Abstract
The Saccharomyces cerevisiae PIS1 gene is essential and required for the final step in the de novo synthesis of phosphatidylinositol. Transcription of the PIS1 gene is uncoupled from the factors that regulate other yeast phospholipid biosynthetic genes. Most of the phospholipid biosynthetic genes are regulated in response to inositol and choline via a regulatory circuit that includes the Ino2p:Ino4p activator complex and the Opi1p repressor. PIS1 is regulated in response to carbon source and anaerobic growth conditions. Both of these regulatory responses are modest, which is not entirely surprising since PIS1 is essential. However, even modest regulation of PIS1 expression has been shown to affect phosphatidylinositol metabolism and to affect cell cycle progression. This prompted the present study, which employed a genomic screen, database mining, and more traditional promoter analysis to identify genes that affect PIS1 expression. A screen of the viable yeast deletion set identified 120 genes that affect expression of a PIS1-lacZ reporter. The gene set included several peroxisomal genes, silencing genes, and transcription factors. Factors suggested by database mining, such as Pho2 and Yfl044c, were also found to affect PIS1-lacZ expression. A PIS1 promoter deletion study identified an upstream regulatory sequence element that was required for carbon source regulation located downstream of three previously defined upstream activation sequence elements. Collectively, these studies demonstrate how a collection of genomic and traditional strategies can be implemented to identify a set of genes that affect the regulation of an essential gene.
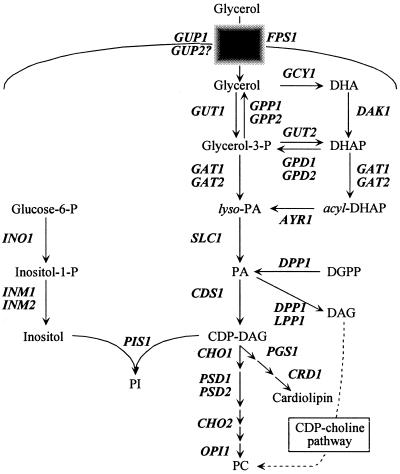
Yeast has been an excellent model for the study of phospholipid biosynthesis (Fig. 1) (11, 12, 28, 33). Phosphatidylinositol (PI) is an essential phospholipid in all eukaryotic cells (3, 11, 12, 28, 33, 38, 54). In yeast, PI is synthesized de novo by the product of the PIS1 gene, PI synthase (18, 23, 24, 36, 53-56), and represents 12 to 27% of the total phospholipid composition (11, 12, 28, 33). In addition to a structural role, PI is a precursor of phosphoinositides, sphingolipids, and inositol polyphosphates (11, 12, 28, 33). PI and these metabolites are required for a diverse set of processes that include glycolipid anchoring of proteins (69), signal transduction (21, 58), mRNA export (57, 64-66), and vesicle trafficking (17). In spite of the importance of PI and its metabolites, relatively little is known about factors that regulate PIS1 expression.
FIG. 1.
Schematic depiction of the S. cerevisiae phospholipid biosynthetic pathway. The CDP-choline pathway, also known as the “Kennedy” and “salvage pathway,” is noted by a broken arrow. Genes are designated in boldface and italic type. Abbreviations: DHA, dihydroxyacetone; DHAP, dihydroxyacetone phosphate; PA, phosphatidic acid; DGPP, diacylglycerol pyrophosphate; DAG, diacylglcerol.
Our understanding of the role of PIS1 expression in regulating PI synthesis is conflicted. One study reported that overexpression of the human PIS1 gene in COS-7 cells yielded a significant increase in PI synthase activity (25-fold) but a modest increase in PI levels (8.2%) (50). However, another report indicated that overexpression of the rat PIS1 gene in NIH3T3 cells yielded elevated levels of PI, PI-4,5-P2, and PI-3,4,5-P3 (19). PIS1 overexpression also decreased the doubling time of transformed cells and accelerated G1 progression (19). Consistent with the effect on G1 progression, cyclin D1 and cyclin E levels were elevated (19). Furthermore, Rous sarcoma virus-infected NIH3T3 cells and activated erbB2-transformed NIH3T3 cells also overexpress PIS1 and have elevated PI levels (37). Finally, specific inhibition of PI synthase activity using inostamycin reduces PI levels and inhibits induction of S phase (18, 36).
PI synthase is a membrane-associated enzyme that catalyzes the condensation of CDP-diacylglycerol and inositol to PI (23, 54) (Fig. 1). Disruption of the PIS1 gene results in lethality (54). Because PIS1 is essential, it is not entirely surprising that yeast cells do not extensively regulate PIS1 expression or PI synthase levels (2, 24, 25). PIS1 gene expression is not coregulated with the other phospholipid biosynthetic genes in response to inositol and choline (2, 11, 12, 28, 33) but is instead regulated by carbon source and oxygen. PIS1 expression is repressed in response to glycerol and aerobic conditions (2, 25). Promoter deletion analysis identified three upstream activation sequence (UAS) elements (UAS1 to UAS3) required for PIS1 gene expression (25). However, the element required for glycerol repression was not identified. The region that includes the UAS3 element also contains an upstream regulatory sequence (URS) that binds Rox1p to exert anaerobic regulation (25). The significance of the anaerobic regulation is evidenced by altered membrane composition. PI levels are elevated in cells grown anaerobically, and phosphatidylcholine (PC) and CDP-diacylglycerol levels are also affected by oxygen (25).
PIS1 gene expression is insensitive to inositol and choline; however, inositol does affect PI synthase activity. High levels of inositol increase the rate of PI synthesis because the Km of PI synthase for inositol (0.21 mM) is ninefold greater than the intracellular concentration of inositol (24 μM) (41). When cells are grown in inositol, PI levels double at the expense of PC synthesis as inositol is also a noncompetitive inhibitor of phosphatidylserine synthase (CHO1 gene product), the first enzyme in the PC branch of phospholipid biosynthesis (Fig. 1) (41).
PIS1 transcript levels are established by an unusual combination of low transcription initiation rates and very stable mRNA. The PIS1 gene is expressed from a weak promoter that is comparable in activity to that of the GAL4, INO2, and INO4 transcription factor genes (1). However, the weakness of the PIS1 promoter is compensated by a stable transcript (half-life, 58 min) (1). In fact, the PIS1 transcript is among the most stable yeast mRNAs (34). The combination of low transcription initiation rates and high transcript stability may ensure that PIS1 transcripts change slowly in response to environmental changes.
In this study, we utilized a genomic strategy to identify genes that affect regulation of PIS1 expression. Analysis of the viable yeast deletion set (VYDS) identified several genes that affect PIS1 expression and regulation. We also examined the role of transcription factors defined by a “ChIP-on-chip” (chromatin immunoprecipitation-on-microarray chip) approach (62) on PIS1 expression. Last, we identified a URS needed for PIS1 repression in response to glycerol.
MATERIALS AND METHODS
Strains, media, and growth conditions. The Saccharomyces cerevisiae strains used in this study were BRS1001 (MATa ade2-1 his3-11,15 leu2-3,112 can1-100 ura3-1 trp1-1), BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), and BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and the complete VYDS in the BY4742 and BY4741 backgrounds. Yeast cultures were grown at 30°C in complete synthetic medium (42) (containing 2% glucose) lacking uracil. Where appropriate, glucose concentration was varied or substituted with 3% glycerol, 3% acetate, 3% ethanol, 3% lactate, or 0.1% oleic acid with 0.2% Tween 80. Where indicated, 75 μM inositol and 1 mM choline were added (I+C+ medium) and/or NaCl was added (0.7 M final concentration).
Genomic studies.
Plasmid pMA107 (2), containing 629 bp of the PIS1 promoter fused to the lacZ gene, was transformed into the BY4742-based VYDS (Res Gen Invitrogen Corp., Carlsbad, Calif.) by a standard lithium acetate yeast transformation procedure (14) in 96-well microtiter plates. Transformants were selected on complete synthetic medium lacking uracil (Ura−), replicated onto Ura− 2% glucose X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and Ura− 3% glycerol X-Gal plates and allowed to grow at 30°C for 48 h. VYDS transformants were screened for altered PIS1-lacZ expression, indicated by a light-blue phenotype on 2% glucose X-Gal plates or a dark-blue phenotype on 3% glycerol X-Gal plates relative to the isogenic wild-type (BY4742) strain. To eliminate false positives, pMA107 transformants that exhibited a phenotype were transformed with plasmids pJH330 (543 bp of the INO1 gene and 132 codons of the INO1 gene fused in frame to the lacZ reporter gene in YEp357R) (22) and YEp357R-TCM1 (TCM1-lacZ) (described below). Mutant alleles in the VYDS were confirmed by sequencing bar codes that marked each mutant.
