Abstract
Stalled bacterial ribosomes are freed by transfer-messenger RNA (tmRNA). With the help of small protein B (SmpB), protein synthesis restarts and tmRNA adds a tag to the stalled protein for destruction. The conformation of a 347 nt long tmRNA from a thermophile and its interactions with SmpB were monitored using structural probes. The RNA is highly folded, including the reading frame, with <30% of unpaired residues. Footprints between SmpB and tmRNA are in the elbow of the tRNA domain, in some pseudoknots including one essential for function and in the lower part of the stem exiting the tRNA domain. The footprints outside the tRNA domain are scattered onto the tmRNA sequence, but form a cluster onto its tertiary structure derived from cryo-EM data. Some footprints flank the first triplet to be translated in tmRNA, suggesting that SmpB participates in the insertion of the tmRNA-encoded reading frame into the decoding center. To discriminate between a conformational rearrangement of tmRNA and independent binding sites, surface plasmon resonance was used and has identified three independent binding sites of SmpB on the RNA, including the site on the tRNA domain. Accordingly, SmpB is proposed to move on the tmRNA scaffold during trans-translation.
INTRODUCTION
In cells, translation failures have to be dealt quickly and efficiently. Defective messages occur during growth, and stalling on such messages sequesters ribosomes and produces incomplete polypeptides. In bacteria, transfer-messenger RNA [(tmRNA) known as SsrA or 10Sa RNA] eliminates such problematic messages, releases stalled ribosomes and targets the incomplete proteins for degradation (1). During protein synthesis, this mechanism, termed trans-translation, ensures the quality control of the intracellular proteins. Bacterial tmRNA is aminoacylated at its 3′ end with alanine by alanyl-tRNA synthetase (AlaRS) (2), followed by a ‘codon–anticodon’ independent transfer of the alanine to the stalled polypeptide chain. Translation resumes at an internal position within tmRNA sequence and the ribosome is freed upon release at a tmRNA-encoded termination codon, with the production of a tagged polypeptide chain. The tagged protein is recognized and degraded by specific proteases and the stalled ribosome is recycled [reviewed in (3,4)]. The recently determined cryo-EM structure visualizing tmRNA entry into a stalled ribosome has provided first detailed insights into the intricate mechanisms involved in trans-translation (5).
In many species, the structure of tmRNA has been deduced by sequence comparison [listed in the tmRNA website, (6)]. For some species, tmRNA folding has been studied by structural probes in solution (7–9). In one-piece tmRNAs, the 5′ and 3′ termini join to form a tRNA-like domain extended by a long interrupted helix abutting in several pseudoknots, with only the one flanking the resume codon being essential for function (10,11). Structural information on full-length tmRNA based on experimental data is restricted to the mesophilic gamma-proteobacteria and cyanobacteria.
The small protein B (SmpB) is an essential cofactor of tmRNA during trans-translation (12). SmpB is universally conserved in bacteria, potentiates tmRNA aminoacylation (13) and is required for interaction of tmRNA with the ribosome (14,15). SmpB can bind the ribosomes directly in the absence of tmRNA (16), suggesting that the protein steps in very early on in the process. SmpB is an antiparallel β-barrel surrounded by three α-helices (17,18), with an unstructured C-terminal domain that probably folds when the protein binds the stalled ribosomes to rescue. SmpB binds to the elbow region of the tRNA domain of tmRNA (19), with sequence-specific interactions at the 3′ end of the D-loop and at a short connector loop (C-loop) that links helix H5 to the T-arm H6 (Figure 1A). In the X-ray structure of the 68 nt long tRNA domain of tmRNA with SmpB, the acceptor stem of tmRNA is disordered. Therefore, the structural basis of the improvement of tmRNA aminoacylation by the protein remains unrevealed. The structural study was performed with a small RNA fragment corresponding only to the tRNA-like domain. However, it is not known whether the protein has additional contacts elsewhere in the tmRNA structure or influences the folding of other functional domains of tmRNA, as the mRNA domain. In Escherichia coli, the few accessible Gs within the RNA structure were monitored by RNase T1 footprints (13), and a truncated version of tmRNA lacking the tag reading frame and three pseudoknots was mapped with enzymes (20). These preliminary studies have identified some footprints of the protein on the tRNA domain of tmRNA (tmRNAΔ), in accordance with its binding as solved by X-ray crystallography (19).
Figure 1.
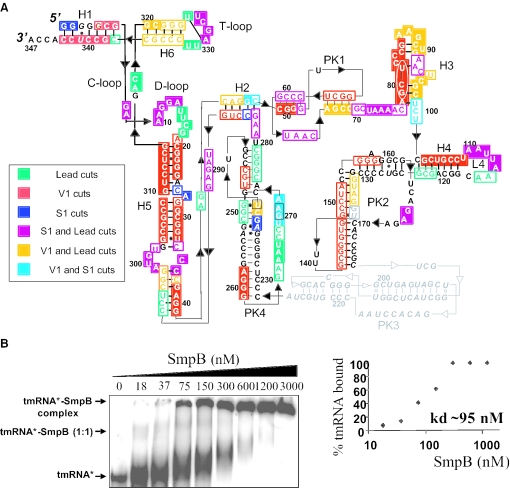
(A) Secondary structure of full-length tmRNA from A.aeolicus deduced by structural probes in solution. The meaning of the colored symbols is detailed in the inset. The intensity of the cleavages is correlated to the intensity of the coloring: open for weak/medium cuts and filled for heavy cuts. Consistently observed degradation sites are indicated by nucleotides squared in black. Structural domains, such as pseudoknots (PK1–PK4), helices (H1–H6) and loops (C-, D- and T-), are indicated. The resume codon is circled. Structural information is unavailable for PK3 (in gray). The black bars and dots are the Watson–Crick (GC or AU) and Wobble (GU) base pairs, respectively. (B) Native gel retardation assay between labeled tmRNA and purified SmpB-His from A.aeolicus, with a graphical representation of the binding.
