Summary
Tuberculosis-diabetes mellitus (TB-DM) is linked to a distinct inflammatory profile, which can be assessed using multi-omics analyses. Here, a machine learning algorithm was applied to multi-platform data, including cytokines and gene expression in peripheral blood and eicosanoids in urine, in a Brazilian multi-center TB cohort. There were four clinical groups: TB-DM(n = 24), TB only(n = 28), DM(HbA1c ≥ 6.5%) only(n = 11), and a control group of close TB contacts who did not have TB or DM(n = 13). After cross-validation, baseline expression or abundance of MMP-28, LTE-4, 11-dTxB2, PGDM, FBXO6, SECTM1, and LINCO2009 differentiated the four patient groups. A distinct multi-omic-derived, dimensionally reduced, signature was associated with TB, regardless of glycemic status. SECTM1 and FBXO6 mRNA levels were positively correlated with sputum acid-fast bacilli grade in TB-DM. Values of the biomarkers decreased during the course of anti-TB therapy. Our study identified several markers associated with the pathophysiology of TB-DM that could be evaluated in future mechanistic investigations.
Subject areas: Bioinformatics, Biological sciences, Health sciences, Internal medicine, Medical informatics, Medical specialty, Medicine, Natural sciences
Graphical abstract
Highlights
-
•
A distinct multi-omic signature characterizes TB regardless of DM status
-
•
Anti-tubercular therapy leads to decreases in expression of the multi-omic signature
-
•
The biomarkers identified may serve as candidates for host-directed therapies
Bioinformatics; Biological sciences; Health sciences; Internal medicine; Medical informatics; Medical specialty; Medicine; Natural sciences
Introduction
Tuberculosis (TB) affects approximately 10 million people annually, and despite being a preventable and curable disease, 1.6 million people died from TB in 2021.1 About one-quarter of the world’s population is estimated to be infected with Mycobacterium tuberculosis (Mtb).2
After infecting persons via the lungs, Mtb can disseminate and affect several other organ systems, resulting in a large potential range of clinical manifestations and varying degrees of severity depending on the interaction between Mtb and host immune responses. Factors that affect immune responses can influence the inflammatory reactions against Mtb; these include infection with HIV,3 and genetic,4 environmental,5 nutritional,6 and metabolic features such as dysglycemia and diabetes mellitus (DM).
DM is one of the most common metabolic diseases worldwide. Currently, 422 million people are living with DM,7 most of whom reside in low and middle-income countries. Approximately 1.5 million deaths have been attributed to DM annually.7 Sustained and uncontrolled hyperglycemia leads to changes in immune regulation8; therefore, people living with DM are considered immunosuppressed hosts.9 The association between TB and DM has important health consequences. Previous studies have revealed that DM increases the risk of incident TB 3-fold and increases the risk of TB unfavorable outcomes 2-fold.10,11
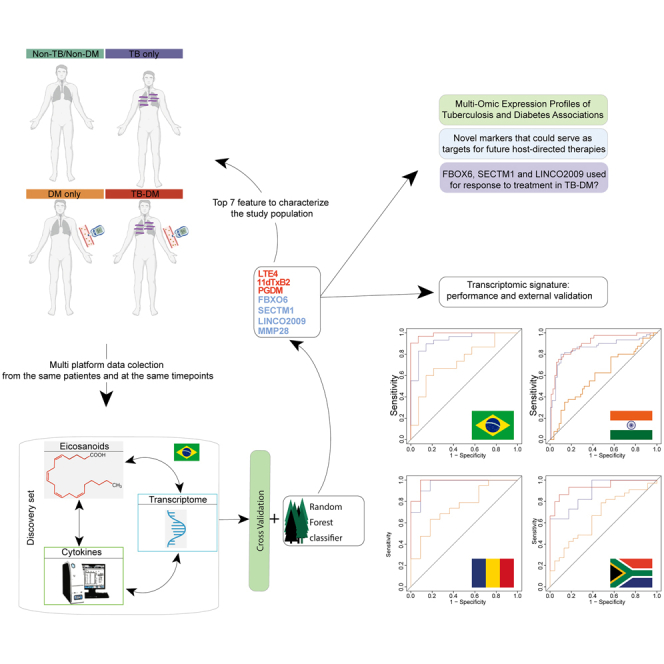
Our group has studied TB-DM interactions, including activation of transcriptional pathways in disease complications,12 persistent inflammation during anti-TB treatment (ATT),13 increased Mtb transmission to close contacts,14 more severe TB clinical presentation,15 and increased risk of unfavorable ATT outcomes.16,17,18 However, while most of the studies used different assays, which were analyzed separately to characterize TB-DM, the use of multiple omics platforms remains poorly explored in the context of TB-DM interactions. Multi-omics provides the opportunity to gain insights into disease pathogenesis, given that it simultaneously explores several components of immune responses through multiple assay platforms. This is a powerful tool to generate hypotheses pertaining to the intricate molecular relationships that may be driving disease. To our knowledge, no previous investigation has examined whether multi-omics analyses can help characterize TB-DM pathogenesis. To fill this knowledge gap, we leveraged samples and data collected in the multi-center prospective cohort of the Regional Prospective Observational Research in Tuberculosis (RePORT)-Brazil (www.reportbrazil.org). We used transcriptomic and cytokine data from peripheral blood and eicosanoids measured in urine, collected at the same time points from persons with TB-DM, TB only, DM only, and non-TB/non-DM close contacts (control group). We applied a random forest model to investigate a possible multi-omic signature that could characterize TB-DM. Furthermore, we tracked the expression of the components of the multi-omic signature in the study groups during the course of ATT. Our results identified novel pathways that may drive disease pathogenesis and serve as targets for future host-directed therapies for TB-DM and as markers of progression to cure after antitubercular therapy initiation. Additionally, the transcriptomic markers identified here have shown a robust accuracy to detect TB-DM in an external validation study using cohorts from India, South Africa, and Romania.
Results
Characteristics of study population
All patients included in our analysis were classified according to their status regarding TB disease and DM. The groups were composed of 24 with TB-DM, 28 TB only, 11 DM only, and 13 non-TB/non-DM (controls). TB-DM and DM only participants were older, with a median age of 46.5 (37–55.7) and 56 (IQR:51–59) years, respectively (p < 0.001; Table S1). In addition, controls and participants with DM were more frequently female than those with TB-DM and TB only. Higher BMI was found in the controls and in DM only groups, with a median (IQR) of 29 (24–33) and 29 (27–32), respectively, compared to 20 (18–22) and 22 (18–26) in TB only and TB-DM patients (p < 0.001) (Table S1). The levels of HbA1c were higher in those with TB-DM (median [IQR] 8.1 [6.6–11.3], followed by DM only 7.1 [6.9–8.4] p < 0.001). Of note, TB only patients were not significantly different regarding AFB (acid-fast bacilli) smear grade and the proportion with cavities on chest X-ray (Table S1).
Multi-omic expression profiles of tuberculosis and diabetes associations
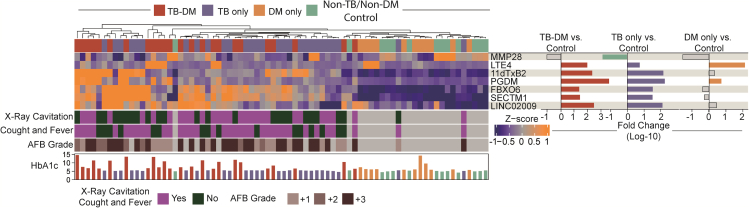
To address the aim of finding a distinct profile that distinguishes TB regardless of DM, our first step was to study the multi-omic alterations in TB and DM using a well-established dimensionality reduction approach (Figure S1). This approach selected 7 parameters that could most contribute to discrimination between all the study groups (Figure S1). The Gini score and mean accuracy for each marker used in our model to discriminate between the clinical groups were provided in Table S2 and Figure S2. To further evaluate the baseline multi-omic profiles of the clinical groups, we first examined the expression of the seven selected features (Figure 1), as described in the STAR methods section. Using a heatmap with log-10 transformed values to assess the overall sample expression, we discovered that TB patients (TB-DM and TB only) had a distinct expression profile compared to non-TB groups, including controls and DM only, with a tendency toward higher expression of 11dTxB2, PGDM, FBXO6, SECTM1, and LINC02009 in TB patients (Figure 1, left panel; Table S3). Of note, the heatmap demonstrated that the impact of TB disease on the biosignature expression values was able to cluster TB-disease participants independent of DM status (Figure 1, left panel; Table S2). The fold-difference of the multi-omic markers between the clinical groups versus control group is summarized in Figure 1, right panel. When the TB only group was compared with controls, the profile was similar to that observed between TB-DM vs. control group, with higher expressions observed in the majority of the markers in both TB groups, except for MMP28, which was higher in the control group when compared with the TB only group. (Figure 1, right panel). Extending the approach to DM only and control differences, we found higher levels of LTE4 (leukotriene E4) and PGDM (prostaglandin D metabolite) in DM only participants (Figure 1, right panel). These findings reveal a higher multi-omic expression of biomarkers capable of clustering TB patients, regardless of DM status, which suggests differential expression triggered by TB disease.