Plasmid construction.
The construction of some of the PIS1-cat reporters has been previously reported (25). Briefly, a nested set of PIS1 promoter deletions was created by PCR using appropriate oligonucleotides (Table 1). The 5′-terminal deletion mutants were created using the reverse primer PIS1-3′ (−1) along with the forward primer set PIS1 (−918) to PIS1 (−127) (Table 1). The individual PCR products were cloned into pGEM-T (Promega, Madison, Wis.). PIS1 promoter fragments were excised from pGEM-T by digestion with BamHI and BglII and inserted into pBM2015 to create a fusion with the cat reporter gene (29). To create PIS1 promoter internal deletions, the regions from position −325 to various downstream points was amplified using forward primer PIS1 (−325) along with the reverse primer set PIS1 (−185) to PIS1 (−51B) (Table 1). These fragments were sequentially cloned into pGEM-T and pBM2015 as described above. The regions downstream of each internal deletion were amplified using the reverse primer PIS1-3′ (−1) along with the forward primer set PIS1 (−101F) to PIS1 (−26F) in addition to PIS1 (−149F) and PIS1 (−127) (Table 1). These fragments were sequentially cloned into pGEM-T and pBM2015 containing the relevant regions above the deletion endpoints as described above. For each plasmid, the name indicates the deletion endpoints. The pBM2015 derivatives were sequenced by the Wayne State University Core Sequencing Facility by using the primer pBM2015-SEQ. Yeast strains containing the promoter-cat plasmids integrated at the GAL4 locus were created by transformation and characterized by Southern blot hybridization as previously described (4).
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotide (position) | Sequence |
|---|---|
| PIS1-3′ (−1) | 5′-GGATCCCTTGTACTATCACAC-3′ |
| PIS1 (−918) | 5′-AGATCTGAATTCCCTGCACGC-3′ |
| PIS1 (−621) | 5′-AGATCTAAGCTTTTAGCCATG-3′ |
| PIS1 (−521) | 5′-AGATCTTAAATCCAAATCTTC-3′ |
| PIS1 (−325) | 5′-AGATCTTGTGCTTGAGGCTCA-3′ |
| PIS1 (−224) | 5′-AGATCTTTTTTTCATTTTATA-3′ |
| PIS1 (−205) | 5′-AGATCTTTCACGTCGCTCCGC-3′ |
| PIS1 (−184) | 5′-AGATCTATCCCTATTACGGAA-3′ |
| PIS1 (−149F) | 5′-AGATCCGCCCCTCCTATTGTT-3′ |
| PIS1 (−138) | 5′-AGATCTGTTTTTTCCGTCTCG-3′ |
| PIS1 (−127) | 5′-AGATCTCTCGAGGATTTTTCA-3′ |
| PIS1 (−185) | 5′-GGATCCTTACGTGCGGAGCGA-3′ |
| PIS1 (−149B) | 5′-GGATCCTTTCCCTATTGAGAA-3′ |
| PIS1 (−126B) | 5′-GGATCCACGGAAAAAACAATA-3′ |
| PIS1 (−101B) | 5′-GGATCCATACTAAAAGTGAAA-3′ |
| PIS1 (−76B) | 5′-GGATCCCTCATGTTTTTTATG-3′ |
| PIS1 (−51B) | 5′-GGATCCTGACGGCAAATAAAC-3′ |
| PIS1 (−101F) | 5′-AGATCTATAAGTAAAACATAA-3′ |
| PIS1 (−76F) | 5′-AGATCTAGGTGGTATGGTTTA-3′ |
| PIS1 (−51F) | 5′-AGATCTCTGTGGAGCCTTCAA-3′ |
| PIS1 (−26F) | 5′-AGATCTTAAGAGGGAAAGTGT-3′ |
| PIS1 URS (−76) | 5′-CTCGAGAGGTGGTATGGTTTA-3′ |
| PIS1 URS (−51) | 5′-CTCGAGTGACGGCAAATAAAC-3′ |
| TCM1 (−572) | 5′-AAGCTTCTTACGTTATCATTC-3′ |
| TCM1 (−1) | 5′-GAATTCGATTGATTGTTGTAG-3′ |
| pBM2015-SEQ | 5′-TCTCGTGCGGAGATGACTGC-3′ |
Plasmid YEp357R-TCM1 contains 572 bp of the sequences upstream of the TCM1 gene fused in frame to the lacZ reporter gene in YEp357R (52). This plasmid was constructed by amplifying 572 bp of the TCM1 promoter using S. cerevisiae genomic DNA (Res Gen Invitrogen Corp.) by using primers TCM1 (−572) and TCM1 (−1) (Table 1). The 572-bp PCR product was cloned into pGEM-T (Promega) and then excised by digestion with HindIII and EcoRI and inserted into YEp357R. Insert orientation and sequence were confirmed by DNA sequencing.
Plasmid pLGΔ312 + PIS1 URSGLY contains a 25-bp fragment of the PIS1 promoter (−76 to −51) inserted upstream of the lacZ reporter gene in pLGΔ312 (32). This plasmid was constructed by amplifying 25 bp of the PIS1 gene promoter (−76 to −51) from plasmid pPIS1(−325) using primers PIS1 URS (−76) and PIS1 URS (−51) (Table 1). The 37-bp PCR product was cloned into pGEM-T (Promega) and then excised by digestion with XhoI and inserted into plasmid pLGΔ312. Insert orientation and sequence were confirmed by DNA sequencing.
Enzyme assays.
β-Galactosidase and chloramphenicol acetyltransferase (CAT) assays were performed as described previously (4, 48). However, some of the β-galactosidase experiments were performed using a microtiter plate platform. Units of β-galactosidase activity were defined as follows: (A420/minute/milligram of total protein) × 1,000. Units of CAT activity were defined as counts per minute in the organic phase and expressed as a percentage of the total counts per minute (percent conversion) divided by the amount of protein assayed (in micrograms) and the time of incubation (in hours). Protein concentration in each extract was determined by using a Bio-Rad (Rockville Center, N.Y.) protein assay kit. A minimum of five independent measurements were made for each data point.
Growth phase assays.
Expression of the PIS1 gene was monitored from BRS1001 transformants harboring the pPIS1-918 (cat) reporter construct. Cells were grown in I−C− and I+C+ media in both the absence and presence of 0.7 M NaCl. Five-milliliter aliquots were removed at several intervals over the span of 100 h and monitored for density (using a Klett-Summerson colorimeter). CAT activity (described above) was determined from all samples simultaneously.
RESULTS
A genome-wide screen for genes that affect PIS1-lacZ expression in glucose and glycerol.
To obtain a comprehensive understanding of the regulation of PIS1 expression, we screened the VYDS (∼4,800 viable mutant strains) for changes in PIS1-lacZ expression of strains grown on different carbon sources. Even though PIS1 is regulated only threefold in response to carbon source, this regulation was clearly discernible on X-Gal medium containing glucose (dark blue) and glycerol (light blue). The PIS1-lacZ reporter (pMA107) (2) was transformed into the BY4742-based VYDS (MATα set), and transformants were screened for light-blue colonies on X-Gal glucose medium and dark-blue colonies on X-Gal glycerol medium. Three rounds of screening identified 178 mutants that yielded reduced expression in glucose and 59 mutants with increased expression in glycerol. To eliminate mutants that nonspecifically affected lacZ expression, we rescreened the mutants using INO1-lacZ (phospholipid biosynthetic gene) (22) and TCM1-lacZ (ribosomal protein gene) reporters. Strains exhibiting altered expression of all three reporters (109 strains on glucose and 8 strains on glycerol) were removed from the data set. The remaining mutants affected PIS1 specifically, PIS1 and INO1, or PIS1 and TCM1 (Table 2).
TABLE 2.
