Abstract
Pseudogenes, in the case of protein-coding genes, are gene copies that have lost the ability to code for a protein; they are typically identified through annotation of disabled, decayed or incomplete protein-coding sequences. Processed pseudogenes (PΨgs) are made through mRNA retrotransposition. There is overwhelming genomic evidence for thousands of human PΨgs and also dozens of human processed genes that comprise complete retrotransposed copies of other genes. Here, we survey for an intermediate entity, the transcribed processed pseudogene (TPΨg), which is disabled but nonetheless transcribed. TPΨgs may affect expression of paralogous genes, as observed in the case of the mouse makorin1-p1 TPΨg. To elucidate their role, we identified human TPΨgs by mapping expressed sequences onto PΨgs and, reciprocally, extracting TPΨgs from known mRNAs. We consider only those PΨgs that are homologous to either non-mammalian eukaryotic proteins or protein domains of known structure, and require detection of identical coding-sequence disablements in both the expressed and genomic sequences. Oligonucleotide microarray data provide further expression verification. Overall, we find 166–233 TPΨgs (∼4–6% of PΨgs). Proteins/transcripts with the highest numbers of homologous TPΨgs generally have many homologous PΨgs and are abundantly expressed. TPΨgs are significantly over-represented near both the 5′ and 3′ ends of genes; this suggests that TPΨgs can be formed through gene–promoter co-option, or intrusion into untranslated regions. However, roughly half of the TPΨgs are located away from genes in the intergenic DNA and thus may be co-opting cryptic promoters of undesignated origin. Furthermore, TPΨgs are unlike other PΨgs and processed genes in the following ways: (i) they do not show a significant tendency to either deposit on or originate from the X chromosome; (ii) only 5% of human TPΨgs have potential orthologs in mouse. This latter finding indicates that the vast majority of TPΨgs is lineage specific. This is likely linked to well-documented extensive lineage-specific SINE/LINE activity. The list of TPΨgs is available at: http://www.biology.mcgill.ca/faculty/harrison/tppg/bppg.tov (or) http:pseudogene.org.
INTRODUCTION
The search for novel functional elements in the human genome is imperative and ongoing (1–3). Pseudogenes (gene copies that have lost their protein-coding ability) are a form of sequence of potential functional utility (4). Substantial progress has been made in the annotation of pseudogenes (5–11). There may be twice as many pseudogenes (derived from protein-coding genes) in the human genome as protein-coding genes (6–10).
Pseudogenes (derived from protein-coding genes) are typically ‘diagnosed’ through searching for the ‘symptoms’ of a lack of protein-coding ability. These symptoms include: frame disablement (from premature stop codons and frameshifts), coding sequence decay (typically detectable through examination of non-synonymous and synonymous substitution rates) or incompleteness (either from sequence truncation or from the loss of essential signals for transcription, splicing and translation) (6–10). Processed pseudogenes (PΨgs) are made through retrotransposition of mRNAs. There is ubiquitous genomic evidence for thousands of PΨgs in mammals (5–10). Similarly, dozens of processed genes (i.e. genes made by retrotransposition of the complete sequence of other genes) have arisen in both the mouse and human genomes (12,13). This mass gene retrotransposition may arise, at least in part, as a by-product of long interspersed element (LINE) retrotransposition (14). Retrotransposition is clearly an active process in mammalian gene evolution (15). Here, we search for an intermediate type of retrotransposed gene sequence: the transcribed processed pseudogene (shortened as TPΨg), which is a PΨg that is disabled but nonetheless transcribed.
Historically, there have been several isolated reports of transcribed pseudogenes, of either the duplicated or the processed form (16–21). Two recent studies have demonstrated that such transcribed pseudogenes can regulate transcription of homologous protein-coding genes. Transcription of a pseudogene in Lymnea stagnalis, that is homologous to the nitric oxide synthase gene, decreases the expression levels for the gene through formation of a RNA duplex; this is thought to arise via a reverse-complement sequence found at the 5′ end of the pseudogene transcript (20). In a second example, transcription of the makorin1-p1 TPΨg in mouse was required for the stability of the mRNA from a homologous gene makorin1 (21). This regulation was deduced to arise from an element in the 5′ areas of both the gene and the pseudogene (21).
In addition to helping to elucidate such regulatory roles, annotation of TPΨgs will further add to our understanding of the dynamics of gene evolution through retrotransposition (15). Also, it is crucial to annotate TPΨgs correctly as a part of the ongoing process of correct cDNA/expressed sequence tag (EST) mapping during genome annotation, and for more accurate interpretation of microarray expression data (22,23). Here, we have performed a data-mining expedition for human TPΨgs using a rigorous method that applies stringent filters to avoid data pollution. TPΨgs have a markedly distinct distribution in the genome when compared with other PΨgs and processed genes. A key result is that TPΨgs are significantly likely to insert near the 5′ and 3′ ends of genes, implying that TPΨgs can be generated by co-option of promoter elements or by intrusion into untranslated regions (UTRs) as ‘molecular passengers’. Also, we find that the vast majority of TPΨgs are human-lineage specific compared with mouse.
Definitions and terms
An mRNA can be reverse transcribed and re-integrated into the genomic DNA, possibly as a by-product of LINE-1 retrotransposition (14). The parent gene of the mRNA need not be on the same chromosome as the retrotransposed copy. Such a retrotransposed mRNA has three possible fates in the present-day genome: (i) formation of a non-transcribed PΨg, (ii) formation of a TPΨg or (iii) formation of a processed gene (or part of a gene).
A PΨg can be defined as any disrupted, decayed or incomplete copy of a gene that has arisen through such retrotransposition. In the process of evolution, PΨgs accumulate disablements (frameshifts and premature stop codons) in their apparent coding sequences. Procedures to annotate PΨgs using disablement detection have been described previously (4,5,7), and serve as the basis for the present analysis.
Operationally, a TPΨg is defined as a PΨg for which an expressed sequence is mappable across any of its coding-sequence disablements, i.e. the disablement occurs in both the expressed sequence and the genomic sequence (see Methods for details).
A processed gene is any undisrupted retrotransposed copy of a gene that also has low Ka/Ks values indicative of selection pressure on coding ability (see Methods for details).
