Abstract
The Rho G protein Cdc42 and its exchange factor Cdc24 are required for hyphal growth of the human fungal pathogen Candida albicans. Previously, we reported that strains ectopically expressing Cdc24 or Cdc42 are unable to form hyphae in response to serum. Here we investigated the role of these two proteins in hyphal growth, using quantitative real-time PCR to measure induction of hypha-specific genes together with time lapse microscopy. Expression of the hypha-specific genes examined depends on the cyclic AMP-dependent protein kinase A pathway culminating in the Efg1 and Tec1 transcription factors. We show that strains with reduced levels of CDC24 or CDC42 transcripts induce hypha-specific genes yet cannot maintain their expression in response to serum. Furthermore, in serum these mutants form elongated buds compared to the wild type and mutant budding cells, as observed by time lapse microscopy. Using Cdc24 fused to green fluorescent protein, we also show that Cdc24 is recruited to and persists at the germ tube tip during hyphal growth. Altogether these data demonstrate that the Cdc24/Cdc42 GTPase module is required for maintenance of hyphal growth. In addition, overexpression studies indicate that specific levels of Cdc24 and Cdc42 are important for invasive hyphal growth. In response to serum, CDC24 transcript levels increase transiently in a Tec1-dependent fashion, as do the G-protein RHO3 and the Rho1 GTPase activating protein BEM2 transcript levels. These results suggest that a positive feedback loop between Cdc24 and Tec1 contributes to an increase in active Cdc42 at the tip of the germ tube which is important for hypha formation.
Polarized growth in response to external signals is essential for both the internal organization of cells and generation of complex multicellular structures. Many different cell types from neurites to fungi form highly elongated protrusions in response to different environmental signals. The Rho family GTPase Cdc42 and its GDP-GTP exchange factors are critical for the cytoskeletal and transcriptional changes underlying such dramatic cell polarization processes. In the budding yeast Saccharomyces cerevisiae and the pathogenic yeast Candida albicans, Cdc42 and its exchange factor Cdc24 are necessary for polarized growth during budding (22, 56). Furthermore, these two proteins have been shown to be specifically required for mating in S. cerevisiae (6, 33, 37, 42) and invasive hyphal growth of C. albicans (7, 48, 49). In S. cerevisiae, these proteins are necessary both for establishing and maintaining cell polarity (8, 21, 36, 51).
During C. albicans hyphal formation, growth is restricted to a small region of the cell surface, resulting in the protrusion of a germ tube which ultimately forms highly elongated filaments. These hyphal filaments can extend to many times the length of the original cell body (blastospore), have one or more septal junctions, and may or may not be branched. Actin and a number of proteins involved in regulation of the cytoskeleton localize to the tips of germ tubes. F-actin patches are clustered at the site of the incipient germ tube just prior to evagination (2), similar to the cytoskeleton-associated polarity protein Spa2 and Cdc42, which are also observed at the site of nascent germ tube formation (20, 59, 60). Furthermore, in hyphal filaments, Cdc42 and actin, but not Spa2, localize to the septal junctions, similar to septin localization (18, 47, 50). Hyphal formation in C. albicans is in some respects similar to S. cerevisiae mating and pseudohyphal formation (20, 46). As is the case during S. cerevisiae mating and pseudohyphal growth (4, 14), F-actin is required for the highly polarized growth that occurs during germ tube formation in C. albicans (1, 20). During this process, F-actin is required for the recruitment to and maintenance of Cdc42 at the tip of the protrusion and partial depolymerization of the actin cytoskeleton inhibits hyphal formation but not bud formation (20). Several C. albicans mutants with mutations in genes that regulate the actin cytoskeleton, including myo5, cdc42, and cdc24, undergo budding growth yet are unable to form invasive hyphal filaments, suggesting that hyphal growth requires increased or sustained actin polarization compared to budding cells (7, 39, 49).
A variety of signals, including elevated temperature, alkaline pH, serum, and N-acetylglucosamine can trigger the C. albicans yeast-to-filamentous growth transition. As is the case for invasive and pseudohyphal growth of S. cerevisiae, both mitogen-activated protein (MAP) kinase and cyclic AMP (cAMP)-dependent protein kinase A pathways are required for the induction of a range of genes that are critical for C. albicans hyphal development (17, 27, 55). These two signaling pathways culminate in specific transcription factors, Cph1 for the MAP kinase pathway and Efg1 for the cAMP-dependent pathway. Efg1 appears to be more important for hyphal growth in response to a range of environmental signals, in particular serum (28). Tec1, a member of the TEA/ATTC transcription factor family (which includes mammalian TEF-1, ScTec1, AnAbaA, and DmScalloped), in part functions downstream of Efg1 (25, 41). In addition to these transcription factors which can activate gene expression, negative transcriptional regulators have also been described, such as Tup1, Nrg1, and Rfg1 (12, 13, 23, 24, 34). A number of hypha-specific genes (HSGs) have been identified whose expression is dramatically increased during hyphal formation. These genes encode cell wall proteins such as ECE1 (9), HWP1 (43, 44), HYR1 (5), and ALS8 (58), a G1 cyclin protein, HGC1 (59), and secreted aspartic proteinases such as those encoded by the SAP genes (32). The induction of these HSGs during hyphal formation is essentially dependent on the cAMP-dependent protein kinase A pathway (19). While some of these proteins are important for virulence in a mouse model of systemic candidiasis, only HGC1 is absolutely required for hyphal formation (59).
Cdc42 and Cdc24 are required for C. albicans hyphal development (7, 48, 49); however, the specific roles and regulation of these two proteins in HSG induction and morphological changes that accompany hyphal formation are less clear. Very recently, C. albicans cdc42 mutants based on S. cerevisiae cdc42 mutants defective in pseudohyphal growth were shown to have reduced expression of several HSGs (49). Here we show that cdc42 and cdc24 mutants which express reduced levels of each respective protein and are defective in hyphal formation in response to serum are able to induce HSGs. However, these mutants cannot maintain elevated levels of these HSGs. Analyses of cell morphology further indicate that these mutants respond to serum but are unable to sustain this morphological response. Overexpression studies indicate that specific levels of Cdc24 and Cdc42 are critical for invasive hyphal growth. In response to serum, CDC24 transcript levels increase transiently in a Tec1-dependent fashion and Cdc24 is recruited to the tip of the incipient germ tube. These results suggest that a positive feedback loop between Cdc24 and Tec1 contributes to an increase in the level of active Cdc42 restricted to the tip of the germ tube, which is important for the polarized growth necessary for hyphal formation.
MATERIALS AND METHODS
General techniques.
Yeast extract-peptone-dextrose (YEPD) or synthetic complete (SC) medium supplemented with 80 mg of uridine (Uri)/liter was used, and strains were grown at 30°C. For hyphal induction in liquid medium, cells were incubated in YEPD containing 50% fetal calf serum (FCS; Dominique Dutscher) at 37°C. In order to reduce fluorescence background in experiments for which results are shown in Fig. 8D, cells were incubated in SC medium. For time lapse microscopy, cells were incubated on 25% YEPD agar-75% FCS pads at 37°C. Where indicated, 2.5 mM methionine and 2.5 mM cysteine were included in the medium. For invasive growth in solid medium, cells were spotted on YEPD agar plates containing 50% FCS and grown at 37°C.
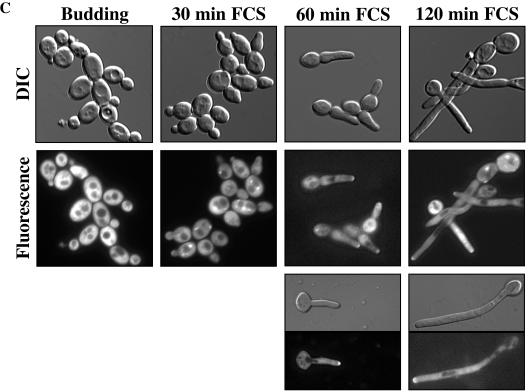
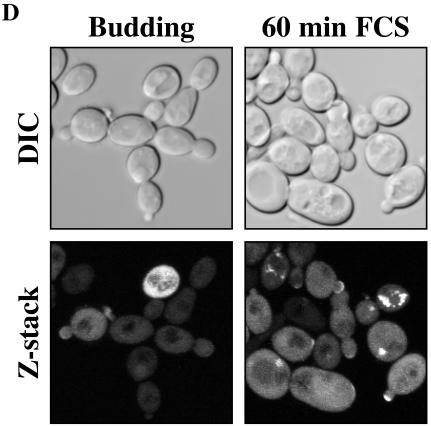
FIG. 8.
Cdc24 is recruited to the tips of germ tubes in response to FCS. (A) Cdc24GFP is functional. The indicated strains were grown on medium which permits (−Met/Cys) or represses (+Met/Cys) MET3-driven CDC24 expression for 2 days at 30°C. (B) Immunoblot analysis of cdc24Δ/PMETCDC24 mutant cells with or without integrated Cdc24GFP. Extracts from cells grown at 30°C were analyzed with anti-GFP polyclonal antibodies. (C) Cdc24GFP localization over 120 min in cells in the presence of FCS. cdc24Δ/PMETCDC24 cells overexpressing Cdc24GFP were grown in YEPD-Uri medium with Met and Cys at 30°C (Budding) and for the indicated times in FCS at 37°C. DIC and fluorescence images are shown. Panels show individual cells after 60 and 120 min in FCS at 37°C. (D) Cdc24GFP localization in the presence or absence of FCS. cdc24Δ/PMETCDC24 cells overexpressing Cdc24GFP grown in SC medium with Met and Cys for 60 min in the presence or absence of FCS were examined by confocal microscopy. DIC images are average intensity projections, and fluorescence images are maximum intensity projections of 15 to 17 0.2-μm z sections.