Genes that affect PIS1-lacZ expression
| Reduced expression in glucose
|
Increased expression in glycerol
|
||||
|---|---|---|---|---|---|
| PIS1 | PIS1 and INO1 | PIS1 and TCM1 | PIS1 | PIS1 and INO1 | PIS1 and TCM1 |
| APT1 | APG17 | ACE2 | AAD4 | ACB1 | NYV1 |
| CPD1 | COQ4 | CIK1 | DCP3 | ADH2 | PAC1 |
| CPT1 | DYN1 | CPS1 | ENT4 | CTF19 | RIF1 |
| CRH1 | ENT3 | DFG10 | HST3 | CVT19 | YDR445C |
| CWH41 | HIR3 | EAP1 | HST4 | EGD1 | YDR521W |
| FAB1 | PDX3 | GSH1 | OGG1 | HSP104 | YGL250W |
| GCS1 | REG2 | LSB3 | PEX3 | NTG2 | YLL032C |
| GPD1 | RMD1 | MUD1 | PEX4 | PEX22 | YLL054C |
| MUM2 | RPB9 | NHX1 | PEX17 | RIM8 | YLL057C |
| NRG2 | RPS25A | PEX7 | PMT3 | THI12 | YPL144W |
| OSM1 | VTA1 | PHO2 | PTC3 | TIR4 | |
| PET130 | YBR053C | PTC1 | ROT2 | VPS33 | |
| PIM1 | YDL119C | RPB4 | RPL8B | VTS1 | |
| POG1 | YDL129W | SIN3 | SMY1 | YTP1 | |
| RDH54 | YDL173W | SIT4 | STM1 | YCR076C | |
| SPO75 | YGL165C | TTR1 | YDR124W | YDR417C | |
| SRT1 | YJL178C | YAL068C | YGR022C | YOL153C | |
| WH14 | YNR025C | YDL025C | YKR043C | YOR315W | |
| YBR250W | YOL107W | YDL057W | YOR029W | YPL162C | |
| YDR332W | YPR123C | YDL063C | YOR322C | YPL166W | |
| YGL050W | YKL147W | YPR151C | |||
| YGR242W | YLR254C | ||||
| YIL028W | YLR366W | ||||
| YMR073C | YNL134C | ||||
| YPL077C | |||||
In strains grown on 2% glucose, 25 mutant strains displayed decreased PIS1-lacZ expression, 20 mutant strains affected PIS1-lacZ and INO1-lacZ expression, and 24 mutant strains yielded altered PIS1-lacZ and TCM1-lacZ expression (Table 2). In strains grown in 3% glycerol, PIS1-lacZ expression was specifically altered in 20 mutant strains, 21 mutant strains had altered PIS1-lacZ and INO1-lacZ expression, and 10 strains yielded altered PIS1-lacZ and TCM1-lacZ expression (Table 2).
The mutant set contains an overrepresentation of mutants that affect a few specific biological processes. The genomic screen identified six genes involved in peroxisome biogenesis (pex3, pex4, pex17, and pex22) and function (acb1 and gpd1). The GPD1 gene is also involved in an early step in phospholipid biosynthesis (Fig. 1). The screen also yielded four genes required for chromatin silencing (rif1, hst3, hst4, and sin3). There is an overrepresentation of mutants in DNA repair (ptc3, ogg1, ntg2, rpb4, and rpb9) which has recently been associated with PI metabolism (84). It is interesting that two subunits of RNA polymerase involved in transcription-coupled repair were also identified (rpb4 and rpb9). The genomic screen also yielded two mutants involved in carbon source regulation that may play a role in glycerol repression (nrg2 and reg2). Last, genes required for PI metabolism were also identified (FAB1 and ACB1 are required for synthesis of phosphoinositides and sphingolipids, respectively). We assume that the genomic screen described here did not identify every gene affecting PIS1-lacZ expression (see below); however, it did provide a wealth of information that would not have been possible if more traditional genetic screens were used.
Peroxisomal biogenesis affects glycerol regulation of PIS1-lacZ expression.
Peroxisome biogenesis is a conserved process among eukaryotes, involving at least 32 known peroxins (20, 60, 70, 72-74, 77, 78, 80, 81). Peroxisomes are membrane-bound organelles that function in metabolic pathways involved in the inactivation of toxic substances (H2O2-based respiration), the regulation of cellular oxygen levels, and the metabolism of lipids, nitrogen bases, and carbohydrates including fatty acid β-oxidation (60, 72, 74, 77, 78). Genomic analysis of the VYDS strain collection identified that PIS1-lacZ expression in cells grown on 3% glycerol X-Gal plates was significantly increased in pex3Δ, pex4Δ, pex17Δ, and pex22Δ mutant strains. However, we assumed that because the screen of the VYDS relied on a relatively modest phenotype, it probably did not identify all genes that regulate PIS1-lacZ expression. Given this possibility, we also tested the other known pexΔ mutants present in the VYDS that were not identified by the genomic plate screen. We performed liquid assays to quantify the effect of all known pexΔ mutants grown in medium containing 2% glucose and 3% glycerol on PIS1-lacZ expression. In addition, we also quantified PIS1-lacZ expression in medium containing 0.1% oleic acid, since this carbon source causes peroxisomes to proliferate (70).
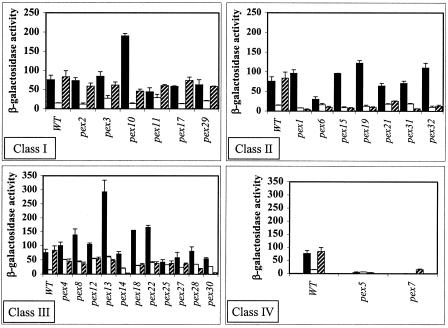
As expected in the wild-type strain, PIS1-lacZ expression was repressed in 3% glycerol but unaffected in 0.1% oleic acid (Fig. 2). The results show that most of the pexΔ mutants affected expression of the PIS1-lacZ reporter under at least one growth condition. This included three of the mutants that were identified in the initial plate screen. That is, PIS1-lacZ gene expression in cultures grown in 3% glycerol was elevated in pex3Δ, pex4Δ, and pex22Δ mutant strains. It is important that we also observed elevated expression in the pex12Δ and pex13Δ mutants grown in 3% glycerol (Fig. 2). These two mutants were present in the original plate screen but eliminated by subsequent screens, suggesting that our screening criteria may have been too stringent. However, we also observed that the pex17Δ mutant identified in the plate screen did not reveal a difference relative to the wild type when quantified by the liquid assay. This discrepancy may be explained by the fact that the plate assay reports the accumulation of β-galactosidase over several phases of growth, whereas the liquid assay quantifies β-galactosidase activity at a single stage of growth.
FIG. 2.
Peroxisomal biogenesis mutants affect PIS1-lacZ gene expression. PIS1-lacZ gene expression was assayed by using plasmid pMA107 containing 629 bp of the PIS1 gene promoter (−629 to −1). Plasmid pMA107 was transformed into a wild-type (WT) strain (BY4742; MATα) and isogenic strains containing pexΔ mutant alleles, and β-galactosidase activity was measured. Cultures were grown in Ura− medium containing 2% glucose (solid bars), 3% glycerol (empty bars), or 0.1% oleic acid with 0.2% Tween 80 (hatched bars). The mutants were divided into four classes described in the text. Note that the y-axis range differs between the bar graphs.
The effect of the pexΔ mutants on PIS1-lacZ expression fell into four classes. Class I contained mutants that generally displayed the same pattern of PIS1-lacZ expression in the three carbon sources as that of the wild-type strain, elevated expression in glucose and oleic acid and decreased expression in glycerol (pex2Δ, pex3Δ, pex10Δ, pex11Δ, pex17Δ, and pex29Δ) (Fig. 2). However, the pex3Δ, pex10Δ, and pex11Δ mutants clearly yielded altered expression levels relative to that of the wild-type control. Class II included several mutants that yielded levels of expression in oleic acid that were below wild-type expression levels in oleic acid (pex1Δ, pex6Δ, pex15Δ, pex19Δ, pex21Δ, pex31Δ, and pex32Δ) (Fig. 2). Class III included mutants that yielded levels of expression in glycerol that were above wild-type expression levels in glycerol and decreased levels of expression in oleic acid (pex4Δ, pex8Δ, pex12Δ, pex13Δ, pex14Δ, pex18Δ, pex22Δ, pex25Δ, pex27Δ, pex28Δ, and pex30Δ) (Fig. 2). Class IV included mutants that generally yielded low levels of expression in all three carbon sources (pex5Δ and pex7Δ) (Fig. 2).
The results clearly show that screening of the VYDS can yield valuable information regarding regulation of gene expression, even for a modestly regulated gene. These pex mutants could not possibly have been identified by more traditional means. Most importantly, these results provide the first evidence of coordination between the synthesis of peroxisomes and phospholipids.
Chromatin silencing genes affect PIS1-lacZ expression.
