Abstract
A new type of DNA transposon, Mutyl, has been identified in the sequenced genome of the yeast Yarrowia lipolytica. This transposon is 7,413 bp long and carries two open reading frames (ORFs) which potentially encode proteins of 459 and 1,178 amino acids, respectively. Whereas the first ORF shows no significant homology to previously described proteins, the second ORF shows sequence similarities with various Mutator-like element (MULE)-encoded transposases, including the bacterial transposase signature sequence. Other MULE features shared by Mutyl include a zinc finger motif in the putative transposase, a 22-bp-long imperfect inverted repeat at each end, and a 9- to 10-bp duplication of its target site in the chromosome. Of the five copies of Mutyl present in the genome, one has a deletion of the first 8 bases, and the others are full length with a single base change in one element. The first potential gene of Mutyl, mutB, was shown to be expressed in exponentially growing cells. Its sequence contains a predicted intron with two 5′ splice sites, a single branch point, and two 3′ splice sites. Its mRNA is alternatively spliced, as judged by reverse transcription-PCR, and generates four mRNAs corresponding to protein-coding sequences of 128, 156, 161, and 190 amino acids. Of the three distinct lineages characterized in Y. lipolytica, strains from the German lineage and the French lineage do not carry Mutyl. A study of the distribution of Mutyl in strains of the French lineage evidenced a recent transposition event. Taken together, these results indicate that Mutyl is still active.
Transposable elements (TEs) are ubiquitous, as they can be found in most genomes. Their multiplication leads to an increased number of repeated sequences in the genomes that can be involved in various processes including genome rearrangements and homologous recombination. TEs are therefore responsible for a large part of the observed genome plasticity. Two types of TEs that differ in their propagation mechanism have been described; the retrotransposons that use an RNA intermediate and a reverse transcription step make up class I, whereas the DNA transposons that can propagate as DNA with the help of a transposase constitute class II. The distribution and copy number of these TEs vary enormously according to the organism as well as the type of element considered.
Among the class II elements, the Mutator (Mu) system, a family of elements discovered in maize, causes a high mutation frequency (33). The Mu system contains several elements which have unrelated internal sequences but possess conserved, similar-sized terminal inverted repeats (TIR) (see reference 40 for a review). These elements can be either autonomous (MuDR) or nonautonomous (Mu). Transposition of Mu elements is dependent on the autonomous MuDR elements. These harbor two open reading frames (ORFs), mudrA and mudrB. The nearly identical ∼215-bp TIRs of MuDR contain the promoters of the mudrA and mudrB genes. The mudrA gene likely encodes a transposase, as its expression is sufficient for somatic excision (24, 31). The mudrA gene product is able to bind to the TIRs specifically (4). Deletion of the mudrA gene abolishes all Mutator activity (24). The transposase encoded by the mudrA gene shows similarity with the transposase of bacterial insertion sequences (12). The role of the mudrB gene product is not yet clearly known, although it seems to be necessary for the integration process. Interestingly, all other Mutator-like elements (MULEs) lack the mudrB gene (23), even those that are able to transpose (38). MuDR, when it integrates in the genome, creates a target site duplication (TSD) of 9 bp. Since the discovery of MuDR, MULEs have been found in various angiosperms and were shown to be widespread in grasses (12, 22, 25, 26, 41). Recent genome sequencing projects like those for Arabidopsis thaliana and Oryza sativa revealed the high diversity of this class of transposons within these genomes (26, 42). The organization of a number of these elements departed from that of the complete MULEs, which contain MURA homologs, long TIRs, and direct repeats at the point of insertion. In particular, some elements carrying small imperfect TIRs or completely lacking inverted repeats at their extremities have been described previously (22, 42). A recent work provided for the first time evidence for the presence of an active MULE in an ascomycete, Fusarium oxysporum (8). The presence of related elements in other filamentous fungi like Magnaporthe grisea, Neurospora crassa, and Aspergillus fumigatus was also reported (8).
In hemiascomycetous yeasts, only retrotransposons have been described. The large majority of yeast TEs are long terminal repeat (LTR) retrotransposons of the Ty1-copia and Ty3-gypsy types (see reference 27 and references therein). Recently, non-LTR retrotransposons have been described in two hemiascomycetous yeasts, the Zorro families in Candida albicans (16) and the Ylli family in Yarrowia lipolytica (6). C. albicans seems to be a reservoir of various transposons, as a number of families of DNA transposons were found but not characterized further (http://biocadmin.otago.ac.nz/retrobase/home.htm).
Y. lipolytica is a dimorphic yeast which can grow on fatty acids and alkanes. In contrast to other yeasts, the Y. lipolytica genome shares several properties with higher eukaryotes, such as dispersion of the 5S RNA genes and the presence of a typical signal recognition particle 7S RNA (2). Y. lipolytica is heterothallic, and although it can display sexuality, most of its isolates are haploid, reminiscent of filamentous fungi. Despite the number of features shared by this yeast with higher eukaryotes and filamentous fungi, rRNA gene sequence phylogeny placed it unambiguously among the hemiascomycetous yeasts (21). Compared to other yeasts, Y. lipolytica seems to have an unusual transposon content. Ylli belongs to the L1 clade and shares most of the properties of known non-LTR retrotransposons (two ORFs, strong conservation of the organization and of the sequence of ORF2, and high copy number of mainly 5′ truncates) (6). Y. lipolytica harbors a Ty3-gypsy LTR retrotransposon, Ylt1, which has an unusually long LTR (37). Y. lipolytica also contains LTRs with no associated transposon (7) and a degenerated Ty3-gypsy-like element in some strains (27).
During the complete sequencing of the yeast Y. lipolytica (11), we detected a repetitive sequence that shares structural features with MULEs. Although transposons are ubiquitous, this is the first MULE as well as the first fully described DNA transposon in hemiascomycetous yeasts that we present here.
MATERIALS AND METHODS
Strains and media.
The Y. lipolytica strains used in this study are listed in Table 1. Cells were routinely grown in YPD medium (1% yeast extracts, 1% peptone, 1% glucose) at 28°C with shaking. Strains were provided by the Collection de Levures d'Intérêt Biotechnologique (http://www.inra.fr/clib/).
TABLE 1.
Yeast strains used in this study
| Strain | CLIB no.a | Originb |
|---|---|---|
| W29 | 89 | Paris sewage; wild isolate |
| H222 | 80 | German soil; wild isolate |
| CBS6124 | 183 | American corn processing plant; wild isolate (type strain) |
| CBS6124-1 | 77 | Spore of type strain CBS6124 |
| CBS6124-2 | 78 | Spore of type strain CBS6124 |
| CBS6125 | 79 | American corn processing plant; wild isolate |
| B204-12D | 257 | Derivative of an initial cross between CBS6124-1 and CBS6124-2 |
| CX161-1B | 153 | Derivative of an initial cross between CBS6124-2 and CBS6125 |
| E150 | 122 | Derivative of an initial cross between CBS6124-2 and W29 |
CLIB, Collection de Levures d'Intérêt Biotechnologique.
