Abstract
Homeodomain proteins are central regulators of development in eukaryotes. In fungi, homeodomain proteins have been shown to control cell identity and sexual development. Cryptococcus neoformans is a human fungal pathogen with a defined sexual cycle that produces spores, the suspected infectious particles. Previously, only a single homeodomain regulatory protein involved in sexual development, Sxi1α, had been identified. Here we present the discovery of Sxi2a, a predicted but heretofore elusive cell-type-specific homeodomain protein essential for the regulation of sexual development. Our studies reveal that Sxi2a is necessary for proper sexual development and sufficient to drive this development in otherwise haploid α cells. We further show that Sxi1α and Sxi2a interact with one another and impart similar expression patterns for two key mating genes. The discovery of Sxi2a and its relationship with Sxi1α leads to a new model for how the sexual cycle is controlled in C. neoformans, with implications for virulence.
Patterning during development in animals is controlled by a special set of coordinately regulated genes, the homeobox genes. These genes encode homeodomain proteins that act in concert with other cellular factors to establish patterns in diverse organisms. For example, in the fruit fly Drosophila melanogaster, where homeobox genes were first discovered, expression of homeodomain proteins specifies identity along the anteroposterior axis (17). In plants, homeodomain proteins regulate, among other things, the patterning of petals in flowers (25). These proteins are also important in controlling sexual differentiation, as has been documented in the mouse (45). The roles of homeodomain proteins have also been studied in fungi. In the yeast Saccharomyces cerevisiae, homeodomain proteins establish cell identity (27), and in the mushroom Coprinus cinereus, myriad homeodomain proteins are involved in mating and sexual development (11).
Sexual reproduction is a central part of eukaryotic life cycles and appears to be important for long-term success. Organisms with strictly clonal life cycles go extinct more rapidly than those with sexual cycles, suggesting that sex confers an advantage, although the reasons for the dominance of sexual reproduction are controversial (7). There are two prevailing hypotheses about how advantage is conferred: sexual recombination either allows the propagation of beneficial mutations in a population or facilitates the purging of deleterious mutations (37, 50). In either case, the majority of eukaryotes utilize a sexual phase in their life cycles. Even in organisms previously thought to not engage in this form of reproduction, cryptic or rarely utilized sexual cycles have been identified, emphasizing the importance of maintaining the machinery to carry out sexual reproduction.
The ascomycetous pathogenic yeast Candida albicans was thought to reproduce only clonally, until a cryptic mating process was identified (24, 36) and the genome sequence revealed conservation of nearly all of the genes required for complete sexual development (47). Another fungus pathogenic for humans, Cryptococcus neoformans, is a divergent basidiomycete that has also maintained the ability to undergo sexual development (31). In this case, although sexual reproduction has not been observed directly in the environment, strains of opposite mating types undergo sexual development in the laboratory (1, 23). This complex process in C. neoformans is initiated by the fusion of two haploid cells of different mating types (a and α). Fusion results in both an altered cell type and a change in growth pattern from yeast cells to dikaryotic filaments. The filamentous dikaryon has the capacity to form specialized meiotic structures and produce haploid spore products (30), suspected to represent the infectious particles inhaled into the lungs of human hosts.
Spores can be analyzed in the laboratory, providing a powerful genetic tool that has been invaluable in the identification of virulence traits that allow C. neoformans to promote disease. C. neoformans is known for causing fungal meningitis in immunocompromised patients, and an important virulence attribute is mating type (9). Almost all isolates of C. neoformans are of a single mating type (α). Animal experiments have shown that in some genetic backgrounds, α cells are more virulent than a cells (32, 33). This difference between a and α cells has prompted numerous studies to determine the molecular differences between the two mating types in an effort to understand the virulence process.
We previously discovered a key α-specific protein that establishes cell identity and controls sexual development in C. neoformans. Sex inducer 1α (Sxi1α) was shown to be necessary for proper sexual differentiation and sufficient to drive sexual development when expressed in a cells (22). Although this α-specific factor can promote sexual development, it was clear from genetic studies that a factor from a cells was also required for appropriate completion of the sexual cycle. Using a combination of molecular genetics and bioinformatics, we identify here a gene in the mating-type locus of a cells, SXI2a, with similarity to homeodomain DNA binding proteins. We show that this a-specific factor is essential for proper sexual development and, like SXI1α in a cells, can change the identity of α cells, causing them to adopt an a/α cell fate. Furthermore, two-hybrid and gene expression studies suggest that Sxi2a functions in concert with Sxi1α through a direct interaction to regulate transcription of key mating genes and induce sexual development. These studies reveal that an interaction between homeodomain proteins of two dissimilar classes governs sexual development in C. neoformans, reminiscent of previous and classic studies in both ascomycetes and basidiomycetes (S. cerevisiae, Ustilago maydis, C. cinereus, and C. albicans) involving homeodomain protein complexes, including a1/α2 and bE/bW. These discoveries establish a foundation upon which further insights into the control of cell identity and cell differentiation in this ubiquitous human fungal pathogen can be derived.
MATERIALS AND METHODS
Constructing sxi2adeletion strains.
To create the sxi2aΔ deletion strain, a PCR overlap approach was used as described previously (13). The 5′ flanking region was amplified with primers JOHE9021 (CAGGCAGCCAGATACAGAGG) and JOHE9023 (GGTCGAGCAACTTCGCTCGGTGATGGTAGAACTGGAGA), the 3′ flanking region was amplified with primers JOHE9024 (CCACCTCCTGGAGGCAAGTAGGAGATTTGTATGCAATAC) and JOHE9026 (GATTGTGTGTAACATTGGAG), and the URA5 selectable marker was amplified with primers JOHE9022 (TCTCCAGTTCTACCATCACCGAGCGAAGTTGCTCGACC) and JOHE9025 (GTATTGCATACAAATCTCCTACTTGCCTCCAGGAGGTGG). The PCR product was introduced into the MATa serotype D ura5 strain JEC34 by biolistic transformation, and transformants were selected on medium lacking uracil (−ura) and containing 1 M sorbitol (14, 46). Transformants were screened by PCR for the proper integration of the deletion construct, positive clones were confirmed by Southern blot analysis, and the resulting independent sxi2aΔ strains were designated CHY766 to CHY770.
PCR amplification.