Strains and plasmids.
The prototrophic wild-type (BWP17 URA+ HIS+ ARG+ [57]), cdc24Δ::HIS1/cdc24::PMETCDC24 (7), and cdc42Δ::HIS1/cdc42::PMETCDC42 (7) strains and the tec1Δ::hisG/tec1Δ::hisG strain CaAs15 (41) were previously described (Table 1). Overexpression constructs were generated by cloning the CDC24 or CDC42 open reading frame into pAW6 (38) followed by subcloning PADH-CDC24-GFP-ADHT or PADH-CDC42-ADHT into pExpArg (7). Initially, an XhoI site was inserted in the URA3 promoter of pAW6 by site-directed mutagenesis with Pfu polymerase (Promega) by the DpnI method (52), resulting in pAW6-X. A sequence encoding a hemagglutinin (HA) epitope was inserted by site-directed mutagenesis just 5′ of the green fluorescent protein (GFP) coding sequence in pAW6-X, resulting in pAW6-HA. For CDC24GFP expression, a unique SacI site was introduced by site-directed mutagenesis just 5′ of the CDC24 stop codon in pExpArgCDC24 (7), and the BamHI/SacI CDC24 open reading frame was then cloned into the BglII/SacI-digested pAW6-HA vector, resulting in pAW6-CDC24. A NotI/XhoI PADH-CDC24HAGFP-ADHT fragment was cloned into pExpArg, resulting in pExpArg-PADHCDC24GFP. For Cdc42 expression a unique BamHI site was introduced 5′ of ATG in pRS202-Cdc42 (7) by site-directed mutagenesis, resulting in pRS202-Cdc42-B. A BamHI/MluI CDC42 fragment was then cloned into BglII/MluI-digested pAW6-X, resulting in pAW6-CDC42, in which the GFP coding sequence has been removed. A NotI/XhoI PADH-CDC42-ADHT fragment was cloned into pExpArg, resulting in pExpArg-PADHCDC42. pExpArg-PADHCDC24GFP or pExpArg-PADHCDC42 was targeted to the RP10 locus after StuI or NcoI digestion, respectively, transformation, and selection for arginine prototrophy. Correct integration of pExpArg-PADHCDC24GFP or pExpArg-PAHDCDC42 was identified by PCR, and protein expression was confirmed by immunoblotting. Two independent integrants for expression of Cdc24 and Cdc42 were subsequently analyzed and gave similar results.
TABLE 1.
Strains used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| BWP17 | ura3Δ::λ imm434/ura3Δ::λ imm434 his1Δ::hisG/his1Δ::hisG arg4Δ::hisG/arg4Δ::hisG | 57 |
| PY47 | cdc42Δ::HIS1/cdc42::URA3-PMETCDC42 arg4::hisG/arg4::hisG | 7 |
| PY82 | ura3Δ::λ imm434/ura3Δ::λ imm434 his1::hisG/HIS1::his1::hisG arg4::hisG/URA3::ARG4::arg4::hisG | 7 |
| PY92 | Same as PY101 with RP10::ARG4 | 7 |
| PY101 | cdc24Δ::HIS1/cdc24::URA3-PMETCDC24 arg4::hisG/arg4::hisG | 7 |
| PY109 | Same as PY101 with RP10::ARG4-PADHCDC24GFP | This study |
| PY113 | Same as PY47 with RP10::ARG4-PADHCDC24GFP | This study |
| PY115 | Same as PY47 with RP10::ARG4-PADHCDC42 | This study |
| PY121 | Same as PY101 with RP10::ARG4-PADHCDC42 | This study |
| PY123 | Same as PY47 with RP10::ARG4 | This study |
| CaAs15 | ura3/ura3 tec1Δ::hisG/tec1Δ::hisG | 41 |
qRT-PCR.
For quantitative real-time PCR (qRT-PCR) analysis of RNA transcript levels, RNA was extracted from cells grown in YEPD-Uri or YEPD-Uri-FCS. RNase-free ultrapure water (Qbiogene) was used in all subsequent steps. RNA was isolated from 20 ml of log-phase cells, using Tri-reagent (Euromedex) following the manufacturer's instructions with an added acid-washed glass bead (Sigma) cell breakage step, using a Ribolyser (Hybaid). RNA (4 μg) was incubated with DNase RQ1 (Promega) in the presence of RNasin (Promega) for 30 min at 37°C and inactivated for 5 min at 75°C. This material was used for cDNA synthesis with Moloney murine leukemia virus reverse transcriptase (Invitrogen) following the manufacturer's instructions.
qRT-PCR was carried out on an ABI Prism 7000 Sequence Detection system (Applied Biosystems) using ABI Prism 7000 SDS Software. Oligonucleotide primers were chosen with ABI primer express software (Applied Biosystems) and were 18 to 25 bases long (Table 2). Primer pairs had melting temperatures between 58 and 60°C with either a 1°C or no difference in melting temperature and resulted in amplicons of 75 to 150 bp. All primer pairs produced products of the expected sizes. For qRT-PCR, qPCR Mastermix plus for SYBR Green I (Eurogentec) was used according to the manufacturer's recommendations except that 25-μl reaction mixtures were used. Prior to transcript quantitation, efficiency values were determined for all primer pairs, and except for RIM101 (efficiency 80%) efficiencies of 90% or greater were observed. Each reaction mixture contained a 300 nM concentration of each primer and 5 μl of cDNA product diluted 1:100 except for CDC24 analyses, where a cDNA product diluted 1:20 was used. Reactions were run in sealed 96-well plates (ABgene) using recommended cycle times and temperatures. For CDC24 and CDC42, values are averages of results with two different primer pairs. Signals from all reactions were substantially higher than for the no-cDNA controls, including signals from all HSGs in the absence of serum, which were at least 10 times higher than those for the no-cDNA controls. RNA transcript levels were calculated relative to the level of actin transcripts, using the equation 2Ctgene/2Ctactin, where Ct is the cycle number at which the amplified target reaches the fixed threshold of 0.2. Therefore, a value of 1 indicates a transcript level equal to that of actin. For hyphal formation time courses, exponentially growing strains (in YEPD-Uri with an optical density at 600 nm of ∼ 0.4) were diluted with an equal volume of prewarmed FCS and incubated for 10, 30, and 120 min at 37°C. Aliquots for RNA isolation and cell imaging were removed at indicated times. Aliquots from zero time points were incubated in the absence of FCS for 30 min at 37°C.
TABLE 2.
Primers used for qRT-PCR analyses
| Primer | Sequence |
|---|---|
| CDC24b+ | GGCCACCAATGCGTCAAC |
| CDC24b− | TAGCTGGTAAAACCCCTGCAGTA |
| CDC24c+ | ACGTGATTCCGACCTGTCATTTA |
| CDC24c− | GATGGCGGGAACTGTGAGA |
| CDC42c+ | TCCCCAATCACCCAGGAA |
| CDC42c− | TGCAGCTACTATAGCCTCGTCAA |
| CDC42d+ | GGGTGAAAAATTGGCTAAGGAA |
| CDC42d− | CCTCTTTGAGTCAATGCAGAACA |
| ACT1+ | ATGTTCCCAGGTATTGCTGA |
| ACT1− | ACATTTGTGGTGAACAATGG |
| ECE1+ | CCAGAAATTGTTGCTCGTGTTG |
| ECE1− | CAGGACGCCATCAAAAACG |
| HGC1+ | AAAGCTGTGATTAAATCGGTTTTGA |
| HGC1− | AATTGAGGACCTTTTGAATGGAAA |
| ALS8+ | TTGGCAACGTGCACCTTTC |
| ALS8− | GTGGTCACGGCGGTAGTACTG |
| HWP1+ | CGGAATCTAGTGCTGTCGTCTCT |
| HWP1− | TAGGAGCGACACTTGAGTAATTGG |
| HYR1+ | CTCAACCTCAGTGCTGCATTAGAA |
| HYR1− | AGCCCAAGTAGCACCAGAATGA |
| SAP5+ | CAATTATTTGCTAACGTTTGGTCTACTAG |
| SAP5− | CTTCGCCGCTTTGAAAACC |
| SAP6+ | TCTCCAGGGTTTGTTACCTTAGACTT |
| SAP6− | TTAGATTCGGCAGTTGGATCATC |
| CPH1+ | GGTGGCGGCAGTGATAGTG |
| CPH1− | GTGTACTCCGGTGACGATTTTTC |
| TEC1+ | TGGATTCATACCGTATTGGTCATTA |
| TEC1− | TCGGGCAATCCTTTGAATAAA |
| EFG1+ | TCCCACCACATGTATCGACAA |
| EFG1− | GAAGTGCTCGAGGCGTTCA |
| RIM101+ | AAGTGACGATTGATGGGTTCAATT |
| RIM101− | GCCAAGAACAGCATGGATCATT |
| CZF1+ | TAGGCGGTCCACCATTTCC |
| CZF1− | GGATAACCAGTGTTTGGCATATACC |
| NRG1+ | ACCAACAACCCATGTACCAAAAC |
| NRG1− | ATTAGCCCTGGAGATGGTCTGA |
| TUP1+ | CTCAAAGGACCGTGGTGTCAT |
| TUP1− | GGCCCTGCAACATCAACAAT |
| BEM2+ | GATTTATTCCCCCGAGAGTCGTA |
| BEM2− | CCCTTCACGATGGGTTAATGTT |
| RHO3+ | AAGGGTGTTAACGAAGCATTTTCT |
| RHO3− | CATTGTCATTGGCACCTTTAGG |
Western blot analyses.