The origin of the strains was described previously by Barth and Gaillardin (3).
Sequencing and sequence assembly.
All the sequences used in this study were generated on the Y. lipolytica strain E150. Most of them were determined by Genoscope/CNS during the Genolevures project (11). Sequence assembly was performed by using the programs phred (version 0.980904.c) and phrap (version 0.960731) with a minscore of 14 and a mismatch of 30 (13, 14). The sequence compilation was visualized with Consed (18). Additional sequences were determined by Genome Express (Meylan, France).
Sequence analysis.
The BLAST search tool (1) was used to screen sequence databases for homology. Sequences were analyzed with various programs in the GCG environment (Genetics Computer Group, Madison, Wis.), including FASTA (30) and Staden. The alignment of the transposase domains described previously by Chalvet et al. (8) was used as a basis for the alignment shown in Fig. 2. Sequence alignments were generated by using CLUSTAL X (39) and were manually adjusted with Genedoc (http://www.psc.edu/biomed/genedoc). Phylogenetic trees were generated by the Phylip maximum-parsimony method (http://evolution.genetics.washington.edu/phylip.html) and the Phyml maximum-likelihood method (19). Trees were visualized with Treeview, version 1.6.5 (29).
FIG. 2.
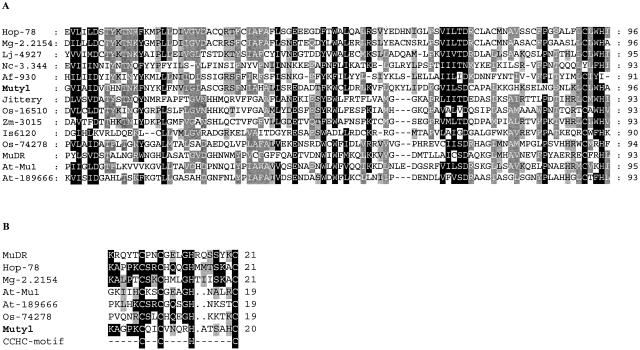
Amino acid alignment of two domains of MutA with related proteins. The sequences are from the transposase of bacterial element IS6120 (Mycobacterium smegmatis [GenBank accession no. M69182]), the MudrA transposase from Zea mays MuDR (accession no. M76978), putative transposases from the plants, Z. mays Zm-Jittery (accession no. AF247646), Z. mays Zm-3015 (accession no. AF466931), A. thaliana At-Mu1 (accession no. AC002883) and At-189666 (accession no. NM_113946), O. sativa Os-74278 (accession no. AC083943.8) and Os-16510 (accession no. AP005198), Lotus japonicus Lj-4927 (accession no. AP004927), and from the fungi Fusarium oxysporum Hop-78 (accession no. AY267761), M. grisea Mg-2.2154 (contig no. 2.2154), N. crassa Nc-3.344 (contig no. 3.344), A. fumigatus Af-930 (contig no. 930) and Mutyl (this work). (A) Comparison of domain 1 of the transposases. Conserved residues in 80% of the sequences are shaded in black, conserved residues in 60% of the sequences are shaded in dark gray, and conserved residues in 40% of the sequences are shaded in light gray. (B) Comparison of the zinc finger of the transposases. Only identical residues are shaded in black. Conserved residues are shaded in gray.
Northern blots, Southern blots, and DNA hybridization.
Total RNA was isolated by using an RNeasy kit (QIAGEN) according to the manufacturer's instructions. About 20 μg of RNA was size separated with 1.2% agarose gel electrophoresis in 1× FA buffer (20 mM morpholinepropanesulfonic acid, 5 mM sodium acetate, 1 mM EDTA [pH 7]) with 1.8% formaldehyde. Genomic DNA was extracted from yeasts as described previously (34). About 500 ng of genomic DNA was digested with ClaI or MluI enzymes according to the manufacturer's recommendations (Biolabs) and was separated on 1% agarose gel in 0.5× Tris-acetate-EDTA. After electrophoresis separation, total RNA and/or digested DNA was transferred onto GeneScreen nylon membranes (Perkin-Elmer Life Sciences) as described previously (43). DNA probes were PCR amplified on genomic DNA of strain E150. PCR products were purified on 1% low-melting agarose gel. DNA probes were labeled with [α-32P]dCTP by using a Megaprime labeling kit (Amersham Biosciences), and hybridizations were performed by using Denhardt buffer at 65°C (36). Final washes were performed at 65°C with 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate.
PCR.
PCR amplifications were run with a Perkin-Elmer 2400 thermocycler in 50 μl with 2.5 U of Ex-Taq DNA polymerase (Takara) in the recommended buffer and about 100 ng of genomic DNA as a template. PCR amplification of full-length elements was performed under the following conditions: 2 min at 94°C and 5 cycles of 30 s at 94°C, 30 s at 51°C, and 7 min at 72°C followed by 25 cycles of 30 s at 94°C, 30 s at 53°C, and 7 min at 72°C with 10-s increments at each cycle, with a final extension step of 5 min at 72°C. All other amplifications were performed as follows: 2 min at 94°C followed by 30 cycles of 30 s at 94°C, 30 s at 50°C, and 3 min at 72°C, with a final extension step of 3 min at 72°C. The oligonucleotides used for PCR amplification are listed in the supplemental material (see Table S1).
Reverse transcription (RT)-PCR was performed with Ready-To-Go RT-PCR beads (Amersham), according to the manufacturer's recommendations, on 1.5 μg of total RNAs prepared as described above. The first step was done under the following conditions: 30 min at 42°C and 5 min at 94°C with the MUTB_L oligonucleotide. This cDNA was used to seed a classical PCR described above with MUTB_U and MutBL oligonucleotides.
Nucleotide sequence accession number.
Sequence data from this article have been deposited in the EMBL and GenBank databases under accession no. AJ621548.
RESULTS
Description of Mutyl, a novel Y. lipolytica transposon.
During the complete genome sequencing project of Y. lipolytica (11), we detected a contig corresponding to a repeated sequence that displayed similarities with MULEs from O. sativa. All the sequences related to this new repeat were screened among all the sequences generated during the sequencing project using reiterated BLASTN and were then assembled as described in Materials and Methods. A total of 10 contigs were generated. The largest of these contigs (8,951 bp) was further shown to be 100% identical over 7,413 bp to other sequences located on different chromosomes of the completely assembled sequence of Y. lipolytica. This 7,413-bp DNA sequence was analyzed further. BLASTX (1) comparison to the sequences in the National Center for Biotechnology Information database revealed weak matches with the Hop element of F. oxysporum (44% similarity over 346 amino acids [aa]), several putative far-red impaired response proteins of O. sativa (43% similarity over 200 aa), and a Mutator-related transposase of A. thaliana (45% similarity over 174 aa).