Figure 1.
Distinct multi-omic expression profiles identified tuberculosis regardless the glycemic status
Right panel. A hierarchical cluster analysis (Ward method with 100 × bootstrap) was employed to test the overall expression of plasma cytokines, gene expression and urinary eicosanoids in the study population. Dendrograms represent Canberra distance. Left panel. Differential expression analysis was used to calculate the fold-changes and show differences in biomarkers levels for each clinical group (TB, TB-DM, and DM) versus control. Differences that reached statistical significance after adjustment for multiple comparisons (adjusted p < 0.05) are represented in colored bars.
Variation in the multi-omic activation according to AFB grade and clinical group
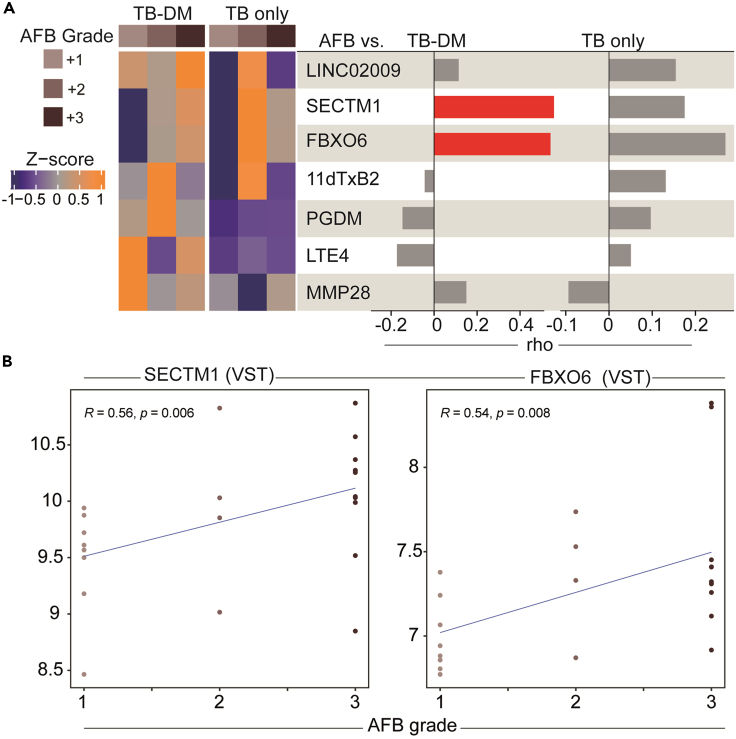
After identifying a multi-omic profile that characterized the TB only and TB-DM groups, our next step was to evaluate whether glycemic status affected the expression of markers according to AFB smear grade (Figure 2). Using hierarchical clustering on the Z score normalized data at baseline, we observed that patients with TB (with and without DM) and AFB smear grade equal to 2+ tended to express higher levels of SECTM1, FBXO6, and 11dTxB2 compared with other AFB grades (Figure 2A). Those marker levels were lower among those with TB with AFB grade 1+ (Figure 2A). On the other hand, the TB-DM group showed higher expression of LTE4 and MMP28 in those with AFB grade 1+, PGDM and 11dTxB2 in AFB grade 2+, and finally FBXO6, SECTM1, and LINCO2009 in AFB grade 3+ (Figure 2A). Next, using Spearman correlation analysis we found that the AFB smear grade was correlated with SECTM1 and FBXO6 expression in TB-DM participants. No significant correlation was observed in the TB only group (Figures 2A and 2B). No significant correlation was observed in the TB only group (Figure 2A). This finding suggests that the presence of TB was associated with changes in multi-omic expression according to AFB smear grade.
Figure 2.
A distinct multi-omic expression and correlation profile between AFB grade and markers among TB groups
(A) Left panel. A hierarchical cluster analysis (Ward method with 100 × bootstrap) was employed to evaluate multi-omic marker expression according to the AFB smear grade in TB and TB-DM, as indicated. Right panel. A Spearman correlation analysis was used to study the influence of the AFB smear grade on multi-omic marker expression. The rho values are shown. Red bars indicate correlation with p value < 0.05.
(B) Spearman correlation plots demonstrating the associations between the expression levels of the indicated genes and the AFB smear grade. Line represent linear regression. R: Spearman rho value.
Changes in multi-omic expression after anti-tuberculosis therapy initiation
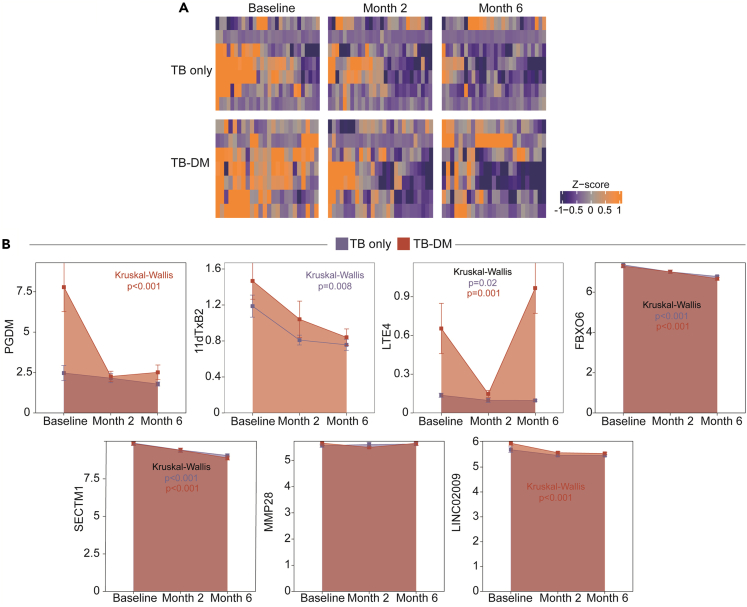
We next evaluated changes in the biomarker expressions or concentrations after ATT initiation (Figure 3). Using a heatmap approach, we were able to show an overall decreasing tendency in marker values, in both the TB and TB-DM groups (Figure 3A). Next, we prospectively assessed marker expressions to evaluate the dynamics of multi-omic expression during ATT (Figure 3B). We discerned a reduced level of 11dTxB2 in the TB group, as well as a reduced level of FBXO6 and SECTM1 expression in both the TB and TB-DM groups, and reduced LINC02009 expression in the TB-DM group.
Figure 3.
Changes in multi-omic expression after anti-tuberculosis therapy initiation
(A) A hierarchical cluster analysis (Ward method with 100 × bootstrap) was employed to evaluate multi-omic marker expression in TB and TB-DM after anti-tuberculosis therapy initiation.
(B) A boxplot was used to test the changes in multi-omic levels in months 2 and 6 after ATT. In the graphs, dots represent median and whiskers represent interquartile range values.
