Abstract
CPEB is an RNA binding protein that interacts with the maturation-type cytoplasmic polyadenylation element (CPE) (consensus UUUUUAU) to promote polyadenylation and translational activation of maternal mRNAs in Xenopus laevis. CPEB, which is conserved from mammals to invertebrates, is composed of three regions: an amino-terminal portion with no obvious functional motif, two RNA recognition motifs (RRMs), and a cysteine-histidine region that is reminiscent of a zinc finger. In this study, we investigated the physical properties of CPEB required for RNA binding. CPEB can interact with RNA as a monomer, and phosphorylation, which modifies the protein during oocyte maturation, has little effect on RNA binding. Deletion mutations of CPEB have been overexpressed in Escherichia coli and used in a series of RNA gel shift experiments. Although a full-length and a truncated CPEB that lacks 139 amino-terminal amino acids bind CPE-containing RNA avidly, proteins that have had either RRM deleted bind RNA much less efficiently. CPEB that has had the cysteine-histidine region deleted has no detectable capacity to bind RNA. Single alanine substitutions of specific cysteine or histidine residues within this region also abolish RNA binding, pointing to the importance of this highly conserved domain of the protein. Chelation of metal ions by 1,10-phenanthroline inhibits the ability of CPEB to bind RNA; however, RNA binding is restored if the reaction is supplemented with zinc. CPEB also binds other metals such as cobalt and cadmium, but these destroy RNA binding. These data indicate that the RRMs and a zinc finger region of CPEB are essential for RNA binding.
In many animals, maternal mRNAs that are synthesized and stored in a translationally dormant form during oogenesis become activated either upon reentry into the meiotic divisions (oocyte maturation) or after fertilization. These mRNAs encode a number of important products such as those that drive the early embryonic cell divisions, establish embryonic polarity, and determine certain cell lineages (15, 40, 42). The translational repression/activation of maternal mRNA appears to be quite complex and involves inhibitory (masking) cis elements (10, 16, 32, 37), some clearly defined inhibitory proteins (4, 31), positively acting cis elements (26, 33, 38, 41), and activator proteins (14). In several cases, translational activation is not direct but instead is mediated by changes in poly(A) tail length. That is, in oocytes, several translationally dormant mRNAs contain relatively short poly(A) tails, usually consisting of fewer than 20 nucleotides. During oocyte maturation or early embryogenesis, several specific mRNAs undergo poly(A) elongation and resulting translational activation (15, 30, 42).
In maturing Xenopus oocytes, two cis elements located in the 3′ untranslated region (UTR) of target RNAs, the cytoplasmic polyadenylation element (CPE) and the hexanucleotide AAUAAA, regulate poly(A) elongation. The CPE, which has the general structure UUUUUAU, usually resides within 30 nucleotides 5′ of the hexanucleotide but can be immediately adjacent to the hexanucleotide, as in c-mos mRNA (UUUUAUAAUAAA), or even overlap with it, as in cyclin A1 mRNA (UUUUUAAUAAA) (11, 39). All of these maturation-type CPEs are bound by the protein CPEB, which is necessary for cytoplasmic polyadenylation (14, 28, 39). Although the precise function of CPEB is unclear, it may recruit or stabilize factors on the hexanucleotide, such as CPSF (cleavage and polyadenylation specificity factor) (3), a group of four polypeptides that is essential for nuclear pre-mRNA cleavage and polyadenylation. At least for nuclear polyadenylation, it is probably CPSF that recruits poly(A) polymerase to catalyze poly(A) addition. During maturation, cytoplasmic poly(A) addition, in a mechanism as yet unknown, induces 5′ cap ribose methylation (i.e., cap II formation) (18). It is cap II formation, then, that is at least partly responsible for translational activation (18, 19).
Xenopus CPEB, a 62-kDa protein, is composed of three regions. The amino-terminal portion (region 1) is conserved in the mouse protein (13) but contains no known functional motif. Region 2 includes two RNA recognition motifs (RRMs) that are conserved in Drosophila ORB (22) and three Caenorhabditis elegans open reading frames (43), as well as in the mouse protein (13). Region 3, which is also conserved in these vertebrate and invertebrate proteins, is composed of a cysteine-histidine repeat that is similar to a metal-coordinating region (2, 35).
In this report, we have examined the primary structure of CPEB that is important for specific interaction with RNA. First, we show that the phosphorylation of CPEB, which takes place during oocyte maturation, enhances about twofold its ability to be UV cross-linked to RNA (28). Using Escherichia coli-expressed protein, we show that CPEB can bind RNA as a monomer and that both RRMs, especially RRM1, are important for RNA binding. In addition, the cysteine-histidine region is essential for RNA binding. Point mutations of specific cysteine and histidine residues within this region abolish binding, thereby demonstrating the importance of these evolutionarily conserved amino acids. Chelation of metal ions also destroys RNA binding by CPEB, but binding is restored by subsequent addition of zinc. Other metals such as cobalt and cadmium can be bound by CPEB, but these inhibit RNA binding, probably by altering the structure of the protein. Therefore, CPEB contains three domains, two RRMs and a zinc finger, that are crucial for RNA binding.
MATERIALS AND METHODS
RNA synthesis.
Cloning and transcription details for the cyclin B1 clones were described previously (39). B1wt was previously referred to as scyclin B1, and B1cpe1 was previously referred to as scyclin B1/CPE1. Cloning and transcription details for embryonic histone B4, previously referred to as sB4, were also described previously (39). RNA transcripts were synthesized in vitro, using a kit from Promega (Madison, Wis.). After transcription, the RNAs were electrophoresed in 6% acrylamide–6 M urea gels and extracted in buffer containing 0.3 M sodium acetate, 1 mM EDTA, and 0.1% sodium dodecyl sulfate (SDS).
Construction of full-length and mutant CPEB cDNAs. (i) Deletion mutations.