All PCR amplifications were performed with an MJ Research DNA engine DYAD thermal cycler and the ExTaq PCR system (Intergen). Primers were used at a concentration of 0.4 μM, and templates for PCRs were titrated and evaluated empirically for each product. For diagnostic PCR to establish linkage of SXI2a with MATa primers to the SXI2a and SXI1α genes and a control sequence were used on genomic DNA templates. The following primers were used to show α-specific segregation: SXI1α, JOHE6710 (CGAAGGGCAAAGTCGAAAACG) and JOHE6711 (CCGAAATAATGGGAACTCC); SXI2a, JOHE9028 (ATGGGCAGCAACCTTGACATC) and JOHE9872 (GGTGAATGCAGCATGTTGGTTG); and for the cell type-independent control fragment, JOHE6712 (CTTACCAGTTTGGCTCCTTA) and JOHE6713 (CCTTCTTGGCTAAACCTTTC). The following primers were used to generate Northern probes: SXI1α, JOHE6710 and JOHE8207 (CGAGGATCCTTAACACGCTAGGCGCGG); SXI2a, JOHE9028 and JOHE9870 (GGATAGATCTTACCCCCTGAGGACTGT); MFa, JOHE6683 (TTCTTCGGCAGCCTCACTAT) and JOHE6684 (GAAAAGAGGTACGAGTAGAT); MFα; JOHE1204 (TTTTACGCTTTTTGCAGATTCCGCCAAA) and JOHE3242 (GACCACTGTTTCTTTCGTTCT); and GPD1 (glyceraldehyde-3-phosphate dehydrogenase), JOHE6524 (CGTCGTTGAATCTACCGGTG) and JOHE6525 (CACCAGCAATGTAAGAGATG). The amplification conditions for PCR were 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s. All PCR samples were visualized by standard DNA electrophoretic techniques (38).
Strain manipulations and media.
All strains used were of the serotype D background and are described in the relevant procedures. All were handled by standard techniques and with standard media as described previously (2, 40). Mating and self-filamentation assays were conducted on V8 medium at room temperature in the dark for 2 to 4 days. Filamentation was evaluated by observing the periphery of test spots on V8 medium. The mating tester strains used were JEC20 (a) and JEC21 (α) (33). For confrontation assays, strains were streaked after 2 days on yeast extract-peptone-dextrose (YPD) agar near one another on solid filament agar plates and incubated at room temperature in the dark for 6 days before they were photographed. Fusion assays were carried out with strains after growth for 2 days on YPD agar. Cells were resuspended in 1 ml of phosphate-buffered saline, quantitated in a spectrophotometer, and diluted to 108 cells/ml. Equal numbers of mating partners were mixed, a V8 agar plate was spotted with 10 μl, and plates were incubated at room temperature in the dark. After 24 h, the cells were scraped off the V8 plates, resuspended in 100 μl of water, and spread on selective plates. Plates were incubated at 30°C for 5 days, and the resulting colonies were counted. Test crosses were as follows: 1, a Ura+ nat− (JEC20) × α Ura− nat+ (CHY621); 2, Ura+ nat− (JEC20) × sxi1αΔ Ura− nat+ (CHY618); 3, sxi2aΔ Ura+ nat− (CHY768) × α Ura− nat+ (CHY621); and 4, sxi2aΔ Ura+ nat− (CHY768) × sxi1αΔ Ura− nat+ (CHY618). Fusants were selected on proline medium (1.7 g of yeast nitrogen base [without amino acids and without ammonium sulfate], 1 g of l-proline, 20 g of dextrose, 20 g of agar per liter) containing 100 μg of ml nourseothricin per ml. A cross between strains known to have fusion defects, a cpk1 Ura+ Ade− (JEC171) × cpk1 Ade+ Ura− (RDC3), resulted in no fusants after growth under selection on SD (synthetic medium plus dextrose [40])−adenine−uracil plates. To test the effects of ectopic expression of SXI2a, the SXI2a open reading frame (ORF) and approximately 300 bp of 3′ untranslated region were liberated from plasmid pCH269 with BglII. The resulting fragment was cloned into the BamHI site of the telomeric, GPD1-containing plasmid pRCD85 to create pCH285. This plasmid was digested with meganuclease I-SceI and transformed via electroporation into JEC43 (α ura5), JEC34 (a ura5), and CHY925 (sxi2aΔ ura5) to create the strains CHY1014, CHY1022, and CHY927, respectively. To test ectopic expression of SXI1α in a/a cells, plasmid pCH258 was transformed into strain CHY640 (a/a ura5/ura5) via electroporation and selected on SD plates without uracil to generate strain CHY815. Transformants were tested for self-filamentous behavior on V8 plates as described above. Spore analysis was carried out on strains CHY1014 and CHY815. Spores from V8 were microdissected and germinated on YPD. Each segregant was then tested for ploidy via fluorescence-activated cell sorter (FACS) analysis as described previously (41) and for auxotrophic markers. Twenty-two spores were evaluated from strain CHY1014, and all were found to be Ura− α haploids, indicating loss of the URA5-marked SXI2a plasmid. Thirty-nine spores were evaluated from CHY815, and all were a haploids whose marker phenotypes are indicated in Fig. 4.
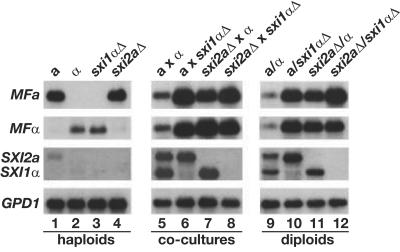
FIG. 4.
Sxi1α and Sxi2a regulate pheromone gene expression. A Northern blot showing the levels of selected transcripts in wild-type, sxi1αΔ, and sxi2aΔ strains after growth on V8 medium in three different contexts, indicated below the lane numbers: haploid strains alone, mixed mating cocultures, and diploid strains alone. The top two panels show RNA probed with the MFa and MFα ORFs. The third panel shows the same blot probed with portions of the SXI1α and SXI2a ORFs. The bottom panel shows the same blot probed with a portion of the GPD1 ORF. Lane 1, wild-type haploid a; lane 2, wild-type haploid α; lane 3, haploid sxi1αΔ; lane 4, haploid sxi2aΔ; lane 5, wild-type a cocultured with wild-type α; lane 6, wild-type a cocultured with sxi1αΔ; lane 7, sxi2aΔ cocultured with a wild-type α; lane 8, sxi2aΔ cocultured with a sxi1αΔ; lane 9, a/α diploid; lane 10, a/sxi1αΔ diploid; lane 11, sxi2aΔ/α diploid; lane 12, sxi2aΔ/sxi1αΔ diploid.
Two hybrid assays.
Constructs for SXI1α and SXI2a were generated with coding regions excised from cDNA-containing plasmids. SXI1α cDNA was cloned as a BamHI fragment from pCH271 into the BamHI site of pGAD-C1 (amino-terminal Gal4 activation domain [AD]), pGBD-C1 (amino-terminal Gal4 DNA binding domain [BD]), and pCDBD2 (carboxy-terminal Gal4 DNA BD). SXI2a cDNA was cloned as a BglII fragment from pCH274 into the BglII sites of pGAD-C1and pGBD-C1 and into the BamHI site of pCDBD2. The truncated versions of SXI2a were generated by PCR with primer JOHE9028.1 (GGATAGATCTATGGGCAGCAACCTTGACATC) in combination with JOHE11774 (GGATAGATCTGCAGAAGACACCAGTTTATC), JOHE11775 (GGATAGATCTAGGGCGAGAGTGCGGGACTTG), or JOHE11776 (GGATAGATCTAAGACCAAGTTCCATCTTTAG) to generate 1,725-, 671-, and 406-bp products, respectively. Products were digested with BglII and cloned into the BglII site of pGAD-C1. All constructs were sequenced across the ligation junctions or PCR-generated regions, and no deviations from the expected sequences were identified. Each pairwise combination of SXI1α and SXI2a fusions in the yeast two-hybrid vectors was transformed into cells of yeast strain PJ69-4a (26). Transformants were selected on medium lacking tryptophan and leucine (SD −leu −trp) and then tested for growth on medium lacking adenine (SD −leu −trp −ade) and medium lacking histidine (SD −leu −trp −his + 3AT). β-Galactosidase assays were performed as described previously (8).