The multi-omic approach resulted in a transcriptomic signature to detect TB-DM
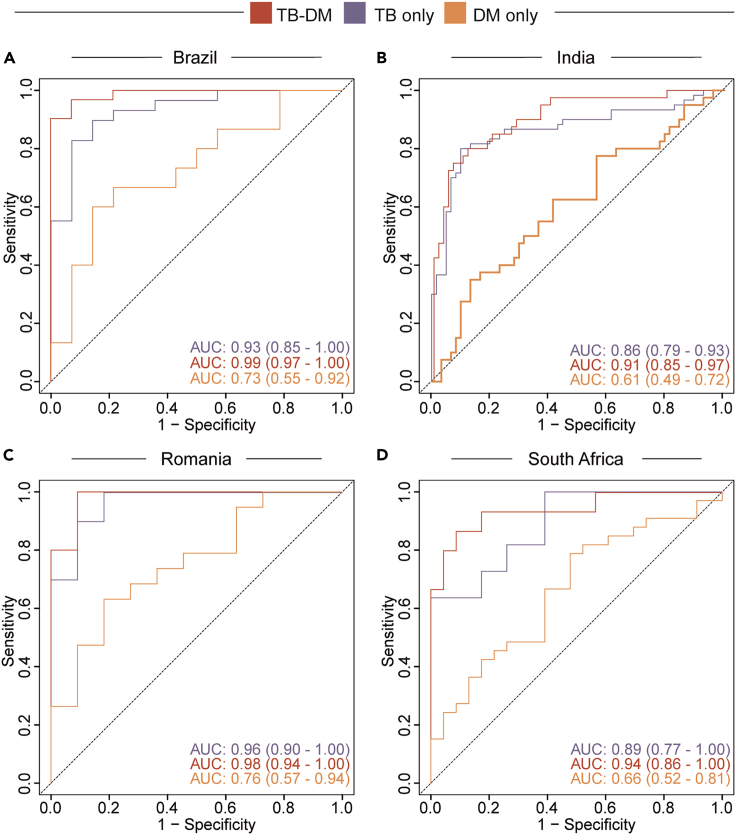
After evaluating the multi-omic profile of TB and TB-DM interaction, we extended our analysis to verify the expression of the transcriptomic markers identified here. Using data from a multi-center, prospective cohort study of our group,19 which assessed transcriptomic data from 290 participants, recruited from two sites in India, one in Brazil and publicity available RNA-seq data from two sites of a TANDEM consortium (Romania and South Africa),20 we test the accuracy of the transcriptomic markers identified here by our machine learning analysis (Figures 4 and S3). First, we used a heatmap approach applied to the four countries, verifying the expression of MMP28, SECTM1, FBOX6, and LINCO2009 (Figure S3). Our results identified that these genes were capable of detecting TB regardless of the glycemic status in Brazil, India, and Romania (Figure S3), marked by higher expression of the markers in TB-DM and TB only (Figure S3). Additionally, we used a fold-change Log2 to study the expression of the markers in each clinical group and in each country (Figure S3). Of interest, our results revealed a similar expression profile in TB-DM and TB only in Brazil and India, with higher expression of SECTM1, FBOX6, and LINCO2009 when compared with non-TB/non-DM controls, whereas MMP28 was higher in the controls (Figure S3). Next, we used the receiver operating characteristic (ROC) curve, employing the genes MMP28, SECTM1, FBOX6, and LINCO2009 to verify the accuracy of these markers, together, to identify TB only, TB-DM, and DM only (Figure 4), shown a robust accuracy in detecting TB-DM, area under curve (AUC) 0.99, 0.91, 0.98, and 0.94 in Brazil, India, Romania, and South Africa, respectively (Figure 4). Our result identifies a transcriptomic signature, composed by expression levels of MMP28, SECTM1, FBOX6, and LINCO2009, and identified using machine learning analysis applied to multi-platform data, with a great accuracy to detect TB-DM.
Figure 4.
Accuracy of a new transcriptomic signature to detect TB-DM
Accuracy of the gene signature detected in the random forest analysis to classify TB, with or without DM in Brazil (A), India (B), Romania (C) and South Africa (D). Receiver operator characteristics (ROC) curve analysis was used to check the accuracy of the signature genes identified by the random forest model to classify the TB, TB-DM, and DM groups in each clinical site as indicated with respect to TB disease.
Discussion
To achieve the World Health Organization targets for TB elimination, new strategies in the TB field are required. The study of omics has been extensively applied to provide insights into the pathophysiological changes induced by Mtb infection. Several mRNA transcriptomic signatures have been proposed in recent years as new tools for the diagnosis, prognosis, and treatment monitoring of Mtb infection and TB disease.21,22,23,24 Nevertheless, advances have been made using isolated omics, and the employment of an integrative analysis could better dissect the changes triggered by Mtb infection. Here, we performed an integrated omics analysis, using transcriptomic data; eicosanoid measurements in urine; and soluble inflammatory biomarkers in blood, to identify a multi-omic profile that characterized TB and DM.
Our first step was to perform dimensionality data reduction. A random forest model was applied to the integrated multi-omic data (Luminex and RNAseq from peripheral blood and eicosanoids from urine) to select the most informative features. The random forest algorithm works based on summarizing multiple decision trees, evaluating the importance of markers in classifying the samples within the data. The variables were evaluated by using the mean decrease of accuracy and Gini indexes. This algorithm has been used by our group in TB and HIV coinfection studies.25,26 This approach was also applied to the conceptualization of predictive models in cardiovascular diseases27 and to predict outcomes in neurosurgery.28 After cross-validation, MMP-28, LTE-4, 11- dTxB2, PGDM, FBXO6, SECTM1, and LINCO2009 were selected. Importantly, despite the crucial role of cytokines in TB inflammatory activation, none of these markers were selected by this approach.
MMP-28 is a protein coding gene that encodes a secreted enzyme associated with casein degradation and is involved in the breakdown of extracellular matrix.29 The role of MMPs in TB pathogenesis has been largely studied and linked with tissue remodeling.30 After Mtb infection, inflammatory activation leads to a host response that increases tissue destruction involving MMPs, and disrupts homeostasis, which is required for Mtb dissemination. Some evidence has shown the expression of MMP28 in normal tissues, indicating its role in the preservation or maintenance of homeostatic conditions.29,31 In accordance, our results identified higher expression of MMP-28 in the control group when compared with TB only patients, which reinforces the marker activity in homeostasis. If confirmed in other cohorts and future studies, the MMP-28 pathway could be used in the future as a target for host-directed therapies, to improve tissue healing orrecovery or limiting the damage caused by the responses against the Mtb.
The role of eicosanoids in TB pathogenesis has been largely explored,32,33 and the changes in the arachidonic acid pathways persist after ATT.34 Here, we identified three eicosanoids which, in TANDEM, could help future studies that develop a host-based therapy. LTE4 is a lipid inflammatory mediator and the unique cysteinyl leukotriene stable and abundant in vivo.35 LTE4 has been described as having a critical role in pulmonary inflammation,35 through leukocyte activation and triggering cytokine production and macrophage necrosis.36 Their action in TB pathogenesis remains unclear, but LTE-4 was associated with mucosal eosinophilia and airway hyperresponsiveness in asthma diseases35 and seems increased in persons with DM.37 In accordance, our results demonstrated higher levels of LTE4 in TB only, TB-DM and DM only when compared with non-TB/non-DM control group. Of note, we showed a decrease in the levels of LTE4 after ATT initiation, mainly in those with TB only, suggesting an indirect attenuation of lung damage after the induction phase of ATT, the first two months. However, in TB-DM participants, the levels of LTE4 returned to higher expression in month 6, corroborating a hypothesis from one of our previous publications that showed persistent inflammation during ATT in TB-DM participants13 (11dTxB2 is a urinary metabolite produced from the breakdown of the thromboxane A2). Their levels, measured in urine, can be used to evaluate the response to aspirin therapy in heart disease,38 asthma,39 and in diseases associated with higher platelet activation40 as an indirect measure of platelet activity. The role of 11dTxB2 in TB pathogenesis remains unclear, but TB patients usually present with higher platelet counts that correlate with disease severity and hypercoagulability.41 Platelets contribute to MMP-mediated tissue damage through their effects on monocytes, leading to upregulation of activation markers, increases in MMP secretion and enhanced phagocytosis.41 Our results reveal higher levels of 11dTxB2 associated with TB, independent of glycemic status, with decreases in the expression after ATT that could be associated with platelet activation and tissue injury consequences. Of note, considering the effects of platelets on TB pathogenesis, some anti-platelet drugs such as aspirin could be a potential target for host-directed therapy in pulmonary TB.42 Finally, PGDM is a urinary metabolite from PGD2, and little is known about its function in TB and in infected alveolar macrophages. A recent study in Histoplasma capsulatum infection demonstrated an immunostimulatory effect of PGD2, contributing to fungicidal mechanisms and controlling the inflammatory damage.43 Our results showed higher levels of PGD2 in all three groups when compared with controls, suggesting lung damage triggered by TB disease, but the real role of the molecule in the pathophysiology of the disease needs to be better evaluated. Our finding highlights the potential role of eicosanoids as candidates for host-directed therapy in TB and TB-DM. The blockage of LTE4 pathways could attenuate the cytokine storm, inflammatory activation,44 and tissue damage. Leukotriene blockers are being used in asthma treatment, and 11dTxB2 has the potential to be used in the future as a marker of platelet activation in TB, as in disease progression, as well as to measure response to the host-directed therapies using anti-platelet agents.