For immunoblot analyses, cells were broken with glass beads by agitation in a Ribolyser. Proteins were then separated on sodium dodecyl sulfate-polyacrylamide (18 or 8%) gels and transferred to a Protran nitrocellulose membrane (Schleicher & Schuell). The membranes were probed with either anti-Cdc42 polyclonal antibodies which were produced against maltose binding protein-ScCdc42 (to be described elsewhere) (1:1,000) or anti-GFP polyclonal antibodies (1:1,000) (36), followed by ECL visualization (Amersham).
Microscopy.
For colony morphology, cells grown on agar-containing medium for 8 days at 30°C were imaged with a Leica MZ6 dissection scope at a magnification of ×20. For cell morphology, cells were grown to exponential phase and then shifted at 37°C for 2 h in the presence of FCS (1:1, vol/vol). Cells were imaged as described previously (7) using a Leica DMR epifluorescence microscope with a numerical aperture (NA) 1.35 × 63 objective. Images were recorded with a Princeton Instruments Micromax charge-coupled device (Roper Scientific), using IPLab (Scanalytics) software. Time lapse differential interference contrast (DIC) microscopy was carried out with a Deltavision deconvolution microscopy system (Applied Precision) on an Olympus IX-70 microscope with an NA 1.4 × 60 objective. Exponentially growing cells were mixed with an equal volume of FCS and spotted on YEPD-FCS agar pads as previously described (3), and an environmental temperature-controlled chamber (Solent Scientific) was used. In each time lapse, five cells were monitored by DIC microscopy. The maximum lengths of cell buds (just prior to cell division) were measured from time lapse movies using ImageJ software. Confocal microscopy was carried out on a Zeiss LSM 510 META Axiovert 200 M microscope with an NA 1.4 × 63 objective and 488-nm LASER excitation. DIC and fluorescence images were captured, and arithmetic average projections (DIC images) or maximum intensity projections (fluorescence images) of z stacks were calculated with ImageJ.
RESULTS
Reduced levels of Cdc24 and Cdc42 in cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants.
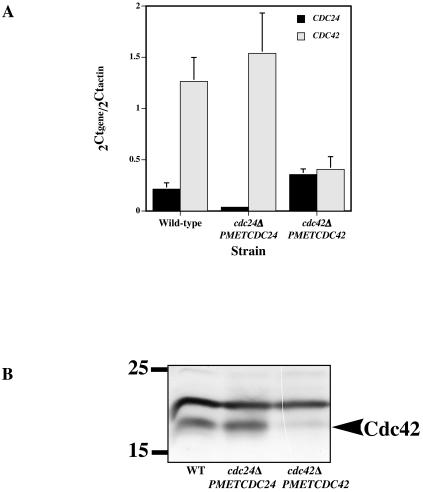
We initially examined the levels of CDC24 and CDC42 RNA transcripts in budding wild-type, cdc24Δ/PMETCDC24, and cdc42Δ/PMETCDC42 strains. To quantitate the transcript levels from cells growing in rich medium at 30°C, we used qRT-PCR with two different primer pairs. Figure 1A shows that in wild-type cells, CDC42 transcripts are approximately six times more abundant than CDC24 transcripts. In cdc24Δ/PMETCDC24 mutant cells, CDC24 transcript levels were reduced sixfold, whereas CDC42 transcript levels were slightly elevated compared to those of a wild-type strain. Similarly, in cdc42Δ/PMETCDC42 mutant cells, CDC42 transcript levels were reduced threefold in contrast to CDC24 transcript levels, which were somewhat increased. Analyses of Cdc42 protein levels by immunoblotting confirmed these qRT-PCR results (Fig. 1B) and were consistent with the difference between Cdc24 levels expressed from the CDC24 and MET3 promoters (7). These results show that each mutant has reduced expression levels of the respective gene when grown in YEPD medium. The modest increase in CDC42 transcript levels in a cdc24Δ/PMETCDC24 mutant and CDC24 transcript levels in a cdc42Δ/PMETCDC42 mutant suggests a compensatory effect between G protein and activator (exchange factor).
FIG.1.
Cdc24 and Cdc42 levels are reduced in cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 strains. (A) Transcript levels of CDC24 and CDC42 in the wild type and ectopic expression mutants. RNA transcript levels normalized to actin transcript levels (2Ctgene/2Ctactin) were determined from budding cells grown at 30°C in YEPD-Uri medium. Values are the means of three or four independent experiments, with bars representing standard deviations. (B) Cdc42 protein levels in the wild type and ectopic expression mutants. Immunoblot analysis of Cdc42 protein levels from the indicated strains grown in YEPD-Uri medium at 30°C. Anti-Cdc42 polyclonal antibodies were used. The upper band is a nonspecific protein cross-reacting with anti-Cdc42 antibody.
Temporal gene induction during hyphal growth.
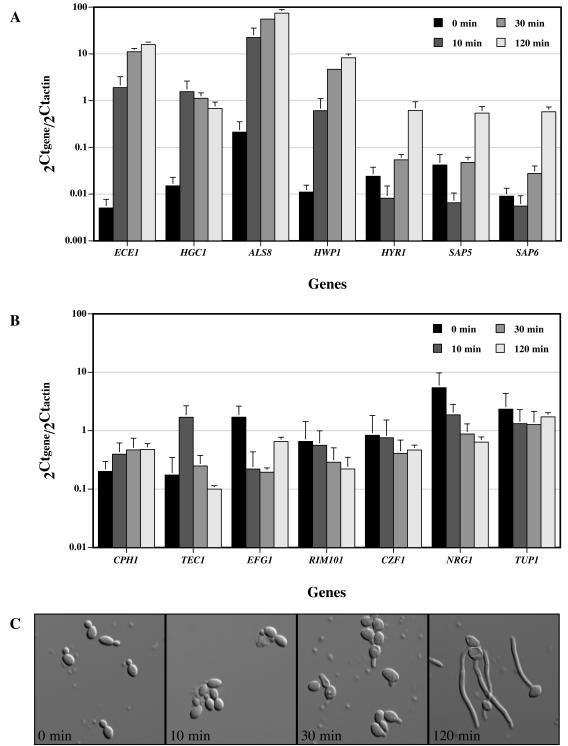
To establish a quantitative measure of hyphal response complementary to morphological changes, we monitored the transcript levels of a variety of different HSGs and transcription factors in cells exposed to serum. We focused on the following HSGs: ECE1, which encodes a cell wall protein whose expression correlates with the extent of cell elongation (9); HGC1, which encodes a G1 cyclin protein required for hyphal formation (59); ALS8, which encodes an agglutinin-like cell wall glycosylphosphatidylinositol (GPI) protein (58); HWP1, which encodes a cell wall GPI mannoprotein involved in cell adhesion (43, 44); HYR1, which encodes a cell wall GPI protein (5); and SAP5 and SAP6, which encode secreted aspartic proteinases (32). These HSGs encode proteins associated with C. albicans virulence, yet of these genes only HGC1 is required for hyphal formation (59). Figure 2A shows the transcript levels of these HSGs in wild-type cells incubated in the presence or absence of FCS at 37°C for 10, 30, and 120 min. These transcript levels were low relative to those of actin (10- to 100-fold lower) in the absence of serum, yet were nonetheless detectable. All of these HSGs were strongly induced in the presence of serum (100- to 1,000-fold increases).
FIG. 2.
Quantitative analyses of hyphal gene and transcription factor expression in wild-type cells exposed to FCS. (A) Normalized transcript levels of HSGs in wild-type cells responding to FCS at 37°C. Relative transcript abundance values (2Ctgene/2Ctactin) are the means of three independent experiments with standard deviations indicated. (B) Normalized transcript levels of transcription factors in wild-type cells responding to FCS at 37°C. Relative transcript abundance values (2Ctgene/2Ctactin) are the means of three independent experiments with standard deviations indicated. (C) Representative images of cells responding to FCS. DIC images of fixed cells from the same experiment were recorded at the indicated times.