In this paper, the 347 nt long full-length tmRNA and SmpB from Aquifex aeolicus were cloned, produced and purified, and their binding capacities were analyzed in vitro. A tmRNA–SmpB complex from a bacterial thermophile was selected because the available crystal structure of a portion (one-fifth) of tmRNA in complex with SmpB is from a thermophile and because thermophilic RNAs are highly folded to adapt to elevated temperatures. In addition, experimentally based structural information on tmRNAs from thermophiles outside the tRNA domain is not available. The solution structure of the RNA and the molecular basis of its interaction with SmpB were monitored in solution. The contacts detected between the tRNA-like domain and the protein are maintained in the context of the full-length RNA. We also detected new ones near the resume codon and on a pseudoknot that is essential for function. This suggests that SmpB also participates in the late events of trans-translation, when translation resumes onto tmRNA reading frame. Surface plasmon resonance (SPR) has identified three independent binding sites of SmpB on the full-length RNA, including the expected site on the tRNA-like domain. This result implies that SmpB moves on the tmRNA scaffold during trans-translation. Indeed, the tmRNA–SmpB complex has to undergo substantial conformational rearrangements when the tmRNA internal coding sequence fits into the decoding center of the small ribosomal subunit.
MATERIALS AND METHODS
Oligodeoxynucleotides and enzymes
All the synthetic DNA oligonucleotides were synthesized by Eurogentech. DNA primers: 5′-GGGAAGCTTAATACGACTCACTATAGGGGGCGGAAAGGAT-3′ and 5′-GGGGATCCTGGTGGAGGCGGCGG-3′ were used for cloning A.aeolicus tmRNA sequence in plasmid pUC19. T7 RNA polymerase was prepared as described by Wyatt et al. (21). Restriction enzymes BamHI, HindIII, BstN1, alkaline phosphatase and T4 polynucleotide kinase were from New England Biolabs (Berverly, MA). Taq DNA polymerase, T4 DNA ligase and T4 RNA ligase were from Gibco-BRL Life Technologies (Cergy-Pontoise, France). RNases S1, V1, U2 and T1, [γ-32P]ATP (3000 mCi/mmol), [α-32P]pCp (3000 mCi/mmol) and l-[3-3H]alanine (74 Ci/mmol) were from Amersham-Pharmacia-Biotech (Orsay, France).
Cloning, transcription and purification of tmRNA from A.aeolicus
tmRNA was produced in vitro and purified as described for E.coli (7). The DNA sequence was cloned downstream of a T7 RNA polymerase promoter in a pUC19 vector, using BamHI and HindIII restriction enzymes (22). The recombinant plasmid was linearized with restriction nuclease BstN1 before transcription, so that in vitro transcribed RNAs will end with the 3′ terminal CCA triplet. In vitro transcription of A.aeolicus tmRNA was performed as described previously (23). Electrophoresis on denaturing gels enabled us to separate the transcribed RNAs from non-incorporated nucleotides and DNA fragments. Synthetic RNAs were electroeluted, and pure in vitro transcribed RNAs were recovered by ethanol precipitation. Labeling at the 5′ end of the RNAs was performed with [α-32P]ATP and phage T4 polynucleotide kinase for 1 h at 37°C after dephosphorylation with alkaline phosphatase (24). Labeling at the 3′ end was carried out overnight at 4°C by ligation of [α-32P]pCp using T4 RNA ligase. After labeling, the RNAs were gel purified, eluted passively and ethanol precipitated. tmRNAΔ was produced as described previously (19).
Overexpression and purification of SmpB and alanyl-tRNA synthetase from A.aeolicus
The pET28-derived plasmid containing the A.aeolicus full-length alanyl-tRNA synthetase sequence was a kind gift from S. Gutmann and N. Ban (ETH, Zurich, Switzerland). The plasmid was transformed into E.coli BL21(DE3)-RIL-Codon-Plus (Stratagen). AlaRS expression was triggered by isopropyl-β-d-thiogalactopyranoside. A 30 min 70°C heat denaturation step followed by centrifugation eliminates most of the E.coli endogenous proteins. Afterwards, the thermophile his-tagged AlaRS was purified on a Nickel column. A.aeolicus AlaRS was then quantified and purity was verified visually by SDS–PAGE. SmpB protein cloning and purification procedure was described previously (19). All of the proteins were at least 98% pure as judged by SDS–PAGE analysis.
Aminoacylation assays
tmRNA was first denatured at 82°C for 2 min and then allowed to cool down for 30 min at room temperature. SmpB-His was then added at various concentrations in an incubation medium containing 50 mM HEPES, pH 7.6, 12 mM MgCl2, 2.5 mM ATP, 0.5 mM Spermine, 50 pmol of tmRNA and 50 μM 3H-labeled alanine (25). SmpB and tmRNA were allowed to bind for 15 min at 55°C. Aminoacylation reactions were then performed at the same temperature by adding 10 μg of purified A.aeolicus AlaRS. The aliquots were spotted onto 3MM Whatman papers at different times, precipitated with 5% trichloroacetic acid, washed three times with 5% trichloroacetic acid, washed once with pure ethanol, dried and quantified on a Wallac 1409 Liquid Scintillation Counter.