FBXO6 is a protein code gene associated with phosphorylation-dependent ubiquitination, largely studied in cancer,45 without robust evidence in infectious diseases. In a murine study of Influenza A infection, deficiencies in FBXO6 expression were associated with decreased pulmonary viral replication, inflammatory responses, and mortality.46 Despite the absence of robust publications involving the gene in the TB field, we found an interesting correlation between the gene expression levels of FBXO6 and the AFB smear grade in TB-DM patients. Additionally, after ATT initiation, we identified a decrease in the expression in both TB and TB-DM, highlighting the possibilities of a new molecule associated with bacilli proliferation and target for host directed therapies. To test this hypothesis, future studies are required to delineate the mechanisms of a potential role for FBXO6 in TB.
SECTM1 is a gene that encodes a transmembrane and secreted protein. In humans, SECTM1 works as a T/NK cell co-stimulatory molecule that has been associated with CD4 and CD8 T cell proliferation and interferon-gamma (IFN-γ) production47 and reported as an IFN early response gene.48 In TB, IFN- γ displays a pivotal role in macrophage activation and immune protection against the bacilli.49,50 Importantly, SECTM1 was identified in a recent study that evaluated host protein features in persons with HIV, as a marker to evaluate Mtb infection prior to TB diagnosis.51 In accordance with the literature, our findings revealed increased expression of SECTM1 in TB participants, independent of the glycemic status. Additionally, we showed a direct correlation between SECTM1 expression level and AFB smear grade in the TB-DM group, highlighting it as a possible marker to evaluate disease severity in TB-DM. Finally, during ATT the levels of SECTM1 decreased, in both the TB and TB-DM groups. Our findings emphasize the role of SECTM1 as a marker of TB, disease progression in TB-DM, and prognosis, as recently proposed.52 The last selected marker by our random forest model was the LINCO2009, a non-coding RNA with unknown function in TB. The non-coding RNA displays a high potential to modulate biochemical pathways, and our results could represent a potential topic of interest for further investigations in TB-DM pathology. Future studies are required to better evaluate the role and potential of LINCO2009 in TB and TB-DM.
The final step of our study was to analyze the accuracy of the transcriptomic markers to detect TB-DM. Several studies around the world have been conducted to identify a transcriptomic profile that characterizes TB-DM interaction, but none of these obtained robust results.12,20 We recently published a multi-center, prospective cohort study of whole blood gene expression in TB-DM (the MSTDI study).19 We applied a random forest model to a whole RNA-seq from two sites in India53,54 and one site in Brazil, as a discovery set, and used public RNA-seq data from two sites of the TANDEM consortium20 as a validation set (Romania and South Africa), to verify the dynamicity across population. We identified four genes as more informative features to detect TB-DM, SMARCD3, BATF2, VAMP5, and ANKRD22, with an accuracy of 0.97 in Brazil and South Africa, 0.92 in India and 0.89 in Romania.19 Here, using a similar machine learning analysis applied to multi-platform data from Brazil, we identified another four genes as more informative features to detect TB-DM, LINCO2009, SECTM1, FBXO6, and MMP28, with a concise and robust accuracy in discrimination TB-DM. We extended our approach and tested this new transcriptomic signature in the sites previously studied in MSTDI and found a better performance in all four sites using the transcriptomic markers identified here, with an accuracy of 0.99, 0.91, 0.98, and 0.94 in Brazil, India, Romania, and South Africa, respectively. This finding highlights the potential applicability of multi-omic platforms and machine learning approaches in the study of inflammatory and/or infectious diseases since the findings obtained through the application of the decision tree on the three platforms were better and more concise than what was found when we applied the model only to transcriptomic data.
Despite some limitations, to the best of our knowledge, this is the first study to evaluate the multi-omic analysis of TB and TB-DM, with data retrieved from samples collected from the same patients and time points. By identifying novel markers that could serve as targets for future host-directed therapies, our results expand the current knowledge regarding TB-DM pathogenesis and that can be useful to improve anti-TB treatment outcomes in this population.
Limitations of the study
This study had some limitations. First, the populations were different regarding age and BMI, factors that could influence inflammatory activation. A validation in an independent cohort is necessary to ensure the robustness of our findings. In the pre-analytical phase of the current study, we defined that all RePORT Brazil participants who had data on all the omics platforms investigated here would be included, and no pre-analytical matching of the study sample was performed. This approach may have introduced bias, which will require further investigation in additional cohorts to clarify. Second, the measurement of HbA1c levels was performed only at baseline, and changes in glycemia could also affect the immune responses after therapy initiation. We are currently developing a prospective investigation to assess HbA1c levels in a novel cohort from the RePORT Brazil attended population.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| Raw RNAseq data from TANDEM | Eckold et al.20 | Bioproject ID: PRJNA470512 |
| Raw RNAseq from RePORT | Kornfeld et al.53; Gupte et al.54; Queiroz et al.19 | GEOncbi ID: GSE181143 |
| Software and algorithms | ||
| R version 4.2.2. | R Core Team | https://cran.r-project.org/ |
| STAR version 2.7.10 | Dobin et al.55 | https://code.google.com/archive/p/rna-star/ |
| DESeq2 version 1.40.2 | Love et al.56 | https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
| randomForest version 4.7–1.1 | Breiman et al.57 | https://cran.r-project.org/web/packages/randomForest/index.html |
| caret version 6.0–94 | Kuhn et al.58 | https://cran.r-project.org/web/packages/caret/index.html |
| complexheatmap | https://doi.org/10.1093/bioinformatics/btw313 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| Hmisc version 5.1–1 | https://cran.rproject.org/web/packages/Hmisc/Hmisc.pdf | https://hbiostat.org/r/hmisc/ |
Resource availability
Lead contact
Further information and requests regarding the packages employed for the analysis performed in this study should be directed to and will be fulfilled, by the lead contact, Bruno B. Andrade (bruno.andrade@fiocruz.br).
Materials availability
-
•
This study did not generate new unique reagents.
Data and code availability
-
•
Data used here were from two previous works. The gene expression from the main population, Salvador site from RePORT Brazil, and Indian population (used only in the validation of the transcriptomic signature) were from the MSTDI study and have been deposited at the GEO ncbi database and is publicly available as of the date of publication (Accession number GSE181143, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181143).
The validation of the transcriptomic signature identified here was performed using an external cohort, with population from South Africa and Romania, previously published by the TANDEM consortium and have been previously deposited at the SRA database and are publicly available as of the date of publication (BioProject ID PRJNA470512).
-
•
All employed packages' references are available at the key resources tables and methodology.
-
•
This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Ethics statement
The study was conducted according to the Declaration of Helsinki. Data and specimens were obtained from the RePORT-Brazil observational cohort study. The study was approved by the Institutional Review Boards of the Instituto Nacional de Infectologia Evandro Chagas, Fundação Oswaldo Cruz, Brazil (CAAE: 25102412.3.1001.5262). Written informed consents were obtained from all voluntary participants.
