Abstract
Nonribosomal peptides, made by nonribosomal peptide synthetases, have diverse biological activities, including roles as fungal virulence effectors. Inspection of the genome of Cochliobolus heterostrophus, a fungal pathogen of maize and a member of a genus noted for secondary metabolite production, revealed eight multimodular nonribosomal peptide synthase (NPS) genes and three monomodular NPS-like genes, one of which encodes a nonribosomal peptide synthetase/polyketide synthase hybrid enzyme presumed to be involved in synthesis of a peptide/polyketide molecule. Deletion of each NPS gene and phenotypic analyses showed that the product of only one of these genes, NPS6, is required for normal virulence on maize. NPS6 is also required for resistance to hydrogen peroxide, suggesting it may protect the fungus from oxidative stress. This and all other nps mutants had normal growth, mating ability, and appressoria. Real-time PCR analysis showed that expression of all NPS genes is low (relative to that of actin), that all (except possibly NPS2) are expressed during vegetative growth, and that expression is induced by nitrogen starvation. Only NPS6 is unfailingly conserved among euascomycete fungi, including plant and human pathogens and saprobes, suggesting the possibility that NPS6 activity provides oxidative stress protection during both saprobic and parasitic growth.
Nonribosomal peptide synthetases are multimodular enzymes that make nonribosomal peptides through a thiotemplate mechanism independent of ribosomes. Nonribosomal peptides can be composed of d- and l-amino acids, protein and nonprotein amino acids, hydroxy acids, ornithine, β-amino acids, and other unusual constituents (36). Nonribosomal peptides can be linear, cyclic, or branched cyclic and may be modified by glycosylation, N-methylation, or acylation. In addition to structural diversity, nonribosomal peptides have a broad spectrum of biological activities, some of which have been useful in medicine, agriculture, and biological research (24, 37, 40, 47). Products made by nonribosomal peptide synthetases or nonribosomal peptide synthetase/polyketide synthase hybrid enzymes include well-known antibiotics (penicillin, erythromycin, and vancomycin), immunosuppressants (cyclosporin and rapamycin), antitumor agents (actinomycin, bleomycin, and epothilone) (39, 47), and toxins involved in pathogenesis (HC-toxin, enniatin, AM-toxin, and probably victorin) (17, 22, 32, 38, 48, 49).
A minimal nonribosomal peptide synthetase module is composed of an AMP-binding adenylation (A) and a thiolation (T, also called peptidyl carrier protein) domain (37). The A domain (500 to 600 amino acid residues) is required for amino acid substrate recognition and activation. The 80- to 100-amino-acid-residue T domain, located downstream of the A domain, is the site for 4′-phosphopantetheine cofactor binding; the holoenzyme then activates aminoacyl substrates to form a thioester bond. A condensation (C) domain (≈450 amino acids) is typically found after each A-T module and functions in peptide bond formation and elongation of the nascent peptide. Generally, the number and order of modules present in a nonribosomal peptide synthetase determine the length and structure of the resulting nonribosomal peptide. In addition to A, T, and C domains, an N-methyl transferase (M) domain that methylates the amino acid specified by the A domain may be inserted between the A and T domains of any given module, and an epimerase (E) domain that changes an amino acid from the l- to the d-form may be inserted between the T and C domains. In some nonribosomal peptide synthetases, a thioesterase domain is found at the C-terminal end of the protein and is thought to release the nonribosomal peptide from the nonribosomal peptide synthetase (7).
Although the thiotemplate mechanism of nonribosomal peptide synthesis is well documented, the physiological significance of the small peptides to the producing organisms is largely unknown. Some proposed activities include roles as signal molecules for coordination of growth and differentiation (16, 21, 28, 35), as aids in the breakdown of cellular metabolic products (9), as defense compounds that kill competing microorganisms (46), as siderophores to assist in iron uptake (5), and as virulence effectors (17, 22, 32, 38).
To date, the majority of characterized nonribosomal peptide synthetases and their nonribosomal peptide products have been from bacteria, especially spore-forming gram-positive soil bacteria such as Bacillus and Streptomyces species (37, 47). Relatively fewer fungal genes encoding nonribosomal peptide synthetases have been fully sequenced and their corresponding peptide structures determined. These include Tolypocladium inflatum (niveum) simA, which controls production of the immunosuppressive drug cyclosporine (50), Penicillium chrysogenum acvA, which controls production of the antibiotic penicillin (2), Ustilago maydis sid2 and Aspergillus nidulans sidC, each of which controls production of a siderophore (55, 12), Claviceps purpurea cpps1 and cpps2, both of which are involved in the synthesis of ergot alkaloids (6, 43), Hypocrea virens tex1, which controls peptaibol synthesis (51), Leptosphaeria maculans SirP, which controls sirodesmin biosynthesis (12), and three genes that control production of peptide virulence effectors: HTS1, which controls production of HC toxin by C. carbonum, a maize pathogen (38), AMT1, which controls production of AM toxin by Alternaria alternata, an apple tree pathogen (22), and Esyn1, which controls production of enniatin by Fusarium scirpi and other Fusarium spp., potato and tomato pathogens (17, 19).
Our study had two goals: first, to identify all the NPS genes in the genome of a filamentous fungus, and second, to determine the phenotype resulting from mutation of each NPS gene identified, with emphasis on discovery of any that might be involved in fungal virulence. C. heterostrophus is an especially appropriate subject for this study because it belongs to a genus renowned for its ability to produce secondary metabolites, including nonribosomal peptides (49, 53). Moreover, it is genetically tractable by both conventional and molecular methods, it undergoes efficient homologous recombination between transforming DNA and target genomic sequences (52), which facilitates functional analysis by site-specific gene deletion, and the genome has been sequenced (see Acknowledgments). Eleven NPS and NPS-like genes (and two pseudogenes) were found in the C. heterostrophus genome, one of which is required for normal virulence of the fungus to maize.
MATERIALS AND METHODS
Data mining and gene annotation.
To identify NPS genes, two strategies were used. First, the C. heterostrophus strain C4 (ATCC 48331) sequence at ≈5X coverage was batch BLASTed (BLASTX) against the NR database and hits to NPS genes were extracted. Second, a database of known NPS genes plus the candidate C. heterostrophus NPS genes (identified in the first BLAST query) was constructed and used to query the C. heterostrophus genome. Among the hits (>100) were genes other than NPSs that also encode proteins with A domains, such as aminopeptidases, ATPases, and H+/K+ transporters/exchangers; these were discarded. The candidate NPS genes on the 28 contigs that remained were evaluated further.
Manual annotation of the NPS sequences and alignment of each against the others and against known NPSs were used to define the structures of these genes. For each gene, the first ATG in a favorable context (11) opening the candidate open reading frame was chosen as the start site. All sequence ambiguities and gaps between contigs were resolved with primers designed to amplify the target region plus about 100 bp on either side. PCR fragments were sequenced and reassembled with the original contigs with the Phred/Phrap program (13, 14). Newly assembled contigs were reannotated to define coding sequences. Putative introns were verified with a 3′ rapid amplification of cDNA ends kit for cDNA amplification (Life Technology, Gaithersburg, Md.). Reverse transcription-PCR products, reverse transcribed from RNA isolated from mycelium grown for 18 h in liquid complete medium (26), were used as the templates for PCR amplification of regions spanning putative introns with primers matching ≈100 bp up- and downstream of the predicted introns. PCR products were sequenced directly or after cloning in TOPO vector PCR0.2.1 (Invitrogen).
Media, strains, and transformation.