Conceptual translation of the studied DNA sequence revealed two potential proteins of 459 and 1,178 aa, in the same orientation, on two different frames and separated by 722 bases, the second ORF displaying sequence similarity with the fungal and plant MULE transposases. The first ORF starts at base 1308. The second ORF ends at 467 bases from the end of the element (Fig. 1). These results suggested that Y. lipolytica harbors a repeated element encoding a protein with similarities with Mutator-like elements. We call this element Mutyl (for Mutator of Y. lipolytica), with the first 459-amino-acid-long ORF designated mutB and the ORF encoding the putative transposase designated mutA. Interestingly, Zea mays MuDR harbors two ORFs that are convergently transcribed from oppositely orientated promoters, whereas the structure of the Y. lipolytica element is closer to that of bacterial DNA transposons with two ORFs transcribed in the same orientation.
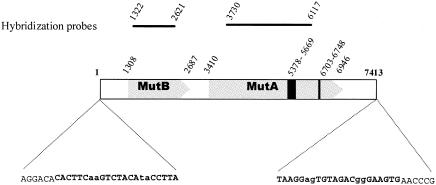
FIG. 1.
Structure of Mutyl. The different parts of Mutyl are depicted. The nucleotide numbers corresponding to the start and the stop of the ORFs and to various features are shown. The sequences of the ends of Mutyl are also shown. Boldface type indicates the TIRs. Bases that are variable between TIRs are in lowercase type. Underlined characters correspond to the element ends. Locations of probes (above), transposase domain 1 (black box), and transposase zinc finger (black line) are also shown.
The first ORF did not show any similarity in the BLAST searches, except for a stretch of 135 aa displaying 44% similarity with the ABF1 homologue from Kluyveromyces marxianus. This result might indicate that MutB, like the transcription factor ABF1, is able to bind DNA. Consistent with this observation, we found two putative zinc finger motifs of the C2HC type, CX10CX1HX2C and CX4CX1HX5C, in the sequence of MutB. These two zinc finger motifs are separated by 112 amino acids.
Mutyl belongs to the MULE class of DNA transposons.
Several structural features characterize the MULEs (23). The regulatory elements, MuDR, encode a transposase, which has a signature sequence homologous to that of bacterial transposases as defined previously by Eisen et al. (12). The presence of a zinc finger domain in the C-terminal part of the transposase was also described, but only in eukaryotic transposases. MULEs also possess inverted repeats (TIR) of variable size at the ends of the element.
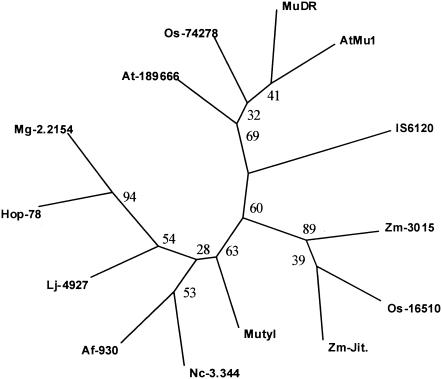
We found a 95-aa-long region in MutA which displayed similarity with the transposase sequence signature; an alignment of the region of different elements, derived previously (8), is shown in Fig. 2A. By further analyzing the sequence of MutA, we detected a zinc finger domain of the C2HC type matching that of various MULEs (Fig. 2B). Phylogenetic analysis was performed on the conserved signature sequence of the transposases of the elements shown in Fig. 2A. The reconstructed phylogenetic tree shown in Fig. 3 was obtained by using maximum parsimony. Essentially similar results and equivalent bootstrap values were obtained by using maximum-likelihood methods (data not shown). Plant elements were split into two different clades, and another clade contained the MULEs from ascomycetes, as reported previously (8). The Y. lipolytica element is distinct from the two plant clades. The phylogenetic tree also indicates that Mutyl is part of the MULE superfamily. A plant element from Lotus japonicus was also associated with the clade of the fungal elements, confirming that phylogenies of the MULEs and those of their hosts are not always congruent (23).
FIG. 3.
Phylogeny of several MULEs. Shown is an unrooted phylogenetic tree of various MULE transposases and transposase of IS6120 derived from parsimony analysis using the amino acid alignment of domain 1, as shown in Fig. 2A. Bootstrap values based on 100 replicates are indicated at the nodes. An essentially similar tree was obtained with the maximum-likelihood method.
We also looked for repeated inverted sequences at the ends of the element (Fig. 1). We found a 22-bp-long stretch 6 bp away from each extremity that is imperfectly repeated. The 5′ imperfect repeat, CACTTC AA GTCTACA T ACCTTA, is found inverted at the 3′ end with the doublets GG and the base G replacing the doublets and the base of the 5′ end, AA and T, giving CACTTC GG GTCTACA G ACCTTA. Initially discovered MULEs were shown to carry a ∼100-bp-long TIR. Since then, several elements from plants including A. thaliana, O. sativa, and various grasses were classified as MULEs even though they lack repeated sequences at their extremities (22) or carry short or imperfect TIRs (GenBank accession no. AJ238507.1 [25]). Comparison of the imperfect TIRs of Mutyl and already-described TIRs did not provide any information on the conservation of the transposase binding site as reported previously (25). Nevertheless, a 7-base-long sequence, CATACCT, common to the putative 5′-end TIR of Mutyl and that of Hop may be significant. Only MuDR seems to carry two ORFs (23), and these are transcribed convergently from the promoters carrying TIRs. In this respect, Mutyl displays a very unusual structure. It carries two ORFs transcribed in the same orientation. Sequences upstream of each ORFs do not display similarity, suggesting that the two corresponding Mutyl promoters are different in structure. Indeed, no peculiar sequence was detected upstream of the mutB gene, whereas two 20-base-long hairpin structures with an energy of formation of −15.8 kcal/mol, separated by 29 bases with a single base change, GTACT/A GTACAGTACTTGTAC, were found in the promoter region of mutA. A third similar but shorter hairpin, GTACAGTACTGTAC (energy of formation of −8.8 kcal/mol), was found 193 bases downstream of the second hairpin and 73 bases downstream of the ATG of mutA. This could imply that if this latter conserved hairpin structure is involved in the regulation of the transcription of mutA, the actual start of MutA is further downstream of the ATG selected in our study.
Strong sequence conservation indicates that Mutyl is a recently active element.