Study design and population
This was a retrospective analysis of a prospective observational study with data and specimens collected within the scope of RePORT-Brazil,59,60,61 at the Salvador site. All participants were enrolled between June 2015 and June 2019 and followed for up to 24 months. Several studies from RePORT-Brazil have explored different aspects of TB-DM.14,16,32,62 However, the evaluation of multi-platform data has not been previously explored. Additionally, to evaluate the accuracy of the transcriptomic signature identified in this study we used public data from a previously published work from the RePORT-India53,54 and TANDEM consortium.20
Participants from the RePORT-Brazil were at least 18 years old, had an Mtb culture-positive sputum at enrollment (for those in the TB groups) and were evaluated at three in-person visits: (i) ATT initiation (baseline), (ii) two months after initiating ATT, and (iii) after completing treatment (approximately month 6). The sputum smears were prepared by the Ziel–Nielsen method using 1% carbol-fuchsin and scored using the International Union Against Tuberculosis and Lung Disease (IUATLD) scale.63 Non-TB/Non-DM controls and DM patients (without TB) were selected from close TB contacts who agreed to participate in the study. These patients were evaluated at two visits: baseline and 6 months after enrollment. We selected those participants who tested negative, at both visits, for Mtb infection by QuantiFERON-TB Gold. The selection of the participants included in the sub-groups was based on availability of databases on each assay platform assessed in the present study. As a result, only participants who had data on all the platforms evaluated were included. This was pre-specified in the analysis plan to allow data integration and multi-omic analyses. Sociodemographic, clinical, and epidemiologic data such as age, sex, race/ethnicity (self-reported), and body mass index (BMI) were collected at baseline and shown in Table S1. Of note, gander variable was not collected. Regarding the biological sex and race/ethnicity (self-reported) our analysis found no differences between the groups. Glycated hemoglobin (HbA1C) was measured in all participants at enrollment.32
Diabetes was defined according to baseline HbA1c, following American Diabetes Association (ADA) guidelines.64 Individuals were classified as having DM (HbA1c ≥ 6.5%) or normoglycemia (HbA1c<5.7%).
Method details
RNA sequencing
Samples from TB patients (with and without DM) were collected at baseline, 2, and 6 months of treatment. Samples for the non-TB/non-DM control and DM only groups were collected at baseline. Total RNA from venous blood (2.5 mL) was collected in PAXgene Blood RNA Tubes (PreAnalytiX), extracted using a PAXgene blood miRNA kit (Qiagen) with the semi-automated QIAcube (Qiagen) and quantified using the LabChip GX HiSens RNA system (PerkinElmer). RNA-seq libraries were prepared using the Bioscientific NEXTflex-Rapid-Directional mRNA-seq sample preparation with the Caliper SciClone. Samples were sequenced using the NextSeq500 High Output kit V2 (Illumina) for 75 cycles. For all samples, RNA was sequenced by Illumina HiSeq 2500 at MedGenome in Bangalore, India.
Collection, processing, and analysis of eicosanoid metabolites
Urine samples from patients were collected at baseline, 2 months, and 6 months of treatment. Urine was stored at −80°C within 1 h of sample collection.65,66 PGE-M and PGD-M are unstable if urine is not stored at −80°C within 90 min of collection. Concentrations of the major urinary prostaglandin (PG)E2 metabolite, PGE-M; the tetranor-PGE1, TN-E, the major urinary metabolite of PGD2, PGD-M; 11-dehydro-thromboxane-B2, dTxB2; the metabolite of PGI2, PGI-M; and leukotriene (LT)E4 were measured in urine at each time point in all study participant. The assays were performed at the Eicosanoid Core Laboratory at Vanderbilt University Medical Center, in the USA. Samples were shipped on dry ice and stored at −80°C until analysis.
[2H6]-PGE-M and [2H11]-TN-E were synthesized as previously described.32 [2H4]-PGI-M and [2H4]-11dTxB2 were purchased from Cayman Chemicals (Ann Arbor, MI USA), and [20,20,20-2H3]-LTE4 from Enzo Life Sciences (Farmingdale, NY USA). Sep-Pak C18 and Oasis HLB (3cc/60mg) extraction cartridges were obtained from Waters Corporation (Milford, MA USA). The organic reagents were of high-performance Liquid Chromatograph (LC) quality and purchased from Sigma Aldrich (St. Louis, MO USA).
Immunoassays
Plasma was stored at −80°C for immunology assays. Using the Bio-Plex Pro Human Cytokine Standard 27-Plex kit (Group I) on the Biorad Bioplex 200 Luminex platform the following analytes were measured: interleukin (IL)-1β, IL-1 receptor antagonist (IL-1Ra), IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p70, IL-13, IL-15 IL-17A, eotaxin, granulocyte colony stimulating factor (G-CSF), granulocyte-macrophage colony stimulating factor (GM-CSF/CSF2), interferon gamma (IFN-γ), monocyte chemoattractant protein-1 (MCP-1)/C-C motif chemokine ligand 2 (CCL2), macrophage inflammatory protein-1 alpha (MIP-1α/CCL3), MIP-1 beta (MIP-1β/CCL4), platelet-derived growth factor-BB (PDGF), regulated on activation, normal T cell expressed and secreted (RANTES/CCL5), tumor necrosis factor-alpha (TNF-α), and vascular endothelial growth factor (VEGF). Biomarkers were chosen based on commercially available pre-mixed kits and included parameters previously described to be involved in TB pathogenesis.34,67
Quantification and statistical analysis
Gene expression analysis
After the quality check, sequences were aligned to the human transcriptome (GRCh38 version 100), comprising mRNA and ncRNA, using STAR.55 Count gene expression matrix was examined using the DESeq2 R package,56 version 4.2.2, to identify differentially expressed genes (DEG) across the four patient groups. Changes in gene expression with false discovery rate (FDR)-adjusted p value <0.05 and log2 fold-difference +1.4 were considered significant.
Feature selection analysis using machine learning
We employed a feature selection approach to identify the most relevant features from the multi-omic factors used here associated with TB-DM. The integrated multi-omic data, composed by all three platforms measured in the study, transcriptomic data, cytokines, and eicosanoids, were used to trained the model and cross validation, applying the random forest algorithm, using the randomForest package.57 1000 trees were used in the model and at each split the number of features was 41.88, the square root of the total of variables in the dataset plus one. Furthermore, a leave-one-out cross-validation with 50-folds, and 5 repetitions was performed to assess the markers’ accuracy, using the caret package.58 The cross-validation allowed the estimation of the model’s accuracy and the No information rate, which was 1(1–0.95) and 0.36 respectively. The p value for accuracy > No information rate was 0.00000000000000022. With this method, we aim to reduce the dimension of data and obtain the most accurate model. Seven features were selected after applied the random forest model and included and the analysis presented here. Figure S1 illustrates the model design. Table S2 and Figure S2 show the variable importance of the top 20 features.
Statistical analysis
The median values with interquartile ranges (IQR) were used as measures of central tendency and dispersion. Features were compared between the study groups using the Kruskal-Wallis test with Dunn’s multiple comparisons. Categorical variables were compared using Chi-square test. Hierarchical cluster analyses (Ward’s method), with 100X bootstrap of Z score normalized data were employed to depict the overall multi-omic expression profile in the study groups. Dendrograms represent Manhattan distance.
Spearman correlation analysis was used to evaluate the interaction between sputum smear AFB grade and multi-omic expression data, and between HbA1c levels and the multi-omic expression data. All univariate comparisons with p values <0.05 after adjustments were considered statistically significant. The statistical analyses were performed using R 4.2.2. The R package used to perform the analyses here is described in Table S4.