Fungal cultures were grown on complete medium (26) with 2% xylose (45) under black lights at 22°C. RNA for expression analysis was prepared from fungal mycelia grown under each of the following conditions: shake liquid complete medium, 30°C, 6 and 20 h (26) for germination and vegetative growth; shake liquid complete medium shifted to liquid minimal medium (26) with nitrogen omitted (30C, 24 h) for nitrogen starvation; liquid Fries medium (33), still culture (22C, 120 h), for high iron (thought to favor secondary metabolite production); and liquid complete medium shifted to solid complete medium with 2% xylose (22C, 72 h) for conidiation.
Sach's medium was used for mating tests (26). Strains C5 (MAT1-1) and C4 (MAT1-2) were used as mating testers. Strain C4 was used to generate all NPS deletion strains except for deletion of NPS11, which was done in near-isogenic strain C5 (ATCC 48332). Fungal protoplast production for transformation was described previously (44), except that 1.5 g of glucanase/100 ml (Sigma, St Louis, Mo.) was substituted for 1.0 g of Novozyme/100 ml. Linearized plasmid DNA (7.5 to 10 μg) or PCR product (2 μg) was used for transformation. Hygromycin B-resistant transformants were purified by isolation of single conidia on complete medium with 2% xylose and hygromycin B-resistant phenotypes were confirmed on complete medium without salts containing 50 μg of hygromycin B/ml. All strains used in this study are described in Table 1.
TABLE 1.
Strains used in this study
| Straina | Genotype |
|---|---|
| C4 | MAT1-2 Tox1 |
| C5 | MAT1-1 tox1 |
| D.C4.pBL8 | MAT1-2 Tox1 nps1-ΔT2 hygB |
| D.C4.pBL33 | MAT1-2 Tox1 nps1-ΔT3 hygB |
| D.C4 pYC1 | MAT1-2 Tox1 nps2-ΔT1 hygB |
| D.C4.pBL11 | MAT1-2 Tox1 nps2-ΔM2 hygB |
| D.C4.pBL16 | MAT1-2 Tox1 nps3-ΔT1 hygB |
| D.C4.pBL14 | MAT1-2 Tox1 nps3-ΔA2 hygB |
| D.C4.pBL12 | MAT1-2 Tox1 nps3-ΔM4 hygB |
| D.C4.pBL34 | MAT1-2 Tox1 nps4-ΔT1 hygB |
| D.C4.pBL13 | MAT1-2 Tox1 nps4-ΔT2 hygB |
| D.C4.pBL38 | MAT1-2 Tox1 nps4-ΔT3 hygB |
| D.C4.pYC2 | MAT1-2 Tox1 nps4-ΔT4 hygB |
| D.C4.pBL36 | MAT1-2 Tox1 nps4-ΔT5 hygB |
| D.C4.pBL30 | MAT1-2 Tox1 nps5-ΔT1 hygB |
| D.C4.pBL21 | MAT1-2 Tox1 nps5-ΔT2 hygB |
| D.C4.pBL15 | MAT1-2 Tox1 nps6-ΔT1 hygB |
| D.C4.356 | MAT1-2 Tox1 nps7-Δ hygB |
| D.C4.pBL29 | MAT1-2 Tox1 nps8-ΔT1 hygB |
| D.C4.1222 b | MAT1-2 Tox1 nps9-ΔM1 hygB |
| D.C4.pBL31 | MAT1-2 Tox1 nps10-ΔT1 hygB |
| D.C5.3HR | MAT1-1 nps11 Δ hygB |
| N6-R-3 | MAT1-1 nps6 hygB |
| N26-R-9 | MAT1-2 nps6 hygB |
| N26-R-10 | MAT1-2 nps6 hygB |
| NR26-R-11 | MAT1-2 NPS6 |
| N26-R-12 | MAT1-1 NPS6 |
| N26-R-13 | MAT1-2 NPS6 |
| N26-R-14 | MAT1-1 nps6 hygB |
| N26-R-15 | MAT1-1 NPS6 |
| N26-R-16 | MAT1-2 NPS6 |
| N26-R-17 | MAT1-2 nps6 hygB |
| N26-R-18 | MAT1-2 NPS6 |
| N26-R-19 | MAT1-1 nps6 hygB |
| N26-R-20 | MAT1-1 nps6 hygB |
C4 = ATCC 48331 (26), C5 = ATCC 48332 (26). The prefix D. refers to a C4 or C5 strain in which a deletion has been made; p refers to deletions that were generated by linearized plasmid DNA; otherwise, fusion PCR was used (4). The prefix N refers to cross number, R indicates random progeny, numeral indicates progeny number. ΔT indicates deletion of a T domain, i.e., ΔT2, for example, indicates deletion of the T domain of module 2; ΔM indicates deletion of an entire module, i.e., ΔM2, indicates deletion of the entire module 2; Δ alone indicates deletion of entire ORF.
Nucleic acid manipulations, PCR, reverse transcription-PCR, and real-time PCR.
Fungal genomic DNA was isolated with the DNeasy Plant minikit or 96-well kits (Qiagen, Valencia, Calif.). Total RNA was isolated with the RNAwiz kit (Ambion, Austin, Tex.). PCR amplification was performed with either Supermix polymerase (Invitrogen, San Diego, Calif.), Hotstart mix (Qiagen, Valencia, Calif.), or Expand long polymerase (Roche. Indianapolis, Ind.). PCR programs were as follows: step 1, 94°C, 3 min, step 2, 94°C, 30 s, step 3, 55°C, 30 s, step 4, 72°C, 1 min/kb, and step 5, 72°C for 10 min. Each cycle was repeated 30 times. Extension time (step 3) was increased to 57°C or decreased to 52°C if the first PCRs did not give specific bands.
For NPS expression analysis, total RNA (50 ng) from samples representing each of the five conditions (above) was reverse transcribed, and amplified by PCR with TaqMan one-step reverse transcription-PCR master mix reagents (Applied Biosystems, Roche, N.J.). Reactions were performed in an ABI Prism 7700 sequence detection system (PE Biosystems) in a MicroAmp Optical 96-well reaction plate (PE Biosystems, Foster City, Calif.) with amplification cycles suggested by the manufacturer. To compare relative abundance of the NPS transcripts, the average cycle value for each NPS was normalized to that of actin (value = 1) for each growth condition. To prepare samples grown under each condition, three flasks were set up and total RNA was extracted from each, then pooled. For each PCR, a different aliquot of the pooled sample representing each condition was used. PCRs were done in triplicate, twice for NPS1-NPS10, three times for NPS6, and once for NPS11 (see Table S1 in the supplemental material).
Fold change, which was used in Table S1 and Fig. 3, was calculated as = 2−ΔCt, where ΔCt = −(CtNPS − Ctactin)threshold 0.2. (23).
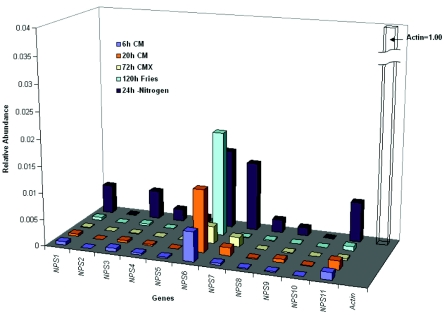
FIG. 3.
Expression of 11 NPS genes in wild-type strain C4 grown under various conditions. Blue/purple: 6 h of germination in liquid complete medium. Orange: 20 h of vegetative growth in liquid complete medium. Yellow: 72 h of asexual development on solid complete medium with 2% xylose. Light blue: 120 h in liquid Fries medium. Maroon: 24 h in liquid minimal medium minus nitrogen. Total RNA from the wild type was tested. Relative abundance was determined by normalizing changes to actin expression (23) (see Table S1).
Construction of NPS deletion strains.