Northern blot analysis.
RNA was prepared from C. neoformans cells with TRIZOL from Invitrogen. Strains were grown on solid V8 medium for 24 h (haploids) or 48 h (cocultures and diploids) at room temperature before they were harvested by scraping off the agar surface. Northern blots were carried out according to standard protocols (4) with 10 μg of total RNA used for each sample. The MFα, MFa, SXI2a, SXI1α, and GPD1 probes were generated by PCR as described above, and radiolabeled probes (Rediprime II kit from Amersham Pharmacia Biotech), were used in hybridization reactions as described previously (12) at 65°C.
Sequence manipulations.
Splice predictions of candidate gene sequences for SXI2a were facilitated with a Softberry algorithm (www.softberry.com). Sequence comparisons were conducted with the BLAST algorithm (3) against the Stanford C. neoformans genome sequence (C. neoformans Genome Project, Stanford Genome Technology Center, and The Institute for Genomic Research). Sequence analyses were conducted and alignments were generated with SeqWeb version 2 (Accelrys).
Nucleotide sequence accession number.
The SXI2a sequence can be accessed in GenBank under accession no. AY911308.
RESULTS
Identification of an a-specific factor in C. neoformans.
Experiments characterizing the role of SXI1α in sexual development revealed that although we had identified a key α-specific factor, an unidentified factor(s) from a cells was also required for sexual development. Ectopic expression of SXI1α in a cells resulted in the initiation and completion of the sexual development process; however, expression of this factor in haploid α or α/α cells did not lead to sexual development, indicating that a signal from a cells was required for the process. To test the hypothesis that predicted a-specific factors controlling sexual development are encoded by the mating-type (MAT) locus in a cells, we constructed a complete deletion of the entire 120,246-bp MATa allele in diploid a/α cells. Cells in which MATa was deleted no longer exhibited the self-filamentous phenotype characteristic of a/α cells, and in mating assays they behaved like haploid α cells (data not shown). These findings confirm that at least one of the factors required to direct formation of mating structures resides in MATa and prompted an intensive bioinformatic analysis of the MATa region to identify genes with similarity to transcription factors found in other fungal MAT loci.
We surmised that a missing a-specific factor might be encoded by a gene with synteny to the SXIIα gene in the α allele of the MAT locus. We utilized the BESTORF gene prediction algorithm from Softberry, Inc., to electronically produce predicted spliced cDNA products encoded by a 10-kb region. A candidate cDNA spanning a novel ORF was identified. This ORF, designated SXI2a, for Sex Inducer 2a, is located in the MATa allele and is the first gene in the locus, residing in the most-telomere-proximal position. This placement is analogous to the position of the SXI1α gene in the MATα allele (Fig. 1A). We also evaluated the MATa allele from another C. neoformans variety (var. grubii) and a sibling species (C. gattii) (16, 35) and found that this ORF was conserved in all three. The ORF is syntenic in only two, and in the third (C. neoformans var. grubii) it now resides at an internal position, likely as a result of gene inversions, which have been shown to be prevalent in the MAT locus (16).
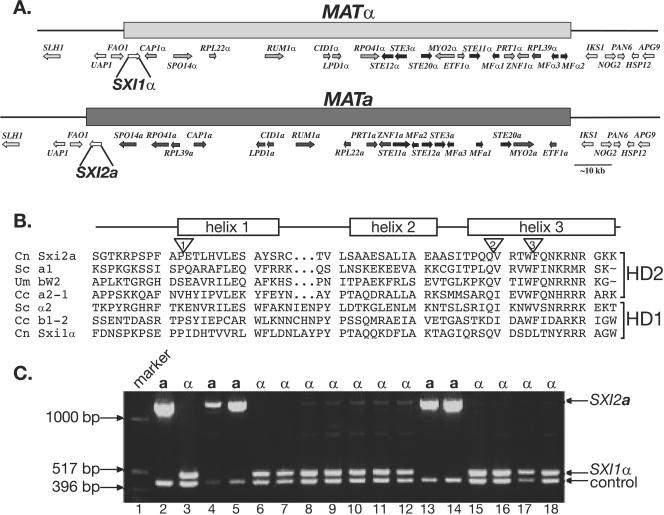
FIG. 1.
MATa contains a gene encoding the predicted homeodomain protein Sxi2a. (A) The SXI2a gene is located in the MAT locus. A schematic representation of the C. neoformans MAT locus shows MATα on the top and MATa on the bottom. The shaded bars represent the region of nonidentical DNA present in MATα and MATa. Genes within the locus are represented by shaded arrows. Each gene within the locus, with the exception of SXI1α and SXI2a, has a counterpart allele that encodes a similar, but not identical, protein in the opposite mating type. SXI1α and SXI2a are unique to their respective mating types. (B) Sxi2a is a homeodomain protein. The predicted homeodomain region of C. neoformans (Cn) Sxi2a is aligned with known homeodomains of other proteins: S. cerevisiae (Sc) a1, U. maydis (Um) bW2, C. cinereus (Cc) a2-1, S. cerevisiae α2, C. cinereus b1-2, and C. neoformans Sxi1α. A schematic representation of the homeodomain region shows the helices of a classic three-helix bundle found in homeodomains above the sequences. C. neoformans Sxi2a, S. cerevisiae a1, U. maydis bW2, and C. cinereus a2-1 fall into the HD2 class of homeodomains, whereas S. cerevisiae α2, C. cinereus b1-2, and C. neoformans Sxi1α are members of the HD1 family containing a 3-amino-acid insertion between helices 1 and 2. The inverted triangles denote the positions of introns in Sxi2a. Introns 1, 2, and 3, are 65, 66, and 294 bases, respectively. The third intron is conserved in many HD2 homeodomain proteins. (C) SXI2a is linked to MATa. Genomic DNA from segregants of a genetic cross was subjected to PCR analysis with primers to the ORF of SXI2a. Lane 1, marker; lanes 2 and 3, DNA from control a cells and α cells, respectively; lanes 4, 5, 13, and 14, DNA from segregants that mate as a cells; lanes 6 to 12 and 15 to 18, DNA from segregants that mate as α cells.