Each of our TPΨgs has ≥1 disablement verified by alignment of the expressed sequences to genomic DNA, in a region of the TPΨg that maps to a known structural protein domain, or to a protein sequence that is conserved in non-mammalian eukaryotes. This three-level verification procedure (genome:transcript:protein) is termed triple alignment. Each verified disablement has an estimated probability of being the result of a sequencing error of ≤10−6, since the error rate for the genomic sequence build is ≤10−4 (24) and the error rate for cDNAs/ESTs is ≤10−2 (25,26). We made a subset of TPΨgs, termed the C set, which has further evidence of lack of coding ability. These have: (i) no continuous segment of sequence that can code for a protein domain (as defined in Methods); (ii) high Ka/Ks values (≥0.5).
As it is possible that a fraction of the TPΨgs that map to introns arise from intron retention in cDNAs or ESTs in the source expressed sequence data, we analyzed all of the data both including and excluding the 67 TPΨgs that map to introns (see Table 3 and below). Our results are unaffected by such potential contamination, as explained below.
Table 3.
Position of TPΨgs, other PΨgs and processed genes relative to annotated genes
| Categories of sequence grouped by position relative to genes | Type of sequence | |||||
|---|---|---|---|---|---|---|
| TPΨgs | Other PΨgs | Processed genes | ||||
| Observed numbera | Expected numberb | Observed numbera | Expected numberb | Observed numbera | Expected numberb | |
| Sequences that overlap gene annotations | 18 (8%) | — | — | — | — | — |
| Sequences mapped to introns of annotated genes | 67 (28%) | 79.7 | 693 (22%) | 1100.0¶¶ | 3 (5%) | 21.2¶¶ |
| Sequences <3000 nt 5′ of start codon of annotated genes | 20 (9%) | 6.8** | 78 (0.7%) | 93.6 | 5 (8%) | 1.9 |
| Sequences <10 000 nt 5′ of start codon of annotated genes | 36 (15%) | 22.3* | 278 (9%) | 307.8 | 7 (11%) | 5.9 |
| Sequences <3000 nt 3′ of translation stop of annotated genes | 22 (9%) | 6.7** | 55 (1.7%) | 92.3¶¶ | 0 (0%) | 1.8 |
| Sequences <10 000 nt 3′ of translation stop of annotated genes | 42 (18%) | 22.2** | 241 (7%) | 306.4¶¶ | 9 (14%) | 5.8 |
| Sequences that are in intergenic DNAc | 109 (47%) | 129.8 | 2109 | 1371.7** | 43 | 31.4 |
aThese categories are not additive, as they are not mutually exclusive, i.e. some TPΨg may be within 10 000 nt of the 5′ end of one gene, and be in the intron of another gene or within 10 000 of the 3′ end of a third gene.
bExpected values are calculated assuming random insertion in the whole genome (without the genomic DNA for annotated genes). For significant over-representation, ** indicates P < 0.001, and * indicates P < 0.01 for a chi-squared test (1 degree of freedom) using Yates correction (similarly, ¶¶ is used for significant under-representation for P < 0.01).
cIntergenic DNA is defined as all of the genomic DNA that does not comprise exons, introns or the regions of genes within 10 000 nt of the translation stop and start of gene coding sequences.
METHODS
Detection of TPΨgs
(i) Mapping expressed sequence data onto existing PΨgs annotations
PΨgs were annotated previously using a method based on the detection of disabled protein homology in genomic DNA (4,5,7). We mapped >6200 of these onto human genome build 34 (from http://www.ensembl.org), through detection of 100% nucleotide sequence matches, removing overlap with coding exons. For each PΨg, the genomic sequence was extracted, both with and without a 6000 nt extension added on to either end to allow for homology matching to ‘pseudo-UTR’ regions. (These sets of genomic DNA are named genPΨg and genPΨg+/−6000.) Three sources of expressed sequences (Refseq mRNAs, Unigene consensuses, and ESTs from dbEST) were downloaded from http://www.ncbi.nih.gov. They were mapped onto genPΨg and genPΨg+/−6000, using BLASTN with low-complexity masking (E-value ≤ 10−10, minimum match length 100 nt) (27,28). From the resulting significant matches, those that align with ≥95% identity were used to generate a second BLASTN search against genPΨg and genPΨg+/−6000, but this time without low-complexity masking, to insure correct sequence identity. Matches to both genPΨg and genPΨg+/−6000 with ≥99% identity over >0.998 of the length of the expressed sequence were then extracted. These expressed sequence matches were filtered to insure that they match more significantly to the PΨg than to any homologous gene. The matching expressed sequences were then re-aligned to the PΨg sequence using FASTY (29), to check that ≥1 disablement (frameshift or premature stop codon) in the PΨg occurs in both the genomic sequence and expressed sequence. Each disablement verified in this way has an estimated probability of being the result of a sequencing error of ≤1 × 10−6; this is because the genomic sequence error rate is ≤1 × 10−4, and the cDNA/EST sequencing error rate is ≤1 × 10−2.
(ii) Extraction of PΨgs that are in Refseq mRNAs
All human Refseq entries corresponding to known mRNAs (total = 20 741) were compiled from data downloaded from the NCBI website (http://www.ncbi.nih.gov). These were compared with all known, non-fragmentary human proteins in the SWISSPROT database (30), using a modification of the disabled protein homology-based procedure developed previously for PΨg annotation (4,5,7,31–33). To insure that all of the candidate TPΨgs in Refseq mRNAs map to a single continuous piece of genomic DNA, we extracted the appropriate mRNA subsequences and mapped them to the human genome using BLASTN. Those segments that matched over their complete length exactly were retained. The resulting TPΨg data were then filtered along with those generated in (i), as detailed in (iii) below.
(iii) Filtering the (transcribed) PΨg data
We applied a set of filters to insure that we were compiling a bona fide list of TPΨgs. All TPΨg data sets were filtered as follows:
Removal of homologies to purely hypothetical proteins or fragmentary proteins: TPΨgs based only on homology to predicted reading frames or reading-frame fragments were removed through BLASTP comparisons (E-value ≤ 10−4) against a library of hypothetical or fragmentary proteins from SWISSPROT (30). These are removed because their disablements may be erroneous (which is inappropriate for the method employed here). Also, they may be inaccurately dated (values for Ks, Ka, etc., may be incorrect).
- Verification that the disablements are in conserved parts of a known protein sequence or domain: We verified that the disablements examined are in known conserved parts of sequences, as detailed below. This list of filters has an ‘if-else-if-else-if’ structure:
- First, we assigned protein structural domains to the TPΨgs, by comparing them with the ASTRALSCOP 95% identity set of protein domains (34), using BLASTP (27) (E-value ≤ 10−4). The total assigned TPΨg subsequence was determined (from the most N-terminal residue that was assigned to a domain, to the most C-terminal). This assigned subsequence was considered disabled, if a frameshift or stop codon occurred >10 residues in from either terminus. This accounts for ∼54% of TPΨgs.