Plasmid pHis-CPEB (encoding H-CPEB) was constructed as described previously (39). Derivatives of pHis-CPEB containing selected portions of CPEB such as the amino-terminal half (N region), RRM1, RRM2, and/or the cysteine-histidine domain were constructed as follows. pN, encoding amino acids 1 to 288 of CPEB, was made by ligation of a BglIIv-BsgI936 (blunt ended) (superscript “v” indicates that the restriction site is present in the vector polylinker; superscript numbers indicate the restriction site location in CPEB cDNA) fragment from pHis-CPEB into the pET15B vector (Novagen, Madison, Wis.) and then cut with BlpI (blunt ended) and BglII. pNc/h contains only the N and cysteine-histidine regions, with the intervening nucleotides encoding RRM1 and RRM2 (amino acids 289 to 511) deleted. This construct was made by amplifying the cysteine-histidine region (encoding amino acids 512 to 568) with 5′ primer 5′GTAGGCCTTGGAAGACTCTGTTTGCCAG3′ and 3′ primer 5′GAGGCCTTAGCTGGAGTCACGACT3′. This amplification product was digested with StuI and ligated, with the BglIIv-BsgI936 (blunt) fragment isolated above, into BamHI (blunt)- and BglII-digested pET15B. pNr2c/h contains the N, RRM2, and cysteine-histidine regions and has a deletion of RRM1, which ranges from amino acid 289 to 413. This clone was made by ligating two fragments from pHis-CPEB, BglIIv-BsgI936 (blunt) with BsgI1313-AatIIv, into pET15B digested with BglII and AatII. pNr1c/h contains the N, RRM1, and cysteine-histidine regions and has a deletion of RRM2, which ranges from amino acid 431 to 511. This clone was made by digesting pHis-CPEB with BamHI, removing the RRM2- and cysteine-histidine-encoding nucleotides, and filling in the BamHI sites of the remaining vector. The cysteine-histidine domain was reintroduced by ligation of the cysteine-histidine domain amplification product (see above) digested with StuI. Clone pNr1r2 contains all of the CPEB coding region (amino acids 1 to 522) except for the carboxy-terminal nucleotides that encode the cysteine-histidine domain (amino acids 523 to 568). ½Nr1r2c/h contains amino acids 140 to 568 of CPEB, with a deletion of nucleotides encoding the first 139 amino acids. This clone was constructed by removing a 410-nucleotide NcoI fragment (NcoI1-NcoI410) from pHis-CPEB.
(ii) Point mutations.
Point mutations within the cysteine-histidine domain were made by using a Chameleon mutagenesis kit (Stratagene, La Jolla, Calif.) as instructed by the supplier. The selection primer was located in the pHis-CPEB polylinker, 5′CGACGACAAGGTCGACGATATGAATATGGCC3′, and changed an XhoI site to a SalI site. This primer was used in conjunction with the following point mutation primers to create new clones: C529/A (5′GGGCCATTCTTCGCCAGAGACCAGG3′), C539/A (5′TTTAAGTATTTCGCGCGCTCCTGTTGGCACTGG3′), and H547/A (TGGCACTGGCAGGCCTCTATGGAAATC3′).
Expression of CPEB in bacteria and their isolation.
Expression of soluble His-CPEB in E. coli and purification on a nickel column were performed as described previously (39). Solubilization and purification of His-CPEB and derivatives in the presence of 6 M urea were performed as described in the Novagene pET system handbook. Eluted His-CPEB and derivatives were dialyzed overnight at 4°C to 4 M urea–10 mM HEPES (pH 8.0)–1 mM dithiothreitol, 100 mM KCl–10% glycerol, aliquoted, and stored at −80°C until use.
Gel shift analyses.
In a 20-μl reaction volume, 50 to 80 fmol of gel-purified, radiolabeled RNA was incubated with 10 to 300 ng of His-CPEB or derivative protein in block mix (10 mM HEPES-KOH [pH 7.7], 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, 10 mM dithiothreitol, 1 μg of tRNA, 2 μg of bovine serum albumin, 8 U of RNasin [Promega]) and 5% glycerol for 20 min at 23°C. RNA gel shifts with proteins isolated in the presence of urea also contained 1 M urea. Heparin was then added to a final concentration of 5 μg/μl, and incubation continued for a further 10 min. Samples were loaded onto a preelectrophoresed 4% polyacrylamide gel (80:1 acrylamide/bisacrylamide) and electrophoresed in 45 mM Tris-borate–1 mM EDTA buffer at 23°C for 15 min at 50 V and then at 200 V until the bromophenol blue dye reached the bottom of the gel. The gel was dried briefly and exposed to a PhosphorImager screen (Molecular Dynamics).
A 1 M stock of 1,10-phenanthroline (Aldrich) was made with 100% ethanol, and a 1 M stock of 4,7-phenanthroline (Aldrich) was made with 50% ethanol. When used in gel shift analyses, reaction mixtures including block mix, protein, and phenanthroline were incubated for 16 h at 23°C. Heat-denatured RNA was then added, and the reaction mixture was incubated and analyzed as described above. The overall percentage of ethanol was equalized in all reactions to control for the phenanthroline vehicle.
UV cross-linking, immunoprecipitation, and Western blotting.
UV cross-linking and Western blot analysis were performed as described previously (14). Immunoprecipitation of UV cross-link labeled CPEB was performed by incubation of a UV cross-linking reaction mixture containing 3.5 × 106 cpm of radiolabeled B4 RNA and 72 μg of oocyte or egg extract with 25 μl of packed protein A-Sepharose beads and 4 μl of crude anti-CPEB polyclonal antiserum in 500 μl of 1× NET buffer (34). This reaction mixture was incubated for 1 h, rotating at 23°C. The beads were washed five times, each time with 1 ml of 1× NET. Fifty microliters of 2× SDS sample buffer was added to the beads; the sample was boiled and then analyzed by polyacrylamide gel electrophoresis (PAGE) and exposure of the dried gel to a PhosphorImager screen.
In other experiments, oocytes were incubated overnight in medium containing [35S]methionine (0.5 mCi/ml) and then induced to mature with progesterone. An extract was prepared by homogenization in 2× PEM buffer [0.2 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES, pH 6.6), 2 mM EGTA, 2 mM MgSO4], which was followed by a brief centrifugation. Potato acid phosphatase was then added as described previously (28), and CPEB was immunoprecipitated and analyzed by SDS-PAGE and fluorography. Other mature oocytes were similarly treated with phosphatase, and CPEB was analyzed by Western blotting.
RESULTS
Phosphorylation modestly increases CPEB cross-linking to RNA.