The predicted protein sequence encoded by the SXI2a ORF contains a domain with similarity to known homeodomain proteins. Identification of SXI2a was challenging because the most conserved region of the homeodomain (helix 3) sequence is interrupted by two introns, resulting in the production of a very small, 4-amino-acid exon (Fig. 1B). The presence of these clustered introns thwarted earlier attempts to identify an a-specific homeodomain factor by using conserved homeodomain sequences from the third helix in BLAST searches against C. neoformans genomic DNA sequence.
Homeodomain proteins from eukaryotes generally fall into two categories: those that conform to a more traditional homeodomain structure designated HD2 (e.g., S. cerevisiae a1 and C. cinereus a2-1) and those that are characterized by a 3-amino-acid insertion between helices 1 and 2 of the homeodomain, designated HD1 (e.g., S. cerevisiae α2 and C. cinereus b1-2) (6, 10). In previous studies, heterodimeric regulatory complexes formed by homeodomain proteins usually include a member from each homeodomain category (11). The well-characterized homeodomain proteins a1 and α2 from S. cerevisiae fall into the HD2 and HD1 classes and interact with one another to form a transcriptional repressor (20). The predicted homeodomain region for Sxi2a is similar to the HD2 class, and the analogous region in Sxi1α is similar to the HD1 class of homeodomains (Fig. 1B). In addition, many HD2 homeobox genes contain a conserved intron in the coding region for the third helix that lies between conserved tryptophan and phenylalanine residues (18). This intron is also conserved in SXI2a. Finally, an analysis of meiotic progeny from a cross between C. neoformans a and α strains revealed that the SXI2a gene is linked to the a mating type, supporting its location within MATa and confirming that SXI2a is an a-specific gene (Fig. 1C).
SXI2a controls sexual development.
Under mating conditions, haploid a and α cells of C. neoformans fuse with one another and undergo sexual development (the formation of filaments, basidia, and spores), as diagrammed in Fig. 2A. We deleted the predicted ORF for the SXI2a gene and tested sxi2aΔ strains in a series of assays to determine the role of Sxi2a in sexual development. In a coculture assay in which the deletion strain was mixed with a wild-type α mating partner under filamentation conditions, there was a dramatic reduction in filament formation (Fig. 2B, top row). This striking result mirrored that of a sxi1α deletion strain cocultured with an a mating partner. In both cases, the sxiΔ strains failed to undergo proper sexual development and produced only rudimentary filaments lacking basidia and spores, which were largely devoid of nuclei and clamp connections. In these respects, the filaments produced most closely resemble conjugation tubes normally produced during the initial stages of mating that facilitate partner recognition and cell-cell fusion.
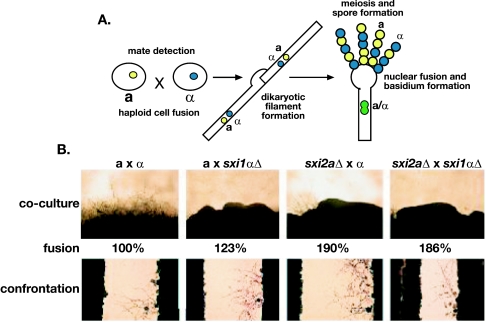
FIG. 2.
SXI2a is required for sexual development. (A) Sexual development in C. neoformans. When haploid a cells encounter haploid α cells at 25°C in the presence of either an unidentified plant factor in V8 medium or nitrogen limitation, the cells initiate a mating response and fuse with one another. The fused cells adopt a filamentous growth pattern in which the haploid nuclei do not fuse with one another. This dikaryotic filament grows in a polar manner, and adjacent filament cells are linked by fused clamp connections. Ultimately, in response to unknown signals, the terminal filament cell ceases extension and forms a rounded compartment at the distal end of the cell. It is in this basidium that nuclear fusion, meiosis, and sporulation occur. Meiotic products are packaged, and spores are extruded in long chains on the surface of the basidium. (B) sxi2aΔ strains can respond to and fuse with a mating partner but fail to form normal filaments. Wild-type, sxi1αΔ, and sxi2aΔ strains were tested in coculture (top row) (48 h), fusion (middle row) (24 h), and confrontation (bottom row) (48 h) assays to assess the nature of their defects in sexual development. Strain combinations are shown above the top row and indicate the strains used in each cross as well as the strains used in the fusion and confrontation assays. There was no significant difference in the extent of the limited rudimentary filaments produced in crosses by the mutant strains.
To further understand the nature of this mating defect, confrontation assays were conducted. In this assay, wild-type cells grown on low-nutrient solid medium respond to one another at a distance: α cells form filamentous projections toward a cells and a cells become enlarged (Fig. 2, bottom row). We tested sxi1αΔ and sxi2aΔ strains for the ability to respond to mating partners. In both cases, the cells responded like wild-type cells, indicating that their defects in filamentation do not reflect deficiencies in sensing or communicating with mating partners via mating pheromones (a-factor and α-factor). This was also the case in assays in which both the sxi1αΔ and sxi2aΔ strains were tested with each other (Fig. 2, bottom row). These observations suggest that the rudimentary filaments produced during coculture of the sxi1α and sxi2a mutants with strains of the opposite mating type are indeed conjugation tubes (rather than true dikaryotic mating filaments or the monokaryotic filaments produced by diploids during self-filamentous growth).
Finally, the sxiΔ strains were tested for the ability to fuse with a mating partner. In this assay, differentially marked strains were cocultured under mating conditions for 24 h and then placed under selection for genetic markers from each parent. The frequency of cell fusion was measured as the number of colonies formed on selective medium. The efficiency of fusion between wild-type a and α cells was designated 100%, and the efficiency of fusion by the deletion strains was evaluated as the percentage of fusion events relative to the expected wild-type frequency (Fig. 2, middle row). By this assay, the sxi1αΔ and sxi2aΔ strains exhibited no defects in cell fusion, while a mating-impaired, fusion-defective mutant strain (cpk1Δ) showed a >1,000-fold decrease in cell fusion, as reported previously (15). Thus, the dramatic defects we observe in sexual development of the sxiΔ strains are not a result of inefficient cell fusion but rather reflect a detrimental effect on subsequent steps in sexual development.
Expression of SXI2a in haploid α cells induces sexual development.
We showed previously that Sxi1α is sufficient to drive filamentation and sporulation in haploid a cells (22). We have carried out a similar analysis with Sxi2a. In this experiment, SXI2a was placed under the control of a constitutive C. neoformans promoter on an autonomously replicating plasmid. The plasmid was transformed into a, α, and a/α cells, and the resulting transformants were tested for the ability to undergo sexual development under inducing conditions. Strikingly, the α cells containing the GPD1-SXI2a plasmid formed filaments, basidia, and spores. A control strain containing an empty vector did not and behaved like a wild-type α strain, showing no filamentation under these conditions (Fig. 3A). This result shows that, like SXI1α, SXI2a is sufficient to induce filamentous behavior that is temporally delayed (data not shown) but morphologically indistinguishable from diploid sexual development. We note that the filaments produced were monokaryotic with unfused clamp connections like those produced by sexual reproduction of a/α diploid cells, rather than those produced by a and α cell mating that are dikaryotic with fused clamp connections (data not shown). In contrast, when the SXI1α and SXI2a genes were introduced into the sxi2aΔ or sxi1αΔ strains, respectively, neither was sufficient to induce self-filamentous growth (data not shown), indicating that the effects of Sxi1α and Sxi2a are dependent on the presence of both factors.
