Abstract
In melanocytes and in melanoma cells, cyclic AMP (cAMP)-elevating agents stimulate melanogenesis and increase the transcription of tyrosinase, the rate-limiting enzyme in melanin synthesis. However, two other enzymes, tyrosinase-related protein 1 (TRP1) and TRP2, are required for a normal melanization process leading to eumelanin synthesis. In B16 melanoma cells, we demonstrated that stimulation of melanogenesis by cAMP-elevating agents results in an increase in tyrosinase, TRP1, and TRP2 expression. cAMP, through a cAMP-dependent protein kinase pathway, stimulates TRP1 and TRP2 promoter activities in both B16 mouse melanoma cells and normal human melanocytes. Regulation of the TRP1 and TRP2 promoters by cAMP involves a M box and an E box. Further, a classical cAMP response element-like motif participates in the cAMP responsiveness of the TRP2 promoter, demonstrating that the TRP2 gene is subjected to different regulatory processes, which could account for its different expression patterns during embryonic development or under specific physiological and pathological conditions. We also found that microphthalmia, a basic helix-loop-helix transcription factor, strongly stimulates the transcriptional activities of the TRP1 and TRP2 promoters, mainly through binding to the M boxes. Additionally, we demonstrated that cAMP increases microphthalmia expression and thereby its binding to TRP1 and TRP2 M boxes. These convergent and compelling results disclose at least a part of the molecular mechanism involved in the regulation of melanogenic gene expression by cAMP and emphasize the pivotal role of microphthalmia in this process.
In mammals, pigmentation results from the synthesis and distribution of melanin in the skin, hair bulbs, and eyes. Melanin synthesis (melanogenesis) takes place in the melanocyte after differentiation of the nonpigmented precursor, the melanoblast (27). Three melanocyte-specific enzymes, tyrosinase, tyrosinase-related protein 1 (TRP1), and TRP2, are involved in this enzymatic process that converts tyrosine to melanin pigments. Although these proteins have similar structures and features, they are expressed by different genes and possess distinct enzymatic activities. Tyrosinase, encoded by the albino locus of the mouse, catalyzes the conversion of tyrosine to 3,4-dihydroxyphenylalanine (DOPA) and of DOPA to DOPA quinone (14, 25, 31). TRP2, encoded by the mouse slaty locus, possesses a Dopachrome tautomerase activity, converting the Dopachrome to 5,6-dihydroxyindole-2-carboxylic acid (DHICA) (3, 19, 42). TRP1, which has been mapped in mouse to the brown locus, catalyzes the oxidation of DHICA to indole-5,6-quinone-2-carboxylic acid (21, 24).
In vivo, melanogenesis is regulated by UVB radiation that can act either directly on melanocytes or indirectly through the release of keratinocyte-derived factors such as interleukins, prostaglandins, and alpha melanocyte-stimulating hormone (α-MSH) (1, 12, 22, 35). Interestingly, α-MSH, one of the most potent activators of melanogenesis, binds to an αs-coupled receptor and increases the intracellular level of cyclic AMP (cAMP) (9, 17, 20, 37). Further, the melanogenic effects of α-MSH can be mimicked by pharmacological cAMP-elevating agents such as forskolin, cholera toxin, and isobutylmethylxanthine (8, 13, 15, 18, 38), indicating that the cAMP pathway plays a pivotal role in the regulation of melanogenesis.