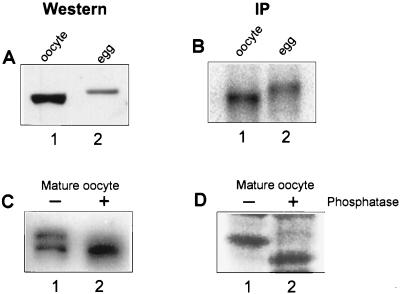
During oocyte maturation, CPEB becomes phosphorylated (14, 28), probably by p34cdc2 (11). Although phosphorylation did not appear to affect the interaction of CPEB with RNA (28), these experiments were performed before antibody to the protein was available, and therefore it was not clear whether possible changes in the level of CPEB could have masked an altered binding affinity. Accordingly, protein extracts prepared from oocytes and ovulated eggs were probed on a Western blot for CPEB (Fig. 1A). As shown previously, egg CPEB had a reduced mobility relative to that from oocytes, which is due to phosphorylation, and commensurately underwent partial degradation (11, 14, 28). To these same extracts was added CPE-containing 32P-labeled RNA, which was followed by UV cross-linking, RNase digestion, and immunoprecipitation with CPEB antibody. Figure 1B shows that CPEB was labeled in both oocyte and egg extracts, which again had a decreased electrophoretic mobility. The quantification of the amount of CPEB in Fig. 1A and the amount of CPEB UV cross-linked to RNA in Fig. 1B demonstrates that the phosphorylation of CPEB results in a 2.3-fold enhancement of UV cross-linking.
FIG. 1.
Phosphorylation of CPEB has a modest effect on RNA binding. (A) A protein extract (30 μg) from stage VI oocytes (lane 1) and shed eggs (lane 2) was resolved by electrophoresis, blotted to nitrocellulose, and probed with rabbit anti-CPEB antisera and then horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G. The horseradish peroxidase was detected by enhanced chemiluminescence. As noted previously (14, 28), CPEB is phosphorylated in the egg, which slows its migration in an SDS-polyacrylamide gel. (B) To these same extracts was added 32P-labeled B4 RNA, which was followed by UV irradiation, RNase digestion, and immunoprecipitation (IP) with CPEB antibody. Radioactive CPEB was detected with a PhosphorImager. (C) One-half of a mature oocyte extract was treated with potato acid phosphatase, and both untreated (lane 1) and treated (lane 2) samples were Western blotted and probed with anti-CPEB antisera. (D) One-half of an extract prepared from [35S]methionine-labeled mature oocytes was treated with phosphatase, and CPEB was immunoprecipitated from both untreated (lane 1) and treated (lane 2) samples and resolved by SDS-PAGE and fluorography.
To determine whether the phosphorylation of CPEB affects Western blotting or immunoprecipitation, two controls were performed. First, one-half of a mature oocyte extract that contained about 50% phosphorylated CPEB was treated with phosphatase, and both treated and untreated samples were analyzed by Western blotting (Fig. 1C). Although a single dephosphorylated CPEB band is evident in the phosphatase-treated sample (lane 2), the amount detected is nearly identical to that in the untreated sample (both phosphorylated and nonphosphorylated CPEB; lane 1). Thus, the phosphorylation of CPEB has no effect on Western blotting. To assess whether phosphorylation of CPEB affects immunoprecipitation, one-half of a different mature oocyte sample containing [35S]methionine-labeled CPEB was treated with phosphatase, which was followed by immunoprecipitation. Figure 1D demonstrates that while the phosphatase correctly dephosphorylated CPEB, this had no effect on the ability of this protein to be immunoprecipitated (compare lanes 1 and 2). Therefore, phosphorylation modestly (2.3-fold) increases the ability CPEB to UV cross-link to RNA.
CPEB can bind RNA as a monomer with a Kd of 130 nM.
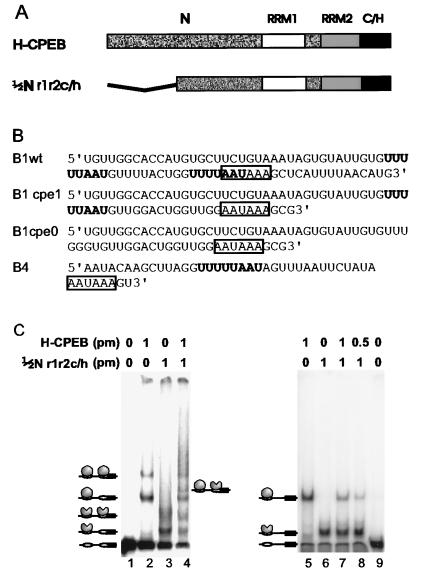
To determine whether CPEB binds RNA as a monomer or a dimer, full-length (H-CPEB) and truncated (½Nr1r2c/h) versions of CPEB were purified from bacteria as histidine-tagged fusion proteins (Fig. 2A) and used in RNA gel shift assays (Fig. 2C). The truncated protein lacked 139 amino-terminal amino acids, which we suspected would not be detrimental for RNA binding (see below and Fig. 3). When used individually in RNA gel shift assays with a small fragment of cyclin B1 RNA which contains two CPEs plus the hexanucleotide (Fig. 2B, B1wt), each protein bound both CPEs, as evidenced by two shifted bands (the nature of complexes in Fig. 2C, lanes 2 and 3, is indicated by a schematic to the left of lane 1). Because the proteins differ in size, the magnitude of the shift was unique for each protein. When the proteins were first mixed and then added to RNA, five shifted bands were evident, four of which corresponded to the original shifts observed with each protein (lane 4). The new band (indicated by a schematic to the right of lane 4) could be due to each protein binding a different CPE on the same RNA or to a heterodimer between the proteins on the same CPE. To distinguish between these possibilities, the same experiments were performed with a small fragment of histone B4 RNA (Fig. 2B, B4), which contains a single CPE plus the hexanucleotide (Fig. 2C, lanes 5 to 9). Again, each protein shifted the RNA uniquely, but in this case only one shifted band was observed (lanes 5 and 6). Addition of both proteins to the RNA resulted in the same shifted bands that were evident when the individual proteins were used (lanes 7 and 8). This result suggests that the new shifted band observed with B1wt RNA only when both proteins were present was due to each protein occupying a different CPE on the same molecule and demonstrates that CPEB can bind RNA as a monomer.
FIG. 2.
CPEB can bind RNA as a monomer. (A) Wild-type CPEB (H-CPEB) and a deletion mutant CPEB (½Nr1r2c/h) lacking 139 amino-terminal amino acids were expressed and purified from bacteria as histidine-tagged fusion proteins. Both proteins contain two RRMs (RRM1 and RRM2) and a cysteine-histidine (C/H) region. About one-half of the N region of CPEB has been removed from the deletion mutant. (B) Sequences of the RNAs used in this study. B1wt is a portion of the cyclin B1 RNA 3′ UTR containing two CPEs; B1cpe1 is a similar portion of the cyclin B1 RNA 3′ UTR containing only one CPE; B1cpe0 is a similar portion of the cyclin B1 3′ UTR containing no CPEs; and B4 is a portion of the 3′ UTR of an embryonic histone containing one CPE. CPEs are indicated in boldface. All RNAs contain a nuclear polyadenylation hexanucleotide (boxed). (C) Wild-type and mutant versions of CPEB were incubated individually or in combination with 50 fmol of radiolabeled B1wt RNA containing two CPEs (lanes 1 to 4) or with 50 fmol of radiolabeled B4 RNA containing one CPE (lanes 5 to 9); the resulting RNA-protein complexes were resolved by electrophoresis in a nondenaturing acrylamide gel and visualized with a PhosphorImager. Schematic representations of RNA protein complexes are shown to the left and right of each panel. Symbols and abbreviations: solid rectangle, AAUAAA; open oval, CPE; shaded oval, H-CPEB; shaded partial oval, ½Nr1r2c/h; pm, picomoles of protein.