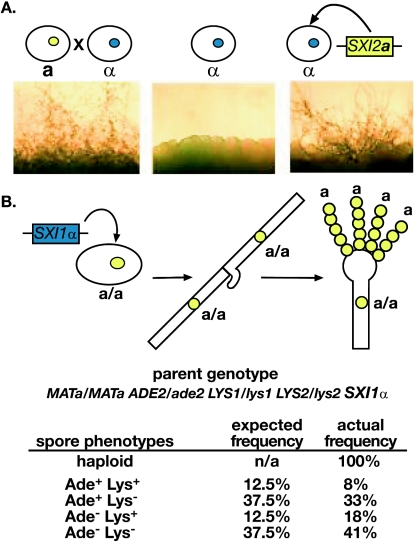
FIG. 3.
SXI2a induces sexual development in α cells. (A) Expression of SXI2a in haploid α cells results in filamentation and spore formation. Panels show the amount of filamentation achieved under mating conditions for each of the strain combinations indicated (from left to right): a wild-type cross between haploid a and α strains (a × α), a haploid α strain (α), and a haploid α strain expressing the a-specific gene SXI2a (α+SXI2a) (B) Schematic representation of filament formation in a/a diploid cells expressing SXI1α and a table showing analysis of spore products. The “parent genotype” is that of the homozygous a/a diploid used in the experiment. The “spore phenotypes” are those combinations expected to result from sporulation of the parent diploid. The frequencies presented result from an analysis of 39 random spores that were micromanipulated, germinated, and analyzed from the a/a+SXI1α strain under filamentation conditions. n/a, not applicable. The structure on the side of the filament cells represents an unfused clamp connection.
One caveat to these heterologous expression studies is that because the test strains were haploid, we could not exclude the possibility that the filamentation and sporulation we observed in the presence of SXI1α or SXI2a were due to the induction of a haploid filamentation and sporulation pathway that has been observed primarily in α cells. Known as monokaryotic fruiting this pathway is generally thought to be strictly mitotic (49). We evaluated this possibility by expressing the SXI1α or SXI2a gene in homozygous diploid cells. In the laboratory, stable, diploid C. neoformans strains have been created that grow as mononucleate, budding yeast (41). We previously created homozygous a/a and α/α diploid strains for ploidy studies and found that they behaved like their haploid counterparts: a/a cells mate with α cells, and α/α cells mate with a cells (22). We used these strains here to evaluate the effects of ectopic expression of SXI1α or SXI2a. We expressed SXI1α in a/a cells heterozygously marked at three genomic loci and observed the induction of filamentation, basidia formation, and sporulation. We analyzed the spore products that were generated and discovered that they were all haploid based on FACS analysis (data not shown), indicating that a chromosomal reduction event had occurred. The spores all mated as a cells and were recombinant for the markers tested. That is, the spores displayed phenotypes consistent with reassortment of the markers from the diploid parent: all of the predicted auxotrophic phenotypes were represented in roughly the expected frequencies for a standard cross between two haploid strains (Fig. 3B). If the expression of SXI1α had activated a mitotic sporulation pathway, we would have expected the production of diploid, prototrophic spores. We observed the same pattern in reciprocal experiments in which SXI2a was expressed in α/α cells (data not shown). Our findings support the hypothesis that the expression of Sxi1α and Sxi2a in the same cell induces complete sexual development, including meiosis and sporulation.
SXI1α and SXI2a impart similar patterns of pheromone gene regulation.
To understand how Sxi1α and Sxi2a control sexual development, we evaluated gene expression in the deletion strains. We analyzed mating conditions in three different settings: haploid cells grown alone, haploid cells grown in coculture with mating partners, and diploid cells grown alone. In each case, RNA was extracted and subjected to Northern blot analysis with probes for five different genes: SXI1α, SXI2a, the pheromone genes MFα and MFa, and the control gene GPD1.
We evaluated the expression patterns of SXI1α and SXI2a in wild-type and deletion strains. Our analysis confirms that, as expected, strains with gene deletions lack detectable levels of transcript from the targeted gene (Fig. 4, lanes 3, 4, 6 to 8, and 10 to 12). The data also show that in wild-type haploid cells, the transcript levels of SXI1α and SXI2a are barely detectable by Northern blotting (Fig. 4, lanes 1 and 2), although these transcripts were detectable by reverse transcription followed by PCR (data not shown). In contrast, both transcripts are up-regulated during coculture and in a/α heterozygous diploids, and they are readily detected by Northern blotting, indicating that gene products supplied by both a and α lead to the induction of SXI1α and SXI2a transcription. This up-regulation was not induced by mating pheromones as has been observed in other basidiomycetes (48) and instead appears to require cell fusion (C. M. Hull and J. Heitman, unpublished data). This regulation is also not dependent upon the presence of SXI1α and SXI2a. That is, strains in which SXI1α has been deleted still exhibit induction of SXI2a and vice versa (Fig. 4, lanes 6, 7, 10, and 11). These data indicate that SXI1α and SXI2a do not work in concert to activate their own transcription, and additional factors specific to a and α cells or regulated by factors specific to a and α cells must be responsible for inducing SXI1α and SXI2a.
We also investigated transcription of the MFα and MFa pheromone genes, which are induced by exposure to cells of the opposite mating type. In haploid cells, the levels of both the a and α pheromone transcripts are consistent with their mating type and do not appear to change in the sxi1αΔ or sxi2aΔ strains (Fig. 4, lanes 1 to 4). Under coculture conditions and also in stable diploid strains, the levels of pheromone transcript change dramatically in response to the deletion of either SXI1α or SXI2a. As was observed previously, transcript levels of both the MFa and MFα pheromone genes increase in dikaryotic and diploid strains in which SXI1α has been deleted, indicating that SXI1α is a formal repressor of pheromone gene transcription (Fig. 4, lanes 6 and 10). This pattern is also observed in the sxi2aΔ strains; the levels of both MFa and MFα pheromone transcript increase in dikaryotic and diploid strains in which SXI2a has been deleted (Fig. 4, lanes 7 and 11). The level of derepression is not substantially different in crosses or strains in which both SXI1α and SXI2a have been deleted (Fig. 4, lanes 8 and 12). These expression data suggest strongly that SXI1α and SXI2a regulate the same target genes and support the hypothesis that Sxi1α and Sxi2a act in concert to repress the transcript levels of mating genes and thereby govern sexual development.
SXI1α and SXI2a interact in a two-hybrid assay.