The temporal induction profiles of these HSGs indicate two types of responses, one which includes the early induced genes ECE1, HGC1, ALS8, and HWP1 (9, 11, 59) and the other comprised of the late induced genes HYR1, SAP5, and SAP6 (29, 40, 41). The level of the former class is maximal or near maximal after 10 min of cell exposure to serum. In contrast, the level of the latter class decreases from 0 to 10 min in serum and subsequently increases progressively with time. Figure 2C shows the morphology of wild-type cells fixed after 0, 10, 30, and 120 min in the presence of serum. Germ tubes start to appear at 30 min and increase in length up to five times the cell body size at 120 min. The early genes are largely induced prior to the observed morphological changes, whereas the late genes are induced concomitant with morphological changes. We also examined the levels of a number of different transcription factors involved in hyphal morphogenesis, including CPH1, TEC1, EFG1, RIM101, CZF1, NRG1, and TUP1 (for a review, see reference 27) (Fig. 2B). TEC1 levels transiently increased in cells exposed to serum for 10 min, whereas EFG1 levels decreased at 10 and 30 min in serum and increased at 120 min, similar to previous observations (41, 45). These results are consistent with a role for TEC1 as a transcriptional activator and EFG1 as a transcriptional repressor during the initial response of cells to serum (41, 45). No significant variation in the levels of CPH1, RIM101, CZF1, and TUP1 transcripts were observed. Consistent with its role as a repressor of hyphal growth, NRG1 levels decreased over time, as previously observed (13).
cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants induce HSGs but are unable to maintain induction.
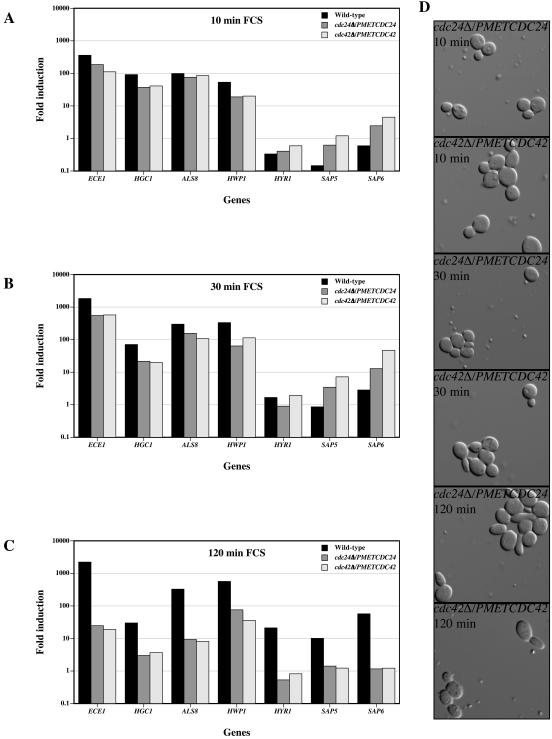
Previously, we demonstrated that cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants are defective in hyphal formation in response to FCS (7). The simplest explanation for this defect is that these mutants do not respond to serum. To test this hypothesis, we examined the levels of HSGs in these mutants in rich medium in the presence of serum. The levels of the HSGs were similar in wild-type, cdc24Δ/PMETCDC24, and cdc42Δ/PMETCDC42 strains prior to the addition of FCS (data not shown). After 10 min in serum, the induction of the HSGs in wild-type, cdc24Δ/PMETCDC24, and cdc42Δ/PMETCDC42 strains was also similar, with the exception of induction of SAP5 and SAP6, which was substantially higher (approximately 10-fold) in the mutants (Fig. 3A). After 30 min in serum, SAP5 and SAP6 levels were still higher in the mutants compared to wild-type cells. In contrast, inductions of ECE1, HGC1, HWP1, HYR1, and ALS8 transcripts were similar or slightly lower in the mutants compared to wild-type cells (Fig. 3B). Strikingly, the early induced genes were substantially affected in the mutants after 120 min in serum relative to wild-type cells. Specifically, ECE1, HGC1, and ALS8 levels were reduced 10- to 30-fold at 120 min compared to the 30-min time point (Fig. 3C), whereas the levels of the transcripts in wild-type cells remained constant between these times. HWP1 and HYR1 transcript levels were approximately 5- and 20-fold lower, respectively, at 120 min in the mutants compared to wild-type cells. Strikingly, the induced levels of SAP5 and SAP6, which were higher in the mutants at the 10- and 30-min time points fell approximately 10-fold between 30 and 120 min, while the levels of these transcripts in wild-type cells increased approximately 10-fold during the same time period. Aliquots of the mutant cells incubated in serum for 10, 30, and 120 min were fixed, and Fig. 3D shows that these cells had buds at all times; however, these buds were somewhat elongated after 30 and 120 min in serum.
FIG. 3.
In response to FCS cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants induce HSGs yet are unable to maintain induced levels. Fold induction of HSGs after incubation of cells for 10 (A), 30 (B), and 120 (C) min in FCS at 37°C is shown. Fold induction is the normalized transcript level at the indicated time divided by the normalized transcript level at time zero. The mean values of two independent experiments are shown. (D) Representative images of cells responding to FCS. DIC images of fixed cells from the same experiment were recorded at the indicated times.
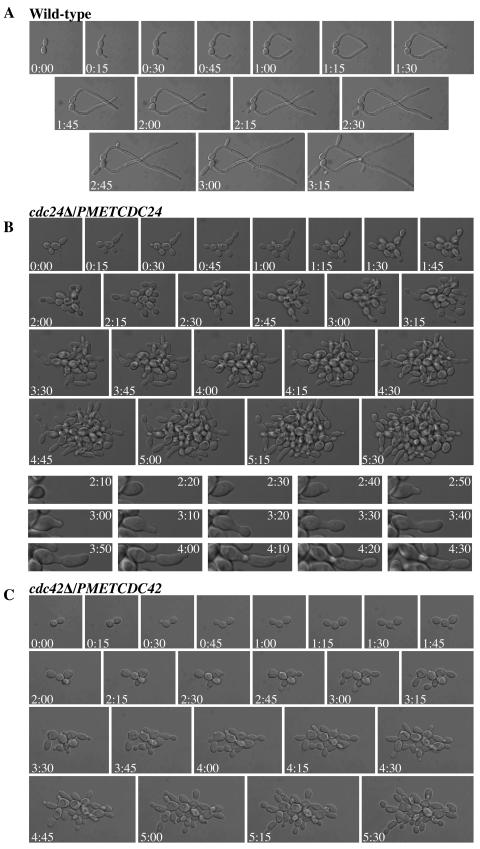
To further investigate the serum-dependent morphology changes and doubling times in cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants, we used time lapse microscopy to follow cells at 37°C either on rich medium agar pads or 25% rich medium agar with 75% FCS. Figure 4A shows that wild-type cells form hyphae in response to serum. The first septum appeared around 100 min, and thereafter they appeared every hour. Hyphal filaments grew to approximately 150 μm in length over the 6.5-h incubation with substantial branching apparent (data not shown). In contrast, both cdc24Δ/PMETCDC24 (Fig. 4B) and cdc42Δ/PMETCDC42 (Fig. 4C) mutants continued to bud in the presence of serum at 37°C. These cells divided every 50 min, similar to the time of septum appearance in wild-type cells under the same conditions and to the doubling time of wild-type and mutant cells in the absence of serum at 37°C (data not shown). Nonetheless, both mutants in the presence of serum were elongated; maximum bud lengths were 6.44 ± 1.00 μm (n = 34 cells) for cdc24Δ/PMETCDC24 mutant cells and 6.46 ± 1.10 μm (n = 40 cells) for cdc42Δ/PMETCDC42 mutant cells compared to the wild type and mutants (data not shown) in the absence of serum; wild-type maximum bud length was 4.83 ± 0.38 μm (n = 40 cells). An example of this morphological change is highlighted in the lower panels of Fig. 4B. Together, HSG induction and morphological changes in cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 strains indicate that these mutants respond to serum but do not maintain this response over time.
FIG. 4.
cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants initiate a morphological response to serum but are unable to form hyphae. (A) Time lapse microscopy of wild-type cells in FCS at 37°C. DIC images were captured every 5 min over 6 h, and images every 15 min are shown over 195 min. (B) Time lapse microscopy of cdc24Δ/PMETCDC24 mutant cells in FCS at 37°C. DIC images were captured every 5 min over 6 h, and images every 15 min are shown during 330 min (top panels). Close-ups of individual cell images (bottom panels) every 10 min from 130 to 270 min are also shown. (C) Time lapse microscopy of cdc42Δ/PMETCDC42 mutant cells in FCS at 37°C. DIC images were captured every 5 min over 6 h, and images every 15 min are shown during 330 min.
Increased Cdc42 levels partially suppress the cdc24Δ/PMETCDC24 invasive hyphal growth defect.