We analyzed the whole genome of Y. lipolytica for the presence of Mutyl in the completed Y. lipolytica E150 sequence by using BLASTN (1). We found three copies of Mutyl identical to that of the sequence compilation of Mutyl. A fourth copy located on chromosome III carried a single-base-pair change at bp 3748 due to a transversion from A to C, resulting in the putative transposase sequence in a replacement of a threonine residue (ACG) by a proline residue (CCG). Another copy of Mutyl was found on chromosome V with an identical sequence, except for a deletion of the first 8 bp of the 5′ end of the element. Overall, the strong sequence similarity of the element indicates that it was recently active or that it is still active. When we analyzed the E150 sequence, we found two sequences with similarity to Mutyl: a 550-bp-long sequence (80% identity at the DNA level) on chromosome III and a 3,352-bp-long sequence (88% identity at the DNA level) on chromosome I.
MULEs are known to create a duplication of their integration site (TSD). We analyzed the sequence in the vicinity of each Mutyl insertion in E150. We found for two full-length copies an exact duplication of 9 bp and for two other copies an exact duplication of 10 bp. For the 5′-truncated copy, we could not find any common sequences because the deletion very likely encompassed more than the Mutyl extremity. Mutyl seemed to have inserted into a very G-rich region, as the probable TSD consists of a stretch of 9 of 10 Gs that bound the 3′ end of the element (Table 2). The G richness may be the cause of the observed rearrangement. The presence of a TSD flanking all the full-length copies of Mutyl and the size of this TSD in all the cases provide more evidence for the classification of Mutyl as a MULE.
TABLE 2.
Characteristics of the Mutyl copies
| Mutyl copy | Length (bp) | TSD | TSD type | Chromosome | Position (bp) | Orientation |
|---|---|---|---|---|---|---|
| 1-1 | 7,413 | ACTTCAATGT | 9-bp perfect | I | 1648460-1655872 | Plus |
| 2-1 | 7,413 | GTAGAAGGG | 9-bp perfect | II | 832949-840361 | Plus |
| 3-1 | 7,413 | GGGGTAAAAC | 10-bp perfect | III | 2113080-2120492 | Minus |
| 4-1 | 7,413 | CAGTATGTAC | 10-bp perfect | IV | 3560088-3567500 | Minus |
| 5-1 | 7,405 | GGGGGGGGGGa | Unknownb | V | 2883442-2890846 | Plus |
Sequence flanking the 3′ end of Mutyl 5-1.
The sequence flanking the 5′ end of Mutyl 5-1 is very likely deleted (see the text).
The genomic environment of these elements was also examined by using TBLASTN and BLASTP to see whether Mutyl was inserted into ORFs. None of the copies of Mutyl were found to be inserted into a detectable ORF. This is not surprising considering the low copy number of Mutyl in the sequenced strain and the overall low gene density of Y. lipolytica, which is below 50% (11). The copy of Mutyl on chromosome II was found 1,002 bp upstream of a full-length Ylt1. None of the other copies were found in the vicinity of any transposon.
Mutyl is not present in all lineages of Y. lipolytica.
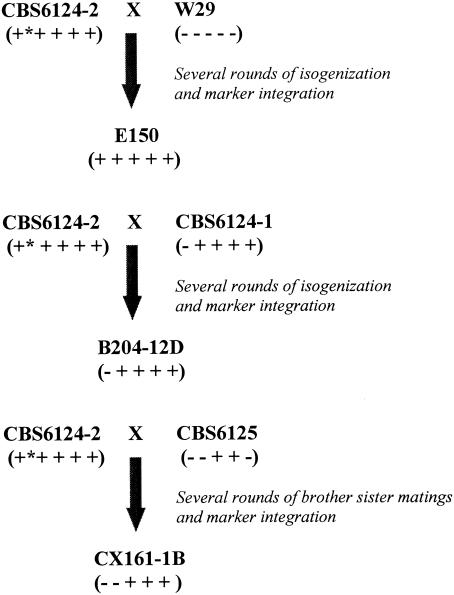
Genetic studies of Y. lipolytica defined three lineages from different geographical origins (3). An American lineage was defined by the type strain CBS6124 (the only diploid strain isolated from the wild), a French lineage was defined by strain W29, and a German lineage was defined by strain H222 (Table 1). Work aiming at the isogenization of strains to allow genetic studies was performed over the years. This work consisted of three initial crosses between a spore of the type strain, CBS6124-2 with W29, with CBS6124-1 (another spore from the type strain) or with CBS6125 (a strain isolated from the same corn processing plant as the type strain). Rounds of back-crosses, mutagenesis, and marker transformation followed which led to the strains we used in this study (Fig. 4) (3).
FIG. 4.
Schematic representation of the various lineages with respect to the presence of Mutyl. The original crosses are depicted. The presence (+) and absence (−) of the E150 Mutyl copies 1 to 5 from left to right are shown within brackets. An * indicates that the element is not full length.
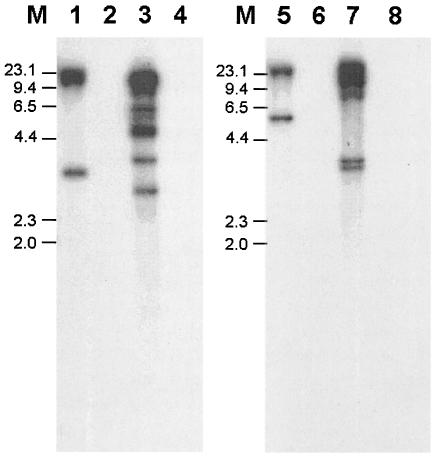
Distribution of transposons in Y. lipolytica was found to vary according to the origin of strains. Indeed, Ylt1, the Ty3-gypsy LTR retrotransposon, was found only in the American lineage and was absent from strains from the French and the German lineages (20). This report led us to perform Southern blots with a 2,370-base-long probe in the mutB gene against genomic DNA from various strains known to belong to the three characterized lineages (3). Analysis of the Southern hybridization shown in Fig. 5 revealed that the Mutyl probe gave strong signals with an isolate from the American lineage (strain CX161-1B) as well as with E150 but not with an isolate from the German lineage (strain H222) or French lineage (strain W29). The presence of Mutyl in E150 is very likely due to the chromosomes inherited from the American strain. Figure 5 also shows that Mutyl copy number is higher in CX161-1B than in E150. This finding is in agreement with the fact that E150 inherited Mutyl carried by chromosomes from the strains of American origin and W29 chromosomes devoid of Mutyl.
FIG. 5.
Hybridization of a Mutyl-specific probe to Y. lipolytica genomic DNA from various strains. Genomic DNA from strains E150 (lanes 1 and 5), W29 (lanes 2 and 6), CX161-1B (lanes 3 and 7), and H222 (lanes 4 and 8) was digested by ClaI (lanes 1 to 4) or MluI (lanes 5 to 8), separated on gel, transferred onto a nylon membrane, and hybridized with 1,282 bp of the mutB gene, as described in Materials and Methods. M indicates lambda DNA digested by HindIII, and the corresponding fragment sizes are indicated.
Mutyl underwent transposition recently.