It has been thought for many years that the regulation of melanin synthesis occurs at the level of tyrosinase, which is the rate-limiting enzyme in melanogenesis. However, TRP1 and TRP2 have been recently shown to play an important role in the control of melanin type (23). Indeed, two types of melanin are produced by melanocytes: pheomelanins, which are red or yellow, and eumelanins, which are brown or black (32). The latter pigments can absorb UV photons and scavenge damaging free radicals generated by UV within the cells, thus preventing DNA damage and sheltering the skin from the harmful effects of UV radiation. TRP1 and TRP2 control distal steps of eumelanin synthesis, thereby fulfilling a key photoprotective function. Noteworthy, the stimulation of melanogenesis by cAMP-elevating agents leads to an increased eumelanin synthesis, suggesting that cAMP regulates TRP1 and TRP2 activity and/or expression. However, the regulation of TRP1 and TRP2 expression by cAMP has not been clearly demonstrated and remains controversial. Interestingly, TRP2 is expressed before tyrosinase and TRP1 during embryogenesis (34). Further, in agouti mice that synthesize only pheomelanins, extinction of TRP1 and TRP2 expression has been reported, while tyrosinase is still expressed (23). These observations suggest that different mechanisms are involved in the regulation of tyrosinase, TRP1, and TRP2 gene expression. Tyrosinase and TRP1 promoters share an 11-bp motif (AGTCATGTGCT) termed the M box located upstream of the TATA box. This motif binds microphthalmia, a basic helix-loop-helix transcription factor that increases tyrosinase and TRP1 promoter activities, thereby playing a key role in the tissue-specific expression of these genes (11, 29, 40). In the TRP2 promoter, a homologous sequence (GTCATGTGCT) is also found upstream of the TATA box (41). However, it has not been clearly established whether microphthalmia binds to and stimulates the TRP2 promoter.
Since a precise control of melanogenic gene expression is crucial for normal melanization, it is essential to study the molecular mechanisms involved in the control of TRP1 and TRP2 gene expression by cAMP. In this study, we demonstrated that cAMP increases the level of TRP1 and TRP2 mRNA. Then, using reporter constructs containing either the 1.1-kb fragment 5′ of the transcriptional start site of the TRP1 gene or the 0.6-kb fragment 5′ of the transcriptional start site of the TRP2 gene, we demonstrated that cAMP-elevating agents, through the cAMP-dependent protein kinase (PKA) pathway, stimulate the transcriptional activity of the TRP1 and TRP2 promoters. Deletion and mutation analysis allowed us to localize the cis-acting elements involved in the cAMP responsiveness of TRP1 and TRP2 promoters. Our data indicate that the M box (GTCATGTGCT) plays a key role in the regulation of TRP1 and TRP2 expression by cAMP. However, we identified in the TRP1 and TRP2 promoters other cis-acting elements involved in the cAMP response, indicating that specific regulatory mechanisms participate in the regulation of TRP1 and TRP2 expression by cAMP. Finally, we showed that microphthalmia binds to the M boxes and transactivates the TRP1 and TRP2 promoters. Further, we observed that cAMP increases microphthalmia expression, thus demonstrating the pivotal role of microphthalmia in the regulation of melanogenic gene expression by cAMP.
MATERIALS AND METHODS
Materials.
MCDB 153, forskolin, bovine serum albumin, hydrocortisone, insulin, phorbol 12-myristate 13-acetate, p-nitrophenyl phosphate (PNPP), 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), aprotinin, and leupeptin were purchased from Sigma Chemical Co. Dispase was from Boehringer, and basic fibroblast growth factor and the basic vector PGL2 were from Promega. Dulbecco’s modified Eagle’s medium, fetal calf serum (FCS), trypsin, and Lipofectamine reagent were from GIBCO. Peroxidase-conjugated anti-rabbit and anti-mouse antibodies were from Dakopatts. Synthetic oligonucleotides were from Oligo Express. Expression vectors encoding the catalytic subunit of PKA and the PKA peptide inhibitor (PKI) were previously described (6, 30).
Cell cultures.
B16-F10 murine melanoma cells and NIH 3T3 fibroblast cells were grown at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% FCS, penicillin (100 U/ml), and streptomycin (50 μg/ml). Epidermal cell suspensions were obtained from foreskins of caucasoid children by overnight digestion in phosphate-buffered saline containing 0.5% dispase grade II at 4°C, followed by a 1-h digestion with 0.05% trypsin–0.02% EDTA in phosphate-buffered saline at 37°C. Cells were grown in MCDB 153 medium supplemented with 2% FCS, hydrocortisone (0.4 μg/ml), insulin (5 μg/ml), 16 nM phorbol 12-myristate 13-acetate, basic fibroblast growth factor (1 ng/ml), penicillin (100 U/ml), and streptomycin (50 μg/ml) in a humidified atmosphere containing 5% CO2 in air at 37°C.