- Otherwise, secondly, TPΨgs not meeting criterion (1) were checked manually for occurrence of disablements in conserved domains using the InterPro (http://www.ebi.ac.uk/interpro) and CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) domain annotation tools.
- Otherwise, thirdly, TPΨgs not meeting criterion (2) (<10% of the sequences) were checked for disablement within a part of the sequence that is conserved in other mammals, and in ≥1 non-mammalian species, using BLASTP (E-value ≤ 10−4).
Removal of candidates with small introns: Putative PΨgs, TPΨgs, and the expressed sequences that match them, were filtered for small intron sequences. A library of introns of <1000 nt was made from genes on human genome build 34. TBLASTN (27) was used to annotate any significant matches to these introns of >0.80 of their length (E-value ≤ 10−4). Any such matches could either be from introns in an aberrant cDNA or be a previously disregarded small intron in the genome. Some additional examples of TPΨgs that map to introns may arise from intron retention in cDNAs or ESTs; however, the main points of the analysis reported in this paper are unaffected by such potential contamination, as explained below.
Removal of possible duplications of single-exon genes and large exons: We wished to insure that there were no single-exon gene duplications in our PΨg and TPΨg sets. To do this, all PΨgs and TPΨgs were compared using BLASTP (E-value ≤ 10−4), to the set of proteins for build 34 (removing those whose genes overlap putative PΨgs) (27). All PΨgs and TPΨgs that had closest-matching homologies to single-exon proteins were removed. Furthermore, we insured that they aligned to their closest-matching homologous human proteins around at least one ‘exon seam’, i.e. a position in a protein sequence that corresponds to an intron–exon boundary. This exon seam filter insures that the pseudogenes considered are processed, and is particularly useful for removing homologies to genes with large exons (e.g. some zinc-finger-containing proteins).
Filtering for processed genes: All TPΨgs were filtered for overlap with annotated processed genes (resulting in the removal of only one putative TPΨg).
After applying these rigorous filters, we had 3418 PΨgs (both transcribed and non-transcribed), and 233 TPΨgs, 218 from mapping expressed sequences to PΨgs and 15 from PΨg extraction from Refseq mRNAs. Almost half (97/233, 42%) of the TPΨg set represent 100% exact matches of expressed sequences to PΨgs. Restricting analysis to just these matches does not affect any of the main trends and results reported here.
Making an obviously decayed C set of TPΨg sequences
We derived a ‘core set’ of TPΨgs that have further evidence of coding-sequence decay. These are dubbed the C set (totaling 177/233, 76%). This set is the union of the following two subsets: (i) TPΨgs without continuous segment of sequence that can code for a protein domain (106/233 TPΨgs, 45%) or (ii) TPΨgs with high Ka/Ks values (>0.50) indicative of lack of coding ability (127/233 TPΨgs, 54%).
(i) Lack of protein domain coding ability
We parsed each TPΨg into subsequences according to the positions of its disablements. If all subsequences could be labeled as ‘unlikely to code for a protein domain’, then the TPΨg was included in the C set. This resulted in inclusion of 106 TPΨgs in the C set. We labeled a subsequence as ‘unlikely to code for a protein domain’ if:
Its length was ≤32 residues. The vast majority (95%) of non-cysteine-rich protein domains in the ASTRALSCOP 40% identity set have sequence lengths >32 residues (34). Cysteine-rich domains (which are likely disulfide-bridged or metal-chelating) are defined as having cysteine concentration <0.077/residue, a value suggested by a bimodality in cysteine concentration, in surveys of cysteine and cystine occurrence in proteins (35,36). Condition (a) was not applied to any fragments that were adjudged cysteine-rich.
It contained a disrupted SCOP domain, as defined in part (iii)(b)(1) above. Such fragments are likely not to constitute a large enough fragment; the reasoning behind this criterion is that evolution has defined and refined the integrity of a body of recurrent folding units (protein domains) (34), and we can therefore use their disruption to evaluate whether a piece of sequence is no longer protein-coding.
(ii) Ka/Ks analysis
We calculated the Ka/Ks values for whole TPΨgs, using the Yang and Nielsen method in PAML (37), using the present-day gene sequence to compare against the pseudogene, as described previously (7). Also, similarly, we calculated Ka/Ks values for subsequences of TPΨgs (≥50 residues) derived by parsing at disablement positions. This parsing allows for the possibility that some of the pseudogene subsequences have coding ability, while others do not, i.e. we can test for a coding ability ‘imbalance’. From these Ka/Ks calculations, we found that only ∼4% of both PΨgs and TPΨgs have two adjacent regions where one is <0.25 (potentially coding) and the other >0.5 (potentially non-coding), indicating that such imbalance is rare. From consulting independent analysis of populations of human genes and PΨgs (6), we ascertained that for a threshold value of Ka/Ks ≥ 0.5, >95% of sequences are predicted to be PΨgs and not genes. We use this as the expectation for the distribution of PΨgs in general. Calculation of Ka/Ks values for gene/pseudogene pairs errs on the side of under-estimation of coding-sequence decay (7).
Conservation of TPΨg in mouse
For each human TPΨg, we searched against potentially orthologous mouse TPΨgs. These ‘moTPΨgs’ were derived by mapping expressed sequences (Refseq mRNAs, Unigene consensus sequences and ESTs) for mouse onto a previously derived set of mouse PΨgs (8), in a similar manner to the human mappings (see above). These were pooled with any existing moTPΨg annotations, and a small number of mouse genes that might be potentially misannotated moTPΨgs. A potentially orthologous moTPΨg was required to match ≥0.5 of the length of the human TPΨg (for BLASTP matches, E-value ≤ 10−4), and to share the same closest-matching human protein with any potential human TPΨg homologs. We did not require that the retrotranspositions be in syntenic positions, since orthologous gene retrotranspositions are not necessarily syntenic (38).
Processed genes
We mapped an independently derived list of processed genes (13) to human genome build 34. In addition to the criteria in (13), we required Ka/Ks values <0.25, and coverage of ≥0.95 of the parent gene's length. Any examples that overlap the TPΨg data set of annotations were removed; vice versa, any TPΨgs that have Ka/Ks < 0.25 and cover ≥0.95 of their parent gene were deleted from the TPΨgs list. Our definitions give two distinct sets of processed genes and TPΨgs; naturally, we miss some sequences that cannot be classified as either a TPΨg or a processed gene.