This observation prompted us to compare the insertion sites between various strains. Oligonucleotidic primers were designed (i) on the flanking regions of each element of E150 to amplify the full-length element and (ii) within the Mutyl sequence to amplify the 5′ end and the 3′ end of Mutyl together with adjacent genomic DNA (Fig. 6A and see Table S1 in the supplemental material). Three PCR amplifications were therefore performed for each of the Mutyl copies detected in E150 on the following strains: E150 (as a control) and its two parental strains W29 and CBS6124-2, CBS6124-1, CBS6125, CX161-1B, and B204-12D. Results of the PCR amplifications are summarized in Table 3.
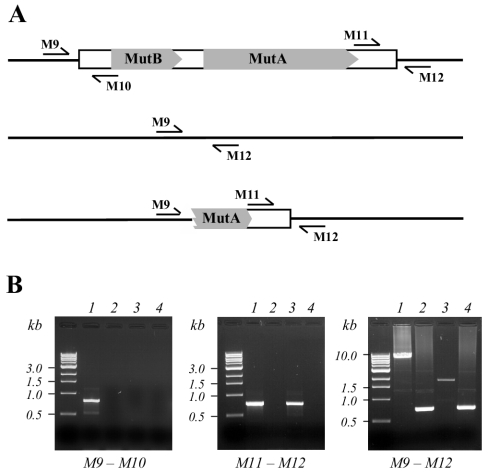
FIG. 6.
PCR amplifications at the Mutyl 1-1 locus in various strains. (A) Schematic representation of the PCR amplifications; top, localization of the primers used for the amplification of the full-length elements and of the junction element-chromosome; middle, no insertion; bottom, predicted structure of the Mutyl 1-1 locus in strain CBS6124-2. (B) PCR amplification on various genomic DNAs with the primer pairs shown in A at the Mutyl 1-1 locus. The PCR products separated on agarose gel as described in Materials and Methods are shown. The primers used for the PCR amplification are under each picture of the gels. Lanes: 1, E150; 2, W29; 3, CBS6124-2; 4, CBS6124.
TABLE 3.
Presence of Mutyl copies in different Y. lipolytica strains
| Strain |
Mutyl copiesa
|
||||
|---|---|---|---|---|---|
| 1-1 | 2-1 | 3-1 | 4-1 | 5-1 | |
| E150 | + | + | + | + | + |
| W29 | − | − | − | − | − |
| CX161-1B | − | − | + | + | + |
| B204-12D | − | + | + | + | + |
| H222 | − | − | − | − | − |
| CBS6124-1 | − | + | + | + | + |
| CBS6124-2 | +b | + | + | + | + |
| CBS6125 | − | − | + | + | − |
A + denotes the presence of Mutyl copies, and a − denotes the absence of Mutyl copies.
Deleted copy.
We found that in the original cross involving CBS6124-2 and CBS6125, which led to CX161-1B, the latter strain received Mutyl 5-1 from CBS6124-2, as no Mutyl is present at the same location in CBS6125. On the other hand, the absence of Mutyl on chromosome II in CX161-1B indicated that the CBS6125 allele was inherited.
Analysis of the presence of Mutyl in the two strains CBS6124-1 and CBS6124-2 (spores of the diploid type strain CBS6124) showed that they have the same content at four of the five tested loci. The case of E150 is also informative: all the Mutyl copies come from CBS6124-2, the sole parent that harbors Mutyl. However, the Mutyl insertion on E150 chromosome I seems to be unique to this strain. Interestingly, we found that the element was partly deleted in the parental strain CBS6124-2, as a PCR amplification of the full-length element led to a 1.8-kb PCR product instead of the expected 8.1-kb PCR product (Fig. 6). In addition, amplification of each end with the adjacent genomic sequences (Fig. 6, primer pairs M9-M10 and M11-M12) led to one PCR product of a size similar to that of E150 at the 3′ end but no detectable PCR product at the 5′ end (Fig. 6B). This result suggests that a genomic rearrangement has recently taken place, i.e., after the original crosses between W29 and CBS6124-2 that ultimately led to E150 (15).
To confirm this result, we performed the PCR amplification with the primers M9 and M12 on the type strain CBS6124 that is predicted to carry two alleles at the Mutyl 1-1 locus. We found one PCR product of around 0.7 kb which corresponded to the expected 749-base product (no insertion) but no PCR product corresponding to a full-length Mutyl insertion (Fig. 6). This result shows that the diploid parental strain that gave the spores CBS6124-2 and eventually E150 does not carry an element on each chromosome I. It thus shows that the presence of an element on CBS6124-2 chromosome I and on E150 chromosome I is the result of a recent transposition event. This transposition event could have taken place at meiosis of CBS6124 to give a spore with a new insertion in chromosome I. This was observed for Tys in Saccharomyces cerevisiae (28, 32). This result clearly indicates that Mutyl is still active; the low number of elements and the few observed transpositions that we observed support a strict regulation of Mutyl propagation.
By using PCR amplification, we also found that the two sequences that show limited homology to Mutyl are present at the same location in the eight strains tested, without detectable size polymorphism (data not shown). This finding indicates that these two sequences could be degenerate copies of an element related to Mutyl.
Mutyl elements are transcriptionally active.
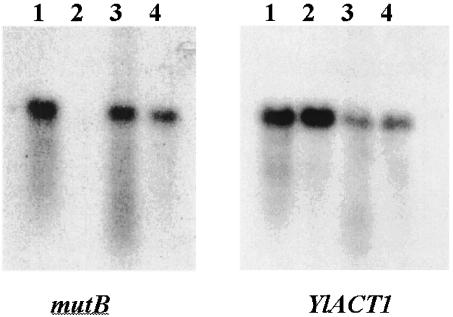
In order to see whether the genes coded by Mutyl are transcribed, we performed a Northern blot with total RNAs isolated from various strains of Y. lipolytica grown in YPD medium at 28°C as described in Materials and Methods. PCR products from part of the mutB and of the mutA genes were used as probes (Fig. 1 and see Table S1 in the supplemental material). Figure 7 shows that when a probe of part of the mutB gene was used, a strong signal was seen in RNA from strains CX161-1B, B204-12D, and E150 but not in control strain W29 as expected. A strong signal was also obtained for all the strains when a probe of the actin gene was used as a control. On the other hand, no clear signal was obtained when a probe covering part of the mutA gene was used in all the strains (data not shown). This result indicates that only the mutB gene is expressed under the conditions tested. This result is reminiscent of the regulation of protein production seen for LTR retrotransposons in which the reverse transcriptase is less abundant than the RNA binding protein. For instance, in yeast Ty1, programmed translational frameshifting reduces the production of TyB to 80% of that of the 5′ TyA (9). Our results indicate that the gene coding for the transposase in Mutyl is under strong genetic regulation; an attractive possibility is that its transcription is under negative control of the mutB gene product.
FIG. 7.