The type of deletion and method of deletion varied for each NPS gene (Table 1). For plasmid constructions, primer pairs were designed and used in PCRs with strain C4 genomic DNA as the template to amplify upstream and downstream sequences flanking the targeted domain. Unique restriction enzyme site sequences (KpnI, SacI, or HindIII) were added to the ends of the primers. The gene hygB, which confers resistance to hygromycin B and was driven by the Aspergillus nidulans trpC promoter (8), was inserted between the two flanking sequences and the product was ligated into a pBluescript KS vector (Stratagene, La Jolla, Calif.). The resulting plasmid was linearized with the appropriate enzyme and used for transformation. For NPS7, NPS9, and NPS11, target sequences were deleted by fusion PCR (4). Briefly, 5 and 3′-flanking sequences and the hygB cassette were amplified and used as templates in a second PCR that fused the 5′ flank to the 5′ two thirds of hygB (HY) and the 3′ flank to the 3′ two thirds of hygB (YG). The resulting PCR products (5′-flank-HY and YG-3′-flank) were purified, concentrated with Qiagen columns (Qiagen, Valencia, Calif.), and used for transformation.
To confirm disruption or deletion of each NPS gene, DNA was isolated from each of eight independent hygromycin B-resistant colonies per deletion. Three pairs of primers were used with genomic DNA (1 μl containing 0.1 to 0.5 μg of DNA) as the template to distinguish homologous from ectopic integration events.
Plant tests.
Strains were grown on complete medium with 2% xylose under black lights at 22°C for 8 to 10 days. Conidia from at least two independent strains/mutant were harvested in 0.05% Tween 20, adjusted to 500 spores/ml, and sprayed on 14-day-old maize plants (W64-N; 15 ml/pot containing six plants) with a pressurized thin-layer chromatography sprayer (Alltech Associates, Deerfield, Ill.). Inoculated plants were placed in a dark mist chamber for 24 h (22°C and 100% relative humidity) after which plants were transferred to a growth chamber (23°C, 12 h of light and 90% relative humidity). Leaves were observed up to 6 days postinfection, and lesion sizes on the fourth-oldest leaves were photographed.
Growth tests.
Fifty conidia from each mutant and from the the wild type were picked onto complete medium with 2% xylose, and germination was monitored for 96 h. The growth rate was determined by measuring diameters of six independent colonies from each mutant and from the the wild type every 24 h.
Mating ability.
To test mating ability, mutant strains were crossed with a wild-type tester of the opposite mating type. Pseudothecial development and fertility were assessed 3 to 4 weeks after mating.
Appressorium formation.
The ability to form appressoria was monitored microscopically after placing 25 μl of conidial suspension on a Teflon-coated slide (Electron Microscopy Sciences, Washington, Pa.) and incubating 4 to 6 h in a humid chamber.
Oxidative stress tests.
Sensitivity to oxidative stress was tested on solid minimal medium with or without hydrogen peroxide in petri dishes (90 mm diameter) (34). Autoclaved minimal medium was cooled to 50°C, then hydrogen peroxide (30%, Sigma, St. Louis, Mo.) was added to a final concentration of 7 mM. Mycelial disks (3 mm diameter) were cut with a cork borer, 1 cm from the edge of an actively growing culture. Disks were inverted and placed in the centers of the plates. Parental strains (wild-type C5 and nps6 mutant N6-R-3), six hygromycin B-resistant progeny (N26-R-9, −10, −14, −17, −19, and −20), and six hygromycin B-sensitive progeny (N26-R-11, −12, −13, −15, −16, and −18) of a cross of the two parents were evaluated. Colony diameters of four replicates per strain were measured after 2 and 3 days of incubation at 22°C in the dark. Growth per 24 h was calculated, and a standard t test was used to determine statistical significance.
AMP-binding adenylation domain genealogy construction and evaluation.
The genealogy of 125 NPS A domains of 48 NPS and NPS-like proteins encoded by NPS genes was inferred by neighbor joining and maximum parsimony analysis in PAUP4.0b8 (Sinauer Associates, Sunderland, Mass.). Accession numbers for all sequences obtained from GenBank are given in Table S2. Alignment was based on 690 amino acids from the A domain; 588 characters were informative, 49 were uninformative, and 53 were constant. Parsimony was performed with 100 random additions, steepest descent on, and tree bisection-reconnection branch swapping. The tree shown in Fig. 7 is one of four most-parsimonious trees of 22,705 steps, consistency index = 0.297, rescaled consistency index = 0.141. The maximum parsimony trees generated by coding gaps as either the 21st amino acid or as missing were not significantly different from each other or from the tree generated by neighbor joining. The genes whose AMP domains grouped with strong bootstrap support in the genealogy were inferred to be orthologs (products of speciation and not gene duplication). Orthology was further supported by identical or nearly identical domain structures.
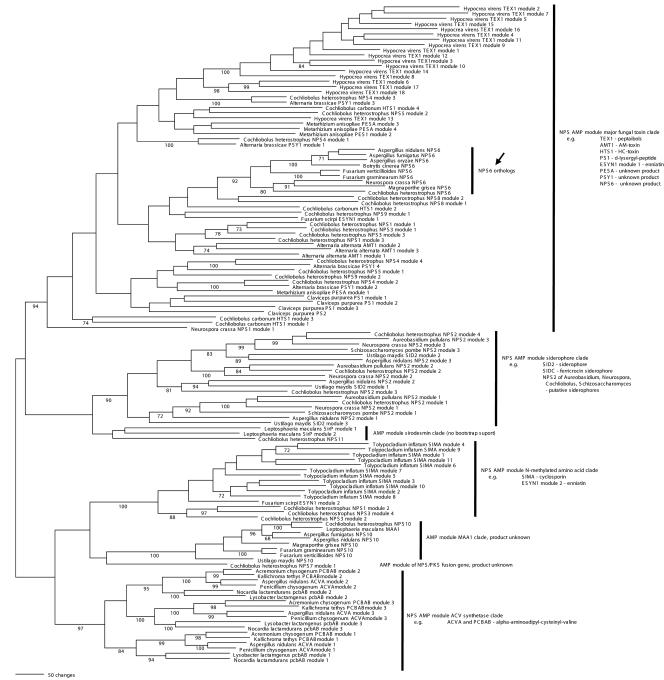
FIG. 7.
Homology of NPS6. Genealogy of 125 NPS AMP-binding adenylation (A) domains of 48 NPS and NPS-like proteins, inferred by maximum parsimony analysis. Vertical bars mark major clades that include functionally similar proteins, and subclades that are comprised of apparently homologous proteins that are predicted to be identical or nearly identical in their basic function. Branch length indicates number of inferred amino acid changes. Numbers below branches indicate percent bootstrap support (when >70%) for each clade (performed with 100 repetitions). Accession numbers for all sequences obtained from GenBank are given in Table S2. Shown is one of four most-parsimonious trees (only the branching order of the NPS6 A domains was ambiguous) (arrow).
Nucleotide sequence accession numbers.
Accession numbers are as follows: C. heterostrophus NPS1 to NPS13, AY884186 to AY884198; G. moniliformis NPS6, AY928085; B. cinerea NPS6, AY928086; A. fumigatus NPS6, AY928087; M. grisea NPS6, AY928088; G. moniliformis NPS10, AY928029; and A. fumigatus NPS10, AY928090.
RESULTS
Identification and annotation of NPS and NPS-like genes in C. heterostrophus.
Analysis of an assembly derived from 5X sequence coverage revealed 28 contigs containing NPS-like A, T, and C domains which mapped to nine metacontigs. The nine metacontigs carried 11 unique NPS or NPS-like genes, named NPS1 to NPS11. NPS8 and NPS9 were found to be linked (≈500 kb apart) on the same metacontig, and NPS1 and NPS10 were located ≈600 kb apart on a different metacontig. In addition, there were two metacontigs that carried open reading frames with conserved A and T domains, named NPS12 and NPS13, however, stop codons were found in the putative coding sequences and consensus intron boundaries that might eliminate them after splicing were not identified. Therefore, NPS12 and NPS13 were considered pseudogenes and not analyzed further.