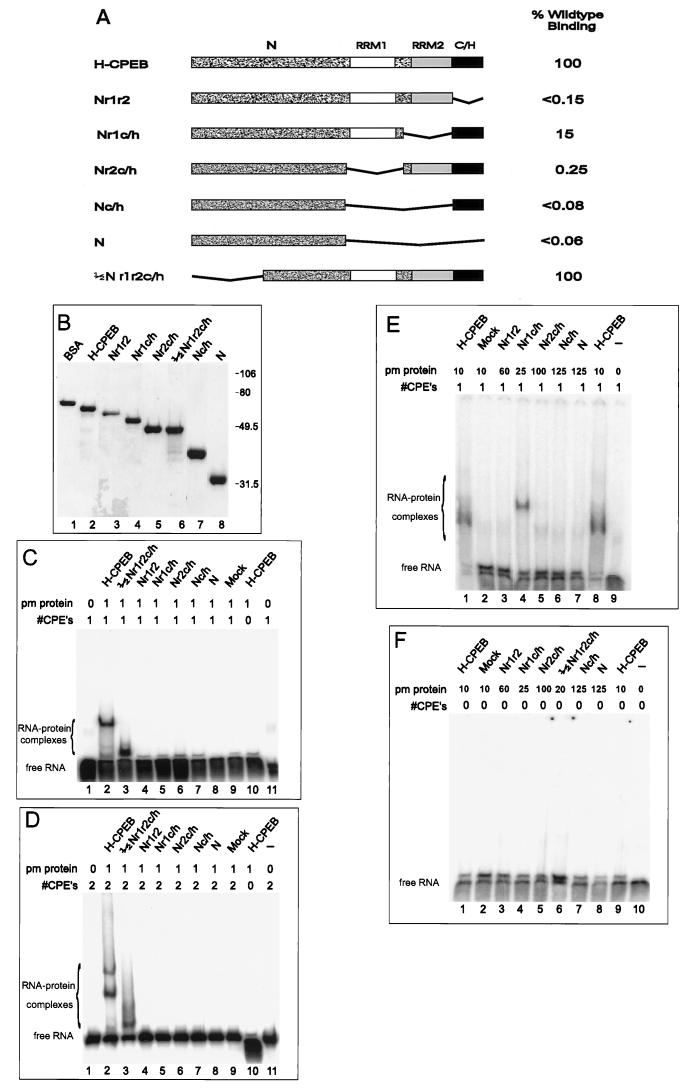
FIG. 3.
Mapping of the RNA binding domain: RNA gel shift analysis of CPEB deletion mutants. (A) Salient features of mutant CPEBs overexpressed in bacteria. Nomenclature of the mutations refers to the regions of CPEB that are retained: N, amino-terminal half; r1, RRM1; r2, RRM2; c/h = cysteine-histidine-rich region. (B) Coomassie blue-stained SDS-polyacrylamide gel of CPEBs used in RNA gel shift assays. Sizes of the protein standards are indicated to the right of the gel. (C) Gel shift analysis of radiolabeled B1cpe1 RNA incubated with 0 or 1 pmol of various CPEBs. (D) Gel shift analysis of radiolabeled B1wt RNA incubated with 0 or 1 pmol of various CPEBs. (E) Gel shift analysis of B1cpe1 RNA incubated with high concentrations (25 to 150 pmol) of various CPEB deletion mutant proteins. (F) Same conditions as in panel E except that B1cpe0 RNA was used. Picomoles of each protein (pm protein) are indicated above each lane. #CPE’s, number of CPEs in the RNA used.
The binding constant of CPEB was determined by gel shift assays using a constant amount of RNA containing either one or two CPEs and increasing amounts of CPEB. CPEB binds to RNA with a Kd of 130 nM (data not shown). Initial Kd determinations with B1wt RNA indicated that CPEB may bind cooperatively to RNA containing two CPEs. However, a Hill plot of these data provided no evidence of cooperative binding (data not shown).
RNA binding by CPEB is controlled by two RRMs and a cysteine-histidine region.
Using mutant recombinant proteins, we analyzed the regions of CPEB required for binding with RNA. Full-length and deletion mutant His-tagged CPEB proteins were expressed from bacteria and purified. Figure 3A depicts the salient features of these proteins visualized on a Coomassie blue-stained SDS-gel (Fig. 3B). Three CPE-containing RNAs, B1wt, B1cpe1, and B4, were chosen for this binding analysis to increase the likelihood that any CPEB-RNA interactions would be detected. Figures 3C and D show that full-length CPEB efficiently bound to both transcripts (lane 2 in each panel), but only when they contained CPEs (lane 10 in each panel). CPEB protein ½Nr1r2c/h, which lacked 139 amino-terminal residues, also bound to both RNAs with an efficiency similar to that of the full-length protein (Fig. 3C and D, lanes 3). However, CPEB proteins that lacked either of the RRMs or the cysteine-histidine region failed to bind either B1cpe1 or B1wt RNA (Fig. 3C and D, lanes 4 to 8).
To determine whether any of these mutant proteins at higher concentrations could bind RNA, additional gel shifts were performed with B1cpe1 and B1cpe0 RNAs. To obtain higher concentrations of CPEB, the protein was solubilized from pelleted material with 6 M urea and the subsequent gel shift assays were performed in the presence of 1 M urea (Materials and Methods). The binding characteristics of CPEB isolated from the soluble and insoluble fractions were virtually identical (compare lanes 2 in Fig. 3D and 4D; also data not shown). CPEB proteins that contained only one RRM (Fig. 3E, lanes 4 and 5) still retained the ability to bind B1cpe1 RNA, but 7- and 400-fold, respectively, less efficiently than full-length H-CPEB (Fig. 3E, lane 2). The RNA-protein complex formed between Nr1c/h and B1cpe1 RNAs migrates more slowly than the full-length H-CPEB–B1cpe1 RNA complex. This is probably due to a difference in the ways these proteins bend RNA and/or their solution structure (21). CPEB protein Nr1r2, which lacked the cysteine-histidine region only, did not bind B1cpe1 RNA even at a comparatively high concentration (Fig. 3E, lane 3). Similarly, other CPEB proteins that lacked both RRMs or the entire carboxy terminus of CPEB failed to bind B1 RNA at high concentrations (Fig. 3E, lanes 6 and 7). None of these proteins bound to B1cpe0 RNA (Fig. 3F). We therefore conclude that RRM2 and the cysteine-histidine region are necessary for the specific binding of CPE-containing RNA. The RNA binding activity of each mutant relative to wild-type CPEB binding is summarized in Fig. 3A.