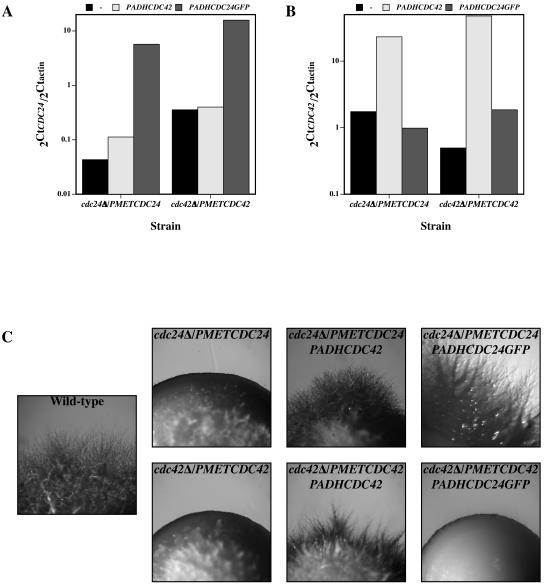
As cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants had reduced levels of the respective proteins, we investigated the effects of overexpression of these proteins on invasive hyphal growth. We overexpressed either Cdc24GFP or Cdc42 using the ADH promoter, and these constructs were integrated into the RP10 locus. Overexpression of either of these proteins in wild-type cells did not perturb budding or invasive hyphal growth (data not shown). Initially, we confirmed that these proteins were overexpressed in the different strains by examining the RNA transcript levels. CDC24 transcript levels were increased 45-fold in cdc42Δ/PMETCDC42 PADHCDC24GFP cells and 30-fold in cdc24Δ/PMETCDC24 PADHCDC24GFP cells compared to wild-type levels, with little to no change in the level of CDC42 (Fig. 5A). CDC42 transcript levels were increased 15-fold in cdc24Δ/PMETCDC24 PADHCDC42 cells and 30-fold in cdc42Δ/PMETCDC42 PADHCDC42 cells compared to wild-type levels (Fig. 5B). Immunoblot analyses confirmed that these strains substantially overexpressed Cdc42 (data not shown). Both cdc24Δ/PMETCDC24 PADHCDC24GFP and cdc42Δ/PMETCDC42 PADHCDC42 cells were viable when grown on medium containing Met and Cys, which repress the MET3 promoter (see Fig. 8A; data not shown). We next examined these strains for invasive hyphal growth on solid rich medium containing FCS. Figure 5C shows that overexpression of CDC42 partially compensates for the invasive hyphal growth defect of cdc24Δ/PMETCDC24 cells. In contrast, overexpression of CDC24GFP, while sufficient to complement the invasive growth defect of cdc24Δ/PMETCDC24 cells, did not enable cdc42Δ/PMETCDC42 cells to invade a solid surface. These results indicate that increased levels of Cdc42 can restore invasive growth in a mutant with reduced Cdc24 levels. However, the increased levels of Cdc24 do not alter the invasive growth defect of the cdc42Δ/PMETCDC42 mutant, perhaps due to a deleterious effect of Cdc24 overexpression in cdc42Δ/PMETCDC42 cells (see below).
FIG. 5.
Overexpression of CDC42 in a cdc24Δ/PMETCDC24 mutant partially suppresses the invasive growth defect. Normalized transcript levels of CDC24 (A) and CDC42 (B) in indicated strains overexpressing CDC42 and CDC24GFP are shown. Relative transcript abundance values (2Ctgene/2Ctactin) are the means of two independent experiments and were determined as for Fig. 3C. (C) Invasive growth of indicated strains overexpressing CDC42 or CDC24GFP. Images were taken after 8 days growth on YEPD-Uri-FCS agar at 37°C.
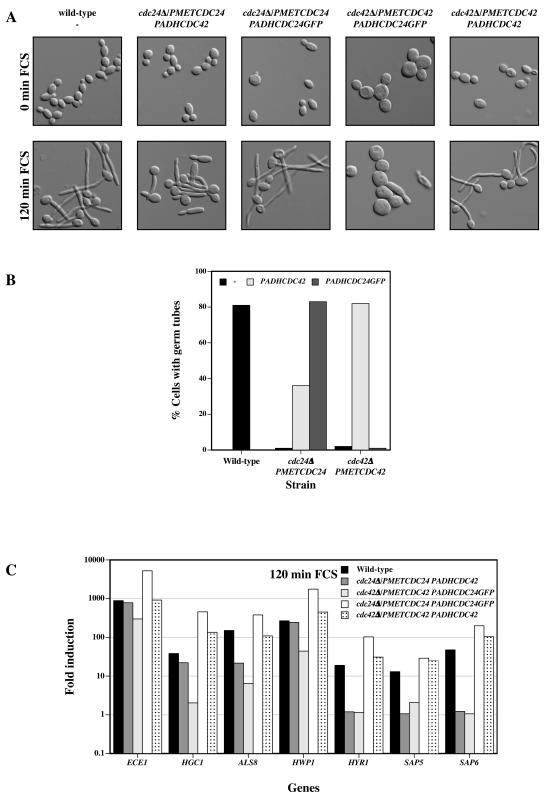
To determine whether cdc24Δ/PMETCDC24 mutant cells that overexpressed Cdc42 were able to form germ tubes and induce HSGs, they were incubated at 37°C over a 120-min period in the presence of serum. We focused primarily on the 120-min incubation time as the defects in the cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants were most pronounced at this time. Figure 6A shows budding cells (0 min with FCS) and cells after 120 min in FCS. All strains appeared normal during budding growth except cdc42Δ/PMETCDC42 PADHCDC24GFP cells, which were substantially larger, indicating that overexpression of Cdc24GFP, while not deleterious to wild-type cells (data not shown) and cdc24Δ/PMETCDC24 mutants, was toxic in cdc42Δ/PMETCDC42 mutant cells. With the exception of these cells, all other strains formed germ tubes. However, closer examination revealed that the lengths of germ tubes varied in the different strains, with germ tubes of cdc24Δ/PMETCDC24 PADHCDC42 cells about 25% shorter and germ tubes of cdc24Δ/PMETCDC24 PADHCDC24GFP cells approximately 25% longer than wild-type and cdc42Δ/PMETCDC42 PADHCDC42 germ tubes. cdc42Δ/PMETCDC42 cells overexpressing Cdc24GFP were also large in the presence of serum. Quantification of the number of cells with germ tubes indicated that Cdc24GFP and Cdc42 overexpression fully compensated the cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants, respectively (Fig. 6B). Strikingly, overexpression of CDC42 resulted in a substantial increase in the percentage of cdc24Δ/PMETCDC24 cells that formed germ tubes. Given that overexpression of Cdc24GFP was deleterious only in cdc42Δ/PMETCDC42 cells grown in rich medium, i.e., with reduced CDC42 levels, we examined the number of cells with germ tubes under conditions in which the MET3 promoter was not repressed (synthetic medium). In these conditions, overexpression of Cdc24GFP resulted in a substantial recovery of the germ tube formation defect (data not shown). These results indicate that the levels of Cdc24 and Cdc42 are critical for invasive hyphal growth and furthermore suggest that an increase in the amount of activated Cdc42 is important in this process.
FIG. 6.
Overexpression of CDC42 in a cdc24Δ/PMETCDC24 mutant partially restores germ tube formation and HSG induction. (A) Representative images of cells responding to FCS. DIC images of fixed cells were recorded at the indicated times. (B) Percentage of cells with germ tubes in indicated strains after 120 min in FCS at 37°C. Values (n = 200 to 250 cells) are from the experiment shown in panel A, with similar results observed in three independent experiments. (C) Fold induction of HSGs after 120 min in FCS at 37°C. Fold induction, determined as for Fig. 3C, of transcript levels from the same experiment for which results are shown in panels A and B.
In the same experiment the induction levels of HSGs in the different strains were determined as an independent assessment of the morphological responses. In cdc24Δ/PMETCDC24 PADHCDC42 cells the induction of the early HSGs ECE1, HGC1, and HWP1 was identical to that in wild-type cells at 120 min (Fig. 6C). In contrast, the induction levels of the late HSGs HYR1, SAP5, and SAP6 in addition to early HSG ALS8 were unaffected by overexpression of Cdc42 in the cdc24Δ/PMETCDC24 mutant, suggesting that Cdc24 is required for maintaining the levels of SAP5, SAP6, and ALS8 and the induction of HYR1. Overexpression of Cdc24GFP in a cdc42Δ/PMETCDC42 mutant had little to no effect on the induction of HSGs, except ECE1, in agreement with the absence of germ tube formation. Consistent with the increased length of germ tubes in cdc24Δ/PMETCDC24 cells overexpressing Cdc24GFP, this strain exhibited increased levels of all HSGs examined relative to wild-type cells, although this effect was less pronounced with SAP5, SAP6, and ALS8 transcripts. HSG levels in cdc42Δ/PMETCDC42 PADHCDC42 cells were similar to those in wild-type cells. These results indicate that increased Cdc42 levels in a cdc24Δ/PMETCDC24 mutant are sufficient for maintenance of early HSG (ECE1, HGC1, and HWP1) expression after 120 min in serum but are not sufficient for the induction of late HSGs (HYR1, SAP5, and SAP6) or the maintenance of ALS8 induction. Together, these results suggest that specific levels of Cdc24 and Cdc42 are critical during invasive hyphal growth. Overexpression of Cdc42 in cdc24Δ/PMETCDC24 mutant cells does not completely suppress the defect in invasive growth, germ tube formation, and HSG induction, suggesting that the CDC24 promoter is important during hyphal growth in response to serum.
Transient increase in Cdc24 levels in response to serum.