Gel-mobility shift assay
The 5′-labeled RNAs (see below) were denatured for 2 min at 80°C and then slowly cooled down to room temperature for 30 min. Standard gel retardation assays contain ∼0.5–1 picomol (100 000 c.p.m.) of labeled tmRNA with the appropriate concentrations of recombinant protein SmpB in a binding buffer (50 mM MES, pH 6.5, 200 mM KCl, 5% glycerol, 5 mM β-mercaptoethanol, 0.01% NP-40 and 0.1 mg/ml BSA), to a final volume of 20 μl (12). A 30 min incubation at room temperature is performed in the presence of 20 U of RNasine, followed by the addition 10 μl of 30% glycerol. The sample is subjected to electrophoresis in a 5% (19/1 acryl/bisacrylamide percentage) non-denaturing PAGE in 90 mM Tris–HCl, pH 8.3, 90 mM boric acid and 2.5 mM EDTA, at 4°C (a 4 h migration at 150 V). The gel is then fixed, dried and the data are analyzed on a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Chemical and enzymatic footprints
Enzymatic digestions or chemical modifications are performed on both 3′- and 5′-labeled tmRNA (100 000 c.p.m./reaction). Labeled tmRNA was gel-purified (5% PAGE), eluted and ethanol-precipitated. Labeled RNA was then heated to 82°C for 2 min and slowly cooled down to room temperature. The labeled RNA was incubated with or without 750 nM of purified protein SmpB for 15 min at room temperature in a final volume of 20 μl. The reaction mixture was then supplemented with 1 μg of total yeast tRNA. Digestions with the various ribonucleases (V1 at 10−4 U, S1 at 5 U, U2 at 2.5 × 10−3 U and T1 at 1.25 U) and probing with lead acetate (at a final concentration of 5 mM) were performed for 15 min at room temperature, except for U2 and T1 (incubations at 50°C). Identical amounts of radioactivity were loaded per lanes. Cleavage or modification sites are detected by gel electrophoresis by direct identification with the T1 and U2 cleavage patterns of the RNA itself. We used a combination of 8, 12 and 15% urea–acrylamide gels in order to scan most of the tmRNA molecules. The denatured gel was fixed, dried and the data were analyzed on a phosphorImager. The quantitation of each fragment is scored manually.
Surface plasmon resonance
The stoichiometry of complex formation was investigated by SPR using BIAcore X biosensor system (BIAcore). For real-time analysis of molecular interactions between poly(A)-RNA and SmpB-His, streptavidin was first immobilized on a C1 sensor chip (BIAcore) by covalent linkage between activated carboxyl groups on the chip matrix and lysine residues in the streptavidin as described by the manufacturer (BIAapplications Handbook, Biacore). A 5′-biotin (dT)20 oligomer at the concentration of 5 μM in 10 mM HEPES, pH 7.4, 150 mM NaCl was bound to the surface of streptavidin-coated sensor chips at a flow rate of 5 μl/min for 10 min. The chips were then washed three times with 20 μl of 50 mM NaOH and two times with 20 μl of 0.1% SDS at a flow rate of 10 μl/min to stabilize the surface prior to the binding of RNA. Chips were equilibrated in the continuous-flow buffer (10 mM HEPES, pH 7.4, 150 mM KCl, 10 mM NH4Cl, 10 mM MgOAc, 3 mM β-mercaptoethanol and 0.05% surfactant P20). The tmRNA and the TLD were polyadenylated using the Poly(A) Tailing Kit as described by the manufacturer (Ambion). Poly(A)-tRNA and Poly(A)-TLD were denaturated by an incubation at 83°C for 3 min in the flow buffer and renatured at room temperature for 20 min. Poly(A)-RNAs (20 nM) were immobilized on one flow cell by an injection at a flow rate of 5 μl/min for 10 min. SmpB-His was injected at the desired concentrations in the running buffer until equilibrium was reached. The Sensor Chip was regenerated by a 60 s injection of 0.1% SDS in 10 mM HEPES, pH 7.5 buffer. Final curves were obtained by subtraction of the signal corresponding to the empty flow cell. Concentration and ratio between SmpB-His (19 410 Da) and immobilized RNA were determined using equation 1
where n is the stoichiometry of the complex, RU is the measured response (units) obtained at binding saturation and Mr is the molecular weight. α = 1 pg of protein bound/RU/mm2 and β = 0.8 pg DNA bound/RU/mm2 (26). These factors arise from the different molar refractive indices of RNA versus proteins, which translate into a different SPR response per unit concentration change. Chips were regenerated by injecting 50 μl of 50 mM NaOH at a flow rate of 10 μl/min.
RESULTS
Structural analysis of tmRNA from a thermophile
Secondary structures of one and two pieces bacterial tmRNAs have been studied experimentally (7–9) and by sequence comparisons (27). Secondary structures of tmRNA from thermophiles, however, have only been inferred from sequence alignments (6). As a starting point for the analysis of the interaction between SmpB and tmRNA in solution, structural probing data were generated to produce a secondary structure model of A.aeolicus tmRNA. A tmRNA transcript was end-labeled, and its solution conformation was probed by chemicals and enzymes. Ribonuclease V1 cleaves double-stranded (ds) RNA or stacked nucleotides, while nuclease S1 cleaves single-stranded (ss) RNA. Lead acetate also cleaves ss RNA but with sensitivity to subtle conformational changes of the RNA chain. The reactivity toward these probes was monitored for the 347 nt long synthetic tmRNA, except for PK3 that is 150 nt away from both ends. Thirty four independent experiments were performed (Figures 2 and 3 are representatives, lanes marked ‘−’). These data are summarized in Figure 1A on a secondary structure model that they, together with the phylogenetic analysis, support.
Figure 2.
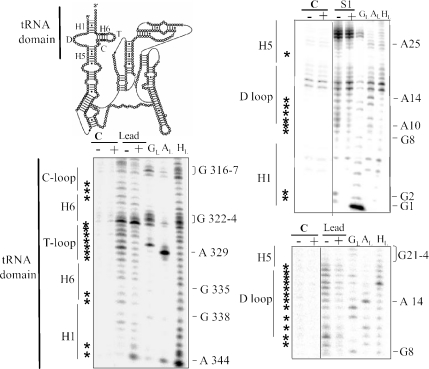
SmpB binds the tRNA-like domain of full-length tmRNA. Autoradiograms of cleavage products of 5′- (right) and 3′- (left) labeled tmRNA by nuclease S1 or lead acetate in the presence (+) or absence (−) of SmpB (750 nM). Lanes C, incubation controls; lanes GL, RNase T1 hydrolysis ladder; lanes AL, RNase U2 hydrolysis ladder; Lanes HL alkaline hydrolysis ladder. The sequence of the RNA is indexed on the right of each panel. Asterisks indicate the nucleotide with reactivity changes in the presence of the protein. The structural domains of the RNA are indicated on the left of each panel and have been indicated on a schematic representation of tmRNA structure (top left).