Western blot assays.
B16 mouse melanoma cells were grown in six-well dishes with or without 20 μM forskolin. After 24 or 48 h of cAMP treatment, cells were lysed in phosphate buffer (pH 6.8) containing 1% (wt/vol) Triton X-100, leupeptin (5 μg/ml), 1 mM AEBSF, and aprotinin (100 IU/ml). Proteins (30 μg) were separated on sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gels and transferred to a nitrocellulose membrane. Tyrosinase, TRP2, and ERK1 proteins were detected with polyclonal antibodies PEP7, PEP8 (23), and C-16, respectively, at a 1/3,000 dilution in saturation buffer and with a secondary peroxidase-conjugated anti-rabbit antibody at a 1/3,000 dilution. TRP1 protein was detected with monoclonal antibody B8G3 (36) at a 1/1,000 dilution in saturation buffer and with a secondary peroxidase-conjugated anti-mouse antibody at a 1/3,000 dilution. Proteins were visualized with the Amersham ECL system. Anti-ERK1 antibody C-16 was from Santa Cruz Biotechnology (Santa Cruz, Calif.).
RNA isolation and reverse transcription-PCR assays.
Total cellular RNA was extracted from control and cAMP-treated cells by a modification of the method of Chomczynski. Ten micrograms of RNA was reverse transcribed by using the Promega reverse transcription system. Twenty-eight cycles of PCR amplification of the resulting cDNA allowed us to quantify tyrosinase, TRP1, and TRP2 mRNAs (53°C, 45 s; 72°C, 1 min; 94°C, 30 s). The 1,191-bp tyrosinase fragment was amplified with primers 5′-CATTTTTGATTTGAGTGTCT-3′ and 5′-TGTGGTAGTCGTCTTTGTCC-3′, the 784-bp TRP1 product was amplified with primers 5′-CTTTCTCCCTTCCTTACTGG-3′ and 5′-TGGCTTCATTCTTGGTGCTT-3′, and the 518-bp TRP2 fragment was amplified with primers 5′-TGAGAAGAAACAAAGTAGGCAGAA-3′ and 5′-CAACCCCAAGAGCAAGACGAAAGC-3′. Specific primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were from Clontech and gave an amplified PCR product of 983 bp. Preliminary trials showed that after 28 cycles of PCR, the reaction remained exponential. The PCR products were electrophoresed on 1% agarose gels and stained with ethidium bromide before visualization under UV light.
Construction of the reporter plasmids.