The modular organization of each of the eleven genes is shown Fig. 1. The numbers of predicted introns per gene are NPS4 (6), NPS5 (2), NPS9 (3), and NPS11 (1); the remainder had none. Interestingly, sequencing the reverse transcription-PCR product did not demonstrate splicing of the predicted intron in NPS11 or one of the predicted introns in NPS5, however, because virtual splicing perfectly preserves the reading frame and domain structure is interrupted if they are not spliced, we suggest this observation is due to incomplete processing of the RNA.
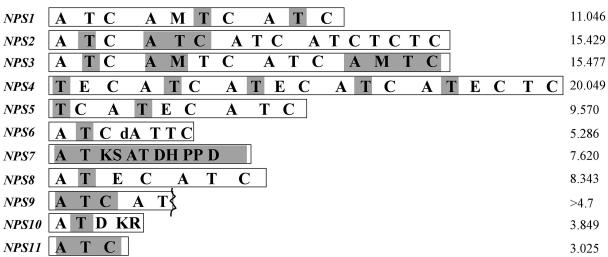
FIG. 1.
Structural organization of C. heterostrophus predicted NPS proteins. Abbreviations: A, adenylation; dA, degenerate adenylation; T, thiolation; C, condensation; E, epimerization; M, N-methyltransferase; D, dehydrogenase; KS, beta-ketoacyl synthase; AT, acyl transferase; DH, dehydratase; KR, ketoreductase. Shaded regions indicate portions of genes that were deleted to make nps mutations. Each box represents an open reading frame, the size of which is indicated on the right in kilobases. The dotted vertical line in NPS9 indicates incomplete sequence. No two predicted proteins are alike in their domain structure.
Consensus A (PF00501), C (PF00668), and E domains for filamentous fungal nonribosomal peptide synthetases are shown in Fig. 2 compared to those of bacterial nonribosomal peptide synthetases. Full-length alignments of the C. heterostrophus NPS A and C domains and their conserved motifs are shown in Fig. S1 and S2.
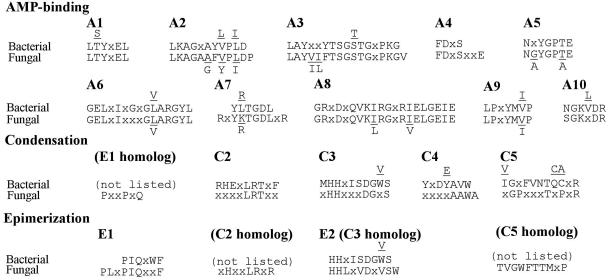
FIG. 2.
Conserved A, C, and E motifs. A comparison of ten A, five C, and two E conserved motifs is shown (41). For each motif, the fungal consensus, based on amino acid sequences encoded by all fungal NPS genes examined in this study (Fig. 7; see Table S2 for complete list) is compared with the bacterial consensus reported (41). The fungal C domains have only C2 to C5 and lack the C1, C6, and C7 domains found in bacterial nonribosomal peptide synthetases. Similarly, the fungal E domains have only E1 and E2 and lack the E3 to E7 domains found in bacterial nonribosomal peptide synthetases.
Predicted protein sequences grouped the 11 genes into two structural categories: NPS1-6, NPS8, and NPS9 are NPS genes encoding multimodular proteins, whereas NPS7, NPS10, and NPS11 are NPS-like genes encoding monomodular proteins, each with a distinctive C-terminal end. NPS7 has a polyketide synthase domain near its C terminus and is thus an NPS/polyketide synthase hybrid (10); it terminates with a dehydrogenase (D) domain of the type found in Lys2 proteins. NPS10 terminates with two of the five domains found in the polyketide synthase portion of NPS7, a D domain followed by a ketoreductase domain. NPS11, although monomodular, has a C domain at its C terminus, which makes the complete protein resemble one unit of a typical multimodular nonribosomal peptide synthetase.
Among the NPS genes, NPS4 is the largest (≈20 kb) encoding a protein with four modules, plus additional domains at the 5′ and 3′ ends. NPS11 is the smallest (≈3 kb) encoding one module. NPS6 encodes a protein with a typical first module, and a second module with a degenerate A and two T domains. E domains were found in NPS4 (upstream of module 1 and in modules 2 and 4), NPS5 (in module 1), and NPS8 (in module 1). M domains were found in NPS1 (in module 2) and NPS3 (in modules 2 and 4). Both NPS4 and NPS5 begin with a T and not an A domain. None of the predicted proteins has a thioesterase domain.
Expression of C. heterostrophus NPS genes.
Real-time PCR was used to determine whether or not each NPS gene is expressed, and if so, whether the expression level is affected by growth condition (Fig. 3 and Table S1). No matter what the growth condition, expression of NPSs was much lower than that of actin. NPS6 expression was higher than that of any other NPS gene under all conditions tested. Transcripts of all NPS genes (except NPS2 and NPS10) were detectable after 6 h of growth in complete medium. NPS2 was not expressed under any condition tested. At 72 h on solid complete medium with 2% xylose, expression was comparable to 20 h in complete medium liquid, except NPS11 was repressed, NPS6 was reduced compared to all other conditions, and transcripts of NPS5 and NPS10 were not detectable. After 120 h still culture in Fries medium, which is thought to favor production of secondary metabolites (33), expression levels were comparable to those at 6 h in liquid complete medium, except for that of NPS6, which had highest expression in Fries medium. NPS10 had no detectable expression after 120 h still culture in Fries medium. Highest expression levels among the active NPS genes were seen in cultures grown in complete medium for 24 h then shifted to nitrogen deficient minimal medium, except for NPS6, which was induced by nitrogen starvation but had maximal expression in Fries medium (Fig. 3 and Table S1).
All NPS genes had lower expression than the actin-encoding control gene, based on average cycle values (Table S1), which ranged from 24 to 40 for the NPS genes and 18 to 22 cycles for the actin gene. Based on these parameters, expression of all NPSs (except NPS2 and NPS10, which had cycle values of 40 on complete medium and therefore a calculation could not be made) was induced 3- to 18-fold by nitrogen starvation, compared with expression during growth on complete medium at 6 h (9-, 8-, 6.5-, 5.4-, 2.6-, 3.6-, 18-, 9.3-, and 5.6-fold increase for NPS1, NPS3, NPS4, NPS5, NPS6, NPS7, NPS8, NPS9, and NPS10, respectively.
NPS6 is necessary for virulence to maize.
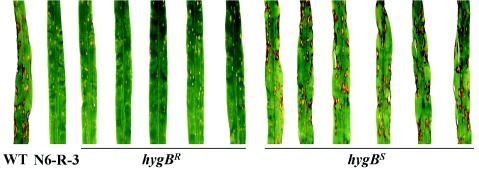
Confirmation of each NPS T domain sequence or module deletion was acquired by PCR, with genomic DNA from each mutant as the template and the primer pairs indicated in Fig. S3. Subsequently, all mutants were screened for altered virulence on maize. Of the 11 NPS genes, only NPS6 was found to be required for normal virulence (Fig. 4). Lesion sizes caused by nps6 mutants were reduced compared to the wild type. One nps6 mutant (D.C4.pBL15) was crossed (cross no. N6) with wild-type strain C5; a hygromycin B-resistant progeny (N6-R-3) was backcrossed to C5 (cross no. N26) and six hygromycin B-resistant (N26-R-9, N26-R-10, N26-R-14, N26-R-17, N26-R-19, and N26-R-20) and six sensitive (N26-R-11, N26-R-12, N26-R-13, N26-R-15, N26-R-16, and N26-R-18) progeny were tested on plants. The small lesion phenotype cosegregated with loss of NPS6 and resistance to hygromycin B (Fig. 4), indicating that the product of NPS6 is necessary for high virulence of C. heterostrophus on maize.