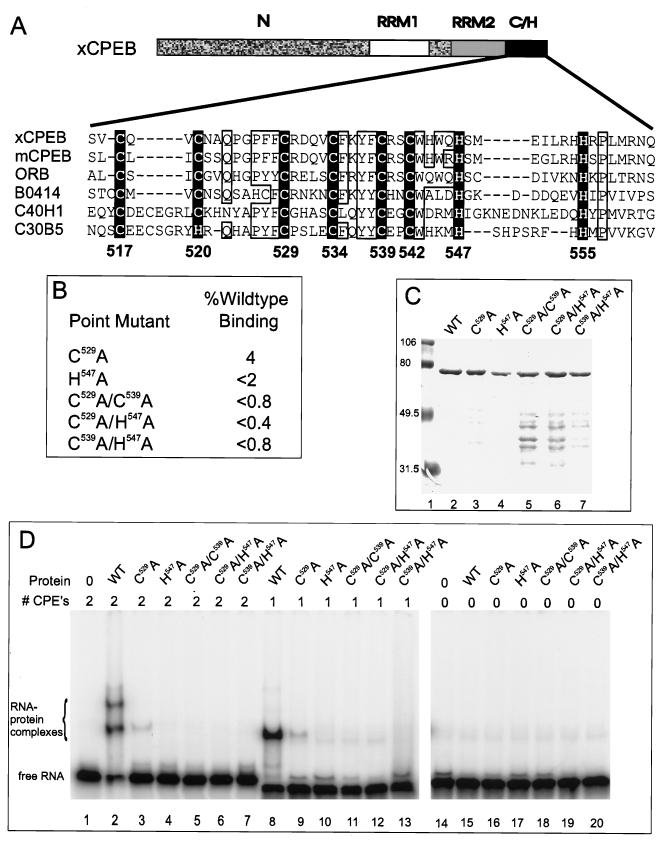
FIG. 4.
Mapping the RNA binding domain: effect of substituting cysteine and histidine residues with alanine. (A) Amino acid sequence alignment of the cysteine-histidine region among CPEB homologs. xCPEB, Xenopus CPEB (accession no. u14169); mCPEB, mouse CPEB (accession no. Y08260); ORB, Drosophila ORB (accession no. X64412); three open reading frames from C. elegans with unknown function: B0414 (accession number AF003145), C40H1 (accession no. Z19154), and C30B5 (accession no. U23450). Conserved cysteine and histidine residues are in white inside a black box. Numbers below the boxes denote positions of amino acids in xCPEB. Note the additional conservation (open boxes) of glutamine (Q), proline (P), aromatic residues (phenylalanine [F], tyrosine [Y], and tryptophan [W]), and basic residues (arginine [R] and lysine [K]). (B) Point mutant clones containing alanine substitutions for specific residues and effects of these mutations on RNA binding expressed as a percentage of wild-type CPEB binding. C529A indicates that C529 was replaced with an alanine, etc. (C) Coomassie blue-stained SDS-polyacrylamide gel of mutant CPEBs used in RNA gel shift analyses. Protein standards are indicated to the left. WT, wild type. (D) RNA gel shift of radiolabeled B1wt RNA (lanes 1 to 7), B1cpe1 RNA (lanes 8 to 13), or B1cpe0 (lanes 14 to 20) incubated with 1 pmol of various CPEBs.
The cysteine-histidine region is remarkably conserved in CPEB-like proteins from mouse, frog, fly, and worm cells. Its primary structure resembles that of a zinc finger in that it contains regularly spaced cysteine and histidine residues surrounded by conserved aromatic and hydrophobic residues (2, 8) (Fig. 4A). To examine more closely the contribution of this region to RNA binding, several CPEB proteins with amino acid substitutions (Fig. 4B and C) were used in RNA gel shifts (Fig. 4D). These substitutions included alanine for C529 or H547 alone or combinations of substitutions for C529 and C539, C529 and H547, and C539 and H547 (Fig. 4B). Figure 4D shows RNA gel shifts with B1wt (lanes 1 to 7), B1cpe1 (lanes 8 to 13), and B1cpe0 (lanes 14 to 20). While wild-type H-CPEB bound RNA with two CPEs or one CPE (lanes 2 and 8), it did not bind RNA with no CPE (lane 15). Importantly, all of the substitutions destroyed RNA binding. Only the C529A substitution showed trace binding activity (4% of wild-type activity; Fig. 4D, lanes 3 and 9). These results (summarized in Fig. 4B) clearly show that the targeted cysteine and histidine residues are critical for RNA binding.
CPEB requires zinc for RNA binding.
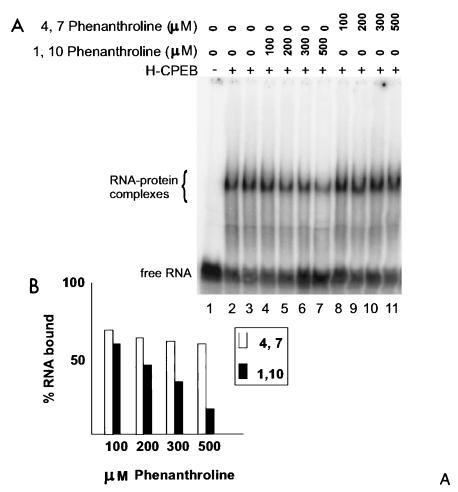
Because the cysteine-histidine region is reminiscent of a zinc finger, we sought to determine whether CPEB requires zinc for RNA binding. To do this, we performed RNA gel shifts with full-length CPEB that was incubated with either 1,10-phenanthroline or 4,7-phenanthroline. The former is a chelator of divalent cations, while the latter, its analog, cannot act as a chelator (1, 20, 21, 27). 1,10-Phenanthroline inhibited RNA binding by about 60% at 500 μM, whereas the analog had no effect at this concentration (Fig. 5A [lanes 7 and 11] and B), indicating that CPEB interaction with RNA requires a metal ion.
FIG. 5.
