were subjected to Edman degradation. The first cycle resulted in an aminoacid with altered retention time. The next four cycles contained Isoleusine, Phenylalanine, Threonine, Lysine and ‘blank’, respectively. When this sequence was compared to LdTOP2 sequence, it was found that the pattern corresponded exactly to that of Tyr775-Ile776-Phe777-Thr778-Lys779, with Tyr775 being the point of covalent attachment to DNA (thus came at an altered retention time). Furthermore, aminoacid 774 of LdTOP2 is Arginine; and trypsin would be expected to cleave after this position as well as after Lys779, to give five aminoacids linked to DNA.
DISCUSSION
Being an essential enzyme, DNA topoisomerase II is conserved among species and has retained the domain organization and motifs important for catalysis and function. However, there must be specific differences in the catalytic properties and efficiency of the enzyme from different sources. The enzyme activity has to be optimized for physiological function and this aspect of the parasite protein has not been addressed so far. In the present study, we have carried out the characterization of four truncation mutants of LdTOP2: LdΔC1058, a 116kDa protein having 178 aminoacids deleted from the C-terminus (14), an 89 kDa protein (LdΔN429) lacking the N-terminal 429 amino acid, a 70 kDa core domain, which has the hydrophilic C-terminus as well as 429 aminoacids from the N-terminus also removed and a 40 kDa protein, which lacks the N-terminal as well as the C-terminal ends. In order to determine whether the individual domains of LdTOP2 still retain activities, we analyzed the capabilities of the deletion mutants to hydrolyze ATP. Only the 116 kDa enzyme (1–1058 aminoacids residues) retained its DNA-stimulated ATPase activity (Figure 2A), which was comparable to the full length LdTOP2 protein. This can be attributed to the fact that only this protein retains the N-terminal homolog of GyrB having an intrinsic ATPase activity. Studies with fragments of DNA gyrase or eukaryotic topoisomerase II or with full-length enzymes have shown a stimulatory effect of DNA on topoisomerase II catalyzed ATP-hydrolysis (12,23,24). Wang and co-workers have recently shown that the stimulatory effect of DNA on the ATPase activity of topoisomerase II fragments increases if the enzyme fragment besides the N-terminal ATPase domain also contains the B' region of the enzyme (25). This correlates with our previous observation that ATP hydrolysis by the N-terminal 1–385 fragment fails to be stimulated by DNA (Sengupta et al., unpublished data), while the present study shows that the stimulation by DNA increases 2-fold as the length of the enzyme increases. Further, the DNA-stimulated ATPase activity of the E.coli holoenzyme varies from 2- to 100-fold, while yeast and calf thymus topoisomerase II show 20- and 4-fold stimulation of ATP hydrolysis by DNA (26).
It has been shown earlier in the case of drosophila that deletion of ATPase domain and C-terminal tail of topoisomerase II does not affect its DNA breakage and rejoining activity (13). Consistent with this, our results also show that all the truncation mutants except the 40 kDa protein retain DNA-binding and cleavage activity. The 40 kDa protein fail to show DNA-binding and cleavage activities, probably due to the inability of the protein to form a dimer as the putative dimer interface was earlier mapped between the amino acid residues 1022–1037; and the deletion of this region results in a protein which cannot complement temperature-sensitive mutant yeast strain (14). Cleavage assay by various truncation mutants reveals an optimum cleavage at 200 mM KCl concentration but the enzymes are active even at 300 mM KCl concentration. This is in contrast to the results obtained in case of yeast and human topoisomerase II, where maximum cleavage activity is observed at 100–150 mM KCl concentration but no cleavable complexes are apparent above 225 mM KCl concentration (27). It is well established that the strength of a given protein-DNA interaction is inversely related to salt concentration (28). The Kd of LdΔC1058 is 3.3 nM and that of the core and LdΔN429 is 4.2 nM and 4 nM, respectively, while the Kd of the full length enzyme is 3.0 nM. This value is about ∼7-fold higher than E.coli gyrase but ∼3-fold lower than Mycobacterium segmentis (26). This value is much lower than the dissociation constant of human topoisomerase II for fourway junction DNA (29 nM) and linear DNA (130 nM) as shown by gel retardation assay (29). L.donovani topoisomerase II truncation mutants can perform cleavage of DNA at high salt. It may be attributed to the fact that this enzyme has high affinity for DNA and a large amount of salt is required to break the interaction between the core domain of LdTOP2 with DNA. This also correlates with the fact that LdΔC1058, which has highest activity at 200 mM salt concentration, has the lowest Kd value among the mutants.