Northern blot of Mutyl-specific probes to separation of total RNA from Y. lipolytica. Total RNAs from E150 (lane 1), W29 (lane 2), B204-12D (lane 3), and CX161-1B (lane 4) were separated on gel, transferred onto a nylon membrane, and hybridized with 1,282 bp of the mutB gene or 382 bp of the actin-encoding gene from strain E150 as described in Materials and Methods.
The mutB mRNA is alternatively spliced.
Interestingly, the ORFs encoded by Mutyl are very large. MutA is much bigger than bacterial or eukaryotic transposases and similar in size to putative far-red impaired response proteins of O. sativa. With 459 aa, MutB of Mutyl is much larger than maize MURB (207 aa). The size of MutB and the presence of introns in the ORFs of MuDR prompted us to look for intronic sequences in the ORFs of Mutyl. Interestingly, two putative 5′ splice sites GUGAGU separated by five nucleotides were detected in mutB upstream of a conserved branch point AUCUAAC at an expected distance from two possible 3′ splice sites, CAG and UAG, themselves separated by one nucleotide, as defined previously (5).
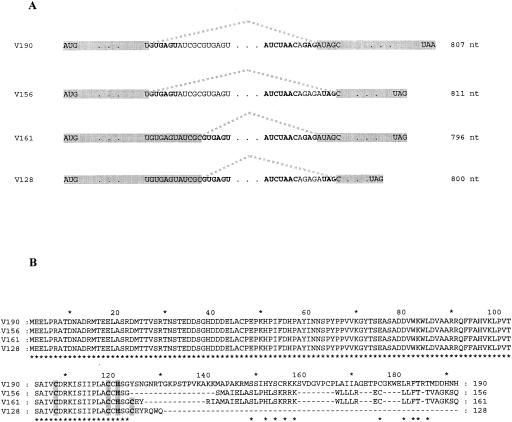
RT-PCR was therefore performed in order to determine whether both genes were transcribed and whether the predicted intron within mutB was spliced. No PCR product was obtained with various oligonucleotides specific for the mutA gene, indicating that this gene is transcribed at a low level or not at all. On the other hand, a ca. 500-bp PCR fragment was obtained for mutB, whose size was consistent with a spliced form of the mutB mRNA. Seven PCR fragments resulting from RT-PCR were cloned into pBluescript and sequenced on both strands. Surprisingly, four different sequences that correspond to all the possible alternative combinations of the two predicted 5′ splice sites and the two predicted 3′ splice sites (introns of 811, 807, 800, and 796 nucleotides) (Fig. 8A) were obtained. The multiple mRNAs thus generated would lead to four protein variants of 128, 156, 161, and 190 amino acids, as shown in Fig. 8B. Interestingly, only two of these proteins, V156 and V161, retained a putative zinc-binding element of the type CX10CX1CX2H (Fig. 8B). The same situation was seen in S. cerevisiae with YML034, which, once spliced, loses a putative zinc-binding site.
FIG. 8.
Alternatively spliced mutB mRNAs. (A) Representation of the four possible combinations of splicing of the mutB mRNA. RT-PCR was performed on total RNAs extracted from E150. The conserved splice sites used in each of the combinations are in boldface type. The variant proteins (left) and the sizes of the introns (right) are indicated. nt, nucleotides. (B) Conceptual translation of each of the four mutB mRNAs and CLUSTAL X multiple alignment. The amino acids which form putative zinc-binding domains are shaded. *, common amino acids.
Presence of metazoan-like SR splicing factors in Y. lipolytica.
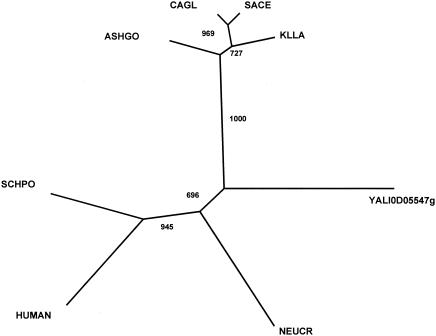
A search for serine/arginine-rich splicing factors (SR splicing factors) involved in the regulation of alternative splicing was conducted in the genome sequence of Y. lipolytica. Putative orthologues of Srp1 of Schizosaccharomyces pombe and human SFRS2 were found in Saccharomyces kluyveri, Kluyveromyces lactis, K. marxianus, Candida tropicalis, and C. albicans (5). We found an SRP1 putative orthologue in Y. lipolytica (see Fig. S1 in the supplemental material) which was not present in the partial genome sequence generated by the Genolevures I project (7). A search for S. pombe SRP2 orthologues led us to find an orthologue in Y. lipolytica and other yeasts like S. cerevisiae, K. lactis, Candida glabrata, and Ashbya gossypii. We performed a CLUSTAL analysis on the proteins of the hemiascomycetous yeasts, S. pombe, N. crassa, and a member of the human SFR family (see Fig. S1 in the supplemental material). Interestingly, a phylogenetic tree revealed two clades, which indicates that the Y. lipolytica orthologue belongs to the clade containing the members of the metazoan family (Fig. 9).
FIG. 9.
Phylogeny of several Srp2-like proteins. Shown is an unrooted phylogenetic tree of various orthologues of S. pombe Srp2 protein derived from parsimony analysis using the amino acid alignment shown in the supplemental material. Bootstrap values based on 100 replicates are indicated at the nodes. The sequences used were Y. lipolytica (Q6CA64, YALI0D05547g), C. glabrata (Q6FRZ2), S. cerevisiae (Q01560), K. lactis (Q6CWW5), A. gossypii (Q759U0), N. crassa (Q7S6F4), Homo sapiens (Q13247), and S. pombe (P78814).
DISCUSSION
By analyzing the recently sequenced genome of Y. lipolytica, we discovered a novel TE that resembles the MULEs from plants and fungi but which also displays unusual features for this type of transposon. Like Z. mays MuDR, the Y. lipolytica Mutyl encodes two ORFs, the second of which shares organization and sequence similarity with putative transposases from MULEs identified in various organisms including ascomycetes and plants. The Y. lipolytica element also carries imperfect TIRs of 22 bp that are located 6 bp away from each end of the element. Mutyl also shares with the MULEs the ability to generate a 9- to 10-bp duplication of the insertion site. To our view, these properties classify Mutyl as a MULE.
Although Mutyl carries two ORFs, the organization of the TE is different from that of MuDR, as unlike MuDR, Mutyl ORFs are predicted to be transcribed in the same orientation. Whereas the promoters of both ORFs of MuDR are nearly identical, as they are part of the TIRs, the promoters in Mutyl do not share any similarity. This finding implies that regulation of the expression of both genes is unlikely to be similar. In this respect, we were able to show that under the growth conditions used here, i.e., exponential growth in rich medium, only the mutB gene is expressed. Expression of Z. mays MuDR genes is regulated developmentally (35). Y. lipolytica is a dimorphic yeast that switches from yeast form to filament in response to environmental and nutritional signals. It would be of interest to determine whether Mutyl is regulated differentially according to the morphology of the cells or in response to environmental and nutritional stress. Ylt1 transposition was shown to be induced in cells grown in medium with acetate as the sole carbon source (G. Barth, personal communication).