FIG. 4.
nps6 mutant has reduced virulence on maize. Leaves were inoculated with progeny of a cross between an nps6 mutant and the wild type. All hygromycin B-resistant progeny are nps6 mutants (from left, leaves 3 to 7). Hygromycin B-sensitive progeny are the wild type (from left, leaves 8 to 13). The photo was taken 4 days postinoculation.
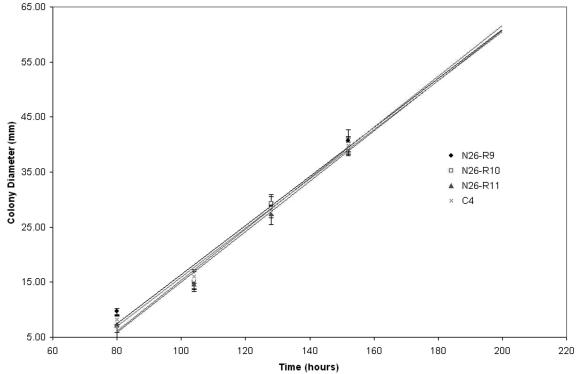
C. heterostrophus NPS genes are not involved in growth, morphology, or reproduction.
Two nps6 mutant progeny (N26-R-9 and N26-R-10), a wild-type (NPS6+) progeny (N26-R-11), and the wild type were monitored for germination and colony radial growth. All 50 conidia from each strain germinated normally; the radial growth rates and colony morphologies of mutants and the wild type were identical (Fig. 5). In addition, nps6 mutants formed normal appressoria and mated normally with the wild type (data not shown). These results suggest that the low-virulence phenotype of the nps6 mutant is not due to defects in morphology, growth, or germination. Strains carrying mutations in all other NPS genes (NPS1-5 and NPS 7-11) were like the wild type in their ability to grow and germinate on complete medium with 2% xylose medium, make appressoria, and mate (data not shown).
FIG. 5.
Evaluation of growth of nps6 mutant strains versus wild-type strains. nps6 mutants grow normally. Symbols: diamond = nps6 (N26-R-9); square = nps6 (N26-R-10); triangle = NPS6 (N26-R-11); and circle = NPS6 (C4). Measurements were taken at 12-h intervals.
nps6 mutants are sensitive to oxidative stress.
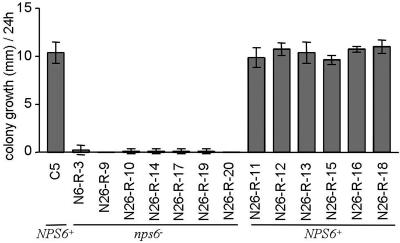
Wild-type strains grew equally fast (P > 0.05) on medium with or without hydrogen peroxide. Growth rates of nps6 mutants were significantly reduced (P < 0.05) in the presence of hydrogen peroxide, compared to the wild type (Fig. 6 and Fig. S4). Hydrogen peroxide sensitivity cosegregated with resistance to hygromycin B and low virulence, indicating that the product of NPS6 activity is involved in protection of the fungus against oxidative stress, and associating oxidative stress tolerance with fungal virulence.
FIG. 6.
Colony diameter increase in 24 h of colonies measured on 7 mM H2O2 amended medium, at 2 and 3 days after setting up the assay. Parents and progeny segregate 1:1 for three phenotypes: sensitivity to hydrogen peroxide, resistance to hygromycin B, and reduced virulence. For strain genotypes, see Table 1 and Materials and Methods. Bar, standard deviation.
NPS6 is conserved in filamentous ascomycetes.
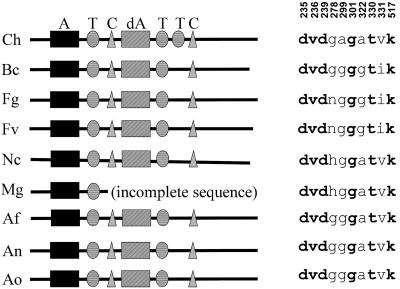
An ortholog of NPS6 was found in every filamentous ascomycete genomic database searched, including Aspergillus fumigatus (TIGR), Aspergillus nidulans (MITCGR), Botryotinia fuckeliana/Botrytis cinerea (TMRI), Gibberella zeae/Fusarium graminearum (MITCGR, TMRI), G. moniliformis/Fusarium verticillioides (TMRI), Magnaporthe grisea (MITCGR), and Neurospora crassa (MITCGR). Orthology was inferred by a phylogenetic analysis of A domains that grouped the module 1 A domains of the predicted NPS proteins of these genes from each genome, with strong bootstrap support (92%, Fig. 7). Orthology was further supported by a full-length alignment of all NPS6 predicted amino acid sequences, which indicated that each has the same general arrangement of two modules, the second of which has a degenerate A domain (Fig. 8). The only protein that varies in its domain structure is C. heterostrophus NPS6, which has two T domains in the second module; only one T domain is found in this module of the other NPS6 proteins.
FIG. 8.
Cartoon of the organization of proteins encoded by nine NPS6 genes and the amino acid specificity code (42) of A module 1 for each of nine homologous NPS6 proteins. Note conservation of specificity residues; numbers correspond to specificity residue positions, (42). The homolog from M. grisea was inferred to have the same modular arrangement; the contig containing this gene terminates prematurely after the first module, and the second module was not found elsewhere in the genomic sequence. Ch, C. heterostrophus; Fg, F. graminearum; Bc, B. cinerea; Nc, N. crassa; Mg, M. grisea; Fv, F. verticillioides; Af, A. fumigatus; An, A. nidulans; Ao, A. oryzae; A, T, C, and dA, see the legend to Fig. 1.
An analysis of the A domain of module 1 indicated that NPS6 proteins are highly conserved for the 10 nonconsecutive amino acids that specify which amino acid is adenylated (42) (Fig. 8 and Fig. S1). Although the specified amino acid could not be determined by a close match to previously characterized A domains (42, 49), the results suggest that all NPS6 genes are not only phylogenetic homologs, but also their products are functionally homologous, at least in terms of the small molecule that is made as a result of NPS6 activity.
In contrast, results of the phylogenetic analyses indicated most of the C. heterostrophus NPS and NPS-like genes have not yet been found in other fungi. A similar observation has been made for fungal PKS genes in fungi (25). Besides NPS6, only two additional homologous matches were identified: NPS4 (the gene product and resulting peptide are not characterized) is found in Alternaria brassicae PSY1 (16), and NPS10 (which encodes a monomodular NPS-like protein) is found in A. fumigatus, A. nidulans, F. graminearum, F. verticillioides, M. grisea, L. maculans (maa1), and Ustilago maydis, but not in N. crassa or B. cinerea.
DISCUSSION
Whole-genome inventory of NPS genes.
It is clear from our genome-wide analysis of C. heterostrophus and from similar analyses with additional fungal genomes (54) that NPSs are abundant in filamentous ascomycetes. C. heterostrophus, for example, has 11, each of which was mutated in this study, plus two pseudogenes. This number rivals the numbers reported for Streptomyces and Mycobacterium spp., bacterial genera renowned for their pharmaceutically important secondary metabolite output (31). Of the 11 C. heterostrophus NPSs, only one had a detectable phenotype when mutated, suggesting that the presumed small peptide products of nonribosomal peptide synthetase proteins usually do not have obvious biological activity. This fact increases the difficulty of detecting both the peptide and the corresponding NPS gene in the absence of genome sequence. This difficulty is exacerbated by the fact that most of the C. heterostrophus NPS genes appear to be transcribed at low levels (≤2% that of the actin gene control) under standard laboratory culture conditions. Low expression suggests that the small molecule biosynthetic pathways represented by the C. heterostrophus NPS genes may be cryptic under common laboratory culture conditions and that special measures may be necessary to detect biological activity of the small molecules predicted by the presence of these genes in the genome. Furthermore, there may be little correlation between RNA and protein levels, or with factors relating to enzyme activity and secretion of the final metabolite.