The type II A subfamily of DNA topoisomerases, to which LdTOP2 belongs, shares an important catalytic property with type IA DNA topoisomerase in their common requirement for Mg2+ in catalysis. In contrast, the type IB subfamily enzymes (represented by human topoisomerase I, vaccinia virus topoisomerase I and even L.donovani topoisomerase I) do not require divalent ions in their catalytic mechanism (30). Although cleavage assay by topoisomerase II is supported by a number of divalent cations including Mg2+ and Co2+ (31), we have also used Ca2+ for two reasons. First, all known eukaryotic topoisomerase II generate higher cleavage in the presence of Ca2+ ions than in the presence of Mg2+ ions (32,33). Calcium-promoted, drug independent DNA cleavage sites induced by yeast and human wild-type topoisomerase II and E.coli gyrase have been reported (33,34). Apart from the increased cleavage, the major properties of DNA scission including salt requirements and nucleotide specificity are retained in Ca2+ containing reactions. Second, Ca2+ inhibits a low level of exonuclease activity sometimes evident in Mg2+ containing reactions. It was observed that DNA cleavage activity was supported by a broad range of Mg2+ and Ca2+ concentration (Figure 5B and C). However, the optimal level of cleavage was observed at 6 and 4 mM divalent cation concentration, respectively.
We have tested the sensitivity of the truncation constructs towards the drug etoposide. In our study, we observed that etoposide does not enhance the formation of the cleavable complex by the mutant proteins. Earlier studies have shown that hypo phosphorylation of human topoisomerase II at the C- terminus render cells resistant to etoposide and other cleavage enhancing drugs (35). Several point mutations or truncations at the C-terminus of topoisomerase II that confer resistance to drugs (36) have been identified. It has been shown for many other multidomain proteins that truncation of the protein in many cases renders it incapable of giving full activity or in other cases makes it drug resistant (37,38). In the present study, we report that the full length Ld TOP2 is inhibited by etoposide. We have also observed that etoposide inhibits ATP hydrolysis by the N-terminal 1–385 aminoacids of LdTOP2 (T. Sengupta, M. Mukherjee, A. Das, C. Mandal, R. Das, T. Mukherjee and H. K. Majumder, unpublished data). But here we have shown that etoposide fails to enhance the formation of the cleavable complex by truncated LdTOP2, whereas the cleavable complex formation is enhanced by the wild-type enzyme. We propose that a full-length enzyme is required for interaction with a drug. This study also provides a clue that besides containing regulatory sequences like nuclear localization signals and dimerization domain, the C-terminus is involved in drug interaction but this proposition requires further experimental validation.
The direct sequencing of the DNA-linked core fragment of LdTOP2 identified Tyr775 as the active site residue of the parasite protein. This tyrosine is conserved in all known type II DNA topoisomerases. A clear expectation is that the homologous tyrosine in each of the type II DNA topoisomerases including tyrosine 785, 781, 783 and 804 of Drosophila melanogaster, Schizosaccharomyces pombe, Saccharomyces cerevisiae, and human, respectively, (39–42) are functionally identical to Tyr 775 of L.donovani topoisomerase II. As noted previously in each of the above mentioned cases, the active site tyrosine in LdTOP2 is also preceded by an arginine residue likely to facilitate the participation of the tyrosine residue in the trans-esterification reaction.