Acknowledgments
We thank the study participants. We recognize the important contributions of the RePORT-Brazil Consortium, who participated at different stages of the consortium and helped to build the databases explored in the present study: Vanessa Nascimento and Michael S. Rocha (Instituto Brasileiro para Investigação da Tuberculose, Fundação José Silveira, Salvador, Brazil, Multinational Organization Network Sponsoring Translational and Epidemiological Research Initiative, Salvador, Brazil); Saulo R. N. Santos and André Ramos (Instituto Brasileiro para Investigação da Tuberculose, Fundação José Silveira, Salvador, Brazil); Juan Manuel Cubillos-Ângulo (Multinational Organization Network Sponsoring Translational and Epidemiological Research Initiative, Salvador, Brazil); Jéssica Rebouças-Silva (Faculdade de Medicina, Universidade Federal da Bahia, Salvador, Brazil, Laboratório de Inflamação e Biomarcadores, Instituto Gonçalo Moniz, Fundação Oswaldo Cruz, Salvador, Brazil); Pedro Brito (Multinational Organization Network Sponsoring Translational and Epidemiological Research Initiative, Salvador, Brazil, Laboratório de Inflamação e Biomarcadores, Instituto Gonçalo Moniz, Fundação Oswaldo Cruz, Salvador, Brazil); Carolina A. S. Schmaltz, Adriano Gomes-Silva, Francine P. Ignácio, Maria C. Lourenço, Aline Benjamin, Jamile G. de Oliveira, Solange Cavalcante, and Betina Durovni (Laboratório de Pesquisa Clínica em Micobacteriose, Instituto Nacional de Infectologia Evandro Chagas, Fiocruz, Rio de Janeiro, Brazil); Alysson G. Costa (Fundação Medicina Tropical Dr Heitor Vieira Dourado, Manaus, Brazil, Programa de Pós-Graduação em Medicina Tropical, Universidade do Estado do Amazonas, Manaus, Brazil, Universidade Federal do Amazonas, Manaus, Brazil); Leandro Sousa Garcia, Brenda K. de Sousa Carvalho, Bruna P. de Loiola, and Alexandra B. Souza (Fundação Medicina Tropical Dr Heitor Vieira Dourado, Manaus, Brazil, Programa de Pós-Graduação em Medicina Tropical, Universidade do Estado do Amazonas, Manaus, Brazil); Elisangela C. Silva, Mayla Mello, and Adriana S. R. Moreira (Programa de Pós-Graduação em Medicina Tropical, Universidade do Estado do Amazonas, Manaus, Brazil); José R. Lapa-e-Silva (Programa Acadêmico de Tuberculose da Faculdade de Medicina, Universidade Federal do Rio de Janeiro, Rio de Janeiro, Brazil; Megan Turner, John R Koethe, and Carlos Henrique Serezani (Division of Infectious Diseases, Department of Medicine, Vanderbilt University School of Medicine, Nashville, TN, United States). A special thanks to Elze Leite (FIOCRUZ, Salvador, Brazil), Eduardo Gama (FIOCRUZ, Rio de Janeiro, Brazil), Elcimar Junior (FMT-HVD, Manaus, Brazil), and Hilary Vansell (VUMC, Nashville, USA) for administrative and logistical support. Some data used here were originated from publicly available databases previously reported by the RePORT India and the TANDEM consortium, and thus we thank the research teams that generated the primary data. This work was supported by the Departamento de Ciência e Tecnologia (DECIT)—Secretaria de Ciência e Tecnologia (SCTIE)—Ministério da Saúde, Brazil [25029.000507/2013-07 to V.R.], the National Institute of Allergy and Infectious Diseases [U01-AI069923] at NIH, and by The Civilian Research and Development Foundation (CRDF) Global #DAA3-17-63144. The study was partially supported by the Intramural Research Program of the Fundacaao Oswaldo Cruz and the Intramural Research Program of the Fundação José Silveira. B.B.A., V.C.R., A.K., and M.C.-S. are senior scientists from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). M.B.A. is Researcher Level I from The Consejo Nacional de Ciencia, Tecnología e Innovación Tecnológica (CONCYTEC) and is supported by Project Number 5 R01-AI147765-04 from NIH. G.C.S. is a scientific initiation fellow from CNPq. M.A.-P. and B.B.-D. received a research fellowship from the Coordenaçaão de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, finance code: 001). The funders had no role in study design, data collection, and analysis, the decision to publish, or the preparation of the manuscript.
Author contributions
A.T.L.Q., B.B.A., T.S., M.C.-S., A.K., V.R., and M.F. designed the study and mentored the work; M.B.A., B.B.-D., M.A.-P., A.A., and B.B.A. performed the experiments and data collections; C.L.V., E.R.F., G.C.S., B.B.A., and A.T.L.Q. analyzed the data; M.B.A., M.M.B., B.B.-D., M.A.-P., V.C.R., A.L.K., M.C.-S., and T.R.S. helped with data interpretation; C.L.V., G.C.S., B.B.A., and A.T.L.Q. wrote the manuscript. All authors contributed to the article and approved the submitted version.
Declaration of interests
The authors declare no competing interests.
Published: February 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109135.
Supplemental information
References
- 1.World Health Organization . 2022. Global Tuberculosis Report. [Google Scholar]
- 2.Houben R.M.G.J., Dodd P.J. The Global Burden of Latent Tuberculosis Infection: A Re-estimation Using Mathematical Modelling. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell L.C.K., Noursadeghi M. Pathogenesis of HIV-1 and Mycobacterium tuberculosis co-infection. Nat. Rev. Microbiol. 2018;16:80–90. doi: 10.1038/nrmicro.2017.128. [DOI] [PubMed] [Google Scholar]
- 4.Kathirvel M., Mahadevan S. The role of epigenetics in tuberculosis infection. Epigenomics. 2016;8:537–549. doi: 10.2217/epi.16.1. [DOI] [PubMed] [Google Scholar]
- 5.Murray M., Oxlade O., Lin H.H. Modeling social, environmental and biological determinants of tuberculosis. Int. J. Tuberc. Lung Dis. 2011;15:64–70. doi: 10.5588/ijtld.10.0535. [DOI] [PubMed] [Google Scholar]
- 6.Gupta K.B., Gupta R., Atreja A., Verma M., Vishvkarma S. Tuberculosis and nutrition. Lung India. 2009;26:9–16. doi: 10.4103/0970-2113.45198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization . 2016. Global Report on Diabetes. [Google Scholar]
- 8.Mantovani A., Garlanda C. Humoral Innate Immunity and Acute-Phase Proteins. N. Engl. J. Med. 2023;388:439–452. doi: 10.1056/NEJMra2206346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berbudi A., Rahmadika N., Tjahjadi A.I., Ruslami R. Type 2 Diabetes and its Impact on the Immune System. Curr. Diabetes Rev. 2020;16:442–449. doi: 10.2174/1573399815666191024085838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Restrepo B.I. Diabetes and Tuberculosis. Microbiol. Spectr. 2016;4:1–11. doi: 10.1128/microbiolspec.TNMI7-0023-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeon C.Y., Murray M.B. Diabetes mellitus increases the risk of active tuberculosis: a systematic review of 13 observational studies. PLoS Med. 2008;5:e152. doi: 10.1371/journal.pmed.0050152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prada-Medina C.A., Fukutani K.F., Pavan Kumar N., Gil-Santana L., Babu S., Lichtenstein F., West K., Sivakumar S., Menon P.A., Viswanathan V., et al. Systems Immunology of Diabetes-Tuberculosis Comorbidity Reveals Signatures of Disease Complications. Sci. Rep. 2017;7:1999. doi: 10.1038/s41598-017-01767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar N.P., Fukutani K.F., Shruthi B.S., Alves T., Silveira-Mattos P.S., Rocha M.S., West K., Natarajan M., Viswanathan V., Babu S., et al. Persistent inflammation during anti-tuberculosis treatment with diabetes comorbidity. Elife. 2019;8 doi: 10.7554/eLife.46477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arriaga M.B., Rocha M.S., Nogueira B.M.F., Nascimento V., Araújo-Pereira M., Souza A.B., Andrade A.M.S., Costa A.G., Gomes-Silva A., Silva E.C., et al. The Effect of Diabetes and Prediabetes on Mycobacterium tuberculosis Transmission to Close Contacts. J. Infect. Dis. 2021;224:2064–2072. doi: 10.1093/infdis/jiab264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil-Santana L., Almeida-Junior J.L., Oliveira C.A.M., Hickson L.S., Daltro C., Castro S., Kornfeld H., Netto E.M., Andrade B.B. Diabetes Is Associated with Worse Clinical Presentation in Tuberculosis Patients from Brazil: A Retrospective Cohort Study. PLoS One. 2016;11 doi: 10.1371/journal.pone.0146876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arriaga M.B., Araújo-Pereira M., Barreto-Duarte B., Nogueira B., Freire M.V.C.N.S., Queiroz A.T.L., Rodrigues M.M.S., Rocha M.S., Souza A.B., Spener-Gomes R., et al. The Effect of Diabetes and Prediabetes on Antituberculosis Treatment Outcomes: A Multicenter Prospective Cohort Study. J. Infect. Dis. 2022;225:617–626. doi: 10.1093/infdis/jiab427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barreda N.N., Arriaga M.B., Aliaga J.G., Lopez K., Sanabria O.M., Carmo T.A., Fróes Neto J.F., Lecca L., Andrade B.B., Calderon R.I. Severe pulmonary radiological manifestations are associated with a distinct biochemical profile in blood of tuberculosis patients with dysglycemia. BMC Infect. Dis. 2020;20:139. doi: 10.1186/s12879-020-4843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calderon R.I., Arriaga M.B., Aliaga J.G., Barreda N.N., Sanabria O.M., Barreto-Duarte B., Franco J.P.D., Lecca L., Andrade B.B., Carvalho A.C.C., Kritski A.L. Persistent dysglycemia is associated with unfavorable treatment outcomes in patients with pulmonary tuberculosis from Peru. Int. J. Infect. Dis. 2022;116:293–301. doi: 10.1016/j.ijid.2022.01.012. [DOI] [PubMed] [Google Scholar]