Metal chelation inhibits RNA binding by CPEB. (A) Wild-type CPEB was incubated overnight with various amounts of 1,10-phenanthroline or 4,7-phenanthroline (100 to 500 μM) and then used in a gel shift with radiolabeled B4 RNA. (B) Quantification of the binding data, plotted as percent RNA bound versus concentration of phenanthroline. Closed bars, reaction mixtures containing 1,10-phenanthroline; open bars, reaction mixtures containing 4,7-phenanthroline.
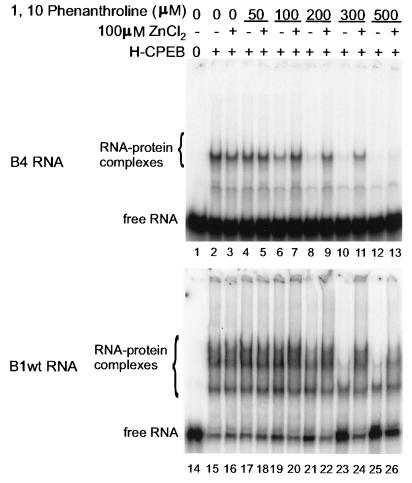
To determine whether zinc can restore RNA binding by CPEB, gel shift mixtures containing various amounts of 1,10-phenanthroline were supplemented with 100 μM ZnCl2. In these experiments, as little as 100 μM phenanthroline resulted in a loss of B4 RNA binding. (Note that this agent was freshly prepared for these experiments, whereas that used for Fig. 5 was older and partly inactive. The 4,7-phenanthroline, however, was freshly prepared.) Zinc very clearly restored RNA binding in the presence of up to 300 μM chelating agent (Fig. 6, lanes 5 to 13). Similar observations were made with B1wt RNA. In this case, a minimum of 100 μM phenanthroline was necessary to reduce binding. Zinc restored RNA binding in the presence of up to 500 μM chelating agent (Fig. 6, lanes 15 to 26). This result demonstrates that CPEB requires a metal cofactor for RNA binding, which is most likely coordinated by the cysteine and histidine residues noted above.
FIG. 6.
Zinc restores the ability of 1,10-phenanthroline-treated CPEB to bind RNA. H-CPEB was mixed with 50 to 500 μM 1,10-phenanthroline and 0 or 100 μM ZnCl2, incubated with radiolabeled B4 (top) or B1wt (bottom) RNA, and analyzed by RNA gel shift.
We also note that it might appear that 1,10-phenanthroline inhibited CPEB binding to two of the CPEs but not to the third (compare lanes 15 and 23 in Fig. 6). However, it should be borne in mind that this agent probably had not chelated all of the zinc; hence, the protein still contained enough of the metal to be partially active and thus occupied different CPEs in the population of RNA molecules, producing mostly a single shifted band. This is also similar to the case with mutant C259A, which retained some ability to bind RNA even in the presence of excess zinc (Fig. 4B). Thus, it, too, could induce a gel shift of RNA, but mostly interacted with a single CPE per RNA.
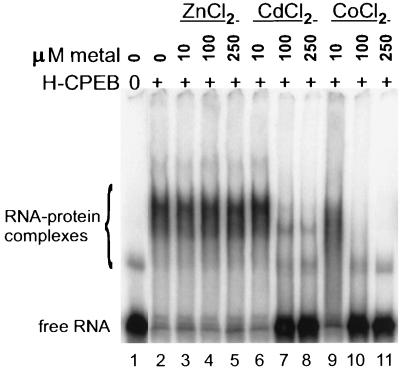
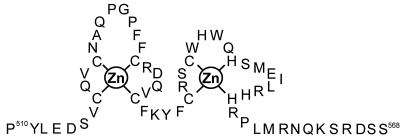
Metal-coordinating regions bind not only zinc but also other metals (29). To assess whether this is the case with the metal coordination region of CPEB, full-length protein (without prior treatment with 1,10-phenanthroline) was supplemented with several concentrations of ZnCl2, CdCl2, or CoCl2 and then used in a gel shift assay with B1wt RNA (Fig. 7). Although CPEB shifted RNA in the presence of ZnCl2 (lanes 3 to 5), it lost the ability to do so with either CdCl2 (lanes 6 to 8) or CoCl2 (lanes 9 to 11). These data suggest that Cd2+ and Co2+ can compete with Zn2+ and that they may alter the structure of the protein in such a way as to destroy RNA binding. We therefore conclude that Zn2+ is the likely physiological cofactor that is required for CPEB to bind RNA. Based on this finding, and on a general comparison with other zinc finger proteins (2), Fig. 8 shows a hypothetical structure of two zinc fingers in CPEB where the zinc ions are coordinated by four cysteine residues in finger 1 and two cysteine and two histidine residues in finger 2.
FIG. 7.
Other metals cannot substitute for zinc in promoting RNA binding by CPEB. Wild-type CPEB (H-CPEB) was incubated overnight with 10 to 250 μM ZnCl2, CdCl2, or CoCl2 and then analyzed for activity in an RNA gel shift with radiolabeled B1wt RNA.
FIG. 8.
Hypothetical structure of the zinc finger region of CPEB. CPEB is predicted to have two zinc fingers: the first Zn2+ coordinated by cysteines 517, 520, 529, and 534, and the second Zn2+ coordinated by cysteines 539 and 542 and histidines 547 and 555.
DISCUSSION
We show that the binding of CPEB to its specific recognition sequence, the UUUUUAU-type CPE, is mediated through two RRMs and a novel zinc finger domain. Although the RRMs, especially RRM1, contribute significantly to RNA binding, it is the metal coordination region, together with a Zn2+ cofactor, that is essential for this to occur.
Recombinant CPEB binds to RNA with a Kd of 130 nM, a value that is similar to those for other RNA binding proteins (5) and only slightly higher than the oocyte concentration of CPEB (32 nM [14]). A Hill plot of the kinetics of the binding of CPEB with the two-CPE-containing cyclin B1wt RNA presented no evidence of binding cooperatively (data not shown). In addition, the experiment presented in Fig. 2, which is based on a procedure used to determine whether GAL4 binds DNA as a monomer or dimer (7, 21), demonstrates that CPEB binds RNA as a monomer. Previous evidence indicates that RNAs containing multiple CPEs bind multiple CPEBs and are polyadenylated more efficiently, presumably due to an enhanced ability to attract polyadenylation complexes (39).
CPEB RRMs greatly enhance RNA binding.