The results presented in this study with the truncated LdTOP2 proteins enhance our understanding in the following ways: (i) they indicate a clear distinction between the region of LdTOP2 required for ATP hydrolysis and trans-esterification reaction, (ii) they demonstrate that the catalytic domain per se is capable of recognizing the target sequence for trans-esterification reaction and (iii) ATP hydrolysis, which is crucial for topoisomerase II function in vivo, is not required for DNA-binding or cleavage. In summary, the present study demonstrates that the core domain of LdTOP2 is involved in the formation of a covalent complex with DNA, with Tyr 775 being the active site residue. This study also brings forth the fact that the ATPase, DNA cleavage and the C-terminal domain of LdTOP2 are relatively independent in topoisomerase structure, with all the domains working in conjunction for full topoisomerase activity. Our study also brings to light the fact that in addition to containing NLS and dimerization domain, the C-terminus of LdTOP2 contains elements that are implicated in interaction with etoposide. Thus, a future challenge will be the identification of residues in the core, C-terminal and ATPase domain of LdTOP2 that could aid in the design of novel treatment strategies for drug-resistant parasites.
Acknowledgments
This work was supported by a grant from the Department of Biotechnology, Government of India (BT/PR2643/BRB/10/250/2001). We are grateful to Swapan Pramanik for his help with thin layer chromatography and Benu Brata Das for his help with purification of LdTOP2 protein. T.S and R.D were supported by fellowship from Council of Scientific and Industrial Research. Funding to pay the Open Access publication charges for this article was provided by Indian Institute of Chemical Biology, Kolkata, India.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang J.C. Cellular roles of DNA topoisomerases: a molecular perspective. Nature Rev. Mol. Cell. Biol. 2002;3:429–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 3.Burden D.A., Osheroff N. Mechanism of action of eukaryotic topoisomerase II and drugs targeted to the enzyme. Biochim. Biophys. Acta. 1998;1400:139–154. doi: 10.1016/s0167-4781(98)00132-8. [DOI] [PubMed] [Google Scholar]
- 4.Li T.K., Liu L.F. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol. Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 5.Fortune J.M., Osheroff N. Topoisomerase II as a target for anticancer drugs: when enzymes stop being nice. Prog. Nucleic Acid Res. Mol. Biol. 2000;64:221–253. doi: 10.1016/s0079-6603(00)64006-0. [DOI] [PubMed] [Google Scholar]
- 6.Corbett A.H., Osheroff N. When good enzymes go bad: conversion of topoisomeraseII to a cellular toxin by antineoplastic drugs. Chem. Res. Toxicol. 1993;5:585–597. doi: 10.1021/tx00035a001. [DOI] [PubMed] [Google Scholar]
- 7.Froelich-Ammon S.J., Osheroff N. Topoisomerase poisons: harnessing the dark side of enzyme mechanism. J. Biol. Chem. 1995;270:21429–21432. doi: 10.1074/jbc.270.37.21429. [DOI] [PubMed] [Google Scholar]
- 8.Liu L.F. New York: Academic press; 1994. DNA topoisomerases: Topoisomerase targeting drugs. [Google Scholar]
- 9.Das A., Dasgupta A., Sengupta T., Majumder H.K. Topoisomerases of kinetoplastid parasites as potential chemotherapeutic targets. Trends Parasitol. 2004;20:381–387. doi: 10.1016/j.pt.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro T.A., Showalter A.F. In vivo inhibition of trypanosome mitochondrial topoisomerase II: effects on kinetoplast DNA maxicircles. Mol. Cell Biol. 1994;14:5891–58977. doi: 10.1128/mcb.14.9.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A., Dasgupta A., Sharma S., Ghosh M., Sengupta T., Bandopadhyay S., Majumder H.K. Characterisation of the gene encoding type II DNA topoisomerase from Leishmania donovani: a key molecular target in antileishmanial therapy. Nucleic Acids Res. 2001;29:1844–1851. doi: 10.1093/nar/29.9.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardiner L.P., Roper D.I., Hammonds T.R., Maxwell A. The N- terminal domain of human topoisomerase IIalpha is a DNA-dependent ATPase. Biochemistry. 1998;37:6997–7004. doi: 10.1021/bi9818321. [DOI] [PubMed] [Google Scholar]