In the past, the strategies used to isolate NPS genes from fungi have included cross-hybridization with bacterial NPS sequences (40) and purification of nonribosomal peptides for antibody production followed by screening of expression libraries (38). In addition, several laboratories have identified NPSs with PCR primers to the more conserved nonribosomal peptide synthetase motifs (1, 22, 30). Identification of a specific NPS with this strategy is difficult given that use of degenerate primers is required and there are many candidates in any given genome. In most cases, detection of a particular biological activity preceded gene identification, the former being the motivation for the latter. For example, the NPS genes for biosynthesis of three known virulence factors, HC toxin from Cochliobolus carbonum HTS1 (38), AM toxin from Alternaria alternata (22), and enniatin from Fusarium scirpi esyn1 (17), were identified relatively recently compared to the recognition of their biological activities.
NPS6 and virulence.
NPS6 from C. heterostrophus is different from the previously reported NPS genes involved in fungal virulence because it is unfailingly conserved among fungi whereas the others are not. To date, HTS1 has been found only in race 1 of Cochliobolus carbonum (32), AMT1 only in the apple pathotype of Alternaria alternata (22), and Esyn1 only in Fusarium scirpi and certain other Fusarium spp.: F. sambucinum, F. lateritium, and F. avenaceum (19). Since C. heterostrophus NPS6 is conserved in every filamentous ascomycete genome examined, it is predicted to have a conserved function in these fungi. Preliminary data indicate that NPS6 products from the closely related rice pathogen Cochliobolus miyabeanus and the distantly related cereal pathogen Gibberella zeae are also virulence determinants (Oide and Turgeon, unpublished data). NPS6 is present in both pathogenic and saprobic fungi, it is expressed in standard laboratory culture, and it is induced under low-nitrogen and high-iron conditions; these diverse observations suggest that NPS6 may play multiple roles in cells and be involved in defense of fungi against a variety of stresses.
How might we imagine a small peptide to be involved in general virulence? A clue is provided by the observation that nps6 mutants have enhanced sensitivity to hydrogen peroxide, hinting that the small peptide may be involved in protecting the fungus from oxidative stress. If so, this would be consistent with the observation that an oxidative burst commonly occurs when plants are attacked by microbes. For a pathogen to be successful, it would require means for survival in the presence of reactive oxygen species generated by the plant; perhaps the NPS6 peptide contributes in this way. The fact that NPS6 homologs are found not only in pathogens, but also in saprobes, suggests that the NPS6 peptide can help any euascomycete fungus deal with the oxidative stresses it encounters, even if it is not a pathogen. It will be informative, therefore, to investigate the function of NPS6 in saprobic fungi.
Mutations in NPS6 did not cause complete loss of virulence, indicating that additional gene products are required for full virulence. One such factor is CPS1, the product of a gene encoding an acyl-AMP ligase-like enzyme that is required for full virulence by C. heterostrophus, and like NPS6, is ubiquitous among euascomycetes (27). Deletion of CPS1 reduces but does not eliminate virulence on maize; the degree of reduction is less than that of nps6 mutants. The phenotype of the double mutant (cps1 nps6) is the same as that of the nps6 single mutant, which may indicate that NPS6 acts downstream of CPS1 in the same pathway (Lee, unpublished data).
Characteristics of C. heterostrophus NPS and NPS-like genes.
The eight multimodular NPSs consist of typical A-T-C modules (Fig. 1). One peptide, NPS2, terminates with four additional domains (T-C-T-C) that are predicted to function in closing the nascent tetrapeptide into a ring. In the case of U. maydis sid2, a tripeptide is closed into a ring by the same four domains (55). Two putative peptides, NPS4 and NPS5, have additional domains preceding the first A-T-C module. NPS5 begins with two domains (T-C) that may serve to produce a peptide longer than predicted by the number of A-containing modules. A similar arrangement and mechanism occurs in Fusarium scirpi esyn1, which has a C domain that precedes two A-containing modules and results in a cyclic heptapeptide (17). NPS4 begins with three domains (T-E-C); the T and C domains may serve to make the enzyme act in an iterative fashion, while the E domain likely epimerizes the amino acid specified and condensed by the last (fourth) A-containing module.
Interestingly, none of the predicted C. heterostrophus NPS proteins has a thioesterase domain, which is thought in bacteria to be involved in release of peptide products from enzyme templates and from bacterial and fungal ACV-type nonribosomal peptide synthetases (29) (which do not occur in C. heterostrophus). Thus, there is likely an alternative mechanism for peptide release from the fungal enzymes.
Three C. heterostrophus NPS-like genes encode proteins that have a single module, each of which ends in a different C-terminal domain. NPS7 follows its A and T domains with ketosynthase, acyltransferase, dehydratase, ketoreductase, and phosphopantetheine domains that comprise a polyketide synthase module, and terminates with a D domain of the type that also terminates monomodular lysine synthase (lys2), which is found in all fungi and is required for lysine synthesis. NPS7 PKS24 accession no. AY495665 (25) is predicted to synthesize a polyketide with an addition of a single amino acid. This is the only protein in the C. heterostrophus genome that consists of all nonribosomal peptide synthetase- and polyketide synthase-type domains necessary to function as nonribosomal peptide synthetase and polyketide synthase proteins. In contrast, M. grisea encodes no NPS/polyketide synthase hybrid proteins, but instead encodes three complete polyketide synthase/NPS hybrids, including the ACE1 polyketide synthase, whose biosynthetic activity is required for virulence (3), and two polyketide synthase/NPS lovastatin nonaketide synthase-like hybrids (3) whose NPS components lack A and T domains and consist only of a C domain.
A recent comparative genomic survey that focused on PKS genes (25) identified three complete PKS/NPS hybrids each in the genome sequences from B. cinerea (AY495608, AY495610, and AY495612) and F. verticillioides (AY495591, AY495599, and AY495600), but none in N. crassa or C. heterostrophus. Among these four genomes, C. heterostrophus has the only lovastatin nonaketide synthase-type PKS/NPS gene (PKS17; AY495658), which was not considered in this study, as it encodes a protein lacking a complete NPS-type domain. NPS10 terminates with two domains: a D domain followed by a ketoreductase domain more commonly found in polyketide synthases and fatty acid synthetases (20). NPS11 terminates with a C domain and thus resembles a multimodular NPS, even though it contains only a single module. Both NPS10 and NPS11 are predicted to add a single amino acid to an unknown moiety.
Conservation of NPS genes.
C. heterostrophus NPS6 is the only gene that has an ortholog in all other ascomycetes examined (Kroken et al., unpublished data). Orthologs of C. heterostrophus NPS10 have been found in most fungi, except B. cinerea and N. crassa. An ortholog of C. heterostrophus NPS4 has been found only in Alternaria brassicae (16). A comparison of each of these sets of orthologous NPS genes suggests that their homologous A domains all specify the same amino acid, based on an alignment of the 10 nonconsecutive amino acids that confer specificity (42). All remaining C. heterostrophus NPSs appear to be paralogs with no matches in any filamentous fungus examined (Kroken et al., unpublished data) and as such are likely to be responsible for biosynthesis of small peptides or peptide-containing small molecules unique to C. heterostrophus. The A domains of the proteins encoded by these NPSs are divergent in the sequence of the specificity-conferring amino acids. A complete treatment of the phylogenetic relationships of the NPS genes and their modules from all filamentous fungi examined here will be reported elsewhere (Kroken et al., unpublished data).