We have previously described the reporter plasmid containing the 2.2-kb fragment of the mouse tyrosinase promoter (pTyro; −2236 to +59) (5). A 1.1-kb fragment 5′ of the transcriptional start site of the mouse TRP1 gene and a 0.6-kb fragment 5′ of the transcriptional start site of the human TRP2 gene were isolated from genomic DNA by PCR using primers 5′-TGAAGCCACAGAGAATAAGG-3′ (−1247 to −1228) and 5′-CCAGACAGTAAATCCCAAGC-3′ (+82 to +63) for TRP1 and primers 5′-GGTTCCAGTGCCTTCCATAC-3′ (−585 to −566) and 5′-TTTCAGTCTTTTCTTTTCAG-3′ (+359 to +340) for TRP2. Promoter fragments were cloned into the unique SmaI-BglII sites of the PGL2-basic vector (PGL2B) upstream of the luciferase coding sequence (pTRP1 [−1247 to +82] and pTRP2 [−585 to +359]). All deletions and mutations of TRP1 and TRP2 were constructed with a Transformer site-directed mutagenesis kit (Clontech) and were verified by plasmid sequencing. Briefly, the deletions of the TRP1 and TRP2 promoters were obtained with primers selected to hybridize to the multiple cloning sites of PGL2B in the 5′ half and to the 5′ end of the selected deletion (indicated in boldface) in the 3′ half, resulting in the following constructions: Δ1pTRP1 (5′-GGTACTGTAACTGAGCCTTAAGACTTTAACC-3′; −83 to +82), Δ2pTRP1 (5′-GGTACTGTAACTGAGCGTTGGGGCAGGGGGG-3′; −864 to +82), Δ3pTRP1 (5′-CTGTAACTGAGCTAACATAACAGGCATCTTATATCAAGCA-3′; −608 to +82), Δ4pTRP1 (5′-GTACTGTAACTGAGCTAACGTAGAGTAATCATGTATTC-3′; −363 to +82), and Δ5pTRP1 (5′-GGTACTGTAACTGAGCTAACGAGTTTTCAACTTCCAGGAG-3′; −304 to +82) for the TRP1 promoter and Δ1pTRP2 (5′-ACTGAGCTAACATAAACTTTGGGTCATGTG-3′; −144 to +349), Δ2pTRP2 (5′-GGTACTGTAACTGAGCTAACGAGTAAGTTATTATTTGGAG-3′; −475 to +349), Δ3pTRP2 (5′-CTGTAACTGAGCTAACGAGCTCACTGCATC-3′; −273 to +349), and Δ4pTRP2 (5′-CTGTAACTGAGCTAACATAAGGAGCACATGAGCCCAGATA-3′; −220 to +349) for the TRP2 promoter. Mutations were introduced with primers carrying point mutations in the core motif of target sequences. In the TRP1 promoter, the M box (GTCATGTGCT) located between bp −44 and −33 upstream from the initiation start site was mutated in GTCGGATCCT (5′-GGAGGGAGTCGGATCCTGCCTAGTAG-3′) and the E box (CAAGTG) located between bp −238 and −233 was mutated in CGGATC (5′-CAGAAAATACGGATCTGACATTGGCC-3′), giving mMBOXpTRP1, mEBOXpTRP1, or the double mutant mM/ EBOXpTRP1. In the TRP2 promoter, the M box (GTCATGTGCT) located between bp −135 and −129 upstream from the initiation start site was mutated in GTCGGATCCT (5′-CACTTTGGGTCGGATCCTAATGATGA-3′), the E box (CACATG) between bp −346 and −340 was mutated in CGGATC (5′-GGTCTTTTTTGCACGGATCTCAGAAAGC-3′), and the cAMP response element (CRE; TGAGGTCA) located between bp −239 and −232 was mutated in TGTGTTCG (5′-CCAGGATGTCTGTGTTCGCAAGTTTGGC-3′), giving mMBOXpTRP2, mEBOXpTRP2, and mCREpTRP2, respectively.
Transfections and luciferase assays.
B16 melanoma cells were seeded in 24-well dishes, and transient transfections were performed the following day, using 2 μl of Lipofectamine and 0.55 μg of total plasmid DNA in a 200-μl final volume as indicated in figure legends. pCMVβGAL was transfected with the test plasmids to control the variability in transfection efficiency. At 48 h after transfection, soluble extracts were harvested in 50 μl of lysis buffer and assayed for luciferase and β-galactosidase activities. All transfections were repeated at least five times with different plasmid preparations. NIH 3T3 fibroblast cells were seeded in six-well dishes and transiently transfected with 8 μl of Lipofectamine and 2 μg of an expression vector encoding microphthalmia in a 800-μl final volume. At 24 h after transfection, cells were labeled with [35S]methionine-cysteine mix. Human melanocytes were transiently transfected by electroporation. Two million melanocytes were resuspended in 400 μl of MCDB 153. Cell suspensions were placed in a 0.4-cm-gap cuvette with 30 μg of total plasmid DNA. Electroporation was performed with simple electric shock (280 V; 1,050 μF) by using an Easyject electroporator system (Eurogentec). Cells were incubated for 48 h, and then luciferase and β-galactosidase activities were assayed as described for B16 cells.