- 13.Chang S., Hu T., Hsieh T.S. Analysis of a core domain in Drosophila DNA topoisomerase II. Targeting of an antitumor agent ICRF-159. J. Biol. Chem. 1998;273:19822–19828. doi: 10.1074/jbc.273.31.19822. [DOI] [PubMed] [Google Scholar]
- 14.Sengupta T., Mukherjee M., Mandal C.N., Das A., Majumder H.K. Functional dissection of the C-terminal domain of type II DNA topoisomerase from the kinetoplastid hemoflagellate Leishmania donovani. Nucleic Acids Res. 2003;30:5305–5316. doi: 10.1093/nar/gkg727. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Andersen A.H., Christiansen K., Zechiedrich E.L., Jensen P.S., Osheroff N., Westergaard O. Strand specificity of the topoisomerase II mediated double- stranded DNA cleavage reaction. Biochemistry. 1989;28:6237–6244. doi: 10.1021/bi00441a015. [DOI] [PubMed] [Google Scholar]
- 16.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 17.Ostergaard V.H., Giangiacomo L., Bjergbaek L., Knudson B.R., Anderson A.H. Hindering the strand passage reaction of human topoisomerase Iialpha without disturbing DNA cleavage, ATP hydrolysis, or the operation of the N-terminal clamp. J. Biol. Chem. 2004;279:28093–28099. doi: 10.1074/jbc.M402120200. [DOI] [PubMed] [Google Scholar]
- 18.Singh R., Dutta C., Majumder H.K. Analysis of sequences of two different classes of kinetoplast DNA minicircles of Leishmania species. J. Biosci. 1994;19:117–182. [Google Scholar]
- 19.Dasgupta S., Adhya S., Majumder H.K. A simple procedure for the preparation of pure kinetoplast DNA network free of nuclear DNA from the kinetoplast hemoflagellate Leishmania donovani. Anal. Biochem. 1986;158:189–194. doi: 10.1016/0003-2697(86)90608-1. [DOI] [PubMed] [Google Scholar]
- 20.Bodley A.L., Chakraborty A.K., Xie S., Burri A., Shapiro T.A. An unusual typeIB topoisomerase from African trypanosomes. Proc. Natl Acad. Sci. USA. 2003;100:7539–7544. doi: 10.1073/pnas.1330762100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tse Y.C., Kirkegaard K., Wang J.C. Covalent bonds between protein and DNA. Formation of phosphotyrosine linkage between certain DNA topoisomerases and DNA. J. Biol. Chem. 1980;255:5560–5565. [PubMed] [Google Scholar]
- 22.Skoufias D.A., Lacroix F.B., Andreassen P.R., Wilson L., Margolis R.L. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol. Cell. 2004;15:977–990. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 23.Lindsley J.E., Wang J.C. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J. Biol. Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 24.Campbell S., Maxwell A. The ATP-operated clamp of human DNA topoisomerase IIalpha: hyperstimulation of ATPase by ‘piggy-back’ binding. J. Mol. Biol. 2002;320:171–188. doi: 10.1016/S0022-2836(02)00461-8. [DOI] [PubMed] [Google Scholar]
- 25.Olland S., Wang J.C. Catalysis of ATP hydrolysis by two NH(2)- terminal fragments of yeast DNA topoisomerase II. J. Biol. Chem. 1999;274:21688–21694. doi: 10.1074/jbc.274.31.21688. [DOI] [PubMed] [Google Scholar]
- 26.Manjunatha U.H., Dalal M., Chaterjee M., Radha D.R., Visweswariah S.S., Nagaraja V. Functional characterisation of mycobacterial DNA gyrase: an efficient decatenase. Nucleic Acids Res. 2002;30:2144–2153. doi: 10.1093/nar/30.10.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scala D., Escargueil A.E., Couprie J., Larsen A.K. The catalytic activities of DNA topoisomerase II are most closely associated with the DNA cleavage/religation steps. Biochimie. 1999;81:771–779. doi: 10.1016/s0300-9084(99)80136-9. [DOI] [PubMed] [Google Scholar]