Figure 3.
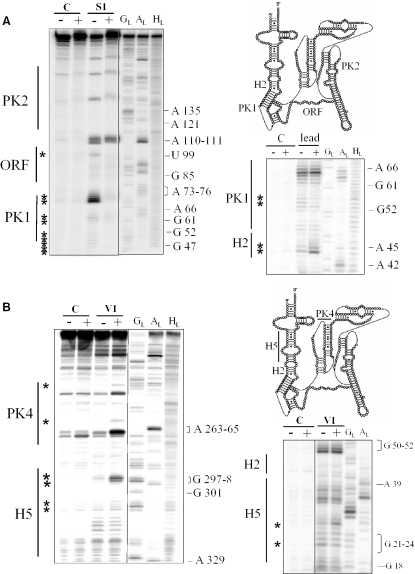
SmpB modifies the conformation or interacts with domains outside the tRNA-like domain. Emphasis to the structural changes induced by the protein on H2, PK1, the internal ORF (A) and on H5 and PK4 (B). The other indications are as in Figure 2.
Double-stranded-specific cuts suggest that H1 (the acceptor stem), H2, the 5′-strand of H4, H5 (heavily cleaved by RNase V1), both stems of PK1, PK2 and PK4 form in solution. Surprisingly, H6 (the T-stem) is cleaved by lead (Figure 2), but precise assignments of the cuts is difficult due to band compressions. Loop 5 (L5, the D-loop), loop 6 (L6, the T-loop) and Loop 4 (L4, encompassing the termination codon of the internal tag reading frame) are heavily cleaved by probes specific for ss RNA, as expected. The nucleotide sequence between PK1 and the resume GCU codon (nt U72–C77) is heavily cleaved by lead and nuclease S1, suggesting that this region is highly accessible within the RNA structure, probably for optimal access by the translational machinery during re-registration. Within H5, there is a C35/A299–U302 internal bulge supported by nuclease S1 cleavages. Strikingly, the coding sequence (from G79 to A111) is heavily cleaved by RNase V1, and some pairings have been proposed (Figure 1A). Spontaneous degradation sites have been mapped in PK4 (U240, G270–A271).
Association constant between SmpB and tmRNA from A.aeolicus
SmpB and tmRNA have been cloned and purified from a thermophile because structural data of the complex between the tRNA domain of tmRNA and SmpB and between full-length tmRNA and SmpB in a stalled ribosome are from thermophiles (5,19). To assess the optimal concentration of purified SmpB required for the enzymatic and chemical footprint assays, interaction between synthetic tmRNA and recombinant SmpB, both from the thermophilic organism A.aeolicus, was analyzed by gel-mobility shift assays (Figure 1B). The observed binding is saturable with half-maximal binding at a free SmpB concentration of ∼95 nM. This value has to be compared with that derived from a complex between tmRNA and SmpB from E.coli, of ∼20 nM (12). At low concentration of SmpB (from 18 to 37 nM), a retarded band is observed (Figure 1B), corresponding to an RNA–protein complex, probably at a one-to-one stoichiometry. Increasing the concentration of SmpB further (from 75 to 3000 nM) leads to the aggregation between the RNA and the protein, in agreement with more than one SmpB per tmRNA, as indicated by the SPR analysis (see below).
Footprints between tmRNA and SmpB from A.aeolicus
Footprint assays were used to monitor the interaction between tmRNA and purified SmpB, an approach that is instrumental to map the contacts between RNAs and proteins (19). The alanyl residue of Ala-tmRNA does not influence the interaction between tmRNA and SmpB (13). End-labeled uncharged tmRNA was incubated with purified SmpB (seven times over the apparent dissociation constant). A total of 14 experiments were performed with nuclease S1, 10 with RNase V1 and 10 with lead acetate, half with 5′-labeled RNA and the other half with 3′-labeled RNA (Figures 2 and 3 are representatives, lanes marked ‘+’). Among them, lead, nuclease S1 and RNase V1 induced cleavages of selected positions within the ribose-phosphate chain of tmRNA and have revealed differences in the reactivity of some nucleotides in the presence or absence of SmpB. These data are summarized in Figure 4. The enzymatic and chemical footprints of the tRNA domain of full-length tmRNA are very similar, if not identical, to those collected for tmRNAΔ, tRNA mimic domain of tmRNA (19), demonstrating that those contacts are maintained in the context of full-length tmRNA. The protein protects nucleotides from both the D- and connector (C) loops as well as 3 nt at the base of the T-stem (H6) from being cleaved by ss-specific probes (Figure 2). These three domains interact with SmpB in the crystal structure of the complex containing only the tRNA domain of tmRNA [tmRNAΔ, (19)]. As for tmRNAΔ, nucleotides from the T-loop are less accessible toward lead, whereas there are no direct contacts between the protein and the T-loop. SmpB binds the elbow region of the tRNA-like domain and probably modifies the conformation of the T-loop indirectly. However, we noticed additional structural changes within the tRNA domain of full-length tmRNA: the reactivity of nucleotide from the first three pairs of the acceptor stem (H1) is modified in the presence of SmpB (Figure 2). SmpB protects the first 2 nt at the very 5′ end of tmRNA and also destabilizes the 3′ end of the acceptor stem. In the X-ray structure, tmRNAΔ acceptor stem was not included in the refined model because the electron density map was poorly defined in this area of the molecule. This is most likely because the conformation of H1 is altered when SmpB binds the tRNA domain.
Figure 4.