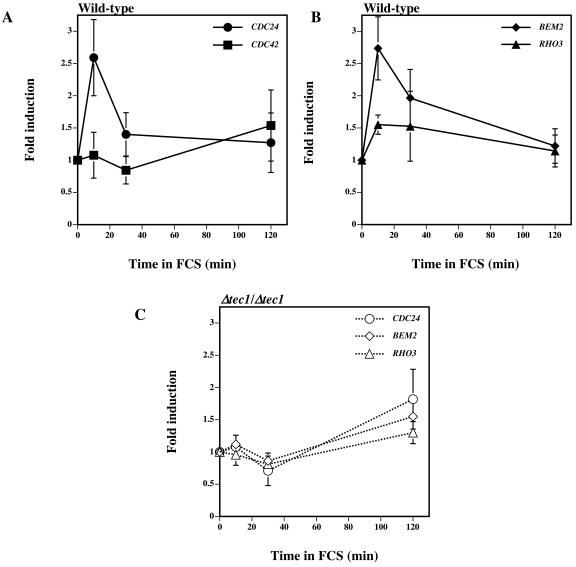
The RNA transcript levels of CDC24 and CDC42 were quantified at 10, 30, and 120 min in serum at 37°C. Figure 7A shows that the CDC24 transcript level increases, with a peak apparent at 10 min subsequent to exposure of cells to serum. This 2.5-fold increase in CDC24 transcript level was not simply due to the temperature shift as an identical experiment without serum revealed no increase in CDC24 message (data not shown). The increase in CDC24 transcripts was transient, with levels at 120 min similar to those observed prior to serum addition. In contrast, CDC42 RNA levels were constant in cells incubated for up to 30 min in the presence of serum and thereafter increased 1.5-fold, consistent with previous observations (31). These results suggest that a specific level of Cdc24 is required for germ tube formation, either for maintaining initial hyphal tip restricted growth or for preventing the apical-to-isotropic growth switch that normally occurs during budding.
FIG. 7.
In response to FCS, CDC24 transcript levels transiently increase in a Tec1-dependent fashion. (A) Levels of CDC24 and CDC42 transcripts in wild-type cells incubated with FCS at 37°C. Fold induction was determined as for Fig. 3C, and values are the averages of five independent experiments, with the bars indicating standard deviations. (B) Levels of BEM2 and RHO3 transcripts in wild-type cells incubated with FCS at 37°C. Fold induction was determined as above, and values are the averages of five independent experiments, with bars indicating standard deviations. (C) Levels of CDC24, BEM2, and RHO3 transcripts in tec1Δ/tec1Δ cells incubated with FCS at 37°C. Fold induction was determined as above, and values are the averages of three independent experiments, with bars indicating standard deviations.
The transient increase in CDC24 transcript levels was reminiscent of the responses of a class of genes in a genome-wide transcription profiling study (35). Intriguingly, two members of this class of transiently upregulated genes are involved in cell polarity, the Rho1 GTPase activating protein (GAP) BEM2 gene and the GTPase RHO3 gene. Using qRT-PCR, we confirmed that the transcript levels of these two genes transiently increase in response to serum at 37°C (Fig. 7B), suggesting a common regulatory mechanism. The transient induction of these genes in cells treated with serum at 37°C was similar to the induction of TEC1 transcripts under the same conditions. Examination of the upstream sequences (700 bp upstream of the ATG) of CDC24, BEM2, and RHO3 revealed a potential Tec1 transcription factor binding site (CATTCY) in each gene. Despite the relatively frequent occurrence of this potential Tec1 binding site, we did not find these sites in the upstream regions of CDC42 or RSR1. To determine if the transient increase in CDC24, BEM2, and RHO3 transcript levels required Tec1, we examined these transcripts in a tec1 deletion mutant under the same conditions. The tec1 deletion mutant was defective in hyphal formation and did not induce the expression of SAP4, SAP5, and SAP6 genes (41). Furthermore, induction of other HSGs examined (ECE1, HGC1, HWP1, HYR1, and ALS8) was substantially reduced in this strain compared to a wild-type strain (data not shown), indicating that expression of these HSGs is Tec1 dependent. Figure 7C shows that the transcripts of the three polarity genes are not transiently increased in a Δtec1/Δtec1 strain in serum at 37°C. However, we did observe that the levels of these transcripts increased after 120 min in serum, similar to the increase in HSGs in this tec1 mutant at this time (data not shown). These results indicate that the Tec1 transcription factor is necessary for the transient induction of several cell polarity proteins. It is attractive to speculate that a transient increase in cell polarity proteins is important for germ tube formation.
Cdc24 is recruited to the site of germ tube formation.
In addition to the specific regulation of Cdc24 levels, and by extrapolation activated Cdc42, during hyphal growth, it is likely that localization of Cdc24 is also crucial during this highly polarized growth process. We used a Cdc24GFP fusion driven by the ADH promoter as we were unable to detect a fluorescence signal in budding cells and cells with germ tubes expressing a Cdc24GFP fusion at endogenous levels. This Cdc24 fusion complemented both budding growth (Fig. 8A) and invasive hyphal growth (Fig. 5C and 6B and C) of the cdc24Δ/PMETCDC24 mutant. Analysis of cells expressing this fusion protein by immunoblotting revealed a protein of the expected size (Fig. 8B) with little to no free GFP observed (data not shown). As shown in Fig. 5A, RNA transcript levels indicated that this fusion was expressed at a level approximately 30-fold higher than the chromosomal wild-type CDC24. Initially, we examined Cdc24GFP localization in a cdc24Δ/PMETCDC24 strain in rich medium containing Met and Cys to repress CDC24 expression. Figure 8C shows budding cells with an observable fluorescence signal; however, very few cells had a polarized or clustered distribution of Cdc24GFP, irrespective of their cell cycle stage. After 30 min in the presence of serum at 37°C, Cdc24GFP could be seen at the nascent germ tube tip, in addition to foci, which appeared adjacent to the vacuole. This localization to the nascent germ tube tip is similar to the distribution of F-actin (2). By 60 and 120 min in serum Cdc24GFP was predominantly restricted to germ tube tips with no observable signal at the cell septum, whereas Cdc42 is localized to the germ tube tip and septum (20 and our unpublished data). Virtually all cells after 1 h in serum had Cdc24GFP localized to the germ tube tips (95%; n = 100 cells). Furthermore, after longer exposure to serum Cdc24GFP was observed at germ tube tips irrespective of the presence or absence of a septum. These results suggest that Cdc24 is recruited to the germ tube tip and persists there during hyphal formation. In contrast to the localization of Cdc24 to the nuclei of S. cerevisiae haploid cells, we did not observe accumulation of Cdc24GFP in the nuclei of budding or hyphal C. albicans cells. To confirm this initial recruitment of Cdc24GFP to the germ tube tips, we examined by confocal microscopy the same cells grown in synthetic medium containing Met and Cys in order to reduce background (Fig. 8D). Sufficient optical sections were taken to observe the entire cell in the z axis, and Fig. 8D (left panels) shows that during budding growth very few cells exhibited a localized or clustered Cdc24GFP distribution. Germ tube formation occurs more slowly in such a synthetic medium compared to rich medium, and hence we examined the initial recruitment of Cdc24GFP after 60 min in serum (Fig. 8D, right panels). At this time a substantial number of cells had Cdc24GFP localized to the nascent germ tubes. Quantitation of the percentage of cells with a patch of Cdc24GFP signal from z-stack images revealed that only 7% of budding cells (n = 150 cells) had clustered Cdc24GFP whereas after 60 min in serum, when few if any germ tubes were observable, 37% of cells (n = 400 cells) exhibited a restricted and localized Cdc24GFP signal. These results indicate that Cdc24 is recruited to and stabilized at the apical site of germ tube growth. Together our results suggest that in response to serum there is an increase in the amount of active Cdc42 that is restricted to the germ tube tip and is important for this highly polarized growth process.
DISCUSSION
Cdc42 and its exchange factor Cdc24 are required for invasive hyphal growth of C. albicans. Strains with reduced Cdc24 or Cdc42 levels are specifically defective in invasive hyphal growth yet induce HSGs in response to serum. Five different HSGs were similarly induced in the wild type and cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants after 30 min in serum at 37°C. Strikingly, after 2 h in serum these mutants had significantly lower HSG levels than wild-type cells. cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutant cells had elongated buds in serum, indicating that these mutants respond to serum but are unable to maintain this response. Overexpression studies indicate that specific levels of Cdc24 and Cdc42 are important for invasive hyphal growth. Furthermore, in response to serum, CDC24 transcript levels increase transiently in a Tec1 transcription factor-dependent fashion. These results suggest a positive feedback loop between Cdc24 and the Tec1 transcription factor; i.e., this exchange factor is required for optimal Tec1-dependent gene induction, and in turn Tec1 is required for the transient induction of CDC24 transcript. In addition to these effects, Cdc24 is recruited to the incipient germ tube tip. Taken together our results suggest that an increase in the level of active Cdc42 restricted to the tip of the germ tube is critical for the highly polarized growth that occurs during hypha formation.
The determination of gene transcript levels using qRT-PCR methods during the yeast-to-hyphal transition is important for the quantitative assessment of hyphal formation. We have examined in wild-type cells the induction of seven different HSGs in addition to seven transcriptional activators or repressors critical for hyphal responses. HSG transcript levels increased 100- to 1,000-fold in response to serum, whereas transcription factor levels either increased or decreased maximally 10-fold. The early induced HSGs, including ECE1, HGC1, ALS8, and HWP1, are already predominantly induced after cell exposure for 10 min in serum, prior to visible morphological changes. This temporal induction profile correlates with the time course of TEC1 transcript induction and EFG1 transcript repression. In contrast, the levels of the late induced HSGs, including HYR1, SAP5, and SAP6, decrease somewhat in the first 10 min of cell incubation in serum and then progressively increase concomitantly with germ tube elongation.