RESULTS AND DISCUSSION
Number of TPΨgs
In total, we found 233 human TPΨgs (Table 1). These TPΨgs form a subset of 3418 previous PΨg annotations that were mapped to build 34 of the human genome (7). These PΨgs were filtered in the same way as the TPΨgs (from a starting total of ∼6200), to remove predicted reading frames, retained introns and potential duplications of single-exon genes or large exons. Using these data, we can estimate that ∼6% (218/3418) of PΨgs are TPΨgs. An additional 15 TPΨgs were derived from a reciprocal process of searching for PΨgs in known Refseq mRNAs, followed by subsequent mapping to the genome.
Table 1.
Summary of numbers of TPΨgs
| Set or subset of TPΨgs | Total number | Total number (without those mapped to introns) |
|---|---|---|
| Mappings to existing pseudogene annotations | 218 | 154 |
| Pseudogene extraction from Refseq mRNAs | 15 | 12 |
| Total TPΨgs | 233 | 166 |
| Expressed sequence support | ||
| TPΨgs that are supported by Refseq mRNAs | 18 (8%) | 16 (10%) |
| TPΨgs that are supported by Unigene consensus sequences | 74 (32%) | 50 (30%) |
| TPΨgs that are supported by dbEST expressed sequence tags | 167 (72%) | 111 (67%) |
| TPΨgs that are supported by dbEST expressed sequence tags and by either a Refseq mRNA or a Unigene consensus | 38 (16%) | 25 (16%) |
| TPΨgs that are additionally supported by oligonucleotide microarray data | 75 (32%) | 53 (32%) |
| Further evidence of decay | ||
| TPΨgs that have no continuous segment likely to code for a protein domain | 106 (45%) | 70 (42%) |
| TPΨgs that have Ka/Ks ≥ 0.5 | 127 (54%) | 88 (53%) |
| C set (TPΨgs that have no continuous segment likely to code for a protein domain or Ka/Ks ≥ 0.5) | 177 (76%) | 123 (74%) |
A small fraction of the TPΨgs (8%) corresponds to known Refseq mRNAs (Table 1). About a third are supported by Unigene consensus sequences, with a large fraction (71%) matching individual ESTs [of this last group, about a quarter (∼23%) are supported by a Refseq mRNA or a Unigene consensus; Table 1]. We sought additional expression verification from a series of high-density oligonucleotide microarrays, composed of ∼52 million 36mers (23). These microarrays were applied to probe the transcriptionally active regions of the human genome, in a strand-sensitive way. Using the same data and statistical method (i.e. a sign test) for scoring the genes' transcriptional activity (22,23), we found that 75/233 (32%) TPΨgs were transcriptionally active in liver (P < 0.05) (Table 1). In comparison, 64% of genes from RefSeq mRNAs, 57% of Ensembl annotated genes (39), and 35% of genes predicted with the program GENSCAN (40), were found to be transcribed in liver.
The C set of more obviously decayed TPΨgs comprises 76% of the total population; 45% of TPΨgs having no continuous segment likely to code for a protein domain, and 54% of TPΨgs having Ka/Ks ≥ 0.5 (Table 1). Obvious degradation of the coding sequences is demonstrated for this set from analysis of protein-domain mapping and Ka/Ks (see Methods). Additionally, other factors (not examined in the present analysis) are expected to cause lack of coding ability in TPΨgs or arise as further consequences. It is likely that TPΨgs will not have appropriate start codon context (41), therefore leading to little or no efficient translation initiation. Also, those TPΨgs that are inserted into 3′-UTRs of mRNAs will be unlikely to become protein-coding through being downstream of a clearly defined coding sequence (although it is conceivable that they may be translatable in the 5′-UTR). Furthermore, a consequence of any frameshift in a sequence is the likelihood of an additional 20 residues or so of non-coding DNA, added onto the end of the sequence truncation (on average, in randomly picked, conceptually translated intergenic DNA, a stop codon will appear ∼20 residues downstream of any starting point); such additional sequence may lead to aggregation or misfolding in the cell.
The proportions of TPΨgs break down in a similar fashion to that just described above for the total data set, when the 67 examples that map to introns are removed (Table 1).
Closest matching human proteins for TPΨgs
TPΨgs were grouped according to their closest-matching human protein (Table 2). Each table entry represents a single ‘parent gene’. The total counts are also shown for the TPΨgs that do not map to introns (in square brackets, Table 2). There are 4 human proteins that have ≥4 homologous TPΨgs. The highest number of TPΨgs (5) occur for cyclophilin A, which is required for cis-peptide isomerization (42). All of these proteins arise from highly expressed mRNAs. They also occur in the top 20 proteins when apportioning all PΨgs, in the same way (7).
Table 2.
Human proteins with four or more homologous TPΨgs
| Numbera | Name of human proteinb |
|---|---|
| 5 [4] | Peptidyl-prolyl cis-trans isomerase A (Cyclophilin A) [P62937] |
| 4 [3] | Prohibitin [P35232] |
| 4 [3] | 40S ribosomal protein S12 [P25398] |
| 4 [3] | Actin, cytoplasmic 2 (Gamma-actin) [P63261] |
| Glyceraldehyde-3-phosphate dehydrogenase [P04406, P00354] |
aThe totals in square brackets are for when those mapping to introns are removed.
bThe Swissprot accession numbers are given in square brackets.
TPΨg position relative to genes and the implications for their expression mechanisms
A number of mechanisms for TPΨg expression are plausible. First, TPΨgs may co-opt nearby promoter elements of protein-coding genes. Secondly, they may intrude into the UTRs of another mRNA, as a sort of ‘molecular passenger’. Thirdly, they may make use of cryptic promoter elements in the intergenic DNA; such promoter elements may have originated from transposable elements, or from genomic duplication of genic promoter regions, or sporadically (de novo).
Such mechanisms for TPΨg expression may have a bearing on their overall positional distribution in the genome relative to genes. To investigate this, we classified the TPΨgs into those that: (i) overlap existing coding-sequence exons; (ii) appear inserted in introns; (iii) are inserted in a 3000 or 10 000 nt region 5′ to annotated genes; (iv) are inserted in a 3000 or 10 000 nt region 3′ to annotated genes.