Interestingly, Mutyl carries a transposase whose size is larger than the known MULE transposases, although it has a size similar to that of the members of the far-red impaired response protein family found in O. sativa. Interestingly, these are not encoded on a transposable element, but they are thought to originate from them (23). It must be pointed out that most Y. lipolytica transposon ORFs tend to be larger overall than their corresponding homologues in yeasts or filamentous fungi. Ylt1 encodes the largest ORF among ascomycetous LTR retrotransposons (our unpublished observation), and Ylli carries the largest ORF1 among the known long interspersed nuclear elements. This is not linked to the overall average coding sequence size in Y. lipolytica which is similar to that of other yeasts (11).
Two hypotheses that explain the presence of Mutyl in only one of the Y. lipolytica lineages tested can be put forward. The first explanation is horizontal gene transfer (HGT), while the other is loss from some lineages. The origin of MULEs is still debated, and HGT is frequently evoked to explain some of the discrepancies between the phylogeny of the TEs and that of their hosts. A number of cases of HGT for Y. lipolytica genes with a bacterial origin were suspected during analysis of the genome (11). In this respect, the similar structures of Mutyl and bacterial DNA transposons which do carry two ORFs are striking, despite the fact that MutA is more related to fungal and plant transposases. Our observation of the absence of Mutyl in at least two lineages of Y. lipolytica could mean that Mutyl has recently colonized Y. lipolytica. Consistent with this hypothesis, the GC content of Mutyl is 45.32%, whereas that of Ylli, the non-LTR retrotransposon, is 52.46% and that of Ylt1, the LTR retrotransposon, is 53.84%. The overall GC content of Y. lipolytica varies from 48.59 to 49.30% according to the chromosomes (11).
The other hypothesis is that some lineages of Y. lipolytica are losing Mutyl, consistent with the observed low copy number. The same observation was made concerning Ylt1 that was not present in the strains of French and German lineages. This is also reminiscent of the non-LTR retrotransposons that have been found in only two hemiascomycetous yeast species to date, C. albicans and Y. lipolytica, and in a basidiomycete, Cryptococcus neoformans (17), despite the increasing number of yeast genome sequences available. This finding indicates that because of the vertical transmission of this type of TE, the species belonging to the class of hemiascomycetes are in the process of losing non-LTR retrotransposons. At the moment, we cannot favor either of these hypotheses. In any case, Y. lipolytica is, like C. albicans, a reservoir of various unusual TEs for yeasts (C. Neuvéglise and S. Casaregola, unpublished data) and as such is a precious source of information on the variety of transposons once existing in hemiascomycetous yeasts.
We have shown that the mutB gene is expressed and that its transcript is alternatively spliced to give four mRNA variants (and hence four different proteins). So far, alternative splicing in yeast has only been shown to occur in S. cerevisiae on two genes (10), and this is the first report of such a phenomenon in Y. lipolytica. Interestingly, one of the S. cerevisiae genes, YKL186c/MTR2, was associated with six splice variants, similar to what is observed here for mutB. It is noteworthy that the alternative sites are close to each other, as five nucleotides separate the two 5′ splice sites and one nucleotide separates the two 3′ splice sites. Our search for conservation of genes involved in the regulation of alternative splicing may, however, indicate that a different regulation mechanism exists for each yeast. Y. lipolytica harbors the putative orthologues of two metazoan SR factors, whereas S. cerevisiae does not seem to possess an srp1-like gene (5). In addition, hemiascomycete Srp2-like proteins form a clade distinct from that of the metazoan and fungal proteins to which the Y. lipolytica orthologue belongs. This could reflect a functional difference that has to be experimentally tested. This is reminiscent of a number of processes of Y. lipolytica that are closer to that of higher eukaryotes in contrast to S. cerevisiae (2).
The very strong sequence conservation between the various copies indicates that Mutyl is still active. Consistent with this idea, we showed that at least one of the two encoded ORFs of Mutyl was expressed. We also observed that a transposition event has taken place, very likely at meiosis in the type strain. This transposition event seems to be a rare event that points towards a tight regulation of the propagation of Mutyl. The existence of isolates free of endogenous Mutyl and the fact that Y. lipolytica is genetically amenable render it possible to study of Mutyl's mode of propagation.
Supplementary Material
Acknowledgments
Sophie Oztas from Genoscope (Evry) is gratefully acknowledged for providing us with some of the Mutyl sequences. Sylvie Blanchin-Roland kindly provided the primers for PCR amplification of the YlACT1 gene. We are thankful to Colin Tinsley for the reading of the manuscript.
This work was financially supported by Centre National de la Recherche Scientifique, Institut National de la Recherche Agronomique, and the Groupe De Recherche CNRS 2354 “Génolevures.”
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, G., and C. Gaillardin. 1997. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol. Rev. 19:219-237. [DOI] [PubMed] [Google Scholar]
- 3.Barth, G., and C. Gaillardin. 1996. Yarrowia lipolytica, p. 313-388. In K. Wolf (ed.), Nonconventional yeasts in biotechnology. Springer-Verlag, Berlin, Germany.