Both cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants respond to serum by HSG induction. With the exception of SAP5 and SAP6 there is little difference in HSG induction levels in these mutants after 10 and 30 min in serum, suggesting that at these early times either CDC24 and CDC42 are not required for HSG induction or the low levels of this GTPase module are sufficient for HSG induction. Yet after 120 min they were unable to maintain the elevated levels of HSG transcripts. The serum-dependent induction of all HSGs examined requires the cAMP-dependent protein kinase A signaling pathway (19, 59). Therefore, our results indicate that Cdc24 and Cdc42 are required for the cAMP-dependent signaling pathway which converges on the EFG1 and TEC1 transcription factors (25, 28, 41). Our results contrast with those obtained when Cdc42 was depleted by FLP-mediated CDC42 gene deletion (30). Such Cdc42-depleted cells were unable to form hyphae after 4 h in 10% serum; however HWP1, HYR1, and SAP4, -5, and -6 transcript levels were induced similar to that in wild-type cells. Very recently, C. albicans cdc42 mutants homologous to S. cerevisiae cdc42 pseudohyphal growth mutants which were defective in the yeast-to-hyphal transition were examined for hyphal gene induction. After 2 h in 10% serum containing minimal medium substantial decreases in HWP1 and SAP6 RNA transcript levels were observed (49), consistent with our results at this incubation time. Furthermore, little to no effect was observed on the induction of the GAP1 transcript, which is the only gene thus far identified that is strictly dependent on MAP kinase cascade signaling mediated by the CPH1 transcription factor (10). Together our results and results with cdc42 point mutants indicate that this GTPase module is critical for maintenance of cAMP-dependent gene induction subsequent to prolonged exposure to serum. However, they do not reveal at which level in the cAMP pathway this GTPase module functions during hyphal formation.
The inability of cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 strains to maintain HSG induction suggests a fundamental difference in the requirement for this GTPase module at 30 and 120 min in serum. These mutant cells elongate at early times in serum, consistent with the initiation of hyphal formation. The simplest explanation is that without sufficient Cdc24 or Cdc42, these mutants are unable to restrict growth to a single location (the tip of the incipient germ tube) and instead undergo an apical-to-isotropic growth switch resulting in even growth over the bud surface, as normally occurs during budding growth. In addition to the normal induction of a number of HSGs at early incubation times in serum this apical-to-isotropic growth switch is likely to be delayed in these mutants, resulting in an elongated morphology. Interestingly, the defect in HSG induction and hyphal formation in cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 cells is reminiscent of the defects observed with cln1Δ and crk1Δ (Cdc2-related kinase) cells (16, 29). The former cyclin mutant can form hyphae but is defective in maintaining hyphal growth and HSG induction (29). The latter crk1 mutant does not form hyphae in response to serum and exhibits an approximately 10-fold reduction in HSG induction (16). These phenotypic similarities as well as the striking effects of cdc24Δ/PMETCDC24 and cdc42Δ/PMETCDC42 mutants on HGC1 transcript levels suggest a link between this GTPase module and these cyclins/cyclin-dependent kinases required for hyphal formation.
Our results demonstrate that the levels of Cdc24 and Cdc42 are critical for hyphal formation. In a strain in which the sole copy of CDC24 is expressed from a MET3 promoter, overexpression of Cdc42 is sufficient to partially compensate for the hyphal growth defect. While overexpression of Cdc24GFP in both wild-type and cdc24Δ/PMETCDC24 strains is not deleterious, overexpression of Cdc24GFP in cdc42Δ/PMETCDC42 cells with reduced Cdc42 levels was somewhat toxic and in the presence of serum, cells were osmotically sensitive and lysed frequently. However, Cdc24 overexpression suppressed the germ tube formation defect of cdc42Δ/PMETCDC42 cells grown under nonrepressing conditions. Similarly, overexpression of fast-cycling Cdc42 mutants, both an F28L mutant which is spontaneously activated by virtue of its enhanced intrinsic GTP/GDP exchange rate yet has normal GTPase activity (26) and a G60A mutant which mimics Cdc24 activation in budding yeast (15), resulted in toxicity (unpublished data). Hence, it is difficult to assess the effect of Cdc24 overexpression in cdc42Δ/PMETCDC42 cells with reduced Cdc42 levels. Also consistent with the role of Cdc24 in hyphal formation and HSG induction, cdc24Δ/PMETCDC24 cells which overexpressed Cdc24GFP formed longer germ tubes and had higher levels of all HSGs examined, suggesting that septum formation was delayed in this strain.
While specific Cdc24 and Cdc42 levels are crucial for hyphal formation, the CDC24 promoter appears particularly important for efficient hyphal growth. We previously observed that strains with the sole copy of CDC24 expressed from a MET3 promoter were defective in hyphal formation even under nonrepressing conditions (7). CDC24 transcripts transiently increase after cell incubation for 10 min in the presence of serum, suggesting that such an increase in Cdc24 levels is important for serum-induced germ tube formation. This increase could result in more activated Cdc42 as overexpression of Cdc42 in a cdc24Δ/PMETCDC24 mutant partially compensates for the hyphal growth defect. Using Cdc42/Rac interactive binding (CRIB) domain pulldown methods, we examined the level of activated Cdc42 in cells over time in the presence of serum; however, we have been unable to reproducibly detect differences in the amount of activated Cdc42.
The Cdc42 GTPase module is necessary for optimal cAMP signaling pathway-dependent gene induction mediated by the Efg1 and Tec1 transcription factors. The transient increase in CDC24 transcript levels in wild-type cells exposed to serum at 37°C is comparable to that observed with TEC1. A previous transcription profiling study defined a class of genes which are transiently upregulated in serum-treated cells at 37°C, including two genes involved in cell polarity, BEM2 and RHO3 (35). The two encoded proteins are critical for restricting polarized growth to the apical portion of the hyphae in the filamentous fungus Ashbya gossypii (53, 54). We have confirmed that BEM2 and RHO3 are transiently upregulated in cells incubated with serum and furthermore identified a potential Tec1 binding site in the upstream region of each of these genes as well as in CDC24. In a tec1Δ/tec1Δ strain the transcript levels of BEM2, RHO3, and CDC24 remain constant for at least 30 min in the presence of serum at 37°C, suggesting that Tec1 is responsible for the transient induction of the three polarity proteins. The increase in the transcript levels of these genes along with other HSGs examined at 120 min in serum at 37°C in a tec1Δ/tec1Δ strain is similar to the increase in the EFG1 transcription factor levels at this time point, suggesting that EFG1 has a role in this late transcript upregulation. We speculate that an increase in the levels of a number of polarity proteins is important for the yeast-to-hyphal transition, in particular with regard to restricting the position of the growth site. These proteins serve to appropriately localize activated small G proteins such as Rho3 and Cdc42 to the germ tube tip.
Cdc24 localizes to the tip of the germ tube and appears to persist there throughout the cell cycle. In contrast to Cdc42, which in addition localizes to the septum of dividing hyphae, we did not observe Cdc24 at the cell septum. Interestingly, the cytoskeleton-associated protein Spa2 is also localized at the nascent germ tube tip, but not at the septum in C. albicans cells undergoing hyphal growth (60). Surprisingly, Cdc24 does not show a particularly polarized distribution in budding cells, compared to Cdc42 localization (20 and our unpublished data). We did not observe Cdc24GFP accumulate in the nucleus in either budding cells or cells with germ tubes. This is similar to the localization of Cdc24 in S. cerevisiae diploid cells and suggests that nuclear export of Cdc24 is not important for cell polarity establishment during hyphal formation. Strikingly, Cdc24 is strongly recruited to nascent germ tubes in cells treated for 30 min in rich medium with serum at 37°C, similar to the distribution (clustering) of F-actin (2). This germ tube tip localization is likely to result in an increase in activated Cdc42 at the germ tube tip as Cdc42 also localizes to the nascent germ tube tip (20).
In summary, our results indicate that the level of the Cdc42 GTPase module is critical for the yeast-to-hyphal transition, both with respect to cell morphology changes and HSG induction. Particular levels of this GTPase module are important for maintaining polarized growth and HSG induction. Our results suggest that an exchange factor-transcription factor positive feedback loop mediates an increase in the level of active Cdc42 restricted to the tip of the germ tube which is critical for the highly polarized growth that occurs during hypha formation.
Acknowledgments
This work was supported by the Centre National de la Recherche Scientifique, Fondation pour la Recherche Médicale-BNP-Paribas, and La Ligue Contre le Cancer. The ABI Prism 7000 Sequence Detection System was financed in part by Association pour la Recherche sur le Cancer grant 7696, the Zeiss LSM 510 META confocal microscopy was financed in part by Association pour la Recherche sur le Cancer grant 7830, and Applied Precision Deltavision deconvolution microscopy system was financed by Fondation pour la Recherche Médicale.
We thank D. Libri for advice on qRT-PCR and C. Matthews for aid with microscopy. We thank N. Dean and K. Schröppel for plasmids and strains.