Both RRMs of CPEB, each of which is composed of two canonical RNP domains (6), strongly contribute to RNA binding. The Kd of CPEB increases about 400-fold in the absence of RRM1 and nearly 7-fold in the absence of RRM2. It is not surprising that these RRMs differ in affinity for RNA given that these motifs, even among highly related proteins, can have different binding specificities. For example, HuD, a human RNA binding protein related to Drosophila ELAV, which regulates alternative splicing in neurons (17), contains three RRMs, two of which, RRM1 and RRM2, are necessary for specific RNA binding (9). However, Hel-N1, a close relative of HuD which contains virtually the same three RRMs, employs RRM3 for specific RNA binding (24). Moreover, even within the same protein, two RRMs can have different binding specificities. The yeast poly(A) binding protein contains four RRMs, yet only the two most amino-terminal ones bind poly(A) (5). Another, perhaps more extreme example is the U1 snRNP A protein, whose two RRMs bind entirely different RNAs: RRM1 binds to U1 snRNA, while RRM2 binds to pre-mRNA (25).
CPEB contains a novel zinc finger that is essential for RNA binding.
Several lines of evidence indicate that CPEB is a novel zinc finger-containing RNA binding protein. First, the remarkable conservation of the cysteine and histidine residues in the carboxy region of the proteins compared in Fig. 4 argues that it is a functionally important region. The distinct con- served motif CXNVCX2QX2P(F/Y)FCX4CFXY(F/Y)CX2 CWX3HXNHXPX2(R/K) has not been found in any previously characterized zinc-coordinating proteins to date. Second, mutations of putative zinc coordination residues, C529, C539, and H547, obliterate RNA binding. Third, chelation of metal ions by phenanthroline abolishes binding, and this is reversed by the addition of zinc ions. Fourth, addition of Cd2+ or Co2+ ions abolishes RNA binding, probably by competing with zinc for interaction with CPEB.
We propose that CPEB has two zinc fingers arranged in the structure shown in Fig. 8. In this model, one zinc atom is coordinated by the first four cysteine residues, and a second is coordinated by a subsequent two cysteine and two histidine residues. This structure is based on the fact that the eight putative metal coordinating residues are conserved from mammals to invertebrates and that alanine substitution of C529 or H547 greatly lowers, or completely destroys, RNA binding (Fig. 5). The most straightforward explanation of the effects of these alanine substitutions is that C529 resides in finger 1 and H547 resides in finger 2 and that both fingers are important for RNA binding. In addition, loop 1 (the residues between C517 and C529) contains five hydrophobic residues (A, P, P, F, F) that could stabilize the geometry of the metal binding center (2), although only the second proline is conserved in mouse, Drosophila, and C. elegans proteins. In loop 2 (the residues between C537 and H555), the two tryptophans may supply hydrophobic stability; the first tryptophan is conserved among all the animal groups mentioned above, while the second is absent only from the C. elegans protein. Although other structures are possible (2), this one is the most consistent with the available data.
The chelating agent 1,10-phenanthroline has been used under a variety of conditions to determine the necessity of metal cofactors for the binding of other proteins to nucleic acids (20, 23, 29). 1,10-Phenanthroline, but not its nonchelating 4,7 analog, inhibited CPEB RNA binding, which demonstrates a requirement for a metal cofactor. The fact that zinc restores RNA binding indicates that it is the physiological cofactor (Fig. 6 and data not shown). This is underscored by the observation that other metals such as Cd2+ and Co2+ actually destroy RNA binding (Fig. 7). This is probably caused by a competition between zinc and these other metals for coordination by the cysteine or cysteine-histidine tetrad, with the result that they introduce an altered geometry of the protein that prevents RNA recognition. Similar disruption of nucleic acid binding by proteins induced by various metals has been observed (20).
RNA binding proteins that utilize both RRMs and a zinc finger for binding specificity seem to be relatively rare. Perhaps the reason CPEB does so is that spurious binding to other sequences may lead to cellular catastrophe. The CPEB interaction sequence is UUUUAU, UUUUUAAU, or UUUUUUAU, depending on the RNA (39). However, eukaryotic 3′ UTRs tend to be UA rich, and thus CPE-like sequences are not uncommon in mRNAs that do not undergo cytoplasmic poly(A) elongation. If these mRNAs should erroneously become polyadenylated, they might be prematurely translated, which could abort normal development. Alternatively, a competition for CPEB between functional CPEs and sequences that are merely U rich might inhibit the polyadenylation and translation of, for example, c-mos mRNA, which would block oocyte maturation (12, 36). Highly selective recognition of appropriate CPEs by CPEB may therefore require a complex and discerning RNA recognition domain.
ACKNOWLEDGMENTS
L.E.H. was supported by a Charles A. King Trust administered through The Medical Foundation. This work was supported by grants from the NIH.
We thank B. Stebbins-Boaz for critical reading of the manuscript and other members of the Richter laboratory for useful discussions.
REFERENCES
- 1.Auld D S. Use of chelating agents to inhibit enzymes. Methods Enzymol. 1988;158:110–114. doi: 10.1016/0076-6879(88)58051-5. [DOI] [PubMed] [Google Scholar]
- 2.Berg J M, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 3.Bilger A, Fox C A, Wahle E, Wickens M. Nuclear polyadenylation factors recognize cytoplasmic polyadenylation elements. Genes Dev. 1994;8:1106–1116. doi: 10.1101/gad.8.9.1106. [DOI] [PubMed] [Google Scholar]
- 4.Bouvet P, Wolffe A P. A role for transcription and FRGY2 in masking maternal mRNA within Xenopus oocytes. Cell. 1994;77:931–941. doi: 10.1016/0092-8674(94)90141-4. [DOI] [PubMed] [Google Scholar]
- 5.Burd C G, Matunis E L, Dreyfuss G. The multiple RNA-binding domains of the mRNA poly(A) binding protein have different RNA-binding activities. Mol Cell Biol. 1991;11:3419–3424. doi: 10.1128/mcb.11.7.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 7.Carey M, Kakidani H, Leatherwood J, Mostashari F, Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989;209:423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 8.Cavaloc Y, Popielarz M, Fuchs J-P, Gattoni R, Stevenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35 kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung S L, Jiang S, Cheng S, Furneaux H. Purification and properties of HuD, a neuronal RNA-binding protein. J Biol Chem. 1996;271:11518–11524. doi: 10.1074/jbc.271.19.11518. [DOI] [PubMed] [Google Scholar]
- 10.Dalby B, Glover D M. Discrete sequence elements control posterior pole accumulation and translation repression of maternal cyclin B in Drosophila. EMBO J. 1993;12:1219–1227. doi: 10.1002/j.1460-2075.1993.tb05763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Moor L, Richter J D. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol Cell Biol. 1997;17:6419–6426. doi: 10.1128/mcb.17.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gebauer F, Xu W, Cooper G M, Richter J D. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO J. 1994;13:5712–5720. doi: 10.1002/j.1460-2075.1994.tb06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebauer F, Richter J D. Mouse cytoplasmic polyadenylylation element binding protein: an evolutionarily conserved protein that interacts with the cytoplasmic polyadenylylation elements of c-mos mRNA. Proc Natl Acad Sci USA. 1996;93:14602–14607. doi: 10.1073/pnas.93.25.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hake L E, Richter J D. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 15.Hake L E, Richter J D. Translational regulation of maternal mRNA. Biochim Biophys Acta. 1997;133:M31–M38. doi: 10.1016/s0304-419x(96)00039-x. [DOI] [PubMed] [Google Scholar]
- 16.Kim-Ha J, Kerr K, MacDonald P M. Translational regulation of oskar mRNA by bruno, an ovarian RNA binding protein. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 17.Koushika S P, Lisbin M J, White K. ELAV, a Drosophila neuron-specific protein, mediates the generation of an alternatively spliced neural protein isoform. Curr Biol. 1996;6:1634–1648. doi: 10.1016/s0960-9822(02)70787-2. [DOI] [PubMed] [Google Scholar]
- 18.Kuge H, Richter J D. Cytoplasmic 3′ poly(A) addition induces 5′ cap ribose methylation: implications for translational control of maternal mRNA. EMBO J. 1995;14:6301–6310. doi: 10.1002/j.1460-2075.1995.tb00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuge, H., G. G. Brownlee, P. D. Gershon, and J. D. Richter. Submitted for publication.