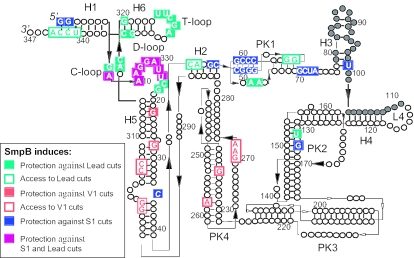
Mapping the structural changes induced by SmpB on tmRNA from A.aeolicus. Colored symbols were used and their meaning is detailed in the inset. The open circles are the nucleotides with no reactivity changes when tmRNA is in complex with SmpB. The structural domains of tmRNA are indicated. Nucleotides filled with gray belong to the internal coding sequence.
SmpB also interacts outside the tRNA domain of tmRNA
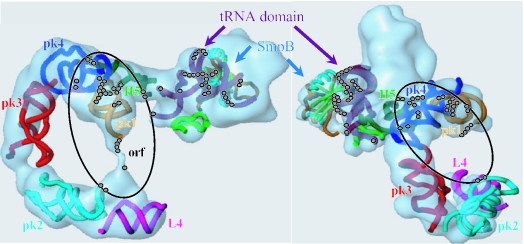
Biochemical and structural data about putative interactions between SmpB and domains outside the tRNA mimic of tmRNA have been collected (Figure 3) and summarized on the experimentally supported structure of tmRNA from A.aeolicus (Figure 4). SmpB modifies the reactivity of 10 nt (out of 16) in both stems of pseudoknot PK1 as well as the reactivity of 2 nt in the loop crossing the shallow groove. Also, nt G70–A73, downstream from PK1, are protected from being cleaved by nuclease S1. The accessibility of nucleotide U99 from tmRNA internal open reading frame (ORF), as well as U134 and G136 from PK2, toward ss-specific probes decreases in the presence of SmpB (data not shown). Those nucleotides, with the exception of A110, are paired suggesting that the protein influences the folding of these domains. Also, SmpB modifies the reactivity of nucleotide in the lower portion of H5 (G28, C35, G297–G298, C305–C306), in H2 (C44–A45, G47–C48) and PK4 (G236, A258, G270–A272). There are no reactivity changes in H4, in PK2 (two exceptions, data not shown) and PK3 (mapping of PK3 is difficult from the ends of tmRNA, but overall we could not detect any differences in the presence of SmpB). The reactivity changes of nucleotide from tmRNA induced by SmpB have been placed onto tmRNA in complex with SmpB (Figure 5) derived from the cryo-EM map obtained for Thermus thermophilus 70S·tRNA·EF-Tu·tmRNA·SmpB with GTP and kirromycin (5). The footprints in the tRNA domain of tmRNA are clustered on the interface between SmpB and tmRNA. Also, whereas the set of footprints away from the tRNA domain are scattered on the secondary structure of tmRNA (Figure 4), they form a cluster when placed onto the 3D model (Figure 5, black circle). The footprints located outside the tRNA domain can result from a conformational rearrangement of tmRNA structure after the binding of SmpB to its elbow region. Alternatively, during trans-translation, SmpB may bind tmRNA at other locations. In order to test these hypotheses, SPR was used to study the stoichiometry of the binding of SmpB to tmRNA.
Figure 5.
Location of the reactivity changes induced when SmpB binds tmRNA onto a 3D model (two views of the model are shown). The coordinates are derived from a cryo-EM study visualizing tmRNA entry into a stalled ribosome from T.thermophilus (5). For clarity, the docking of EF-Tu into the density was omitted. SmpB (α-helices in cyan and β-barrels in light green) binds the tRNA-like domain (in dark purple) of tmRNA. H5 is in green, PK1 in orange, PK2 in light blue, PK3 in red, PK4 in dark blue and L4 in violet. The location of the reactivity changes induced when SmpB binds tmRNA are the beads. The structural alterations outside the tRNA-like domain are circled. The two figures differ by rotating the macromolecular complex along the x and y axes.
Stoichiometry of the interaction between SmpB and tmRNA
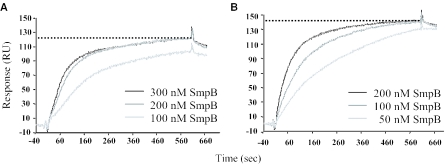
The stoichiometry of SmpB–tmRNA binding was determined by SPR analysis described in Materials and Methods. A biotinylated (dT)20 oligomer was bound to a streptavidin sensor chip. tmRNA or tmRNAΔ was polyadenylated and coupled by their poly(A) tails to a flow chip. The stoichiometry between SmpB and tmRNAΔ was accessed by the injection of increasing concentrations of purified SmpB-His over the Sensor Chip functionalized with 151 RU of polyadenylated tmRNAΔ until saturation of the ligand. Three independent experiments were performed (Figure 6A is representative). A maximum of 122 RU of SmpB-His can bind tmRNAΔ. The amount of protein and RNA can be determined using the conversion factors: 1 RU of protein bound = 1 pg of protein and 1 RU of RNA = 0.8 pg of RNA/mm2 flow cell surface area (26). Given that SmpB-His and poly(A)-tmRNAΔ have a calculated molecular weight of 19 410 and 30 215 Da, respectively, the SmpB-His:tmRNAΔ ratio was estimated from Equation 1 (see Materials and Methods). A stoichiometry of one molecule of SmpB-His per tmRNAΔ was deduced. Three independent experiments were performed and the calculated stoichiometry is 0.87 ± 0.13 SmpB protein for one tmRNAΔ. This stoichiometry is in agreement with the detection of a single molecule of SmpB on tmRNAΔ by X-ray crystallography (19), which argues for the specificity of the interaction in our experimental conditions. To determine the stoichiometry between SmpB and full-length tmRNA, the experiment was repeated using polyadenylated tmRNA as the ligand. A total of 233 RU of coupled tmRNA bind a maximum of 142 RU of SmpB (Figure 6B). Using Equation 1 and a calculated Mr of 129 925 Da for poly(A)-tmRNA, a stoichiometry of 3.26 molecules of SmpB per tmRNA was deduced. Five independent experiments (Figure 6B is representative) performed with various amounts of immobilized tmRNA lead to a stoichiometry of 2.87 ± 0.4 molecules of SmpB per tmRNA.