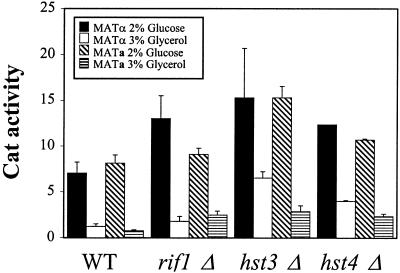
Genomic analysis of the VYDS also identified rif1Δ, hst3Δ, and hst4Δ mutant strains with elevated PIS1-lacZ expression on 3% glycerol X-Gal plates. All three mutant strains are involved in chromatin silencing at telomeres. The HST3 and HST4 genes are homologs of SIR2 and are also involved in regulating short-chain fatty acid metabolism (8, 71). Hst3p is also required for silencing at the origin of replication from the endogenous 2μm plasmid (31). To determine if the PIS1-lacZ plate phenotype was due to genuine effects on transcription and not an effect on plasmid copy number, we assayed a PIS1-cat reporter stably integrated in single copy at the GAL4 locus. We also determined if mating type affected expression by comparing CAT activity in MATa (BY4741 background) and MATα (BY4742 background) transformants. PIS1-cat expression in all three mutant strains was elevated in both glucose and glycerol regardless of mating type, although the hst3Δ and hst4Δ mutants had the more obvious effect (Fig. 3). These results also show that the plate phenotype was not due to an indirect effect of plasmid copy number. This is the first evidence of chromatin silencing affecting PIS1 expression.
FIG. 3.
Chromatin silencing gene mutants affect PIS1-cat expression. A PIS1 promoter fragment (pPIS-325) was fused to the cat reporter gene and integrated in single copy at the GAL4 locus in strains BY4742 (MATα) and BY4741 (MATa) and isogenic rif1Δ, hst3Δ, and hst4Δ strains. PIS1-cat expression was assayed in transformants grown in medium containing either 2% glucose (MATα, solid bars; MATa, hatched bars) or 3% glycerol (MATα, empty bars; MATa, horizontal bars). WT, wild type.
Mutants in glycerol utilization affect regulation of PIS1-lacZ expression.
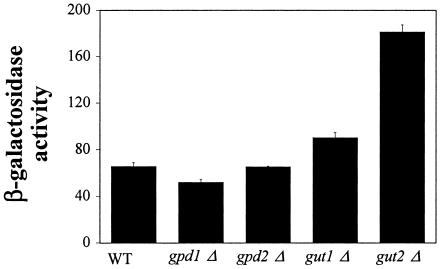
Genomic analysis of the VYDS also identified the gpd1Δ mutant strain with decreased PIS1-lacZ expression on 2% glucose X-Gal plates. GPD1 encodes an NADH-dependent cytosolic glycerol-3-phosphate dehydrogenase, a key enzyme in the initial stages of glycerol metabolism (Fig. 1) (15, 61). This was an interesting result given that PIS1 gene expression is repressed by glycerol. This led us to determine if additional enzymes involved in the early stages of glycerol utilization also affect PIS1-lacZ expression. Thus, we quantified β-galactosidase activity in a gpd1Δ mutant strain as well as gpd2Δ, gut1Δ, and gut2Δ mutant strains grown in 2% glucose. GPD2 encodes an isoform of glycerol-3-phosphate dehydrogenase (Fig. 1) (59), while GUT1 encodes glycerol kinase, and GUT2 encodes mitochondrial glycerol-3-phosphate dehydrogenase (Fig. 1) (26, 27).
PIS1-lacZ expression increased substantially in the gut2Δ strain (176%) and to a much lesser extent in the gut1Δ mutant strain (37%) when cells were grown in 2% glucose (Fig. 4). As expected from the VYDS screen, the gpd1Δ strain yielded a 20% decrease in β-galactosidase activity (Fig. 4). However, the gpd2Δ mutant strain was indistinguishable from the wild-type control (Fig. 4).
FIG. 4.
Mutants in glycerol utilization affect regulation of PIS1-lacZ expression. PIS1-lacZ expression was assayed by using plasmid pMA107 containing 629 bp of PIS1 gene promoter (−629 to −1). Plasmid pMA107 was transformed into a wild-type (WT) strain (BY4742; MATα) and isogenic strains containing gpd1Δ, gpd2Δ, gut1Δ, and gut2Δ mutant alleles, and β-galactosidase activity was measured. Cultures were grown in Ura− medium containing 2% glucose.
PIS1 promoter binding proteins identified by ChIPs are required for PIS1 expression.
In an effort to identify additional factors that regulate PIS1 expression, we mined a ChIP-on-chip database containing information on yeast DNA-binding proteins (47). This analysis identified Pho2, Yfl044c, and Ste12 as weak candidate PIS1 promoter binding proteins (47). Pho2 is involved in the response to phosphate starvation (5, 39, 40, 43, 51, 67, 68), while Ste12 is involved in pheromone and pseudohyphal regulation (16, 35, 62, 83). YFL044c encodes a hypothetical open reading frame (ORF) with unknown function. Interestingly, the pho2 mutant was identified in our plate screen as yielding reduced expression on 2% glucose X-Gal medium (Table 2). Previously, three UAS elements (UAS1 [−224 to −205], UAS2 [−184 to −149], and UAS3 [−149 to −127]) required for PIS1 gene expression were identified (25). Consistent with our results, potential Ste12p and Pho2p binding sites were identified in PIS1 UAS1, while PIS1 UAS2 contains an additional potential Pho2p binding site.
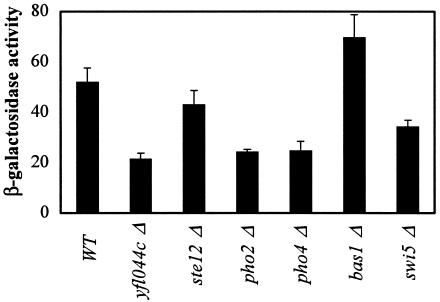
These results prompted us to quantify PIS1-lacZ expression in pho2Δ, yfl044cΔ, and ste12Δ mutant strains. PIS1-lacZ expression generally decreased in the three mutant strains, but the effect was most obvious in the pho2Δ and yfl044cΔ mutants (Fig. 5). The homeodomain transcription factor Pho2 is known to interact with Swi5, Pho4, and Bas1 in vivo and activate transcription of the HO gene (7, 9), phosphate utilization genes (7, 82), and genes involved in purine and histidine biosynthesis (7), respectively. Because Pho2 associates with these three proteins, we also quantified PIS1-lacZ expression in swi5Δ, pho4Δ, and bas1Δ mutant strains. PIS1-lacZ expression increased 34% in the bas1Δ strain, while pho4Δ and swi5Δ mutant strains exhibited lower levels of PIS1-lacZ expression (53% and 34%, respectively) relative to that of the wild-type strain (Fig. 5). Because Pho4 and Pho2 are required for induction of target genes in response to low phosphate concentrations (5, 39, 40, 43, 51, 67, 68), we also quantified the effect of phosphate on PIS1-lacZ expression. However, we found that PIS1-lacZ expression was not significantly affected by phosphate concentration (data not shown).
FIG. 5.
Transcription factor mutants affect PIS1-lacZ gene expression. PIS1-lacZ expression was assayed by using plasmid pMA107 containing 629 bp of PIS1 gene promoter (−629 to −1). Plasmid pMA107 was transformed into a wild-type (WT) strain (BY4742; MATα) and isogenic strains containing yfl044Δ, ste12Δ, pho2Δ, pho4Δ, bas1Δ, and swi5Δ mutant alleles, and β-galactosidase activity was measured. Cultures were grown in Ura− medium containing 2% glucose.
The ste12Δ mutant did not dramatically affect PIS1-lacZ expression under the condition we tested or in response to pheromone induction (data not shown). However, Ste12 may affect PIS1 expression under conditions we have not yet tested since Ste12 is known to regulate a diverse set of genes in response to at least two different signals (16, 35, 62, 83).
The effect of the yfl044cΔ mutant on PIS1-lacZ expression is the first evidence of regulation of gene expression by this putative transcription factor. While the role of Yfl044c was defined by the ChIP-on-chip database, the only other evidence to suggest that it regulates PIS1 expression was our β-galactosidase liquid assay. To corroborate the role of Yfl044c in PIS1 expression, we transformed a pRS200 derivative containing the YFL044c ORF (along with 5′- and 3′-flanking sequences) into the yfl044c mutant strain and assayed it for PIS1-lacZ expression. The presence of YFL044c on the pRS200 plasmid restored PIS1-lacZ expression to a level ∼3-fold higher than that of the wild-type strain (data not shown). This result suggests that Yfl044c may be limiting for PIS1 expression in a wild-type strain under the conditions tested here.
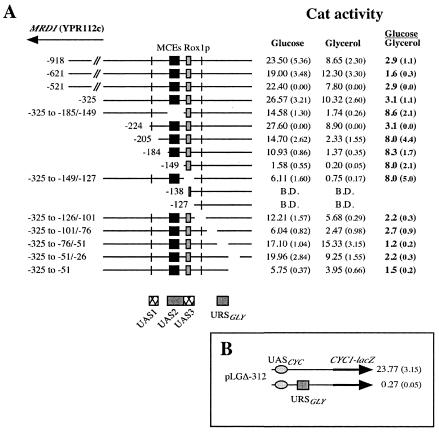
PIS1 promoter deletion analysis identifies a regulatory element required for glycerol repression.