- 20.Labbé S, Larouche L, Mailhot D, Seguin C. Purification of mouse MEP-1, a nuclear protein which binds to the metal regulatory elements of genes encoding metallothionein. Nucleic Acids Res. 1993;21:1549–1554. doi: 10.1093/nar/21.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane D, Prentki P, Chandler M. Use of gel retardation to analyze protein-nucleic acid interactions. Microbiol Rev. 1992;56:509–528. doi: 10.1128/mr.56.4.509-528.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lantz V, Ambrosio L, Schedl P. The Drosophila orb gene is predicted to encode sex-specific germline RNA-binding protein and has localized transcripts in ovaries and early embryos. Development. 1992;115:75–88. doi: 10.1242/dev.115.1.75. [DOI] [PubMed] [Google Scholar]
- 23.Lee S P, Han M K. Zinc stimulates Mg2+-dependent 3′ processing activity of human immunodeficiency virus type 1 integrase in vivo. Biochemistry. 1996;35:5837–5844. doi: 10.1021/bi952056p. [DOI] [PubMed] [Google Scholar]
- 24.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lutz C, Alwine J. Direct interaction of the U1 snRNP-A protein with the upstream element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 26.McGrew L L, Dworkin-Rastl E, Dworkin M B, Richter J D. Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev. 1989;3:803–815. doi: 10.1101/gad.3.6.803. [DOI] [PubMed] [Google Scholar]
- 27.Navaratnam N, Morrison J R, Bhattacharya S, Patel D, Funahashi T, Giannoni F, Teng B-B, Davidson N O, Scott J. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993;268:20709–20712. [PubMed] [Google Scholar]
- 28.Paris J, Swenson K, Piwnica-Worms H, Richter J D. Maturation-specific polyadenylation: in vitro activation by p34cdc2 and phosphorylation of a 58-kD CPE-binding protein. Genes Dev. 1991;5:1697–1708. doi: 10.1101/gad.5.9.1697. [DOI] [PubMed] [Google Scholar]
- 29.Predki P F, Sarkar B. Effect of replacement of “zinc finger” zinc on estrogen receptor DNA interactions. J Biol Chem. 1992;267:5842–5846. [PubMed] [Google Scholar]
- 30.Richter J D. Dynamics of poly(A) addition and removal during development. In: Hershey J, Mathews M, Sonenberg S, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 481–503. [Google Scholar]
- 31.Richter J D, Smith L D. Reversible inhibition of translation by Xenopus oocyte-specific proteins. Nature. 1984;309:378–380. doi: 10.1038/309378a0. [DOI] [PubMed] [Google Scholar]
- 32.Robbie E, Peterson M, Amaya E, Musci T. Temporal regulation of the Xenopus FGF receptor in development: a translation inhibitory element in the 3′ untranslated region. Development. 1995;121:1755–1785. doi: 10.1242/dev.121.6.1775. [DOI] [PubMed] [Google Scholar]
- 33.Salles F, Lieberfarb M, Wreden C, Gergen J, Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994;266:1996–1999. doi: 10.1126/science.7801127. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Sanchez-Garcia I, Rabbits T H. The LIM domain: a new structural motif found in zinc-finger-like proteins. Trends Genet. 1994;10:315–320. doi: 10.1016/0168-9525(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 36.Sheets M D, Wu M, Wickens M. Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature. 1995;374:511–516. doi: 10.1038/374511a0. [DOI] [PubMed] [Google Scholar]
- 37.Standart N, Dale M, Stewart E, Hunt T. Maternal mRNA from clam oocytes can be specifically unmasked in vitro by antisense RNA complementary to the 3′ untranslated region. Genes Dev. 1990;4:2157–2168. doi: 10.1101/gad.4.12a.2157. [DOI] [PubMed] [Google Scholar]
- 38.Stebbins-Boaz B, Richter J D. Multiple sequence elements and a maternal mRNA product control cdk2 RNA polyadenylation and translation during early Xenopus development. Mol Cell Biol. 1994;14:5870–5880. doi: 10.1128/mcb.14.9.5870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stebbins-Boaz B, Hake L E, Richter J D. CPEB controls the cytoplasmic polyadenylation of cyclin, cdk2, and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 40.St. Johnston D, Nusslein-Volhard C. The origin of pattern and polarity in the Drosophila embryo. Cell. 1992;68:210–219. doi: 10.1016/0092-8674(92)90466-p. [DOI] [PubMed] [Google Scholar]
- 41.Strickland S, Huarte J, Belin D, Vassalli A, Rickles R J, Vassalli J-D. Antisense RNA directed against the 3′ noncoding region prevents dormant mRNA activation in mouse oocytes. Science. 1988;241:680–684. doi: 10.1126/science.2456615. [DOI] [PubMed] [Google Scholar]
- 42.Wickens M, Kimble J, Strickland S. Translational control of developmental decisions. In: Hershey J, Mathews M, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 411–450. [Google Scholar]
- 43.Wilson R, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]