REFERENCES
- 1.Akashi, T., T. Kanbe, and K. Tanaka. 1994. The role of the cytoskeleton in the polarized growth of the germ tube in Candida albicans. Microbiology 140:271-280. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, J. M., and D. R. Soll. 1986. Differences in actin localization during bud and hypha formation in the yeast Candida albicans. J. Gen. Microbiol. 132:2035-2047. [DOI] [PubMed] [Google Scholar]
- 3.Arkowitz, R. A., and N. Lowe. 1997. A small conserved domain in the yeast Spa2p is necessary and sufficient for its polarized localization. J. Cell Biol. 138:17-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ayscough, K. R., and D. G. Drubin. 1998. A role for the yeast actin cytoskeleton in pheromone receptor clustering and signalling. Curr. Biol. 8:927-930. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, D. A., P. J. Feldmann, M. Bovey, N. A. Gow, and A. J. Brown. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J. Bacteriol. 178:5353-5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barale, S., D. McCusker, and R. A. Arkowitz. 2004. The exchange factor Cdc24 is required for cell fusion during yeast mating. Eukaryot. Cell 3:1049-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bassilana, M., J. Blyth, and R. A. Arkowitz. 2003. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot. Cell 2:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bidlingmaier, S., and M. Snyder. 2004. Regulation of polarized growth initiation and termination cycles by the polarisome and Cdc42 regulators. J. Cell Biol. 164:207-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birse, C. E., M. Y. Irwin, W. A. Fonzi, and P. S. Sypherd. 1993. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 61:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas, S., M. Roy, and A. Datta. 2003. N-acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology 149:2597-2608. [DOI] [PubMed] [Google Scholar]
- 11.Braun, B. R., W. S. Head, M. X. Wang, and A. D. Johnson. 2000. Identification and characterization of TUP1-regulated genes in Candida albicans. Genetics 156:31-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun, B. R., and A. D. Johnson. 1997. Control of filament formation in Candida albicans by the transcriptional repressor TUP1. Science 277:105-109. [DOI] [PubMed] [Google Scholar]
- 13.Braun, B. R., D. Kadosh, and A. D. Johnson. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20:4753-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cali, B. M., T. C. Doyle, D. Botstein, and G. R. Fink. 1998. Multiple functions for actin during filamentous growth of Saccharomyces cerevisiae. Mol. Biol. Cell 9:1873-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caviston, J. P., S. E. Tcheperegine, and E. Bi. 2002. Singularity in budding: a role for the evolutionarily conserved small GTPase Cdc42p. Proc. Natl. Acad. Sci. USA 99:12185-12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, J., S. Zhou, Q. Wang, X. Chen, T. Pan, and H. Liu. 2000. Crk1, a novel Cdc2-related protein kinase, is required for hyphal development and virulence in Candida albicans. Mol. Cell. Biol. 20:8696-8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst, J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763-1774. [DOI] [PubMed] [Google Scholar]
- 18.Gale, C., M. Gerami-Nejad, M. McClellan, S. Vandoninck, M. S. Longtine, and J. Berman. 2001. Candida albicans Int1p interacts with the septin ring in yeast and hyphal cells. Mol. Biol. Cell 12:3538-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harcus, D., A. Nantel, A. Marcil, T. Rigby, and M. Whiteway. 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 15:4490-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazan, I., and H. Liu. 2002. Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot. Cell 1:856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irazoqui, J. E., A. S. Gladfelter, and D. J. Lew. 2003. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5:1062-1070. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, D. I. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 63:54-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadosh, D., and A. D. Johnson. 2001. Rfg1, a protein related to the Saccharomyces cerevisiae hypoxic regulator Rox1, controls filamentous growth and virulence in Candida albicans. Mol. Cell. Biol. 21:2496-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khalaf, R. A., and R. S. Zitomer. 2001. The DNA binding protein Rfg1 is a repressor of filamentation in Candida albicans. Genetics 157:1503-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane, S., C. Birse, S. Zhou, R. Matson, and H. Liu. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988-48996. [DOI] [PubMed] [Google Scholar]
- 26.Lin, R., S. Bagrodia, R. Cerione, and D. Manor. 1997. A novel Cdc42Hs mutant induces cellular transformation. Curr. Biol. 7:794-797. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H. 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4:728-735. [DOI] [PubMed] [Google Scholar]
- 28.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 29.Loeb, J. D., M. Sepulveda-Becerra, I. Hazan, and H. Liu. 1999. A G1 cyclin is necessary for maintenance of filamentous growth in Candida albicans. Mol. Cell. Biol. 19:4019-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michel, S., S. Ushinsky, B. Klebl, E. Leberer, D. Thomas, M. Whiteway, and J. Morschhauser. 2002. Generation of conditional lethal Candida albicans mutants by inducible deletion of essential genes. Mol. Microbiol. 46:269-280. [DOI] [PubMed] [Google Scholar]
- 31.Mirbod, F., S. Nakashima, Y. Kitajima, R. D. Cannon, and Y. Nozawa. 1997. Molecular cloning of a Rho family, CDC42Ca gene from Candida albicans and its mRNA expression changes during morphogenesis. J. Med. Vet. Mycol. 35:173-179. [PubMed] [Google Scholar]
- 32.Monod, M., G. Togni, B. Hube, and D. Sanglard. 1994. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol. Microbiol. 13:357-368. [DOI] [PubMed] [Google Scholar]
- 33.Moskow, J. J., A. S. Gladfelter, R. E. Lamson, P. M. Pryciak, and D. J. Lew. 2000. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:7559-7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murad, A. M., P. Leng, M. Straffon, J. Wishart, S. Macaskill, D. MacCallum, N. Schnell, D. Talibi, D. Marechal, F. Tekaia, C. d'Enfert, C. Gaillardin, F. C. Odds, and A. J. Brown. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20:4742-4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nern, A., and R. A. Arkowitz. 2000. G proteins mediate changes in cell shape by stabilizing the axis of polarity. Mol. Cell 5:853-864. [DOI] [PubMed] [Google Scholar]
- 37.Nern, A., and R. A. Arkowitz. 1998. A GTP-exchange factor required for cell orientation. Nature 391:195-198. [DOI] [PubMed] [Google Scholar]
- 38.Nishikawa, A., J. B. Poster, Y. Jigami, and N. Dean. 2002. Molecular and phenotypic analysis of CaVRG4, encoding an essential Golgi apparatus GDP-mannose transporter. J. Bacteriol. 184:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oberholzer, U., A. Marcil, E. Leberer, D. Y. Thomas, and M. Whiteway. 2002. Myosin I is required for hypha formation in Candida albicans. Eukaryot. Cell 1:213-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schroppel, K., K. Sprosser, M. Whiteway, D. Y. Thomas, M. Rollinghoff, and C. Csank. 2000. Repression of hyphal proteinase expression by the mitogen-activated protein (MAP) kinase phosphatase Cpp1p of Candida albicans is independent of the MAP kinase Cek1p. Infect. Immun. 68:7159-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schweizer, A., S. Rupp, B. N. Taylor, M. Rollinghoff, and K. Schroppel. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol. Microbiol. 38:435-445. [DOI] [PubMed] [Google Scholar]
- 42.Simon, M. N., C. De Virgilio, B. Souza, J. R. Pringle, A. Abo, and S. I. Reed. 1995. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature 376:702-705. [DOI] [PubMed] [Google Scholar]
- 43.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]
- 44.Staab, J. F., and P. Sundstrom. 1998. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast 14:681-686. [DOI] [PubMed] [Google Scholar]
- 45.Stoldt, V. R., A. Sonneborn, C. E. Leuker, and J. F. Ernst. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 47.Sudbery, P. E. 2001. The germ tubes of Candida albicans hyphae and pseudohyphae show different patterns of septin ring localization. Mol. Microbiol. 41:19-31. [DOI] [PubMed] [Google Scholar]
- 48.Ushinsky, S. C., D. Harcus, J. Ash, D. Dignard, A. Marcil, J. Morchhauser, D. Y. Thomas, M. Whiteway, and E. Leberer. 2002. CDC42 is required for polarized growth in human pathogen Candida albicans. Eukaryot. Cell 1:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VandenBerg, A. L., A. S. Ibrahim, J. E. Edwards, Jr., K. A. Toenjes, and D. I. Johnson. 2004. Cdc42p GTPase regulates the budded-to-hyphal-form transition and expression of hypha-specific transcripts in Candida albicans. Eukaryot. Cell 3:724-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warenda, A. J., and J. B. Konopka. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13:2732-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wedlich-Soldner, R., S. C. Wai, T. Schmidt, and R. Li. 2004. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 166:889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiner, M. P., G. L. Costa, W. Schoettlin, J. Cline, E. Mathur, and J. C. Bauer. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151:119-123. [DOI] [PubMed] [Google Scholar]
- 53.Wendland, J., and P. Philippsen. 2001. Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics 157:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wendland, J., and P. Philippsen. 2000. Determination of cell polarity in germinated spores and hyphal tips of the filamentous ascomycete Ashbya gossypii requires a rhoGAP homolog. J. Cell Sci. 113:1611-1621. [DOI] [PubMed] [Google Scholar]
- 55.Whiteway, M. 2000. Transcriptional control of cell type and morphogenesis in Candida albicans. Curr. Opin. Microbiol. 3:582-588. [DOI] [PubMed] [Google Scholar]
- 56.Whiteway, M., and U. Oberholzer. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7:350-357. [DOI] [PubMed] [Google Scholar]
- 57.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, X., S. H. Oh, G. Cheng, C. B. Green, J. A. Nuessen, K. Yeater, R. P. Leng, A. J. Brown, and L. L. Hoyer. 2004. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology 150:2415-2428. [DOI] [PubMed] [Google Scholar]
- 59.Zheng, X., and Y. Wang. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng, X. D., Y. M. Wang, and Y. Wang. 2003. CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol. Microbiol. 49:1391-1405. [DOI] [PubMed] [Google Scholar]