A nested set of deletions of the PIS1 promoter fused to the cat reporter gene in plasmid pBM2015 was transformed into a wild-type strain (BRS1001) targeting integration at the GAL4 locus in single copy and in a single orientation. We have previously reported the analysis of a deletion subset grown in glucose that identified three UAS elements at positions −224 to −205 (UAS1), −184 to −149 (UAS2), and −149 to −138 (UAS3) (Fig. 6A) (25). Deletion of UAS1 decreased PIS1-cat expression by half (Fig. 6A, compare pPIS-224 and pPIS-205), while deletion of UAS2 exacerbated the decrease in PIS1-cat expression by another 90% (Fig. 6A, compare pPIS-205 and pPIS-149). PIS1-cat expression was below detectable levels when UAS3 was also deleted (Fig. 6A, compare pPIS-225 and pPIS-127). However, the data showed that none of these UAS elements were required for the response to glycerol. Curiously, deleting any one of the three UAS elements increased the degree of repression caused by growth in glycerol (from ∼3-fold to ∼8-fold [Fig. 6A]). One explanation for this result is that when the UAS elements are deleted, the glycerol repression factor is more effective in repressing transcription because promoter activity is lower.
FIG. 6.
A. Identification of a PIS1 promoter element required for glycerol-mediated repression. BRS1001 (wild-type) transformants containing various promoter deletions were grown in synthetic medium lacking uracil and containing either 2% glucose or 3% glycerol. Yeast extracts from the transformants were prepared and assayed for CAT activity. MRD1 is a divergent ORF. The locations of two MCEs (Mcm1p binding sites) (black box), a Rox1p binding site (gray box), potential TATA boxes (bars), three UAS elements, and a URS element are shown. B.D., below detection. B. PIS1 URSGLY element functions as a URS element. Plasmid pLGΔ312 + URSGLY, containing 25 bp of PIS1 gene promoter (−76 to −51), was transformed into a wild-type strain (BRS1001), and β-galactosidase activity was measured. Cultures were grown in Ura− 2% glucose medium.
Because the three UAS elements are required for expression, it was necessary to generate internal deletions in order to analyze promoter sequences downstream of the three UAS elements. Analysis of the PIS1 promoter downstream of the three UAS elements identified a repressor site (URSGLY), located promoter proximal, required for glycerol repression. The data identified a 25-bp region (−76 to −51) that, when deleted, abolishes PIS1 glycerol repression (Fig. 6A). This organization of regulatory elements is reminiscent of GAL4 gene expression which is repressed fourfold by growth in glucose and requires a URS element located downstream of a UAS element (29, 30). A heterologous system was also employed to further examine PIS1 URSGLY function. The 25-bp region (−76 to −51) defined by the deletion analysis was inserted into plasmid pLGΔ312, previously utilized to test URS function (32, 49). Plasmid pLGΔ312 contains the CYC1 promoter fused to the lacZ reporter gene. The 25-bp PIS1 URSGLY caused CYC1-lacZ expression to decrease dramatically (Fig. 6B). This result indicates that the PIS1 URSGLY sequence functions as a URS element. The URSGLY effect on CYC1-lacZ expression in cells grown in glycerol could not be assessed because the CYC1 promoter is regulated by a carbon source.
Glycerol repression of PIS1 expression is reversed by glucose.
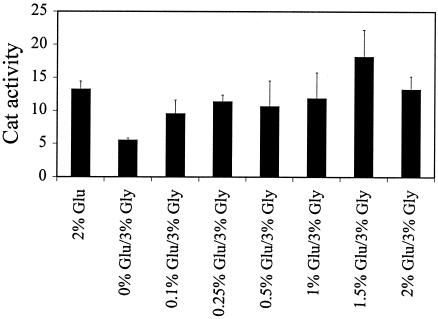
Glycerol-mediated repression of PIS1 transcription is unusual since it does not appear to involve any of the previously identified regulators of carbon source regulation (1, 2). To determine if glycerol-mediated repression is reversible, PIS1-cat activity from a wild-type strain (BRS1001) containing pPIS-325 was assayed from culture grown in medium containing 2% glucose or 3% glycerol with various concentrations of glucose (range, 0.1 to 2%). The data show that glucose concentrations as low as 0.1% were able to partially reverse glycerol repression and that concentrations from 1 to 2% completely reversed repression of PIS1-cat expression (Fig. 7).
FIG. 7.
Glycerol repression of PIS1-cat expression is reversed by glucose. BRS1001 transformants containing pPIS-325 were grown in Ura− synthetic medium containing 3% glycerol and 0, 0.1, 0.25, 0.5, 1.0, 1.5, or 2% glucose, and CAT activity was assayed.
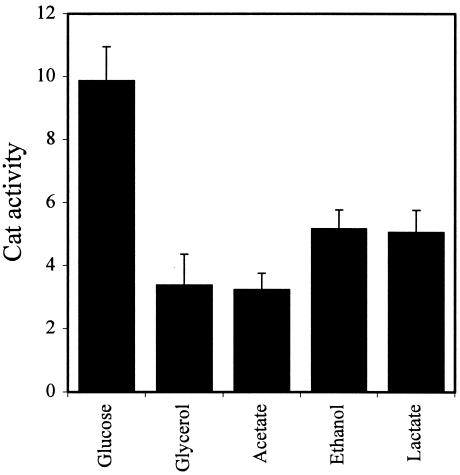
PIS1 expression is elevated in fermentable carbon sources (glucose and galactose) relative to a nonfermentable carbon source (glycerol) (2). We quantified the effect of other nonfermentable carbon sources on PIS1-cat expression by using the pPIS-325 construct. In addition to glycerol, PIS1-cat expression is reduced ∼50% when cells are grown with acetate, ethanol, or lactate as a carbon source (Fig. 8).
FIG. 8.
Analysis of PIS1-cat expression in response to different carbon sources. A PIS1 promoter fragment (pPIS-325) created by PCR was fused to the cat reporter gene and integrated in single copy at the GAL4 locus in strain BRS1001 (wild type). PIS1-cat expression (pPIS-325) was assayed in BRS1001 transformants grown in Ura− medium containing 2% glucose, 3% glycerol, 3% acetate, 3% ethanol, or 3% lactate.
PIS1 expression is not affected by growth phase or hyperosmotic stress.
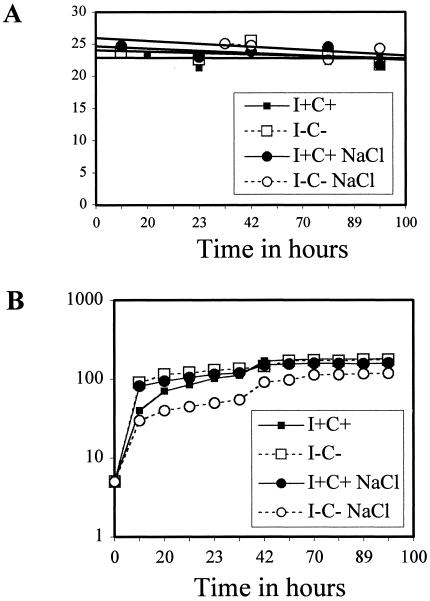
It has been reported that several phospholipid biosynthetic genes are regulated in response to growth phase (46). For example, CHO1-lacZ expression levels in I−C− medium are low during lag phase and increase throughout log phase, peaking at the beginning of stationary phase (46, 63). Once in stationary phase, expression levels decrease precipitously until they reach the initial expression levels seen in lag phase. This expression pattern is similar in I+C+ medium; however, because expression is repressed in this medium, the overall expression levels are lower than those observed in the I−C− medium (46, 63). We also discovered that growing cells in hyperosmotic medium (0.7 M NaCl) eliminates the growth phase regulation without affecting the inositol-choline-mediated regulation (63). These observations prompted an examination of the effect of growth phase and hyperosmotic conditions on PIS1 expression. This examination was done by quantifying CAT activity from pPIS1-918 transformants of BRS1001. These experiments revealed that neither growth phase nor hyperosmotic medium affected expression of the PIS1-cat gene (Fig. 9).
FIG. 9.
A. PIS1-cat expression is not regulated by growth phase or hyperosmotic stress. BRS1001 transformants containing pPIS1-918 were grown in Ura− synthetic medium in the presence of inositol and choline (I+C+) (solid squares), in the absence of inositol and choline (I−C−) (open squares), in the presence of 0.7 M NaCl (solid circles), or in the presence of inositol, choline, and 0.7 M NaCl (open circles). Yeast extracts from the transformants were prepared and assayed for CAT activity. B. Growth patterns of cultures used as described above (A).