Table 3 summarizes these data. A minor proportion (8%) of TPΨgs entail gene coding-sequence annotations, i.e. they are erroneously annotated reading frames. There are 67 TPΨgs that map to introns (Table 3); it is unclear how many of these may arise from intron retention in cDNAs or ESTs. Expectations based on random insertion in the genome were calculated for classes (ii) to (iv). We focus on (iii) and (iv) in particular.
TPΨgs are significantly more likely than random (P < 0.01, chi-squared tests) to be inserted in the regions 5′ and 3′ of annotated genes; this effect is most obvious in the 3000 nt regions 5′ and 3′ to genes, but is still significant up to 10 000 nt in either direction (Table 3). Similar results are observed for the C set of more obviously decayed TPΨgs. The enrichment of TPΨgs observed in the 5′ and 3′ areas of genes can be seen as a simple logical consequence of randomly inserted PΨgs having an increased probability of being transcribed, and is clear support for either co-option of genic promoter elements, or insertion into UTRs as molecular passengers, leading to TPΨg expression. This result is also unaffected by possible contamination from intron retention in cDNAs/ESTs, as, in general, PΨgs are significantly under-represented in introns (Table 3); if one assumed that, in the extreme, all of the TPΨg mappings near the 5′ and 3′ ends of genes were actually mappings to introns, then this would make their over-representation even more significant. The general dearth of PΨgs in introns may be a reflection of an overall genomic tendency for a lack of retroelement insertion in introns (43).
Roughly half of the TPΨgs are located away from genes (>10 000 nt 5′ and 3′ to genes, and overlapping neither an exon nor an intron; Table 3). These thus may be co-opting cryptic promoters of unknown origin in the intergenic DNA, such as those derivable from transposable elements.
In summary, the distribution of TPΨgs in the vicinity of genes is significantly different from that observable for other non-transcribed PΨgs (that have no transcription evidence), and for processed genes in the following ways (Table 3):
TPΨgs are significantly over-represented in the 10 000 nt 5′ and 3′ to genes, whereas other PΨgs and processed genes are not;
Other PΨgs are significantly over-represented in intergenic DNA and significantly under-represented in introns, and processed genes are significantly under-represented in introns; TPΨgs now show such trends for introns or intergenic DNA. In addition, there is a dearth of PΨgs 3′ to genes (Table 3). The reasons for this are unclear; there may be a compositional effect, similar to the relationship between genomic G+C content and ribosomal-protein PΨgs insertion, observed previously (44).
We examined the distribution of 20 TPΨgs that are directly mappable onto known Refseq mRNAs. Thirteen of these overlap an erroneously predicted open reading frame, and two are already annotated as transcribed pseudogenes. None of the five remaining TPΨgs are inserted in the 5′-UTR of a messenger RNA. One explanation for this absence in 5′-UTRs is that a TPΨg would introduce upstream ORFs that interfere with translation initiation (41). The five TPΨgs inserted in the 3′-UTRs of mRNAs are all in the forward direction (i.e. they are all on the same DNA strand as the annotated coding sequence). An example of this is discussed below. In addition, we checked the list of TPΨgs 3′ to annotated genes and within 3000 nt of the end of the coding sequence (Table 3), for additional examples of this ‘passenger’ phenomenon, through manual examination of cDNAs or ESTs for the 5′ genes, but could find no further examples of cDNAs with polyadenylation signals to define the end of the mRNA. Such analysis is complicated by the fact that, in some cases, it may not be possible to distinguish between the original polyadenylation signal of the gene, and an inserted polyadenylation signal arising from the TPΨg.
Distribution on chromosomes
Analysis of the distribution of processed genes in the human and mouse genome has indicated that the X chromosome is a marked outlier, both for processed gene deposition onto the X chromosome and origination from X (13). A similar outlier preference was observed for PΨg deposition onto the X chromosome (but not origination from X) (13). These phenomena may be due to selection pressures to compensate for X-chromosome inactivation during spermatogenesis, in combination with some unaccounted-for mutational biases (13,38).
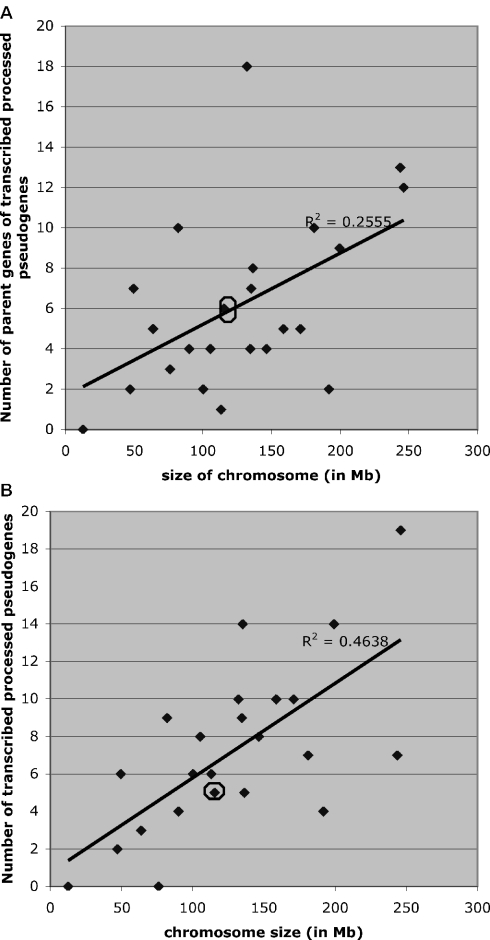
To compare with this previous analysis, we examined the distribution of TPΨg ‘parent genes’ on each chromosome, and also the distribution of the number of TPΨgs per chromosome (Figure 1A and B). Figure 1A indicates the data for origination of TPΨgs, and Figure 1B shows the trend for deposition of TPΨgs onto each chromosome. In each case (origination and deposition), the X chromosome is not an outlier. This may indicate that, in general, TPΨg formation is deleterious, unlike processed gene and non-transcribed PΨg formation, which are arguably, by comparison, beneficial and selectively neutral, respectively. Interestingly, there is some outlier behavior for TPΨg origination from chromosome 12. The same result is obtained, if the 67 TPΨgs that map to introns are removed.
Figure 1.