Figure 6.
Determination of the stoichiometry between SmpB and tmRNA by SPR. The sensorgrams of SmpB binding to either tmRNAΔ (A) or tmRNA (B) were obtained by the injection of increasing amount of SmpB to the flow cell immobilized with either poly(A)-tmRNAΔ (151 RU) (A) or tmRNA (233 RU) (B). The maximum level of the binding of SmpB is indicated by a broken line.
tmRNA aminoacylation in the presence of increasing amounts of SmpB
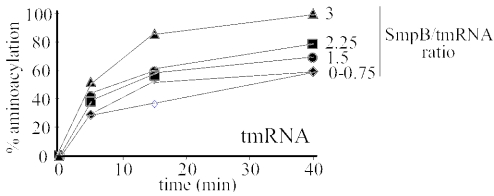
SPR indicates that three molecules of SmpB can potentially bind to one tmRNA. To assess whether or not these results has any physiological relevance during trans-translation, aminoacylation levels of tmRNA were measured in the presence of increasing concentrations of purified SmpB. Therefore, the stoichiometry of SmpB versus full-length tmRNA was gradually increased up to 3:1 and tmRNA aminoacylation with alanine was measured in vitro at 55°C with purified alanyl-tRNA synthetase from A.aeolicus. Aminoacylation plateaus gradually increase up to 100% at a 3:1 SmpB·tmRNA ratio (Figure 7). Lower plateaus mean that a fraction of the RNA is incapable of charging with alanine, probably due to stable non-functional conformations that can be altered by SmpB. For tmRNAΔ, aminoacylation is very efficient and 100% aminoacylation is obtained in the absence of SmpB and also with a one-to-one SmpB·tmRNAΔ ratio. Increasing the SmpB·tmRNAΔ ratio, however, gradually impairs aminoacylation (data not shown).
Figure 7.
Aminoacylation plateau levels of full-length tmRNA with increasing amounts of SmpB. Open diamonds, without SmpB; black diamonds, 0.75 SmpB molecules per tmRNA; black circles, 1.5 SmpB molecules per tmRNA; black squares, 2.25 SmpB molecules per tmRNA; black triangles, 3 SmpB molecules per tmRNA. A beneficial effect on tmRNA aminoacylation was reproducibly observed. Hundred percent aminoacylation plateaus are obtained with three molecules of SmpB per tmRNA.
DISCUSSION
The structure of tmRNA from a thermophile has been mapped in solution and indicates that ∼70% of the nucleotides are paired, to enhance thermostability. The overall structure is consistent with its deduction by sequence comparison (6). However, the internal reading frame is heavily cleaved by ds-specific probes, suggesting internal pairings, as suggested for E.coli tmRNA (7), despite the lack of phylogenetic support. In tmRNA from E.coli, helices and pseudoknots are unstable and breathe in solution (7); whereas in A.aeolicus tmRNA, the paired regions are more stable, as expected for a thermophile. Moreover, this RNA might possess post-transcriptional modifications in vivo that will enhance thermostability further.
Footprints in the tRNA-like domain of full-length tmRNA with SmpB are remarkably similar than those identified between a molecule encompassing only the tRNA portion (tmRNAΔ) and SmpB (19). This indicates that in the full-length RNA, the tRNA domain folds independently and its conformation is not affected by the other domains. Specific protections have been mapped in the T- and D-loops of tmRNA, consistent with the binding of SmpB to the elbow region, as for tmRNAΔ, stabilizing the D-loop in an extended conformation and increasing the angle between the acceptor stem and H5. A single conserved and position-specific G:U base pair in the tRNA acceptor stem is the key identity determinant for aminoacylation of alanine-specific RNAs by AlaRS in vivo (28). SmpB induces structural modifications at the tip of the acceptor stem (the first three pairs), probably to facilitate the exposure of a set of chemical groups of the G:U pair at the third position that is recognized and required for aminoacylation (2). In the X-ray structure between the tRNA domain of tmRNA and SmpB, the electron density map of the acceptor stem is poorly defined, likely induced by this structural perturbation identified in solution. This structural effect induced by SmpB at the vicinity of the G:U pair in tmRNA acceptor stem probably accounts, at least in part, for its stimulation of tmRNA aminoacylation by the AlaRS [(13,14) and this paper].
Additional footprints between SmpB and tmRNA were identified in the lower part of H5, H2, PK1 and PK4 and to a lesser extent in PK2 and the internal ORF. These contact sites are also probably present in E.coli, but may have been missed because one study tested protections by SmpB of the few accessible Gs by RNase T1 footprints (13) and the other has used a truncated version of tmRNA lacking the internal ORF and PK2–PK4 (20). When the footprints are placed onto a 3D model derived from cryo-EM pictures visualizing tmRNA entry into a stalled ribosome in the presence of SmpB (5), those outside the tRNA domain form a compact cluster (Figure 5). This observation implies that these binding sites are not at random onto tmRNA structure, reflecting specific binding sites. These reactivity changes, reflecting either indirect structural changes or direct contacts, are located away from the binding interface between SmpB and the tRNA-like domain. These results suggest that during trans-translation, SmpB could bind tmRNA at different locations. This hypothesis has been strengthened by SPR analysis, indicating that immobilized tmRNA binds three molecules of SmpB. Three molecules of SmpB cannot bind simultaneously tmRNA in vivo, since there is an estimated amount of two molecules of SmpB per molecule of tmRNA, both required to insure the optimal trans-transfer of the alanine of ala-tmRNA to the stalled peptide (16). The three independent recognition sites for SmpB on tmRNA are probably required during the highly dynamic process of trans-translation. Indeed, large conformational changes are required at various steps of trans-translation, including tmRNA accommodation, switching of reading frames and translocation. In particular, SmpB has to move from its location derived from cryo-EM pictures of the early steps of trans-translation, to prevent steric clashes with the P-site tRNA and also to fit the tmRNA–SmpB complex into the P-site of the ribosome after translocation (5). It has been proposed that SmpB may transiently dissociate from tmRNA after translocation to allow an incoming tRNA matching the first codon of the tag to fit into an accessible A-site (19). SmpB remains bound to a ‘ribosome–tmRNA’ complex isolated from cells with stalled translation at various positions within the tag reading frame of tmRNA (29), reinforcing the biological significance of our results and illustrating the essential role of SmpB at early and later stages during trans-translation. Alternatively, during trans-translation, specific steps could require the transient binding of multiple SmpB molecules onto one molecule of tmRNA.