DISCUSSION
Until recently, PIS1 gene expression was not believed to be regulated (2, 25, 45, 75, 76). However, we have previously shown that PIS1 expression is regulated by carbon source (2) and in response to oxygen availability (25, 45, 75, 76). Multiple approaches were applied to identify genes that affect PIS1 expression. The genomic screen of the VYDS identified 69 mutants with reduced PIS1 expression in glucose and 51 mutants with increased PIS1 expression in glycerol. PIS1 promoter deletion analysis coupled with mining of databases of DNA-binding proteins identified three UAS elements and one URS element and putative cognate transcription factors. For example, the promoter deletion analysis identified one UAS element with potential Ste12 and Pho2 binding sites (UAS1 [−224 to −205]), an additional UAS element with a potential Pho2 binding site and two known Mcm1 binding sites (UAS2 [−184 to −149]), and a potential Gcr1 binding site within another UAS element (UAS3 [−149 to −138]), as well as the Rox1 binding site essential for the anaerobic regulation of PIS1 (25). Comparison of promoters from related Saccharomyces yeast species revealed strong sequence similarity among the three UAS regions, including the potential Ste12 binding sites, and known Mcm1 and Rox1 binding sites between S. cerevisiae and S. castellii, S. bayanus, S. kluyveri, S. kudriavzevii, and S. mikatae (25). Mining of the yeast ChIP-on-chip database identified Yfl044c and Mcm1 as candidate PIS1 promoter binding proteins (47).
This study demonstrated that genomic approaches provide an excellent means to identify regulatory mechanisms that control modestly regulated and essential genes such as PIS1. However, while single strategies provide an excellent starting point, it is the combination of multiple genomic approaches that is essential. For example, Rox1 was not identified as a regulator of the PIS1 promoter by the ChIP-on-chip study (P value of 0.19) (47) or by the VYDS screen; however, it was suggested by searches of databases of DNA-binding proteins coupled with promoter deletion (25) and microarray studies (45, 75, 76). Our previous studies have clearly shown that Rox1 binds and bends the PIS1 promoter to exert anaerobic regulation on the PIS1 gene (25). Conversely, the VYDS screen we employed here did not identify Yfl044c, and examination of the PIS1 promoter sequence does not reveal an obvious binding site for Yfl044c (TTCTTKTYYTTTT) (47). However, the ChIP-on-chip study placed Yfl004c on the cusp of the acceptable binding threshold (P value of 0.006) (47), and we have shown here that Yfl044c does regulate PIS1-lacZ expression. The most obvious PIS1 promoter binder defined by the ChIP-on-chip study was Mcm1 (P value of 0.000003), and this protein has previously been shown to bind the PIS1 promoter (44). However, the VYDS screen could not have identified MCM1 since it is an essential gene. The use of multiple genomic approaches should also be expected to generate overlapping information. This was the case with Pho2 (also known as Bas2 and Grf10), which was suggested to be a regulator of PIS1 by the promoter deletion analysis since potential binding sites are found in UAS1 and UAS2 (25). The VYDS screen also identified Pho2 as a regulator of PIS1-lacZ expression. However, the ChIP-on-chip database did not identify Pho2 as a potential binder (P value of 0.5) (47). These results underscore the benefits of employing multiple genomic screens coupled with database mining and more traditional approaches.
This study further demonstrated that genomic approaches might identify biological functions that affect the expression of a gene, thereby illuminating interplay between biological processes. The VYDS screen identified an overrepresentation of mutants that affect three biological processes: peroxisome biogenesis (pex3, pex4, pex17, and pex22) and function (acb1 and gpd1), chromatin silencing (rif1, hst3, hst4, and sin3), and DNA repair (ptc3, ogg1, ntg2, rpb4, and rpb9). Alterations of PIS1 expression in the 26 assayed pex mutants are extremely interesting given the cellular role peroxisomes play, specifically in cellular oxygen regulation, and the metabolism of lipids, nitrogen bases, carbohydrates, and fatty acid β-oxidation (60, 72, 74, 77, 78). This finding suggests that changes within the peroxisome, potentially due to alterations in fatty acid metabolism (60, 72, 74, 77-79), could be responsible for alterations in PIS1 expression, reminiscent of what was seen for CIT2 in cells with altered mitochondrial activity (10, 13). This finding further supports the existence of a pathway for retrograde regulation between peroxisomes and the nucleus and is suggestive of the existence of a novel mechanism for the cell to adjust to changes in peroxisomal activity.
Note the identification of chromatin silencing and DNA repair mutants by the VYDS screen. The dual roles HST3 and HST4 play in chromatin silencing and regulating short-chain fatty acid metabolism again suggest a relationship between PIS1 expression and fatty acid metabolism. Our understanding of the role of Hst3 and Hst4 in silencing is not as well developed as that of other silencing proteins. Moreover, our studies cannot distinguish between direct and indirect effects on PIS1 expression. Additional studies will focus on whether these proteins bind the PIS1 promoter directly. The DNA repair genes defined in the VYDS screen included RPB4 and RPB9. These genes may affect PIS1 expression either because they play a regulatory role in transcription of PIS1 or because of their role in DNA repair. Regardless of the mechanism whereby these two genes affect PIS1 expression, it is clear that other DNA repair genes also affected PIS1 expression. The identification of a link between a damage checkpoint pathway and PI metabolism has been suggested by other genome-wide studies (84). This is not entirely unexpected given that PIS1 and PI have a role in cell cycle progression (18, 19, 36, 37).
The promoter deletion study identified a URS element, downstream of the three UAS elements, that was required for glycerol repression. Examination of the sequence defined by the URSGLY element did not reveal any obvious protein-binding site. This observation is consistent with our findings that many genes involved in carbon source regulation in yeast do not affect PIS1 expression (REG1, GLC7, MIG1, etc. [M. E. Gardocki and J. M. Lopes, unpublished results]). We did, however, find that REG2 and NRG2 affected PIS1-lacZ expression and that these genes have a role in carbon source regulation (6). Nrg2 binds DNA directly, and its levels are decreased in glycerol- or ethanol-grown cells. However, Nrg2 is a repressor and would therefore be expected to increase PIS1 expression in glycerol (6). Moreover, the PIS1 promoter does not contain a binding site for Nrg2. Thus, it seems possible that a novel transcription factor may be involved in glycerol-mediated PIS1 expression. This factor may be present in the collection of genes defined by the VYDS screen.
Our knowledge of PIS1 expression, its regulation, and the various biological processes with which it may be intimately intertwined can only now begin to be discerned as a direct result of the results presented here. Genomic studies coupled with database mining and with more classical strategies can obviously be successfully combined to study modestly regulated genes. Of course, the challenge is in interpreting the wealth of information that these genomic approaches provide. A critical part of this process will be to distinguish between direct and indirect effects. Another important issue that will need to be addressed is to determine which of the genes and processes defined here affect PI metabolism. PI metabolism has been shown to be affected by anaerobic conditions via the Rox1 protein (25) and by carbon source (D. Kamath and J. M. Lopes, unpublished results), suggesting that regulation of PIS1 expression does affect PI metabolism. Furthermore, the long half-life of the PIS1 transcript (57 min) (1) and an AUG codon located upstream of the PI synthase AUG codon (25) may also contribute to the regulation of PIS1 gene expression.
Acknowledgments
We thank Meng Chen, Linan Chen, Leandria Hancock, Ying He, Niketa Jani, Sangeeta Oza, and Jimmy Lo for their efforts in editing the manuscript. We also thank Edward Golenberg and George Brush for helpful discussions.
This work was initially supported by an NSF grant (MCB-0110408) and completed with support from an NSF grant (MCB-0415511) to J.M.L., the William A. Turner, Jr., Memorial Foundation scholarship to M.E.G., and an undergraduate research grant from the Wayne State University Undergraduate Research Council to K.B.
REFERENCES
- 1.Anderson, M. S. 1996. Analysis of PIS1 gene expression in Saccharomyces cerevisiae. Loyola University of Chicago, Maywood, Ill.