Origination and deposition of TPΨgs for different chromosomes. (A) Origination of TPΨgs: this plot shows the number of parent genes of TPΨgs in a chromosome versus the chromosome size (in Mb). (B) Deposition of TPΨgs: this shows the number of TPΨgs per chromosome versus chromosome size (in Mb). Only retrotranspositions from one chromosome to another are considered in each plot. The X chromosome is ringed. Note that for each plot we have corrected for the probability of X and Y chromosome inclusion in gametes [i.e. the size of X is multiplied by 0.75 and Y by 0.25; for comparison see figure 1 in (13)].
Search for potential orthologs in mouse
We investigated mouse/human cross-species conservation of TPΨgs, as an indicator of human-lineage specificity. The 233 human TPΨgs were compared against a set of 215 putative mouse TPΨgs (moTPΨgs) (see Methods for details). We found that 5% (11/233) have potential orthologous TPΨgs. Four of these are for the metabolic enzyme, glyceraldehyde-3-phosphate dehydrogenase, which is ubiquitously and highly expressed, giving this sequence the status of a notable ‘parent gene’ for TPΨgs (see also Table 2). If the human and mouse TPΨgs are not restricted to having the same closest-matching human gene homolog, 28/237 (12%) have potential orthologs.
These results suggest that a minor fraction of TPΨgs could be used in conserved functional roles in mammals. However, given that ∼40% of human PΨgs are conserved in the mouse genome (8), these results imply that TPΨgs are significantly under-conserved between human and mouse (P < 0.001 using binomial statistics) compared with PΨgs in general, and also compared with processed genes (13), which are at most ∼20% lineage-specific. The vast majority of TPΨgs are thus human lineage-specific compared with mouse; indeed, both Alus (which are primate-specific) and PΨgs can be made as by-products of LINE retrotransposition (14), and have similar overall age profiles in the genome (8). These results are also evidence for a general evolutionary selection pressure to delete TPΨgs. This may be because they form a source of transcriptional interference for adjacent genes or homologous genes. However, one must stress that, in the future, increased cDNA coverage for both the mouse and human genomes may modify these statistics somewhat. Such a lack of saturation in current databases of expressed sequences can be demonstrated using some simple sampling analysis. Sampling of TPΨg-matching expressed sequences from random fractional subsets of the total expressed sequence database used in the present analysis (i.e. ESTs + Unigene consensuses + Refseq mRNAs), indicates that we are not near finding all of the TPΨgs in the human genome (or, at least, those discoverable through mapping of expressed sequences). (This sampling analysis is presented in Supplementary Figure 1.)
Examples of TPΨgs
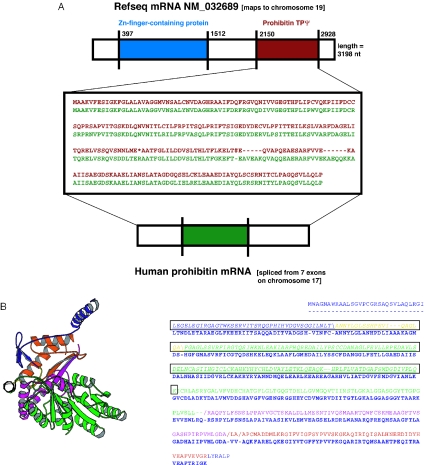
A TPΨg derived from the prohibitin gene is shown in Figure 2A. A prohibitin TPΨg is inserted into the 3′-UTR of a Zn-finger-containing protein. Prohibitin is highly and ubiquitously expressed, and is involved in inhibition of DNA synthesis; its mRNA contains a putative functional RNA element in its own 3′ UTR (45). It is beyond the scope of this present study to ascertain whether this RNA element, in this TPΨg, is intact, as it has not yet been characterized extensively by mutational and biophysical analysis. This TPΨg is one of four that derived from the prohibitin gene (Table 2).
Figure 2.
Examples of TPΨgs. (A) This is a TPΨg derived from the human prohibitin gene. The prohibitin gene contains both a protein-coding region and an RNA in its 3′-UTR (45), but only the segment of the TPΨg corresponding to the protein-coding sequence is shown. In the center is an alignment of the TPΨg (in red) with prohibitin protein (in green). The graphic above it shows the position of the TPΨg (red segment) in the 3′-UTR of an mRNA that codes for a Zn-finger-containing protein (blue segment). (B) An example of a TPΨg that maps to a known globular protein domain. The TPΨg derives from the mRNA for the precursor sequence of mitochondrial 2-amino-3-ketobutyrate coenzyme A. The domain is from the closest-matching protein structure (from E.coli, PDB code 1fc4a). In the Molscript (54) picture, the protein chain trace color changes at the position of each disablement. The alignment of the E.coli domain sequence and the human TPΨg sequence is shown. The part of the sequence that maps to an EST (gi|6138420) is boxed and italicized.
The second example is derived from the precursor sequence of mitochondrial 2-amino-3-ketobutyrate coenzyme A. The crystal structure of the Escherichia coli homolog of this enzyme is known (PDB code 1fc4a). We have indicated how the ‘triple alignment’ of genomic sequence, EST and known protein-domain sequence overlap (Figure 2B). The protein chain is divided into colored segments, with each disablement defining a segment boundary. One can clearly see that the triple alignment covers two disablements in the TPΨg.
CONCLUSIONS
Diverse efforts to map novel elements of potential functional utility in our genome are ongoing (1–3). In the spirit of such endeavors, we have derived a rigorous procedure for annotating a specific novel type of element of potential functional utility, the TPΨg. Applying this method to the human genome, we discovered 166–233 TPΨgs, which represent ∼4–6% of all PΨgs (the lower total arises from setting aside any examples that map to introns). One should point out that we might have missed some TPΨgs; e.g. those without extensive homology to a coding sequence (i.e. those consisting largely of UTR homologies), or TPΨgs formed from single-exon and large-exon genes, or TPΨgs that are transcribed in a low-level beyond detectability through EST/cDNA sequencing.
TPΨgs are significantly more likely in regions close to the 5′ and 3′ ends of genes, compared with both a random insertion model for them throughout the genome, and compared with the distribution observed in general for PΨgs. Furthermore, if one assumes that these 5′ and 3′ regions are actually introns, the significance of the increased 5′ and 3′ density of TPΨgs improves. (This indicates that the increased 5′ and 3′ density is not an artifact of intron retention in cDNA/EST libraries.) This increased density provides evidence that TPΨgs may be expressed through co-option of genic promoter elements or through insertion into UTRs as ‘molecular passengers’. Specific detailed evidence was found for molecular passengers in the 3′-UTRs of known mRNAs; an example of this derived from the prohibitin gene was illustrated (Figure 2A). TPΨgs could thus also have a role as intermediates in protein-coding sequence evolution. A reasonable hypothesis that can be further investigated is that, TPΨgs may represent a source of evolutionary protein novelty, either as ‘molecular passengers’, or as part of alternative splicings (46), through being temporarily released from coding-sequence selection pressures (31,47–50). Use of additional sequence segments may underlie the influence of the [PSI+] prion on phenotypic variability in budding yeast (31,51); analogs of this phenomenon are possible in mammals.