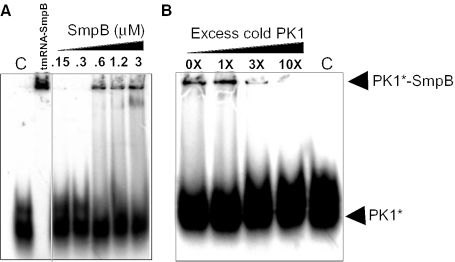
Pseudoknots PK2–PK4 can be replaced with RNA single strands without affecting significantly trans-translation in E.coli cells (11). However, based on extensive mutational analysis (10,15) and phylogenetic conservation among bacterial sequences, pseudoknot PK1 has an important function during trans-translation. Also, the segment between PK1 and the resume codon has been identified by randomization–selection experiments in E.coli tmRNA as an essential locus responsible for precise resume codon selection (30). Twelve footprints have been identified between SmpB and PK1 and four additional ones within the segment connecting PK1 to the resume codon (Figure 4). These data suggest that during trans-translation, SmpB binds at or near PK1 to participate in resume codon selection. In E.coli, attempts to detect the binding of SmpB onto a synthetic fragment corresponding to PK1 by mobility shift assays failed (20). We performed similar experiments between SmpB and synthetic PK1 from A.aeolicus and we have detected the formation of a complex in vitro (Figure 8). The binding is saturable, with half-maximal binding at a free SmpB concentration of ∼750 nM, suggesting that domains in the vicinity of PK1 in tmRNA structure (e.g. H2, the segment connecting PK1 to the resume codon, the reading frame, PK2) enhance binding. These footprints suggest that SmpB might participate in finding the first triplet to be translated in tmRNA, for precise registration of the reading frame. A third binding site may correspond to the footprints identified between the lower part of H5 and pseudoknot PK4.
Figure 8.
Native gel retardation assay between labeled pK1 and purified SmpB-His from A.aeolicus. (A) Increasing amounts of SmpB was incubated with pK1; a full-length tmRNA–SmpB complex is shown as a positive control; (B) a 3- to 10-fold excess of cold PK1 competed labeled PK1 out of the complex. C: Negative control.
Aminoacylation plateau levels are optimal when the stoichiometry between SmpB and tmRNA is 3:1. Similar results were obtained in E.coli (13). This effect on aminoacylation could be due to the saturation of all the potential binding sites of the protein that stabilize tmRNA structure into an active conformation, eliminating a substantial fraction of improperly folded RNAs. In E.coli, using a tmRNA construct containing only the tRNA domain, H5 and PK1, aminoacylation plateau levels are optimal when the stoichiometry between SmpB and tmRNA is 2:1 (13). According to the structural data reported here, this tmRNA fragment has only two independent binding sites for SmpB, the tRNA domain and around PK1.
Translation stalling sequesters ribosomes and produces incomplete polypeptides. In bacteria, the SmpB–tmRNA complex solves these problems. SmpB probably binds the stalled ribosome at first, then could recruit tmRNA and the ribosome switches mRNA templates, leaving the non-stop message and translation resumes on tmRNA reading frame. The independent binding sites of SmpB onto tmRNA might be responsible for the conformational flexibility of the tmRNA–SmpB complex that is required to induce the structural changes involved in the recognition of the stalled ribosomes by the rescue system, the insertion of the tmRNA-encoded reading frame into the decoding center and to thread through the ribosome during trans-translation. Accordingly, SmpB is proposed to move on the tmRNA scaffold during the various steps of trans-translation (initial recognition by the stalled ribosome, codon-independent trans-transfer of the alanine from Ala-tmRNA, translation of the tag reading frame and ribosome recycling).