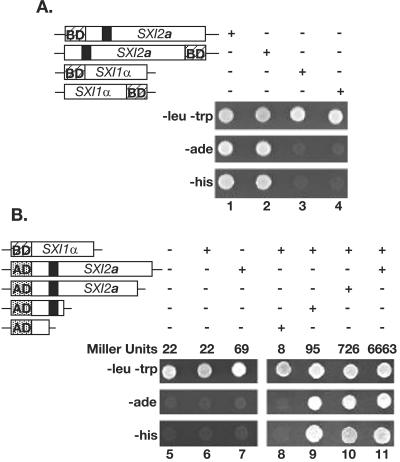
To test the model that Sxi1α and Sxi2a coordinately regulate gene expression by interacting with one another, we evaluated Sxi1α and Sxi2a for the ability to interact, using a yeast two-hybrid assay. The SXI1α and SXI2a ORFs were fused to the GAL4 AD or DNA BD and tested in S. cerevisiae for the ability to transactivate several different reporter genes. All combinations of fusion proteins were constructed: amino-terminal and carboxy-terminal fusions to either the AD or BD for each protein. SXI2a fused to BD results in transactivation in the absence of an AD construct (Fig. 5A, columns 1 and 2). The BD-SXI1α fusion construct did not activate the reporter genes on its own (Fig. 5A, columns 3 and 4) and was used to test for interaction between Sxi1α and Sxi2a. Figure 5B shows that when a DNA BD was fused to the amino terminus of Sxi1α and an AD was fused to the amino terminus of Sxi2a, there was activation of two independent reporter genes required for growth under selection (Fig. 5B, column 11), indicating an interaction between Sxi1α and Sxi2a. Controls (Fig. 5B, columns 5 to 7) confirmed that activation of the reporter was dependent on the presence of both GAL4 fusion proteins, and liquid β-galactosidase assays (reported in Miller units) confirmed the activation of the GAL4-lacZ gene as a third, independent reporter (Fig. 5B). Columns 9 and 10 show that this interaction is dependent on only a small portion of Sxi2a. C-terminal truncations of Sxi2a were tested for the ability to interact with Sxi1α, and the smallest interacting fragment contained the first one-third of the protein, including the homeodomain region (column 9). A smaller fragment in which the homeodomain had been deleted did not interact with Sxi1α (column 8). Although we cannot exclude models in which other proteins might also be present in Sxi1α-Sxi2a complexes, we conclude that the most likely model is that Sxi1α and Sxi2a interact directly, as is the case with other known HD1-HD2 heterodimeric complexes. We propose that this interaction leads to the coordinate regulation by Sxi1α-Sxi2a of sexual development in C. neoformans.
FIG. 5.
Sxi1α and Sxi2a interact with one other. The top panel in each set (−leu −trp) confirms the presence of each two-hybrid plasmid under selection. The middle and bottom panels indicate the presence or absence of activation of a biosynthetic reporter gene (either ADE2 or HIS3). The presence or absence of indicated constructs is represented with + or − over each panel. The fusion proteins tested are indicated in schematic form: the hatched box BD represents the Gal4 DNA BD, the speckled box AD represents the Gal4 AD, and the black box represents the Sxi2a homeodomain region. Growth in a test spot indicates activation of the reporter. (A) Lanes 1 to 4 reveal a transactivating property of Sxi2a in the absence of Sxi1α when fused to a DNA BD. (B) Lanes 5 to 11 confirm an interaction between Sxi1α and Sxi2a and define the region of Sxi2a necessary for interaction. The results of liquid β-galactosidase assays in each column are shown in Miller units.
DISCUSSION
We describe here the discovery of a cell-type-specific homeodomain protein required for sexual development in C. neoformans. Previous work had predicted the presence of an a-specific factor required for sexual development, but sequence analyses had failed to reveal its identity (22). Functional analyses narrowed the region of the genome in which this factor could reside to the a allele of the MAT locus, and bioinformatic approaches identified an a-specific gene located in MATa with similarity to homeobox-containing genes. Deletion of this gene, SXI2a, resulted in a profound defect in sexual development in C. neoformans, and ectopic expression of SXI2a in haploid α cells resulted in the induction of sexual development in the absence of a mating partner. Sxi1α and Sxi2a induce similar patterns of mating gene expression and interact with one another in a two-hybrid assay, supporting a model in which these proteins interact with one another to form a heterodimeric homeodomain transcriptional regulatory complex that specifies the dikaryotic state and thus promotes sexual development. We hypothesize that the absence of either Sxi1α or Sxi2a renders newly fused cells incapable of initiating a new developmental pathway. The factors are both required for specifying the a + α state of the newly formed dikaryon. Without these key proteins, fused cells are arrested and incapable of forming dikaryotic filaments, basidia, or spores. Thus, Sxi1α and Sxi2a are factors that cooperate to change the identities of fused mating partners and direct a new developmental program in C. neoformans.
Model for the regulation of sexual development.
We propose that Sxi1α and Sxi2a carry out the coordinated regulation of sexual development through a direct protein-protein interaction with one another. It has been shown in related fungi that the homeodomain proteins that control sexual development interact with one another to regulate gene expression. A well-studied example of this type of gene regulation is represented by the a1 and α2 proteins from S. cerevisiae (27). In this budding yeast, a1 is produced in a cells and α2 is produced in α cells. When a and α cells mate, the proteins form a heterodimeric complex that alters gene regulation: haploid-specific genes are repressed, and this change allows diploid cells to undergo meiosis and sporulation. This regulatory scheme is also found in U. maydis, where the homeodomain proteins bE and bW form a regulatory heterodimer and promote sexual development (19, 29, 39). In these systems as well as others, the proteins in the heterodimeric complex each fall into a different class of homeodomains and the complexes consist of one HD1 protein and one HD2 protein. In C. neoformans, Sxi1α and Sxi2a are the only predicted cell-type-specific homeodomain DNA binding proteins in the MAT locus required for the control of sexual development, and they fall into the two classes of homeodomains: Sxi1α is an HD1 protein, and Sxi2a is an HD2 protein. These features, along with our data that the proteins interact in a two-hybrid assay, make a compelling argument for a regulatory complex in C. neoformans development. Finally, both factors control expression of the pheromone genes in a similar way, further suggesting that they repress pheromone gene expression in a coordinated manner either directly or indirectly.
Of note regarding Sxi2a was its ability in a two-hybrid assay to activate transcription without a partner protein when fused to the Gal4 DNA BD. Homeodomain partners have been observed to carry out different functions in the heterodimeric complex, suggesting that the strategies for achieving regulation differ between complexes. For example, in C. cinereus, it appears that only one of the heterodimeric partners binds DNA and the other is essential for nuclear localization of the complex, and its DNA BD is not required (42). In S. cerevisiae binding to DNA is primarily mediated by contacts of the a1 protein, and the role of α2 appears to be to enhance DNA binding by a1 (43) and to recruit the Tup1-Ssn6 corepressor complex (28). Perhaps in C. neoformans, the most important role for Sxi1α is to interact with Sxi2a and shuttle it to the DNA of regulated genes to activate transcription. Although the formal roles for Sxi1α and Sxi2a in pheromone gene regulation are as transcriptional repressors, this effect could be indirect, and the complex could be acting upstream to activate repressors of pheromone genes. Understanding the nature of the interaction between Sxi1α, Sxi2a, and their targets will elucidate the mechanisms by which these key regulators regulate transcription and control development.