Metabolic labeling and immunoprecipitation.
B16 mouse melanoma cells and nontransfected or microphthalmia-transfected NIH 3T3 cells were grown in six-well dishes and labeled for 30 h with [35S]methionine-cysteine (0.1 mCi/ml; Amersham) in methionine-cysteine-free medium. Cells were then solubilized at 4°C in radioimmunoprecipitation assay buffer (pH 7.5) containing 10 mM Tris-HCl, 1% sodium deoxycholate, 1% Nonidet P-40, 150 mM NaCl, 0.1% SDS, 5 μg of leupeptin per ml, 1 mM AEBSF, 100 IU of aprotinin per ml, 1 mM NaVO4, 5 mM NaF, 20 mM β-glycerophosphate, and 10 mM PNPP. Total extracts were precleared by incubation with 50 μl of protein A-Sepharose (Pharmacia) and then incubated with 20 μl of antibodies to the C terminus of the microphthalmia (5) complexed to 50 μl of protein A-Sepharose for 1 h at 4°C. The immune complexes were washed five times with radioimmunoprecipitation assay buffer, eluted in SDS-sample buffer at 95°C for 5 min, and analyzed on a 7.5% SDS gel. The specifically bound immune complexes were visualized by autoradiography.
Nuclear extracts and gel mobility shift assay.
B16 cells were stimulated with 20 μM forskolin, and the nuclear extracts were prepared essentially as described previously (7) except that phosphatase inhibitors (1 mM NaVO4, 5 mM NaF, 20 mM β-glycerophosphate, and 10 mM PNPP) were added to the nuclear extraction buffer. Double-stranded synthetic M-BOXTRP1 (5′-GGAGGGAGTCATGTGCTGCCTAG-3′) or E-BOXTRP1 (5′-GAAAATACAAGTGTGACATTG-3′) and M-BOXTRP2 (5′-CTTTGGGTCATGTGCTAATGATG-3′) or E-BOXTRP2 (5′-GTCTTTTTTGCACACATGTCAGAAAGC-3′) were end labeled with T4 polynucleotide kinase and [γ-32P]ATP. Five micrograms of nuclear proteins or 4 μl of the in vitro-translated microphthalmia was preincubated in binding buffer containing 10 mM Tris (pH 7.5), 100 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 4% glycerol, 80 μg of salmon sperm DNA per ml, 0.1 μg of poly(dI-dC), 10% FCS, 2 mM MgCl2, and 2 mM spermidine for 15 min on ice. Then 30,000 to 50,000 cpm of 32P-labeled probe was added to the binding reaction for 10 min at room temperature. DNA-protein complexes were resolved by electrophoresis on a 4% polyacrylamide (37.5:1 acrylamide-bisacrylamide) gel in TBE buffer (22.5 mM Tris-borate, 0.5 mM EDTA [pH 8]) for 2 h at 150 V. When indicated, a 50-fold excess of unlabeled competitor oligonucleotides was added during preincubation. For supershift assays, 0.3 μl of preimmune serum or 0.3 μl of specific antibodies against microphthalmia (5) was preincubated with nuclear extracts or with in vitro-translated microphthalmia in binding reaction buffer before addition of the labeled probe.
RESULTS
Stimulation of tyrosinase, TRP1, and TRP2 expression by cAMP-elevating agents in B16-F10 mouse melanoma cells.