- 2.Anderson, M. S., and J. M. Lopes. 1996. Carbon source regulation of PIS1 gene expression in Saccharomyces cerevisiae involves the MCM1 gene and the two-component regulatory gene, SLN1. J. Biol. Chem. 271:26596-26601. [DOI] [PubMed] [Google Scholar]
- 3.Antonsson, B. 1997. Phosphatidylinositol synthase from mammalian tissues. Biochim. Biophys. Acta 1348:179-186. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner, B. P., and J. M. Lopes. 1995. Autoregulated expression of the yeast INO2 and INO4 helix-loop-helix genes effects cooperative regulation on their target genes. Mol. Cell. Biol. 15:1709-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbaric, S., M. Münsterkötter, C. Goding, and W. Hörz. 1998. Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2. Mol. Cell. Biol. 18:2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkey, C. D., V. K. Vyas, and M. Carlson. 2004. Nrg1 and Nrg2 transcriptional repressors are differently regulated in response to carbon source. Eukaryot. Cell 3:311-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhoite, L. T., J. M. Allen, E. Garcia, L. R. Thomas, I. D. Gregory, W. P. Voth, K. Whelihan, R. J. Rolfes, and D. J. Stillman. 2002. Mutations in the Pho2 (Bas2) transcription factor that differentially affect activation with its partner proteins Bas1, Pho4, and Swi5. J. Biol. Chem. 277:37612-37618. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann, C., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 9.Brazas, R. M., and D. J. Stillman. 1993. The Swi5 zinc-finger and Grf10 homeodomain proteins bind DNA cooperatively at the yeast HO promoter. Proc. Natl. Acad. Sci. USA 90:11237-11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butow, R. A., and N. G. Avadhani. 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14:1-15. [DOI] [PubMed] [Google Scholar]
- 11.Carman, G. M., and S. A. Henry. 1999. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38:361-399. [DOI] [PubMed] [Google Scholar]
- 12.Carman, G. M., and G. M. Zeimetz. 1996. Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 271:13293-13296. [DOI] [PubMed] [Google Scholar]
- 13.Chelstowska, A., and R. A. Butow. 1995. RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J. Biol. Chem. 270:18141-18146. [DOI] [PubMed] [Google Scholar]
- 14.Chen, D.-C., B.-C. Yang, and T.-T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 15.Costenoble, R., L. Adler, C. Niklasson, and G. Liden. 2003. Engineering of the metabolism of Saccharomyces cerevisiae for anaerobic production of mannitol. FEMS Yeast Res. 3:17-25. [DOI] [PubMed] [Google Scholar]
- 16.Crosby, J. A., J. B. Konopka, and S. Fields. 2000. Constitutive activation of the Saccharomyces cerevisiae transcriptional regulator Ste12p by mutations at the amino-terminus. Yeast 16:1365-1375. [DOI] [PubMed] [Google Scholar]
- 17.Czech, M. P. 2000. PIP2 and PIP3: complex roles at the cell surface. Cell 100:603-606. [DOI] [PubMed] [Google Scholar]
- 18.Deguchi, A., M. Imoto, and K. Umezawa. 1996. Inhibition of G1 cyclin expression in normal rat kidney cells by inostamycin, a phosphatidylinositol synthesis inhibitor. J. Biochem. (Tokyo) 120:1118-1122. [DOI] [PubMed] [Google Scholar]
- 19.Deguchi, A., K. Segawa, K. Hosaka, I. B. Weinstein, and K. Umezawa. 2002. Overexpression of phosphatidylinositol synthase enhances growth and G1 progression in NIH3T3 cells. Jpn. J. Cancer Res. 93:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Distel, B., R. Erdmann, S. J. Gould, G. Blobel, D. I. Crane, J. M. Cregg, G. Dodt, Y. Fujiki, J. M. Goodman, W. W. Just, et al. 1996. A unified nomenclature for peroxisome biogenesis factors. J. Cell Biol. 135:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Divecha, N., and R. F. Irvine. 1995. Phospholipid signaling. Cell 80:269-278. [DOI] [PubMed] [Google Scholar]
- 22.Elkhaimi, M., M. R. Kaadige, D. Kamath, J. C. Jackson, J. H. C. Biliran, and J. M. Lopes. 2000. Combinatorial regulation of phospholipid biosynthetic gene expression by the UME6, SIN3, and RPD3 genes. Nucleic Acids Res. 28:3160-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischl, A. S., and G. M. Carman. 1983. Phosphatidylinositol biosynthesis in Saccharomyces cerevisiae: purification and properties of microsome-associated phosphatidylinositol synthase. J. Bacteriol. 154:304-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischl, A. S., M. J. Homann, M. A. Poole, and G. M. Carman. 1986. Phosphatidylinositol synthase from Saccharomyces cerevisiae. J. Biol. Chem. 261:3178-3183. [PubMed] [Google Scholar]
- 25.Gardocki, M. E., and J. M. Lopes. 2003. Expression of the yeast PIS1 gene requires multiple regulatory elements including a Rox1p binding site. J. Biol. Chem. 278:38646-38652. [DOI] [PubMed] [Google Scholar]
- 26.Grauslund, M., J. M. Lopes, and B. Rønnow. 1999. Expression of GUT1, which encodes glycerol kinase in Saccharomyces cerevisiae, is controlled by the positive regulators Adr1p, Ino2p, and Ino4p, and the negative regulator Opi1p in a carbon source-dependent fashion. Nucleic Acids Res. 27:4391-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grauslund, M., and B. Rønnow. 2000. Carbon source-dependent transcriptional regulation of the mitochondrial glycerol-3-phosphate dehydrogenase gene, GUT2, from Saccharomyces cerevisiae. Can. J. Microbiol. 46:1096-1100. [DOI] [PubMed] [Google Scholar]
- 28.Greenberg, M. L., and J. M. Lopes. 1996. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 60:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griggs, D. W., and M. Johnston. 1993. Promoter elements determining weak expression of the GAL4 regulatory gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 13:4999-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griggs, D. W., and M. Johnston. 1991. Regulated expression of the GAL4 activator gene in yeast provides a sensitive genetic switch for glucose repression. Proc. Natl. Acad. Sci. USA 88:8597-8601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grünweller, A., and A. E. Ehrenhofer-Murray. 2002. A novel yeast silencer: the 2μ origin of Saccharomyces cerevisiae has HST3-, MIG1- and SIR-dependent silencing activity. Genetics 162:59-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guarente, L., and T. Mason. 1983. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32:1279-1286. [DOI] [PubMed] [Google Scholar]
- 33.Henry, S. A., and J. L. Patton-Vogt. 1998. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog. Nucleic Acid Res. Mol. Biol. 61:133-179. [DOI] [PubMed] [Google Scholar]
- 34.Herrick, D., R. Parker, and A. Jacobson. 1990. Identification and comparison of stable and unstable mRNAs in Saccharomyces cerevisiae. Mol. Cell. Biol. 10:2269-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 36.Imoto, M., T. Morii, A. Deguchi, and K. Umezawa. 1994. Involvement of phosphatidylinositol synthesis in the regulation of S phase induction. Exp. Cell Res. 215:228-233. [DOI] [PubMed] [Google Scholar]
- 37.Imoto, M., Y. Taniguchi, H. Fujiwara, and K. Umezawa. 1994. Enhancement of CDP-DA:inositol transferase activity in src- and erbB2-transformed cells. Exp. Cell Res. 212:151-154. [DOI] [PubMed] [Google Scholar]
- 38.Jackson, M., D. C. Crick, and P. J. Brennan. 2000. Phosphatidylinositol is an essential phospholipid of mycobacteria. J. Biol. Chem. 275:30092-30099. [DOI] [PubMed] [Google Scholar]
- 39.Kaffman, A., I. Herskowitz, R. Tjian, and E. K. O'Shea. 1994. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science 263:1153-1156. [DOI] [PubMed] [Google Scholar]
- 40.Kaffman, A., N. M. Rank, and E. K. O'Shea. 1998. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kap121. Genes Dev. 12:2673-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelley, M. J., A. M. Bailis, S. A. Henry, and G. M. Carman. 1988. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by inositol. Inositol is an inhibitor of phosphatidylserine synthase activity. J. Biol. Chem. 263:18078-18085. [PubMed] [Google Scholar]
- 42.Kelly, B. L., and M. L. Greenberg. 1990. Characterization and regulation of phosphatidylglycerolphosphate phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1046:144-150. [DOI] [PubMed] [Google Scholar]
- 43.Komeili, A., and E. K. O'Shea. 1999. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science 284:977-980. [DOI] [PubMed] [Google Scholar]