SXI1α and SXI2a are up-regulated during sexual development.
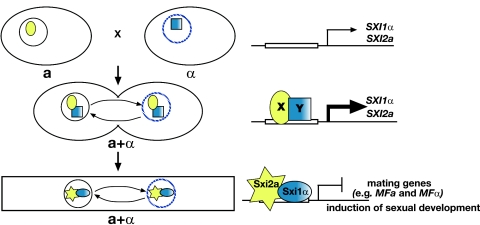
It is clear from our studies that Sxi1α and Sxi2a function after cell fusion. Deletion studies in diploids show that sexual development in these strains cannot take place in the absence of either Sxi1α or Sxi2a, even though both cell and nuclear fusion have taken place. Both proteins are clearly required subsequent to cell fusion for sexual development to continue. This is consistent with the finding that transcription of both SXI1α and SXI2a is up-regulated during mating and in diploids. These findings lead to a model of regulation in which Sxi1α and Sxi2a play very little, if any, role in haploid cells (Fig. 6). However, in response to the fusion of a and α mating partners, SXI1α and SXI2a are up-regulated. Interestingly, this regulation is independent of the presence of both factors, indicating that an Sxi1α-Sxi2a regulatory heterodimer is not responsible for inducing expression of these genes. Instead, other cell-type-specific regulatory factors must participate to induce SXI1α and SXI2a transcription. After induction, a Sxi1α-Sxi2a regulatory complex is then enabled to efficiently repress mating genes (like MFa and MFα) and signal the dikaryotic state. This repression likely prevents additional fusion of dikaryons with haploids and therefore maintains the integrity of the dikaryotic state (34). This altered cell type then has the capacity to undergo sexual development and complete its life cycle.
FIG. 6.
Model for the regulation of sexual development in C. neoformans. Model for the role of Sxi1α and Sxi2a in controlling sexual development. Large ovals represent cells. A solid circle represents the a nucleus, and a hatched blue circle represents the α nucleus. The yellow oval X and shaded blue square Y represent unknown a- and α-specific factors, respectively, that lead to the induction of SXI1α and SXI2a. The blue shaded oval represents Sxi1α, and the yellow star represents Sxi2a. (Top) SXI1α and SXI2a are expressed at low levels in haploid cells. (Middle) Following cell fusion, SXI1α and SXI2a expression is dependent upon and induced by factors from both a and α cells (X and Y). (Bottom) After induction, Sxi1α and Sxi2a form a heterodimeric complex that establishes the dikaryotic state and induces sexual development through the repression of mating genes.
Relationship between sex and virulence.
Although a relationship between sexual development and virulence has been established in some plant fungal pathogens, this link in the human fungal pathogens has been less clear. In the corn smut U. maydis, haploid cells must come into contact with a plant host before sexual differentiation can occur (5), but there is no clear equivalent requirement among any of the human fungal pathogens. To date, there is no evidence for complete sexual development in vivo for any human fungal pathogen, but there are clues that sexual development is an integral part of the virulence process. The clues are most clear in C. neoformans where mating type plays a significant role in how the organism interacts with its environment. Almost all isolates from both patients and the environment are α, suggesting some advantage over a strains (32). In addition, in some backgrounds, α cells are more virulent in animals than a cells, indicating that the factors that specify α cell identity (and thereby control mating and sexual development) also affect prevalence and virulence (33). This relationship is a complex one; however, as Sxi1α does not appear to play a prominent role in virulence (21). Understanding all of the determinants of mating type and the mechanisms of sexual development may ultimately reveal the cell-type-specific properties that contribute to virulence.
Sexual development in C. neoformans is also significant because human infections are thought to be caused by the products of this process, spores. It has been proposed that the small size of the spore makes it a likely candidate for the infectious particle in human infections (9, 44). If the spore is the infectious particle, understanding how sexual development is controlled promises to shed light on how these propagules are created and dispersed in the environment. Our discovery of SXI2a and subsequent studies of the relationship between Sxi1α and Sxi2a answer many questions about sexual development in C. neoformans and afford new opportunities to further explore the process of sexual development in a model human fungal pathogen.
Acknowledgments
We thank Robert Brazas, James Fraser, Alex Idnurm, and Robin Wharton for comments on the manuscript; Cristl Arndt and Joanne Ekena for technical assistance; and members of the Heitman laboratory, especially Rob Davidson, Connie Nichols, and Yen-Ping Hseuh, for their efforts. Special thanks go to Alex Idnurm for his pivotal contributions to the discovery of SXI2a.
This work was supported by an NIAID RO1 grant AI50113 to J.H. and NIAID program project grant AI44975 to the Duke University Mycology Research Unit. C.M.H. was supported by a Damon Runyon Cancer Research Fund Fellowship (DRG-1694) and a Burroughs Wellcome Career Award in the Biomedical Sciences. J.H. is a Burroughs-Wellcome Scholar in Molecular Pathogenic Mycology and an investigator of the Howard Hughes Medical Institute. The C. neoformans Genome Project, Stanford Genome Technology Center, was funded by the National Institute for Allergy and Infectious Diseases, National Institutes of Health [NIAID/NIH], under cooperative agreement U01 AI47087, and The Institute for Genomic Research was funded by the NIAID/NIH under cooperative agreement U01 AI48594.
REFERENCES
- 1.Alspaugh, J. A., R. C. Davidson, and J. Heitman. 2000. Morphogenesis of Cryptococcus neoformans. Dimorphism in human pathogenic and apathogenic yeasts. Contrib. Microbiol. 5:217-238. [DOI] [PubMed] [Google Scholar]
- 2.Alspaugh, J. A., J. R. Perfect, and J. Heitman. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit Gpa1 and cAMP. Genes Dev. 11:3206-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology, vol. 2:13. John Wiley and Sons, Inc., Boston, Mass.
- 5.Banuett, F. 1995. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet. 29:179-208. [DOI] [PubMed] [Google Scholar]
- 6.Burglin, T. R. 1997. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 25:4173-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burt, A. 2000. Perspective: sex, recombination, and the efficacy of selection—was Weismann right? Evol. Int. J. Org. Evol. 54:337-351. [DOI] [PubMed] [Google Scholar]
- 8.Cardenas, M. E., C. Hemenway, R. S. Muir, R. Ye, D. Fiorentino, and J. Heitman. 1994. Immunophilins interact with calcineurin in the absence of exogenous immunosuppressive ligands. EMBO J. 13:5944-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, D.C.