Acknowledgments
D. Goven in the laboratory has helped us with the purification of tmRNA. The clone expressing recombinant A.aeolicus alanyl-tRNA synthetase was provided by Prof. N. Ban (ETH, Zurich). We also thank Dr J. Frank (Albany) for providing the coordinates of the cryo-EM map of tmRNA/SmpB into a stalled ribosome and Dr F. Dardel (Paris V, France) for providing purified PK1. Our laboratory is part of IFR 140 GFAS. This work was supported in part by the Région Bretagne (Programme d'accueil d'équipes en émergence). Funding to pay the Open Access publication charges for this article was provided by Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1.Keiler K.C., Waller P.R., Sauer R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 2.Komine Y., Kitabatake M., Yokogawa T., Nishikawa K., Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc. Natl Acad. Sci. USA. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Withey J.H., Friedman D.I. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 4.Haebel P.W., Gutmann S., Ban N. Dial tm for rescue: tmRNA engages ribosomes stalled on defective mRNAs. Curr. Opin. Struct. Biol. 2004;14:58–65. doi: 10.1016/j.sbi.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Valle M., Gillet R., Kaur S., Henne A., Ramakrishnan V., Frank J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- 6.Gueneau de Novoa P., Williams K.P. The tmRNA website: reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004;32:D104–D108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felden B., Himeno H., Muto A., McCutcheon J.P., Atkins J.F., Gesteland R.F. Probing the structure of the Escherichia coli 10Sa RNA (tmRNA) RNA. 1997;3:89–103. [PMC free article] [PubMed] [Google Scholar]
- 8.Hickerson R.P., Watkins-Sims C.D., Burrows C.J., Atkins J.F., Gesteland R.F., Felden B. A nickel complex cleaves uridine in folded RNA structures: application to E.coli tmRNA and related engineered molecules. J. Mol. Biol. 1998;279:577–587. doi: 10.1006/jmbi.1998.1813. [DOI] [PubMed] [Google Scholar]
- 9.Gaudin C., Zhou X., Williams K.P., Felden B. Two-piece tmRNA in cyanobacteria and its structural analysis. Nucleic Acids Res. 2002;30:2018–2024. doi: 10.1093/nar/30.9.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nameki N., Felden B., Atkins J.F., Gesteland R.F., Himeno H., Muto A. Functional and structural analysis of a pseudoknot upstream of the tag-encoded sequence in E.coli tmRNA. J. Mol. Biol. 1999;286:733–744. doi: 10.1006/jmbi.1998.2487. [DOI] [PubMed] [Google Scholar]
- 11.Nameki N., Tadaki T., Himeno H., Muto A. Three of four pseudoknots in tmRNA are interchangeable and are substitutable with single-stranded RNAs. FEBS Lett. 2000;470:345–349. doi: 10.1016/s0014-5793(00)01349-1. [DOI] [PubMed] [Google Scholar]
- 12.Karzai A.W., Susskind M.M., Sauer R.T. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barends S., Karzai A.W., Sauer R.T., Wower J., Kraal B. Simultaneous and functional binding of SmpB and EF-Tu-TP to the alanyl acceptor arm of tmRNA. J. Mol. Biol. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu Y., Ueda T. The role of SmpB protein in trans-translation. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- 15.Hanawa-Suetsugu K., Takagi M., Inokuchi H., Himeno H., Muto A. SmpB functions in various steps of trans-translation. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hallier M., Ivanova N., Rametti A., Pavlov M., Ehrenberg M., Felden B. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J. Biol. Chem. 2004;279:25978–25985. doi: 10.1074/jbc.M314086200. [DOI] [PubMed] [Google Scholar]
- 17.Dong G., Nowakowski J., Hoffman D.W. Structure of small protein B: the protein component of the tmRNA–SmpB system for ribosome rescue. EMBO J. 2002;21:1845–1854. doi: 10.1093/emboj/21.7.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Someya T., Nameki N., Hosoi H., Suzuki S., Hatanaka H., Fujii M., Terada T., Shirouzu M., Inoue Y., Shibata T., et al. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus. FEBS Lett. 2003;535:94–100. doi: 10.1016/s0014-5793(02)03880-2. [DOI] [PubMed] [Google Scholar]
- 19.Gutmann S., Haebel P.W., Metzinger L., Sutter M., Felden B., Ban N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- 20.Wower J., Zwieb C.W., Hoffman D.W., Wower I.K. SmpB: a protein that binds to double-stranded segments in tmRNA and tRNA. Biochemistry. 2002;41:8826–8836. doi: 10.1021/bi0201365. [DOI] [PubMed] [Google Scholar]
- 21.Wyatt J.R., Chastain M., Puglisi J.D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991;11:764–769. [PubMed] [Google Scholar]
- 22.Perret V., Garcia A., Puglisi J., Grosjean H., Ebel J.-P., Florentz C., Giegé R. Conformation in solution of yeast tRNA(Asp) transcripts deprived of modified nucleotides. Biochimie. 1990;72:735–743. doi: 10.1016/0300-9084(90)90158-d. [DOI] [PubMed] [Google Scholar]
- 23.Felden B., Florentz C., Giegé R., Westhof E. Solution structure of the 3′-end of brome mosaic virus genomic RNAs. Conformational mimicry with canonical tRNAs. J. Mol. Biol. 1994;235:508–531. doi: 10.1006/jmbi.1994.1010. [DOI] [PubMed] [Google Scholar]
- 24.Silberklang M., Prochiantz A., Haenni A.L., Rajbhandary U.L. Studies on the sequence of the 3′-terminal region of turnip-yellow-mosaic-virus RNA. Eur. J. Biochem. 1977;72:465–478. doi: 10.1111/j.1432-1033.1977.tb11270.x. [DOI] [PubMed] [Google Scholar]
- 25.Stepanov V.G., Nyborg J. tmRNA from Thermus thermophilus. Interaction with alanyl-tRNA synthetase and elongation factor Tu. Eur. J. Biochem. 2003;270:463–475. doi: 10.1046/j.1432-1033.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- 26.Van Ryk D.I., Venkatesan S. Real-time kinetics of HIV-1 Rev–Rev response element interactions. Definition of minimal binding sites on RNA and protein and stoichiometric analysis. J. Biol. Chem. 1999;274:17452–17463. doi: 10.1074/jbc.274.25.17452. [DOI] [PubMed] [Google Scholar]
- 27.Williams K.P., Bartel D.P. Phylogenetic analysis of tmRNA secondary structure. RNA. 1996;2:1306–1310. [PMC free article] [PubMed] [Google Scholar]
- 28.McClain W.H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 29.Shpanchenko O.V., Zvereva M.I., Ivanov P.V., Bugaeva E.Y., Rozov A.S., Bogdanov A.A., Kalkum M., Isaksson L.A., Nierhaus K.H., Dontsova O.A. Stepping tmRNA through the ribosome. J. Biol. Chem. 2005 doi: 10.1074/jbc.M409094200. in press. [DOI] [PubMed] [Google Scholar]
- 30.Williams K.P., Martindale K.A., Bartel D.P. Resuming translation on tmRNA: a unique mode of determining a reading frame. EMBO J. 1999;18:5423–5433. doi: 10.1093/emboj/18.19.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]