The diversity of peptides made by nonribosomal peptide synthetases arises from both the specificity of individual A domains and the order of those domains within a multimodular gene. The phylogenetic analyses of A domains suggest that modular rearrangements have played a role in generating novel nonribosomal peptide synthetases. For example, the A domains of modules 1 and 3 of C. heterostrophus NPS1 and C. heterostrophus NPS3 group with a large clade of A modules from nonribosomal peptide synthetases such as AMT1 and HTS1 that make AM toxin and HC toxin, respectively, whereas the A domains from module 2 from C. heterostrophus NPS1 and C. heterostrophus NPS3 and module 4 from C. heterostrophus NPS3 group with the A domains from simA that makes cyclosporine in T. inflatum (Fig. 7). Similarly, the A domain from module 1 of ESYN1 groups with the A domains from AMT1 and HTS1, whereas the A domain from module 2 groups with the A domains from simA.
NPS1, NPS3, and ESYN1 represent novel rearrangements of modules from the same major clades of A domains. The combined processes of rearrangement of A modules and specificity changes of those modules have resulted in a diversity of NPS genes, each of which encodes a protein that is likely to make a unique small peptide. Thus, the total number of nonribosomal peptides made by fungi is undoubtedly large, perhaps greater than the substantial number reported from Streptomyces and Mycobacterium spp. (31).
ADDENDUM IN PROOF
Update: NPS12 (accession no. AY884197) is a gene, not a pseudogene.
Supplementary Material
Acknowledgments
S.K. was supported by a collaborative agreement between UC-Berkeley (N. L Glass and J. W Taylor, Department of Plant and Microbial Biology) and the former Torrey Mesa Research Institute (TMRI)/Syngenta.
B.G.T. did this work at the former Torrey Mesa Research Institute while on a leave of absence from Cornell University.
Sequence data of Aspergillus fumigatus (NIAID U01 A1 48830) is obtained from the Institute for Genomic Research website at http://www.tigr.org. The genomes of B. cinerea, C. heterostrophus, F. graminearum, and F. verticillioides were sequenced by Celera Genomics for TMRI/Syngenta. Intermediate genome assembly of raw shotgun sequences was performed with in-house software (Don Hutchinson, TMRI). Gene predictions were created by Darrell Ricke (TMRI). We thank the Genome Technology and Bioinformatics groups at TMRI for the Cochliobolus sequence database, David Lan for BAC library construction, Natasha Ginzburg for the Phred/Phrap assemblies, Elisabeth Wagner for technical support, and Greg Saenz and Natalie Catlett for reviewing early versions of the manuscript. We thank Steve Briggs, former Director and CEO of TMRI, for his unwavering support.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Bailey, A. M., M. J. Kershaw, B. A. Hunt, I. C. Paterson, A. K. Charnley, S. E. Reynolds, and J. M. Clarkson. 1996. Cloning and sequence analysis of an intron-containing domain from a peptide synthetase-encoding gene of the entomopathogenic fungus Metarhizium anisopliae. Gene 173:195-197. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, J. E., J. M. Blackburn, J. D. Sutherland, and M. C. Wright. 1991. High-level soluble expression of isopenicillin-N synthase isozymes in E. coli. Tetrahedron 47:5991-6002. [Google Scholar]
- 3.Bohnert, H. U., I. Fudal, W. Dioh, D. Tharreau, J. L. Notteghem, and M. H. Lebrun. 2004. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16:2499-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catlett, N. L., B.-N. Lee, O. C. Yoder and B. G. Turgeon. 2003. Split-marker recombination for efficient targeted deletion of fungal genes. Fungal Genet. Newsl. Online 50:9-11. [Google Scholar]
- 5.Challis, G. L., J. Ravel, and C. A. Townsend. 2000. Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7:211-224. [DOI] [PubMed] [Google Scholar]
- 6.Correia, T., N. Grammel, I. Ortel, U. Keller, and P. Tudzynski. 2003. Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea. Chem. Biol. 10:1281-1292. [DOI] [PubMed] [Google Scholar]
- 7.Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema, and D. van Sinderen. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8:821-831. [DOI] [PubMed] [Google Scholar]
- 8.Cullen, D., S. A. Leong, L. J. Wilson, and D. J. Henner. 1987. Transformation of Aspergillus nidulans with the hygromycin-resistance gene, hph. Gene. 57:21-26. [DOI] [PubMed] [Google Scholar]
- 9.Davies, J. 1990. What are antibiotics-archaic functions for modern activities. Mol. Microbiol. 4:1227-1232. [DOI] [PubMed] [Google Scholar]
- 10.Du, L., and B. Shen. 2001. Biosynthesis of hybrid peptide-polyketide natural products. Curr. Opin. Drug Discov. Dev. 4:215-228. [PubMed] [Google Scholar]
- 11.Edelmann, S. E., and C. Staben. 1994. A statistical analysis of sequence features within genes from Neurospora crassa. Exp. Mycol. 18:70-81. [Google Scholar]
- 12.Eisendle, M., H. Oberegger, I. Zadra, and H. Haas. 2003. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC). Mol. Microbiol. 49:359-375. [DOI] [PubMed] [Google Scholar]
- 13.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 14.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 15.Gardiner, D. M., A. J. Cozijnsen, L. M. Wilson, M. S. C. Pedras, and B. J. Howlett. 2004. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol. Microbiol. 53:1307-1318. [DOI] [PubMed] [Google Scholar]
- 16.Guillemette, T., A. Sellam, and P. Simoneau. 2004. Analysis of a nonribosomal peptide synthetase gene from Alternaria brassicae and flanking genomic sequences. Curr. Genet. 45:214-224. [DOI] [PubMed] [Google Scholar]
- 17.Haese, A., M. Schubert, M. Herrmann, and R. Zocher. 1993. Molecular characterization of the enniatin synthetase gene encoding a multifunctional enzyme catalysing N methyldepsipeptide formation in Fusarium scirpi. Mol. Microbiol. 7:905-914. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, J., and D. Dubnau. 1991. Growth stage signal transduction and the requirements for srfA induction in development of competence. J. Bacteriol. 173:7275-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrmann, M., R. Zocher, and A. Haese. 1996. Effect of disruption of the enniatin synthetase gene on the virulence of Fusarium avenaceum. Mol. Plant-Microbe Interact. 9:226-232. [DOI] [PubMed] [Google Scholar]
- 20.Hopwood, D. A. 1997. Genetic contributions to understanding polyketide synthases. Chem. Rev. 97:2465-2498. [DOI] [PubMed] [Google Scholar]
- 21.Horinouchi, S., and T. Beppu. Gene expression in Streptomyces. Tanpakushitsu Kakusan Koso 35:2567-2583, 1990. [PubMed]
- 22.Johnson, R. D., L. Johnson, Y. Itoh, M. Kodama, H. Otani, and K. Kohmoto. 2000. Cloning and characterization of a cyclic peptide synthetase gene from Alternaria alternata apple pathotype whose product is involved in AM-toxin synthesis and pathogenicity. Mol. Plant-Microbe Interact. 13:742-753. [DOI] [PubMed] [Google Scholar]
- 23.Kim, M. K., J. H. Jeon, M. Fujita, L. B. Davin, and N. G. Lewis. 2002. The western red cedar (Thuja plicata) 8-8′ DIRIGENT family displays diverse expression patterns and conserved monolignol coupling specificity. Plant Mol. Biol. 49:199-214. [DOI] [PubMed] [Google Scholar]
- 24.Kleinkauf, H., and H. Von Doehren. 1996. A nonribosomal system of peptide biosynthesis. Eur. J. Biochem. 236:335-351. [DOI] [PubMed] [Google Scholar]
- 25.Kroken, S., N. L. Glass, J. W. Taylor, O. C. Yoder, and B. G. Turgeon. 2003. Phylogenomic analysis of type I polyketide synthase genes in pathogenic and saprobic ascomycetes. Proc. Natl. Acad. Sci. USA 100:15670-15675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leach, J., B. R. Lang, and O. C. Yoder. 1982. Methods for selection of mutants and in vitro culture of Cochliobolus heterostrophus. J. Gen. Microbiol. 128:1719-1729. [Google Scholar]
- 27.Lu, S. W., S. Kroken, B. N. Lee, B. Robbertse, A. C. Churchill, O. C. Yoder, and B. G. Turgeon. 2003. A novel class of gene controlling virulence in plant pathogenic ascomycete fungi. Proc. Natl. Acad. Sci. USA 100:5980-5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marahiel, M. A., W. Danders, M. Krause, and H. Kleinkauf. 1979. Biological role of gramicidin S in spore functions. Studies on gramicidin-S-negative mutants of Bacillus brevis ATCC9999. Eur. J. Biochem. 99:49-55. [DOI] [PubMed] [Google Scholar]
- 29.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2674. [DOI] [PubMed] [Google Scholar]
- 30.Nikolskaya, A. N., D. G. Panaccione, and J. D. Walton. 1995. Identification of peptide synthetase-encoding genes from filamentous fungi producing host-selective phytotoxins or analogs. Gene 165:207-211. [DOI] [PubMed] [Google Scholar]
- 31.Omura, S., H. Ikeda, J. Ishikawa, A. Hanamoto, C. Takahashi, M. Shinose, Y. Takahashi, H. Horikawa, H. Nakazawa, T. Osonoe, H. Kikuchi, T. Shiba, Y. Sakaki, and M. Hattori. 2001. Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc. Natl. Acad. Sci. USA 98:12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panaccione, D. G., J. S. Scott-Craig, J. A. Pocard, and J. D. Walton. 1992. A cyclic peptide synthetase gene required for pathogenicity of the fungus Cochliobolus carbonum on maize. Proc. Natl. Acad. Sci. USA 89:6590-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pringle, R. B., and A. C. Braun. The isolation of the toxin of Helminthosporium victoriae. Phytopathology 47:369-371, 1957.