- 10.Casselton, L. A., and U. Kues. 1994. Mating-type genes in homobasidiomycetes, p. 307-321. In J. Wessels and F. Meinhardt (ed.), The Mycota, vol. 1. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 11.Casselton, L. A., and N. S. Olesnicky. 1998. Molecular genetics of mating recognition in basidiomycete fungi. Microbiol. Mol. Biol. Rev. 62:55-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davidson, R. C., J. R. Blankenship, P. R. Kraus, M. DeJesus-Berrios, C. M. Hull, C. D'Souza, P. Wang, and J. Heitman. 2002. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology 148:2607-2615. [DOI] [PubMed] [Google Scholar]
- 14.Davidson, R. C., M. C. Cruz, R. A. L. Sia, B. M. Allen, J. A. Alspaugh, and J. Heitman. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29:38-48. [DOI] [PubMed] [Google Scholar]
- 15.Davidson, R. C., C. B. Nichols, G. M. Cox, J. R. Perfect, and J. Heitman. 2003. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol. Microbiol. 49:469-485. [DOI] [PubMed] [Google Scholar]
- 16.Fraser, J. A., S. Diezmann, R. L. Subaran, A. Allen, K. B. Lengeler, F. S. Dietrich, and J. Heitman. 9 November 2004, posting date. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2:e384. [Online.] doi:10.1371/journal.pbio.0020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gehring, W. J. 1993. Exploring the homeobox. Gene 135:215-221. [DOI] [PubMed] [Google Scholar]
- 18.Gehring, W. J., M. Affolter, and T. Burglin. 1994. Homeodomain proteins. Annu. Rev. Biochem. 63:487-526. [DOI] [PubMed] [Google Scholar]
- 19.Gillissen, B., J. Borgemann, C. Sandmann, B. Schroeer, M. Bolker, and R. Kahmann. 1992. A two-component regulatory system for self/non-self recognition in Ustilago maydis. Cell 68:647-657. [DOI] [PubMed] [Google Scholar]
- 20.Herskowitz, I., J. Rine, and J. Strathern. 1992. Mating-type determination and mating-type interconversion in Saccharomyces cerevisiae, p. 583-656. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 2. Gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 21.Hull, C. M., G. M. Cox, and J. Heitman. 2004. The α-specific cell identity factor Sxi1α is not required for virulence of Cryptococcus neoformans. Infect. Immun. 72:3643-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hull, C. M., R. C. Davidson, and J. Heitman. 2002. Cell identity and sexual development in Cryptococcus neoformans are controlled by the mating-type-specific homeodomain protein Sxi1α. Genes Dev. 16:3046-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hull, C. M., and J. Heitman. 2002. Genetics of Cryptococcus neoformans. Annu. Rev. Genet. 36:557-615. [DOI] [PubMed] [Google Scholar]
- 24.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in a mammalian host. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 25.Irish, V. F. 1999. Patterning the flower. Dev. Biol. 209:211-220. [DOI] [PubMed] [Google Scholar]
- 26.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, A. D. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5:552-558. [DOI] [PubMed] [Google Scholar]
- 28.Keleher, C. A., M. J. Redd, J. Schultz, M. Carlson, and A. D. Johnson. 1992. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68:709-719. [DOI] [PubMed] [Google Scholar]
- 29.Kronstad, J. W., and C. Staben. 1997. Mating type in filamentous fungi. Annu. Rev. Genet. 31:245-276. [DOI] [PubMed] [Google Scholar]
- 30.Kwon-Chung, K. J. 1976. Morphogenesis of Filobasidiella neoformans, the sexual state of Cryptococcus neoformans. Mycologia 68:821-833. [PubMed] [Google Scholar]
- 31.Kwon-Chung, K. J. 1975. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia 67:1197-1200. [PubMed] [Google Scholar]
- 32.Kwon-Chung, K. J., and J. E. Bennett. 1978. Distribution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. Am. J. Epidemiol. 108:337-340. [DOI] [PubMed] [Google Scholar]
- 33.Kwon-Chung, K. J., J. C. Edman, and B. L. Wickes. 1992. Genetic association of mating types and virulence in Cryptococcus neoformans. Infect. Immun. 60:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laity, C., L. Giasson, R. Campbell, and J. Kronstad. 1995. Heterozygosity at the b mating-type locus attenuates fusion in Ustilago maydis. Curr. Genet. 27:451-459. [DOI] [PubMed] [Google Scholar]
- 35.Lengeler, K. B., D. S. Fox, J. A. Fraser, A. Allen, K. Forrester, F. S. Dietrich, and J. Heitman. 2002. Mating-type locus of Cryptococcus neoformans: a step in the evolution of sex chromosomes. Eukaryot. Cell 1:704-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 37.Rice, W. R., and A. K. Chippindale. 2001. Sexual recombination and the power of natural selection. Science 294:555-559. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schafer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 40.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 41.Sia, R. A., K. B. Lengeler, and J. Heitman. 2000. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet. Biol. 29:153-163. [DOI] [PubMed] [Google Scholar]
- 42.Spit, A., R. H. Hyland, E. J. C. Mellor, and L. A. Casselton. 1998. A role for heterodimerization in nuclear localization of a homeodomain protein. Proc. Natl. Acad. Sci. USA 95:6228-6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stark, M. R., D. Escher, and A. D. Johnson. 1999. A trans-acting peptide activates the yeast a1 repressor by raising its DNA-binding affinity. EMBO J. 18:1621-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sukroongreung, S., K. Kitiniyom, C. Nilakul, and S. Tantimavanich. 1998. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med. Mycol. 36:419-424. [PubMed] [Google Scholar]
- 45.Tilmann, C., and B. Capel. 2002. Cellular and molecular pathways regulating mammalian sex determination. Recent Prog. Horm. Res. 57:1-18. [DOI] [PubMed] [Google Scholar]
- 46.Toffaletti, D. L., T. H. Rude, S. A. Johnston, D. T. Durack, and J. R. Perfect. 1993. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J. Bacteriol. 175:1405-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tzung, K. W., R. M. Williams, S. Scherer, N. Federspiel, T. Jones, N. Hansen, V. Bivolarevic, L. Huizar, C. Komp, R. Surzycki, R. Tamse, R. W. Davis, and N. Agabian. 2001. Genomic evidence for a complete sexual cycle in Candida albicans. Proc. Natl. Acad. Sci. USA 98:3249-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urban, M., R. Kahmann, and M. Bolker. 1996. Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251:31-37. [DOI] [PubMed] [Google Scholar]
- 49.Wickes, B. L., M. E. Mayorga, U. Edman, and J. C. Edman. 1996. Dimorphism and haploid fruiting in Cryptococcus neoformans: association with the α-mating type. Proc. Natl. Acad. Sci. USA 93:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeyl, C., and G. Bell. 1997. The advantage of sex in evolving yeast populations. Nature 388:465-468. [DOI] [PubMed] [Google Scholar]