Two examples of regulation by transcribed pseudogenes of homologous genic transcripts have been observed (20,21). Transcriptional analysis showed that the stability of the makorin1 mRNA in mouse relies upon the expression of its homologous makorin1-p1 TPΨg, through the action of an element at the 5′ end of the makorin1-p1 sequence. However, makorin1-p1 only seems to be conserved in one line of Mus, and has not been found in the rat genome (52). In a second example, transcription of a pseudogene in Lymnea stagnalis, that is homologous to the nitric oxide synthase gene, decreases expression levels for the gene; this is thought to arise via a reverse-complement sequence found at the 5′ end of the pseudogene transcript (20). Alternatively, TPΨgs near genes or in UTRs may also exert a controlling/interfering influence on the genes' transcription and translation, through upstream ORF formation, or the action of other undiscovered elements. Such TPΨgs could exert such effects through co-option as alternative splicings, as has been observed for Alus (53). Also, it is possible that some TPΨgs produce a short peptide that does not misfold or aggregate in the cell, but is still targeted and serves an alternative function as a truncated peptide. Certainly, TPΨgs represent a source of transcriptional ‘noise’, which may have implications for selection pressures on transcription levels, and the degree of variation on which such pressures can act.
Our survey provides evidence for the existence in the human genome of a small population of TPΨgs, which are an intermediate class of retrosequence derived from genes, since they have expression evidence (like genes), but also have evidence of lack of coding ability (like other pseudogenes). The distribution of TPΨgs near the 5′ and 3′ ends of genes indicates that TPΨgs can co-opt genic promoters or intrude into UTRs; furthermore, this is a robust observation that verifies our expression-data mappings. One must also point out, however, that about half of the TPΨgs are located away from genes in intergenic DNA (Table 3), and thus may be co-opting cryptic promoters of undesignated origin. Also, TPΨgs differ from other PΨgs (without transcription evidence) and from processed genes in terms of their distribution per chromosome, and their projected conservation in mouse. Our analysis indicates that, unlike processed genes and other PΨgs, the vast majority (∼95%) of TPΨgs are human lineage-specific. In combination, the chromosomal distribution and mouse conservation for TPΨgs suggests that there is some general evolutionary pressure to delete TPΨgs from the genome. One should point out that the cDNA coverage of both genomes is far from complete (as illustrated here, with some simple sampling analysis), so that the analysis of conservation in mouse should be regarded as tentative.
This TPΨg analysis has important implications for genome annotation. It is still common practice to assume that an mRNA contains one undisrupted open reading frame; however, it is clear that one should routinely check for TPΨgs in the manner described here. Also, this TPΨg annotation is useful for improved interpretation of microarray expression data (22,23). The list of TPΨgs is available at: http://www.biology.mcgill.ca/faculty/harrison/tppg/tppg.tar (or) http://pseudogene.org.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
Thanks to T. Bureau, N. Juretic and D. Hoen (McGill U.) for discussions. This work was supported in part by a Discovery Grant from the National Science and Engineering Council of Canada to P.M.H., and by National Institutes of Health grant # P50 HG02357-01 to M.G. Funding to pay the Open Access publication charges for this article was provided by McGill University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dermitzakis E.T., Reymond A., Scamuffa N., Ucla C., Kirkness E., Rossier C., Antonarakis S.E. Evolutionary discrimination of mammalian conserved non-genic sequences (CNGs) Science. 2003;302:1033–1035. doi: 10.1126/science.1087047. [DOI] [PubMed] [Google Scholar]
- 2.Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 3.Consortium E.P. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 4.Harrison P., Gerstein M. Studying genomes through the aeons: protein families, pseudogenes and proteome evolution. J. Mol. Biol. 2002;318:1155–1174. doi: 10.1016/s0022-2836(02)00109-2. [DOI] [PubMed] [Google Scholar]
- 5.Harrison P.M., Hegyi H., Balasubramanian S., Luscombe N.M., Bertone P., Echols N., Johnson T., Gerstein M. Molecular fossils in the human genome: identification and analysis of the pseudogenes in chromosomes 21 and 22. Genome Res. 2002;12:272–280. doi: 10.1101/gr.207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torrents D., Suyama M., Zdobnov E., Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z., Harrison P., Liu Y., Gerstein M. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 2003;13:2541–2558. doi: 10.1101/gr.1429003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Z., Carriero N., Gerstein M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends Genet. 2004;20:62–67. doi: 10.1016/j.tig.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Consortium M.G.S. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 10.Consortium R.G.S.P. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 11.Chen C., Gentles A.J., Jurka J., Karlin S. Genes, pseudogenes, and Alu sequence organization across human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99:2930–2935. doi: 10.1073/pnas.052692099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 13.Emerson J.J., Kaessmann H., Betran E., Long M. Extensive gene traffic on the mammalian X chromosom. Science. 2004;303:537–540. doi: 10.1126/science.1090042. [DOI] [PubMed] [Google Scholar]
- 14.Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nature Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 15.Brosius J. Genomes were forged by massive bombardments with retroelements and retrosequences. Genetica. 1999;107:209–238. [PubMed] [Google Scholar]
- 16.Fujii G.H., Morimoto A.M., Berson A.E., Bolen J.B. Transcriptional analysis of the PTEN/MMAC1 pseudogene, psiPTEN. Oncogene. 1999;18:1765–1769. doi: 10.1038/sj.onc.1202492. [DOI] [PubMed] [Google Scholar]
- 17.Bristow J., Gitelman S.E., Tee M.K., Staels B., Miller W.L. Abundant adrenal-specific transcription of the human P450c21A ‘pseudogene’. J. Biol. Chem. 1993;268:12919–12924. [PubMed] [Google Scholar]