- 4.Benito, M. I., and V. Walbot. 1997. Characterization of the maize Mutator transposable element MURA transposase as a DNA-binding protein. Mol. Cell. Biol. 17:5165-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bon, E., S. Casaregola, G. Blandin, B. Llorente, C. Neuveglise, M. Munsterkotter, U. Guldener, H. W. Mewes, J. Van Helden, B. Dujon, and C. Gaillardin. 2003. Molecular evolution of eukaryotic genomes: hemiascomycetous yeast spliceosomal introns. Nucleic Acids Res. 31:1121-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casaregola, S., C. Neuvéglise, E. Bon, and C. Gaillardin. 2002. Ylli, a non-LTR retrotransposon L1 family in the dimorphic yeast Yarrowia lipolytica. Mol. Biol. Evol. 19:664-677. [DOI] [PubMed] [Google Scholar]
- 7.Casaregola, S., C. Neuveglise, A. Lepingle, E. Bon, C. Feynerol, F. Artiguenave, P. Wincker, and C. Gaillardin. 2000. Genomic exploration of the hemiascomycetous yeasts: 17. Yarrowia lipolytica. FEBS Lett. 487:95-100. [DOI] [PubMed] [Google Scholar]
- 8.Chalvet, F., C. Grimaldi, F. Kaper, T. Langin, and M. J. Daboussi. 2003. Hop, an active Mutator-like element in the genome of the fungus Fusarium oxysporum. Mol. Biol. Evol. 20:1362-1375. [DOI] [PubMed] [Google Scholar]
- 9.Clare, J. J., M. Belcourt, and P. J. Farabaugh. 1988. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc. Natl. Acad. Sci. USA 85:6816-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis, C. A., L. Grate, M. Spingola, and M. Ares, Jr. 2000. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 28:1700-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, N. Goffard, L. Frangeul, M. Aigle, V. Anthouard, A. Babour, V. Barbe, S. Barnay, S. Blanchin, J. M. Beckerich, E. Beyne, C. Bleykasten, A. Boisrame, J. Boyer, L. Cattolico, F. Confanioleri, A. De Daruvar, L. Despons, E. Fabre, C. Fairhead, H. Ferry-Dumazet, A. Groppi, F. Hantraye, C. Hennequin, N. Jauniaux, P. Joyet, R. Kachouri, A. Kerrest, R. Koszul, M. Lemaire, I. Lesur, L. Ma, H. Muller, J. M. Nicaud, M. Nikolski, S. Oztas, O. Ozier-Kalogeropoulos, S. Pellenz, S. Potier, G. F. Richard, M. L. Straub, A. Suleau, D. Swennen, F. Tekaia, M. Wesolowski-Louvel, E. Westhof, B. Wirth, M. Zeniou-Meyer, I. Zivanovic, M. Bolotin-Fukuhara, A. Thierry, C. Bouchier, B. Caudron, C. Scarpelli, C. Gaillardin, J. Weissenbach, P. Wincker, and J. L. Souciet. 2004. Genome evolution in yeasts. Nature 430:35-44. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, J. A., M. I. Benito, and V. Walbot. 1994. Sequence similarity of putative transposases links the maize Mutator autonomous element and a group of bacterial insertion sequences. Nucleic Acids Res. 22:2634-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 14.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 15.Gaillardin, C. M., V. Charoy, and H. Heslot. 1973. A study of copulation, sporulation and meiotic segregation in Candida lipolytica. Arch. Mikrobiol. 92:69-83. [DOI] [PubMed] [Google Scholar]
- 16.Goodwin, T. J., J. E. Ormandy, and R. T. Poulter. 2001. L1-like non-LTR retrotransposons in the yeast Candida albicans. Curr. Genet. 39:83-91. [DOI] [PubMed] [Google Scholar]
- 17.Goodwin, T. J., and R. T. Poulter. 2001. The diversity of retrotransposons in the yeast Cryptococcus neoformans. Yeast 18:865-880. [DOI] [PubMed] [Google Scholar]
- 18.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 19.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 20.Juretzek, T., M. Le Dall, S. Mauersberger, C. Gaillardin, G. Barth, and J. Nicaud. 2001. Vectors for gene expression and amplification in the yeast Yarrowia lipolytica. Yeast 18:97-113. [DOI] [PubMed] [Google Scholar]
- 21.Kurtzman, C. P., and C. J. Robnett. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek 73:331-371. [DOI] [PubMed] [Google Scholar]
- 22.Le, Q. H., S. Wright, Z. Yu, and T. Bureau. 2000. Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97:7376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisch, D. 2002. Mutator transposons. Trends Plant Sci. 7:498-504. [DOI] [PubMed] [Google Scholar]
- 24.Lisch, D., L. Girard, M. Donlin, and M. Freeling. 1999. Functional analysis of deletion derivatives of the maize transposon MuDR delineates roles for the MURA and MURB proteins. Genetics 151:331-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lisch, D. R., M. Freeling, R. J. Langham, and M. Y. Choy. 2001. Mutator transposase is widespread in the grasses. Plant Physiol. 125:1293-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao, L., T. C. Wood, Y. Yu, M. A. Budiman, J. Tomkins, S. Woo, M. Sasinowski, G. Presting, D. Frisch, S. Goff, R. A. Dean, and R. A. Wing. 2000. Rice transposable elements: a survey of 73,000 sequence-tagged-connectors. Genome Res. 10:982-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuveglise, C., H. Feldmann, E. Bon, C. Gaillardin, and S. Casaregola. 2002. Genomic evolution of the long terminal repeat retrotransposons in hemiascomycetous yeasts. Genome Res. 12:930-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuveglise, C., F. Solano-Serena, P. Brignon, F. Gendre, C. Gaillardin, and S. Casaregola. 2000. Homologous recombination and transposition generate chromosome I neopolymorphism during meiosis in Saccharomyces cerevisiae. Mol. Gen. Genet. 263:722-732. [DOI] [PubMed] [Google Scholar]
- 29.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 30.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raizada, M. N., and V. Walbot. 2000. The late developmental pattern of Mu transposon excision is conferred by a cauliflower mosaic virus 35S-driven MURA cDNA in transgenic maize. Plant Cell 12:5-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ribeiro-dos-Santos, G., A. C. Schenberg, D. C. Gardner, and S. G. Oliver. 1997. Enhancement of Ty transposition at the ADH4 and ADH2 loci in meiotic yeast cells. Mol. Gen. Genet. 254:555-561. [DOI] [PubMed] [Google Scholar]
- 33.Robertson, D. S. 1978. Characterization of a Mutator system in maize. Mutat. Res. 51:21-28. [Google Scholar]
- 34.Romano, A., S. Casaregola, P. Torre, and C. Gaillardin. 1996. Use of RAPD and mitochondrial DNA RFLP for typing of Candida zeylanoides and Debaryomyces hansenii yeast strains isolated from cheese. Syst. Appl. Microbiol. 19:255-264. [Google Scholar]
- 35.Rudenko, G. N., and V. Walbot. 2001. Expression and post-transcriptional regulation of maize transposable element MuDR and its derivatives. Plant Cell 13:553-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Schmid-Berger, N., B. Schmid, and G. Barth. 1994. Ylt1, a highly repetitive retrotransposon in the genome of the dimorphic fungus Yarrowia lipolytica. J. Bacteriol. 176:2477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singer, T., C. Yordan, and R. A. Martienssen. 2001. Robertson's Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1). Genes Dev. 15:591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walbot, V., and G. N. Rudenko. 2002. MuDR/Mu transposable elements of maize, p. 533-564. In R. Craigie, N. L. Craig, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 41.Yoshida, S., K. Tamaki, K. Watanabe, M. Fujino, and C. Nakamura. 1998. A maize MuDR-like element expressed in rice callus subcultured with proline. Hereditas 129:95-99. [DOI] [PubMed] [Google Scholar]
- 42.Yu, Z., S. I. Wright, and T. E. Bureau. 2000. Mutator-like elements in Arabidopsis thaliana. Structure, diversity and evolution. Genetics 156:2019-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zimmermann, M., and P. Fournier. 1996. Electrophoretic karyotyping of yeasts, p. 101-116. In K. Wolf (ed.), Nonconventional yeasts in biotechnology. Springer-Verlag, Berlin, Germany.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.