- 34.Robbertse, B., O. C. Yoder, A. Nguyen, C. L. Schoch, and B. G. Turgeon. 2003. Deletion of all Cochliobolus heterostrophus monofunctional catalase-encoding genes reveals a role for one in sensitivity to oxidative stress but none with a role in virulence. Mol. Plant-Microbe Interact. 16:1013-1021. [DOI] [PubMed] [Google Scholar]
- 35.Schaeffer, P. 1969. Sporulation and the production of antibiotics, exoenzymes, and exotoxins. Bacteriol. Rev. 33:48-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwarzer, D., R. Finking, and M. A. Marahiel. 2003. Nonribosomal peptides: from genes to products. Nat. Prod. Rep. 20:275-287. [DOI] [PubMed] [Google Scholar]
- 37.Schwarzer, D., and M. A. Marahiel. 2001. Multimodular biocatalysts for natural product assembly. Naturwissenschaften 88:93-101. [DOI] [PubMed] [Google Scholar]
- 38.Scott-Craig, J. S., D. G. Panaccione, J. A. Pocard, and J. D. Walton. 1992. The cyclic peptide synthetase catalyzing HC-toxin production in the filamentous fungus Cochliobolus carbonum is encoded by a 15.7-kilobase open reading frame. J. Biol. Chem. 267:26044-26049. [PubMed] [Google Scholar]
- 39.Shen, B., L. Du, C. Sanchez, D. J. Edwards, M. Chen, and J. M. Murrell. 2001. The biosynthetic gene cluster for the anticancer drug bleomycin from Streptomyces verticillus ATCC15003 as a model for hybrid peptide-polyketide natural product biosynthesis. J. Ind. Microbiol. Biotechnol. 27:378-385. [DOI] [PubMed] [Google Scholar]
- 40.Smith, D. J., M. K. Burnham, J. H. Bull, J. E. Hodgson, J. M. Ward, P. Browne, J. Brown, B. Barton, A. J. Earl, and G. Turner. 1990. Beta-lactam antibiotic biosynthetic genes have been conserved in clusters in prokaryotes and eukaryotes. EMBO J. 9:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stachelhaus, T., and M. A. Marahiel. 1995. Modular structure of peptide synthetases revealed by dissection of the multifunctional enzyme GrsA. J. Biol. Chem. 270:6163-6169. [DOI] [PubMed] [Google Scholar]
- 42.Stachelhaus, T., H. D. Mootz, and M. A. Marahiel. 1999. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6:493-505. [DOI] [PubMed] [Google Scholar]
- 43.Tudzynski, P., K. Holter, T. Correia, C. Arntz, N. Grammel, and U. Keller. 1999. Evidence for an ergot alkaloid gene cluster in Claviceps purpurea. Mol. Gen. Genet. 261:133-141. [DOI] [PubMed] [Google Scholar]
- 44.Turgeon, B. G., R. C. Garber, and O. C. Yoder. 1987. Development of a fungal transformation system based on selection of sequences with promoter activity. Mol. Cell. Biol. 7:3297-3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tzeng, T. H., L. K. Lyngholm, C. F. Ford, and C. R. Bronson. 1992. A restriction fragment length polymorphism map and electrophoretic karyotype of the fungal maize pathogen Cochliobolus heterostrophus. Genetics 130:81-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vining, L. C. 1990. Functions of secondary metabolites. Annu. Rev. Microbiol. 44:395-427. [DOI] [PubMed] [Google Scholar]
- 47.von Dohren, H., U. Keller, J. Vater, and R. Zocher. 1997. Multifunctional peptide synthetases. Chem. Rev. 97:2675-2705. [DOI] [PubMed] [Google Scholar]
- 48.Walton, J. D. 1996. Host-selective toxins: agents of compatibility. Plant Cell 8:1723-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walton, J. D., D. G. Panaccione, and H. Hallen. (2004). Peptide synthesis without ribosomes, p. 127-162. In J. Tkacz and L. Lange (ed.), Advances in fungal biotechnology for industry, agriculture, and medicine. Kluwer Academic, New York, N.Y.
- 50.Weber, G., K. Schorgendorfer, E. Schneider-Scherzer, and E. Leitner. 1994. The peptide synthetase catalyzing cyclosporine production in Tolypocladium niveum is encoded by a giant 45.8-kilobase open reading frame. Curr. Genet. 26:120-125. [DOI] [PubMed] [Google Scholar]
- 51.Wiest, A., D. Grzegorski, B. W. Xu, C. Goulard, S. Rebuffat, D. J. Ebbole, B. Bodo, and C. Kenerley. 2002. Identification of peptaibols from Trichoderma virens and cloning of a peptaibol synthetase. J. Biol. Chem. 277:20862-20868. [DOI] [PubMed] [Google Scholar]
- 52.Wirsel, S., B. G. Turgeon, and O. C. Yoder. 1996. Deletion of the Cochliobolus heterostrophus mating type (MAT) locus promotes function of MAT transgenes. Curr. Genet. 29:241-249. [PubMed] [Google Scholar]
- 53.Yoder, O. C., V. Macko, T. J. Wolpert, and B. G. Turgeon. 1997. Cochliobolus spp. and their host-specific toxins, p. 145-166. In G. Carroll and P. Tudzynski (ed.), The mycota, vol. 5. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 54.Yoder, O. C., and B. G. Turgeon. 2001. Fungal genomics and pathogenicity. Curr. Opin. Plant Biol. 4:315-321. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, W. M., G. D. Gentil, A. D. Budde, and S. A. Leong. 2001. Characterization of the Ustilago maydis sid2 gene, encoding a multidomain peptide synthetase in the ferrichrome biosynthetic gene cluster. J. Bacteriol. 183:4040-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.