- 44.Kuo, M.-H., and E. Grayhack. 1994. A library of yeast genomic MCM1 binding sites contains genes involved in cell cycle control, cell wall and membrane structure, and metabolism. Mol. Cell. Biol. 14:348-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwast, K. E., L.-C. Lai, N. Menda, D. T. James III, S. Aref, and P. V. Burke. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184:250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamping, E., J. Lückl, F. Paltauf, S. A. Henry, and S. Kohlwein. 1995. Isolation and characterization of a mutant Saccharomyces cerevisiae with pleiotropic deficiencies in transcriptional activation and repression. Genetics 137:55-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, et al. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 48.Lopes, J. M., and S. A. Henry. 1991. Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 19:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luche, R. M., R. Sumrada, and T. G. Cooper. 1990. A cis-acting element present in multiple genes serves as a repressor protein binding site for the yeast CAR1 gene. Mol. Cell. Biol. 10:3884-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lykidis, A., P. D. Jackson, C. O. Rock, and S. Jackowski. 1997. The role of CDP-diacylglycerol synthetase and phosphatidylinositol synthase activity levels in the regulation of cellular phosphatidylinositol content. J. Biol. Chem. 272:33402-33409. [DOI] [PubMed] [Google Scholar]
- 51.McAndrew, P. C., J. Svaren, S. R. Martin, W. Hörz, and C. R. Goding. 1998. Requirements for chromatin modulation and transcription activation by the Pho4 acidic activation domain. Mol. Cell. Biol. 18:5818-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Myers, A. M., A. Tzagoloff, D. M. Kinney, and C. J. Lusty. 1986. Yeast shuttle and integrative vectors with multiple cloning sites suitable for construction of lacZ fusions. Gene 45:299-310. [DOI] [PubMed] [Google Scholar]
- 53.Nikawa, J.-I., T. Kodaki, and S. Yamashita. 1988. Expression of the Saccharomyces cerevisiae PIS gene and synthesis of phosphatidylinositol in Escherichia coli. J. Bacteriol. 170:4727-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikawa, J.-I., T. Kodaki, and S. Yamashita. 1987. Primary structure and disruption of the phosphatidylinositol synthase gene of Saccharomyces cerevisiae. J. Biol. Chem. 262:4876-4881. [PubMed] [Google Scholar]
- 55.Nikawa, J.-I., and S. Yamashita. 1984. Molecular cloning of the gene encoding CDPdiacylglycerol-inositol 3-phosphatidyltransferase in Saccharomyces cerevisiae. Eur. J. Biochem. 143:251-256. [DOI] [PubMed] [Google Scholar]
- 56.Nikawa, J.-I., and S. Yamashita. 1997. Phosphatidylinositol synthase from yeast. Biochim. Biophys. Acta 1348:173-178. [DOI] [PubMed] [Google Scholar]
- 57.Odom, A. R., A. Stahlberg, S. R. Wente, and J. D. York. 2000. A role for nuclear inositol 1,4,5-triphosphate kinase in transcriptional control. Science 287:2026-2029. [DOI] [PubMed] [Google Scholar]
- 58.Ohanian, J., and V. Ohanian. 2001. Sphingolipids in mammalian cell signalling. Cell. Mol. Life Sci. 58:2053-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Overkamp, K. M., B. M. Bakker, P. Kotter, A. van Tuijl, S. de Vries, J. P. van Dijken, and J. T. Pronk. 2000. In vivo analysis of the mechanisms for oxidation of cytosolic NADH by Saccharomyces cerevisiae mitochondria. J. Bacteriol. 180:2823-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purdue, P. E., and P. B. Lazarow. 2001. Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17:701-752. [DOI] [PubMed] [Google Scholar]
- 61.Remize, F., B. Cambon, L. Barnavon, and S. Dequin. 2003. Glycerol formation during wine fermentation is mainly linked to Gpd1p and is only partly controlled by the HOG pathway. Yeast 20:1243-1253. [DOI] [PubMed] [Google Scholar]
- 62.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, et al. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 63.Robinson, K. A., J. I. Koepke, M. Kharodawala, and J. M. Lopes. 2000. A network of yeast basic helix-loop-helix interactions. Nucleic Acids Res. 28:4460-4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saiardi, A., J. J. Caffrey, S. H. Snyder, and S. B. Shears. 2000. The inositol hexakisphosphate kinase family. J. Biol. Chem. 275:24686-24692. [DOI] [PubMed] [Google Scholar]
- 65.Saiardi, A., J. J. Caffrey, S. H. Snyder, and S. B. Shears. 2000. Inositol polyphosphate multikinase (ArgRIII) determines nuclear mRNA export in Saccharomyces cerevisiae. FEBS Lett. 468:28-32. [DOI] [PubMed] [Google Scholar]
- 66.Saiardi, A., H. Erdjument-Bromage, A. M. Snowman, P. Tempst, and S. H. Snyder. 1999. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr. Biol. 9:1323-1326. [DOI] [PubMed] [Google Scholar]
- 67.Shao, D., C. L. Creasy, and L. W. Bergman. 1998. A cysteine residue in helixII of the bHLH domain is essential for homodimerization of the yeast transcription factor Pho4p. Nucleic Acids Res. 26:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shao, D., C. L. Creasy, and L. W. Bergman. 1996. Interaction of Saccharomyces cerevisiae Pho2 with Pho4 increases the accessibility of the activation domain of Pho4. Mol. Gen. Genet. 251:358-364. [DOI] [PubMed] [Google Scholar]
- 69.Shields, D., and P. Arvan. 1999. Disease models provide insights into post-Golgi protein trafficking, localization and processing. Curr. Opin. Cell Biol. 11:489-494. [DOI] [PubMed] [Google Scholar]
- 70.Smith, J. J., M. Marelli, R. H. Christmas, F. J. Vizeacoumar, D. J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R. A. Rachubinski, and J. D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Starai, V. J., H. Takahashi, J. D. Boeke, and J. C. Escalante-Semerena. 2003. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetics 163:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Subramani, S., A. Koller, and W. B. Snyder. 2000. Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 69:399-418. [DOI] [PubMed] [Google Scholar]
- 73.Tam, Y. Y. C., J. C. Torres-Guzman, F. J. Vizeacoumar, J. J. Smith, M. Marelli, J. D. Aitchison, and R. A. Rachubinski. 2003. Pex11-related proteins in peroxisome dynamics: a role for the novel peroxin Pex27p in controlling peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 14:4089-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Terlecky, S. R., and M. Fransen. 2000. How peroxisomes arise. Traffic 1:465-473. [DOI] [PubMed] [Google Scholar]
- 75.Ter Linde, J. J. M., and H. Y. De Steensma. 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19:825-840. [DOI] [PubMed] [Google Scholar]
- 76.Ter Linde, J. J. M., H. Liang, R. W. Davis, H. Y. De Steensma, J. P. Van Dijken, and J. T. Pronk. 1999. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 181:7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Titorenko, V. I., and R. A. Rachubinski. 2001. The life cycle of the peroxisome. Nat. Rev. Mol. Cell Biol. 2:357-368. [DOI] [PubMed] [Google Scholar]
- 78.van den Bosch, H., R. B. H. Schutgens, R. J. A. Wanders, and J. M. Tager. 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61:157-197. [DOI] [PubMed] [Google Scholar]
- 79.van Roermund, C. W. T., H. R. Waterham, L. Ijlst, and R. J. A. Wanders. 2003. Fatty acid metabolism in Saccharomyces cerevisiae. Cell. Mol. Life Sci. 60:1838-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vizeacoumar, F. J., J. C. Torres-Guzman, D. Bouard, J. D. Aitchison, and R. A. Rachubinski. 2004. Pex30p, Pex31p, and Pex32p form a family of peroxisomal integral membrane proteins regulating peroxisome size and number in Saccharomyces cerevisiae. Mol. Biol. Cell 15:665-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vizeacoumar, F. J., J. C. Torres-Guzman, Y. Y. C. Tam, J. D. Aitchison, and R. A. Rachubinski. 2003. YHR150w and YDR479c encode peroxisomal integral membrane proteins involved in the regulation of peroxisome number, size, and distribution in Saccharomyces cerevisiae. J. Cell Biol. 161:321-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vogel, K., W. Hörz, and A. Hinnen. 1989. The two positively acting regulatory proteins PHO2 and PHO4 physically interact with PHO5 upstream activation regions. Mol. Cell. Biol. 9:2050-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zeitlinger, J., I. Simon, C. T. Harbison, N. Hannett, T. L. Volkert, G. R. Fink, and R. A. Young. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signalling. Cell 113:395-404. [DOI] [PubMed] [Google Scholar]
- 84.Zewail, A., M. W. Xie, Y. Xing, L. Lin, P. F. Zhang, W. Zou, J. P. Saxe, and J. Huang. 2003. Novel functions of the phosphatidylinositol metabolic pathway discovered by a chemical genomics screen with wortmannin. Proc. Natl. Acad. Sci. USA 100:3345-3350. [DOI] [PMC free article] [PubMed] [Google Scholar]