- 18.Zhou B.S., Beidler D.R., Cheng Y.C. Identification of antisense RNA transcripts from a human DNA topoisomerase I pseudogene. Cancer Res. 1992;52:4280–4285. [PubMed] [Google Scholar]
- 19.Olsen M.A., Schechter L.E. Cloning, mRNA localization and evolutionary conservation of a human 5-HT7 receptor pseudogene. Gene. 1999;227:63–69. doi: 10.1016/s0378-1119(98)00588-5. [DOI] [PubMed] [Google Scholar]
- 20.Korneev S.A., Park J.H., O'Shea M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is suppressed by an antisense RNA transcribed from an NOS pseudogene. J. Neurosci. 1999;19:7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirotsune S., Yoshida N., Chen A., Garrett L., Sugiyama F., Takahashi S., Yagami K., Wynshaw-Boris A., Yoshiki A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- 22.Rinn J.L., Euskirchen G., Bertone P., Martone R., Luscombe N.M., Hartman S., Harrison P.M., Nelson F.K., Miller P., Gerstein M., et al. The transcriptional activity of human Chromosome 22. Genes Dev. 2003;17:529–540. doi: 10.1101/gad.1055203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertone P., Stolc V., Royce T.E., Rozowsky J.S., Urban A.E., Zhu X., Rinn J.L., Tongprasit W., Samanta M., Weissman S., et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 24.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. International Human Genome Sequencing Consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 25.Pruitt K.D., Maglott D.R. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler D., Church D., Edgar R., Federhen S., Helmberg W., Madden T., Pontius J., Schuler G., Schriml L., Sequeira E., et al. Database resources of the National Center for Biotechnology Information: update. Nucleic Acids Res. 2004;32:D32–D40. doi: 10.1093/nar/gkh073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wootton J.C., Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 29.Pearson W.R. Flexible sequence similarity searching with the FASTA3 program package. Methods Mol. Biol. 2000;132:185–219. doi: 10.1385/1-59259-192-2:185. [DOI] [PubMed] [Google Scholar]
- 30.Apweiler R., Bairoch A., Wu C., Barker W., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison P.M., Kumar A., Lan N., Echols N., Snyder M., Gerstein M. A small reservoir of disabled ORFs in the sequenced yeast genome and its implications for the dynamics of proteome evolution. J. Mol. Biol. 2002;316:409–419. doi: 10.1006/jmbi.2001.5343. [DOI] [PubMed] [Google Scholar]
- 32.Harrison P.M., Carriero N., Liu Y., Gerstein M. A ‘polyORFomic’ analysis of prokaryote genomes using disabled-homology filtering reveals conserved but undiscovered short ORFs. J. Mol. Biol. 2003;333:885–892. doi: 10.1016/j.jmb.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y., Harrison P.M., Kunin V., Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandonia J., Hon G., Walker N., Lo Conte L., Koehl P., Levitt M., Brenner S. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrison P.M., Sternberg M.J.E. Analysis and classification of disulphide connectivity in proteins. The entropic effect of cross-linkage. J. Mol. Biol. 1994;244:448–463. doi: 10.1006/jmbi.1994.1742. [DOI] [PubMed] [Google Scholar]
- 36.Harrison P.M., Sternberg M.J.E. The disulphide beta-cross: from cystine geometry and clustering to classification of small disulphide-rich protein folds. J. Mol. Biol. 1996;264:603–623. doi: 10.1006/jmbi.1996.0664. [DOI] [PubMed] [Google Scholar]
- 37.Yang Z., Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 38.Bradley J., Baltus A., Skaletsky H., Royce-Tolland M., Dewar K., Page D. An X-to-autosome retrogene is required for spermatogenesis in mice. Nature Genet. 2004;36:872–876. doi: 10.1038/ng1390. [DOI] [PubMed] [Google Scholar]
- 39.Birney E., Andrews D., Bevan P., Caccamo M., Cameron G., Chen Y., Clarke L., Coates G., Cox T., Cuff J., et al. Ensembl 2004. Nucleic Acids Res. 2004;32:D468–D470. doi: 10.1093/nar/gkh038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 41.Lu P.D., Harding H.P., Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell. Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haendler B., Hofer E. Characterization of the human cyclophilin gene and of related processed pseudogenes. Eur. J. Biochem. 1990;190:477–482. doi: 10.1111/j.1432-1033.1990.tb15598.x. [DOI] [PubMed] [Google Scholar]
- 43.Semon M., Duret L. Evidence that functional transcription units cover at least half of the human genome. Trends Genet. 2004;20:229–232. doi: 10.1016/j.tig.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Z., Harrison P., Gerstein M. Identification and analysis of over 2000 ribosomal protein pseudogenes in the human genome. Genome Res. 2002;12:1466–14482. doi: 10.1101/gr.331902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manjeshwar S., Branam D.E., Lerner M.R., Brackett D.J., Jupe E.R. Tumor suppression by the prohibitin gene 3′untranslated region RNA in human breast cancer. Cancer Res. 2003;63:5251–5256. [PubMed] [Google Scholar]
- 46.Modrek B., Lee C.J. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 47.Letunic I., Copley R.R., Bork P. Common exon duplication in animals and its role in alternative splicing. Hum. Mol. Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 48.Kondrashov F.A., Koonin E.V. Evolution of alternative splicing: deletions, insertions and origin of functional parts of proteins from intron sequences. Trends Genet. 2003;19:115–119. doi: 10.1016/S0168-9525(02)00029-X. [DOI] [PubMed] [Google Scholar]
- 49.Trabesinger-Ruef N., Jermann T., Zankel T., Durrant B., Frank G., Benner S.A. Pseudogenes in ribonuclease evolution: a source of new biomacromolecular function? FEBS Lett. 1996;382:319–322. doi: 10.1016/0014-5793(96)00191-3. [DOI] [PubMed] [Google Scholar]
- 50.Balakirev E.S., Ayala F.J. Pseudogenes: Are they ‘junk’ or functional DNA? Annu. Rev. Genet. 2003;37:123–151. doi: 10.1146/annurev.genet.37.040103.103949. [DOI] [PubMed] [Google Scholar]
- 51.True H.L., Berlin I., Lindquist S.L. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 52.Podlaha O., Zhang J. Non-neutral evolution of the transcribed pseudogene Makorin1-p1 in mice. Mol. Biol. Evol. 2003;21:2202–2209. doi: 10.1093/molbev/msh230. [DOI] [PubMed] [Google Scholar]
- 53.Dagan T., Sorek R., Ast G., Graur D. AluGene: a database of Alu elements incorporated within protein-coding genes. Nucleic Acids Res. 2004;32:D489–D492. doi: 10.1093/nar/gkh132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kraulis P.J. Molscript: a program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.