- 28.Record M.T., Jr, Ha J.H., Fisher M.A. Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 1991;208:291–343. doi: 10.1016/0076-6879(91)08018-d. [DOI] [PubMed] [Google Scholar]
- 29.West K.L., Austin C.A. Human DNA topoisomerase II beta binds and cleaves four-way junction DNA in vitro. Nucleic Acids Res. 1999;27:984–992. doi: 10.1093/nar/27.4.984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das B.B., Sen N., Ganguly A., Majumder H.K. Reconstitution and functional characterization of the unusual bi-subunit type I DNA topoisomerase from Leishmania donovani. FEBS Lett. 2004;565:81–88. doi: 10.1016/j.febslet.2004.03.078. [DOI] [PubMed] [Google Scholar]
- 31.Baldwin E.L., Byl J.A., Osheroff N. Cobalt enhances DNA cleavage mediated by human topoisomerase II alpha in vitro and in cultured cells. Biochemistry. 2004;43:728–735. doi: 10.1021/bi035472f. [DOI] [PubMed] [Google Scholar]
- 32.Osheroff N., Zechiedrich E.L. Calcium-promoted DNA cleavage by eukaryotic topoisomerase II: trapping the covalent enzyme-DNA complex in an active form. Biochemistry. 1987;26:4293–4313. doi: 10.1021/bi00388a018. [DOI] [PubMed] [Google Scholar]
- 33.Strumberg D., Nitiss J.L., Dong J., Kohn K.W., Pommier Y. Molecular analysis of yeast and human type II topoisomerases. Enzyme-DNA and drug interactions. J. Biol. Chem. 1999;274:28246–28255. doi: 10.1074/jbc.274.40.28246. [DOI] [PubMed] [Google Scholar]
- 34.Capranico G., Guano F., Moro S., Zagotto G., Sissi C., Gotto B., Zunino F., Menta E., Palumbo M. Mapping drug interactions at the covalent topoisomerase II-DNA complex by bisantrene/amsacrine congeners. J. Biol. Chem. 1998;273:12732–12739. doi: 10.1074/jbc.273.21.12732. [DOI] [PubMed] [Google Scholar]
- 35.Chikamori K., Grabowski D.R., Kinter M., Willard B.B., Yadav S., Aebersold R.H., Bukowski R.M., Hickson I.D., Anderson A.H., Ganapathi M.K., et al. Phosphorylation of serine 1106 in the catalytic domain of topoisomerase II alpha regulates enzymatic activity and drug sensitivity. J. Biol. Chem. 2003;278:12696–12702. doi: 10.1074/jbc.M300837200. [DOI] [PubMed] [Google Scholar]
- 36.Vassetzsky Y.S., Alghisi G.C., Gasser S.M. DNA topoisomerase II mutations and resistance to anti-tumor drugs. Bioessays. 1995;17:767–774. doi: 10.1002/bies.950170906. [DOI] [PubMed] [Google Scholar]
- 37.Brandt P.A., Jacobs B.L. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 2001;75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khoo K.H., Douglas E., Azadi P., Inamine J.M., Besra G.S., Mikusova K., Brennan P.J., Chaterjee D. Truncated structural variants of lipoarabinomannan in ethambutol drug-resistant strains of Mycobacterium smegmatis. Inhibition of arabinan biosynthesis by ethambutol. J. Biol. Chem. 1996;271:28682–28690. doi: 10.1074/jbc.271.45.28682. [DOI] [PubMed] [Google Scholar]
- 39.Wortland S.T., Wang J.C. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:4412–4416. [PubMed] [Google Scholar]
- 40.Uemura T., Morikawa K., Yanagida M. The nucleotide sequence of the fission yeast DNA topoisomerase II gene: structural and functional relationships to other DNA topoisomerases. EMBO J. 1986;5:2355–2361. doi: 10.1002/j.1460-2075.1986.tb04504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wycoff E., Natalie D., Nolan J., Lee M., Hsieh T.S. Structure of the Drosophila DNA topoisomerase II gene. Nucleotide sequence and homology among topoisomerases II. J. Mol. Biol. 1989;505:1–13. doi: 10.1016/0022-2836(89)90361-6. [DOI] [PubMed] [Google Scholar]
- 42.Tsai-Pflugfelder M., Liu L.F., Liu A.A., Tewey K.M., Whangpeng J., Knutsen T., Croce C.M., Wang J.C. Cloning and sequencing of cDNA encoding human DNA topoisomerase II and localization of the gene to chromosome region 17q21-22. Proc. Natl Acad. Sci. USA. 1988;85:7177–7181. doi: 10.1073/pnas.85.19.7177. [DOI] [PMC free article] [PubMed] [Google Scholar]