- 19.Queiroz A.T.L., Vinhaes C.L., Fukutani E.R., Gupte A.N., Kumar N.P., Fukutani K.F., Arriaga M.B., Sterling T.R., Babu S., Gaikwad S., et al. A multi-center, prospective cohort study of whole blood gene expression in the tuberculosis-diabetes interaction. Sci. Rep. 2023;13:7769. doi: 10.1038/s41598-023-34847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckold C., Kumar V., Weiner J., Alisjahbana B., Riza A.L., Ronacher K., Coronel J., Kerry-Barnard S., Malherbe S.T., Kleynhans L., et al. Impact of Intermediate Hyperglycemia and Diabetes on Immune Dysfunction in Tuberculosis. Clin. Infect. Dis. 2021;72:69–78. doi: 10.1093/cid/ciaa751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamada Y., Penn-Nicholson A., Krishnan S., Cirillo D.M., Matteelli A., Wyss R., Denkinger C.M., Rangaka M.X., Ruhwald M., Schumacher S.G. Are mRNA based transcriptomic signatures ready for diagnosing tuberculosis in the clinic? - A review of evidence and the technological landscape. EBioMedicine. 2022;82 doi: 10.1016/j.ebiom.2022.104174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singhania A., Wilkinson R.J., Rodrigue M., Haldar P., O'Garra A. The value of transcriptomics in advancing knowledge of the immune response and diagnosis in tuberculosis. Nat. Immunol. 2018;19:1159–1168. doi: 10.1038/s41590-018-0225-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burel J.G., Babor M., Pomaznoy M., Lindestam Arlehamn C.S., Khan N., Sette A., Peters B. Host Transcriptomics as a Tool to Identify Diagnostic and Mechanistic Immune Signatures of Tuberculosis. Front. Immunol. 2019;10:221. doi: 10.3389/fimmu.2019.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warsinske H., Vashisht R., Khatri P. Host-response-based gene signatures for tuberculosis diagnosis: A systematic comparison of 16 signatures. PLoS Med. 2019;16 doi: 10.1371/journal.pmed.1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan S., Queiroz A.T.L., Gupta A., Gupte N., Bisson G.P., Kumwenda J., Naidoo K., Mohapi L., Mave V., Mngqibisa R., et al. Integrative Multi-Omics Reveals Serum Markers of Tuberculosis in Advanced HIV. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.676980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulkarni V., Queiroz A.T.L., Sangle S., Kagal A., Salvi S., Gupta A., Ellner J., Kadam D., Rolla V.C., Andrade B.B., et al. A Two-Gene Signature for Tuberculosis Diagnosis in Persons With Advanced HIV. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.631165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Wu H., Jin X., Zheng P., Hu S., Xu X., Yu W., Yan J. Study of cardiovascular disease prediction model based on random forest in eastern China. Sci. Rep. 2020;10:5245. doi: 10.1038/s41598-020-62133-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanko M., Grendár M., Snopko P., Opšenák R., Šutovský J., Benčo M., Soršák J., Zeleňák K., Kolarovszki B. Random Forest-Based Prediction of Outcome and Mortality in Patients with Traumatic Brain Injury Undergoing Primary Decompressive Craniectomy. World Neurosurg. 2021;148:e450–e458. doi: 10.1016/j.wneu.2021.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Ra H.J., Parks W.C. Control of matrix metalloproteinase catalytic activity. Matrix Biol. 2007;26:587–596. doi: 10.1016/j.matbio.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amaral E.P., Vinhaes C.L., Oliveira-de-Souza D., Nogueira B., Akrami K.M., Andrade B.B. The Interplay Between Systemic Inflammation, Oxidative Stress, and Tissue Remodeling in Tuberculosis. Antioxid. Redox Signal. 2021;34:471–485. doi: 10.1089/ars.2020.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabir N., Hussain T., Mangi M.H., Zhao D., Zhou X. Matrix metalloproteinases: Expression, regulation and role in the immunopathology of tuberculosis. Cell Prolif. 2019;52 doi: 10.1111/cpr.12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arriaga M.B., Karim F., Queiroz A.T.L., Araújo-Pereira M., Barreto-Duarte B., Sales C., Moosa M.Y.S., Mazibuko M., Milne G.L., Maruri F., et al. Effect of Dysglycemia on Urinary Lipid Mediator Profiles in Persons With Pulmonary Tuberculosis. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.919802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M., Divangahi M., Gan H., Shin D.S.J., Hong S., Lee D.M., Serhan C.N., Behar S.M., Remold H.G. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J. Exp. Med. 2008;205:2791–2801. doi: 10.1084/jem.20080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vinhaes C.L., Oliveira-de-Souza D., Silveira-Mattos P.S., Nogueira B., Shi R., Wei W., Yuan X., Zhang G., Cai Y., Barry C.E., 3rd, et al. Changes in inflammatory protein and lipid mediator profiles persist after antitubercular treatment of pulmonary and extrapulmonary tuberculosis: A prospective cohort study. Cytokine. 2019;123 doi: 10.1016/j.cyto.2019.154759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paruchuri S., Tashimo H., Feng C., Maekawa A., Xing W., Jiang Y., Kanaoka Y., Conley P., Boyce J.A. Leukotriene E4-induced pulmonary inflammation is mediated by the P2Y12 receptor. J. Exp. Med. 2009;206:2543–2555. doi: 10.1084/jem.20091240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salimi M., Stöger L., Liu W., Go S., Pavord I., Klenerman P., Ogg G., Xue L. Cysteinyl leukotriene E(4) activates human group 2 innate lymphoid cells and enhances the effect of prostaglandin D(2) and epithelial cytokines. J. Allergy Clin. Immunol. 2017;140:1090–1100.e11. doi: 10.1016/j.jaci.2016.12.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rafnsson A., Bäck M. Urinary leukotriene E4 is associated with renal function but not with endothelial function in type 2 diabetes. Dis. Markers. 2013;35:475–480. doi: 10.1155/2013/370461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lordkipanidzé M., Pharand C., Schampaert E., Turgeon J., Palisaitis D.A., Diodati J.G. A comparison of six major platelet function tests to determine the prevalence of aspirin resistance in patients with stable coronary artery disease. Eur. Heart J. 2007;28:1702–1708. doi: 10.1093/eurheartj/ehm226. [DOI] [PubMed] [Google Scholar]
- 39.Oosaki R., Mizushima Y., Mita H., Shida T., Akiyama K., Kobayashi M. Urinary leukotriene E4 and 11-dehydrothromboxane B2 in patients with aspirin-sensitive asthma. Allergy. 1997;52:470–473. doi: 10.1111/j.1398-9995.1997.tb01032.x. [DOI] [PubMed] [Google Scholar]
- 40.Catella F., Healy D., Lawson J.A., FitzGerald G.A. 11-Dehydrothromboxane B2: a quantitative index of thromboxane A2 formation in the human circulation. Proc. Natl. Acad. Sci. USA. 1986;83:5861–5865. doi: 10.1073/pnas.83.16.5861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirwan D.E., Chong D.L.W., Friedland J.S. Platelet Activation and the Immune Response to Tuberculosis. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.631696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cubillos-Angulo J.M., Nogueira B.M.F., Arriaga M.B., Barreto-Duarte B., Araújo-Pereira M., Fernandes C.D., Vinhaes C.L., Villalva-Serra K., Nunes V.M., Miguez-Pinto J.P., et al. Host-directed therapies in pulmonary tuberculosis: Updates on anti-inflammatory drugs. Front. Med. 2022;9 doi: 10.3389/fmed.2022.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pereira P.A.T., Assis P.A., Prado M.K.B., Ramos S.G., Aronoff D.M., de Paula-Silva F.W.G., Sorgi C.A., Faccioli L.H. Prostaglandins D(2) and E(2) have opposite effects on alveolar macrophages infected with Histoplasma capsulatum. J. Lipid Res. 2018;59:195–206. doi: 10.1194/jlr.M078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tobin D.M., Roca F.J., Oh S.F., McFarland R., Vickery T.W., Ray J.P., Ko D.C., Zou Y., Bang N.D., Chau T.T.H., et al. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell. 2012;148:434–446. doi: 10.1016/j.cell.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merry C., Fu K., Wang J., Yeh I.J., Zhang Y. Targeting the checkpoint kinase Chk1 in cancer therapy. Cell Cycle. 2010;9:279–283. doi: 10.4161/cc.9.2.10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cen M., Ouyang W., Lin X., Du X., Hu H., Lu H., Zhang W., Xia J., Qin X., Xu F. FBXO6 regulates the antiviral immune responses via mediating alveolar macrophages survival. J. Med. Virol. 2023;95 doi: 10.1002/jmv.28203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang T., Huang C., Lopez-Coral A., Slentz-Kesler K.A., Xiao M., Wherry E.J., Kaufman R.E. K12/SECTM1, an interferon-gamma regulated molecule, synergizes with CD28 to costimulate human T cell proliferation. J. Leukoc. Biol. 2012;91:449–459. doi: 10.1189/jlb.1011498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huyton T., Göttmann W., Bade-Döding C., Paine A., Blasczyk R. The T/NK cell co-stimulatory molecule SECTM1 is an IFN "early response gene" that is negatively regulated by LPS in human monocytic cells. Biochim. Biophys. Acta. 2011;1810:1294–1301. doi: 10.1016/j.bbagen.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 49.Khan T.A., Mazhar H., Saleha S., Tipu H.N., Muhammad N., Abbas M.N. Interferon-Gamma Improves Macrophages Function against M. tuberculosis in Multidrug-Resistant Tuberculosis Patients. Chemother. Res. Pract. 2016;2016 doi: 10.1155/2016/7295390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flynn J.L., Chan J., Triebold K.J., Dalton D.K., Stewart T.A., Bloom B.R. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J. Exp. Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singer S.N., Ndumnego O.C., Kim R.S., Ndung'u T., Anastos K., French A., Churchyard G., Paramithiothis E., Kasprowicz V.O., Achkar J.M. Plasma host protein biomarkers correlating with increasing Mycobacterium tuberculosis infection activity prior to tuberculosis diagnosis in people living with HIV. EBioMedicine. 2022;75 doi: 10.1016/j.ebiom.2021.103787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Young B.L., Mlamla Z., Gqamana P.P., Smit S., Roberts T., Peter J., Theron G., Govender U., Dheda K., Blackburn J. The identification of tuberculosis biomarkers in human urine samples. Eur. Respir. J. 2014;43:1719–1729. doi: 10.1183/09031936.00175113. [DOI] [PubMed] [Google Scholar]
- 53.Kornfeld H., West K., Kane K., Kumpatla S., Zacharias R.R., Martinez-Balzano C., Li W., Viswanathan V. High Prevalence and Heterogeneity of Diabetes in Patients With TB in South India: A Report from the Effects of Diabetes on Tuberculosis Severity (EDOTS) Study. Chest. 2016;149:1501–1508. doi: 10.1016/j.chest.2016.02.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gupte A., Padmapriyadarsini C., Mave V., Kadam D., Suryavanshi N., Shivakumar S.V.B.Y., Kohli R., Gupte N., Thiruvengadam K., Kagal A., et al. Cohort for Tuberculosis Research by the Indo-US Medical Partnership (CTRIUMPH): protocol for a multicentric prospective observational study. BMJ Open. 2016;6 doi: 10.1136/bmjopen-2015-010542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breiman L. Random Forest. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 58.Kuhn M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008;28:26. doi: 10.18637/jss.v028.i05. [DOI] [Google Scholar]
- 59.Arriaga M.B., Amorim G., Queiroz A.T.L., Rodrigues M.M.S., Araújo-Pereira M., Nogueira B.M.F., Souza A.B., Rocha M.S., Benjamin A., Moreira A.S.R., et al. Novel stepwise approach to assess representativeness of a large multicenter observational cohort of tuberculosis patients: The example of RePORT Brazil. Int. J. Infect. Dis. 2021;103:110–118. doi: 10.1016/j.ijid.2020.11.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Geadas C., Stoszek S.K., Sherman D., Andrade B.B., Srinivasan S., Hamilton C.D., Ellner J. Advances in basic and translational tuberculosis research: Proceedings of the first meeting of RePORT international. Tuberculosis (Edinb) 2017;102:55–67. doi: 10.1016/j.tube.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Heijden Y.F., Abdullah F., Andrade B.B., Andrews J.R., Christopher D.J., Croda J., Ewing H., Haas D.W., Hatherill M., Horsburgh C.R., Jr., et al. Building capacity for advances in tuberculosis research; proceedings of the third RePORT international meeting. Tuberculosis (Edinb) 2018;113:153–162. doi: 10.1016/j.tube.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arriaga M.B., Araújo-Pereira M., Barreto-Duarte B., Sales C., Miguez-Pinto J.P., Nogueira E.B., Nogueira B.M.F., Rocha M.S., Souza A.B., Benjamin A., et al. Prevalence and Clinical Profiling of Dysglycemia and HIV Infection in Persons With Pulmonary Tuberculosis in Brazil. Front. Med. 2021;8 doi: 10.3389/fmed.2021.804173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broekmans J.F., Migliori G.B., Rieder H.L., Lees J., Ruutu P., Loddenkemper R., Raviglione M.C., World Health Organization International Union Against Tuberculosis and Lung Disease and Royal Netherlands Tuberculosis Association Working Group. Lung D., Royal Netherlands Tuberculosis Association Working Group European framework for tuberculosis control and elimination in countries with a low incidence. Recommendations of the World Health Organization (WHO), International Union Against Tuberculosis and Lung Disease (IUATLD) and Royal Netherlands Tuberculosis Association (KNCV) Working Group. Eur. Respir. J. 2002;19:765–775. doi: 10.1183/09031936.02.00261402. [DOI] [PubMed] [Google Scholar]
- 64.American Diabetes Association Professional Practice Committee. Draznin B., Aroda V.R., Bakris G., Benson G., Brown F.M., Freeman R., Green J., Huang E., Isaacs D., et al. 6. Glycemic Targets: Standards of Medical Care in Diabetes-2022. Diabetes Care. 2022;45:S83–S96. doi: 10.2337/dc22-S006. [DOI] [PubMed] [Google Scholar]
- 65.Song W.L., Wang M., Ricciotti E., Fries S., Yu Y., Grosser T., Reilly M., Lawson J.A., FitzGerald G.A. Tetranor PGDM, an abundant urinary metabolite reflects biosynthesis of prostaglandin D2 in mice and humans. J. Biol. Chem. 2008;283:1179–1188. doi: 10.1074/jbc.M706839200. [DOI] [PubMed] [Google Scholar]
- 66.Murphey L.J., Williams M.K., Sanchez S.C., Byrne L.M., Csiki I., Oates J.A., Johnson D.H., Morrow J.D. Quantification of the major urinary metabolite of PGE2 by a liquid chromatographic/mass spectrometric assay: determination of cyclooxygenase-specific PGE2 synthesis in healthy humans and those with lung cancer. Anal. Biochem. 2004;334:266–275. doi: 10.1016/j.ab.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 67.Oliveira-de-Souza D., Vinhaes C.L., Arriaga M.B., Kumar N.P., Cubillos-Angulo J.M., Shi R., Wei W., Yuan X., Zhang G., Cai Y., et al. Molecular degree of perturbation of plasma inflammatory markers associated with tuberculosis reveals distinct disease profiles between Indian and Chinese populations. Sci. Rep. 2019;9:8002. doi: 10.1038/s41598-019-44513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data used here were from two previous works. The gene expression from the main population, Salvador site from RePORT Brazil, and Indian population (used only in the validation of the transcriptomic signature) were from the MSTDI study and have been deposited at the GEO ncbi database and is publicly available as of the date of publication (Accession number GSE181143, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE181143).
The validation of the transcriptomic signature identified here was performed using an external cohort, with population from South Africa and Romania, previously published by the TANDEM consortium and have been previously deposited at the SRA database and are publicly available as of the date of publication (BioProject ID PRJNA470512).
-
•
All employed packages' references are available at the key resources tables and methodology.
-
•
This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.