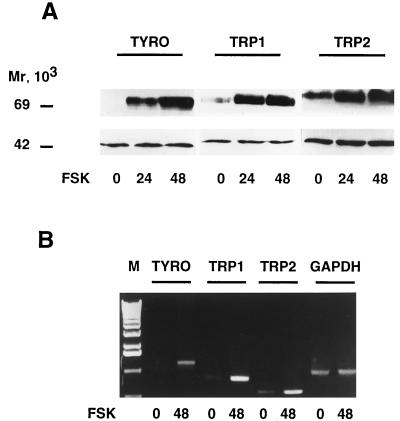
Since conflicting data concerning the effects of cAMP-elevating agents on TRP1 and TRP2 gene expression have been reported, we investigated the effects of forskolin on the amount of tyrosinase, TRP1, and TRP2 proteins and mRNAs in B16-F10 mouse melanoma cells. Western blot experiments showed that 24 or 48 h of treatment with forskolin markedly increased the amount of tyrosinase protein. Forskolin also increased the amount of TRP1 and TRP2 proteins (Fig. 1A, upper panel). The detection of the ERK1 protein at 44 kDa ensured even loading of lanes (Fig. 1A, lower panel).
FIG. 1.
Forskolin treatment increases tyrosinase, TRP1, and TRP2 gene expression in B16 mouse melanoma cells. (A) Thirty micrograms of proteins from control cells and cells treated for 24 or 48 h with 20 μM forskolin (FSK) was subjected to Western blot analysis using antibodies PEP7 for tyrosinase detection, B8G3 for TRP1, and PEP8 for TRP2 (upper panel). The detection of the 44-kDa ERK1 protein ensured even loading of lanes (lower panel). (B) Ten micrograms of total RNA from control B16 cells or cells treated for 48 h with 20 μM forskolin was reverse transcribed. The resulting cDNAs were subjected to 28-cycle PCRs using specific primers that gave amplified products of 1,191 bp for tyrosinase (TYRO), 784 bp for TRP1, and 518 bp for TRP2. A control of PCR amplification with specific primers of GAPDH transcript showed a 983-bp fragment. PCR products were electrophoresed on a 1% agarose gel and stained with ethidium bromide before UV light visualization. DNA molecular weight markers (lane M), from bottom to top: 510, 1,018, 1,636, 2,036, 3,054, and 4,072 bp.
In addition, tyrosinase, TRP1, and TRP2 mRNAs were analyzed by reverse transcription-PCR experiments. Using specific primers, we amplified PCR fragments of 1,191, 784 and 518 bp, corresponding to fragments of the tyrosinase, TRP1, and TRP2 cDNAs, respectively (Fig. 1B). The amounts of the PCR fragments were increased when we used cDNA from B16 cells treated with forskolin for 48 h, reflecting an augmentation of tyrosinase, TRP1, and TRP2 messengers. A control of PCR amplification showed that the GAPDH transcripts were not modified following forskolin treatment.
These results indicate that cAMP-elevating agents increase the amount of tyrosinase, TRP1, and TRP2 proteins and mRNAs, suggesting a coordinated regulation of the expression of these melanogenic enzymes by cAMP in B16-F10 mouse melanoma cells.
Regulation of tyrosinase, TRP1, and TRP2 transcriptional activities by cAMP-elevating agents and PKA.
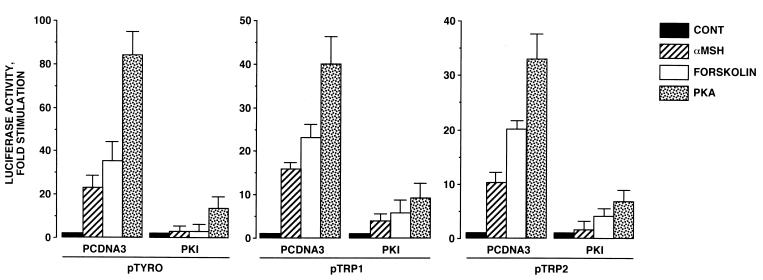
To study the effects of cAMP on TRP1 and TRP2 gene transcription, we constructed two luciferase reporter plasmids, containing either TRP1 (pTRP1; −1247 to +82) or TRP2 (pTRP2; −585 to +349) promoter fragments that contain the elements accountable for melanocyte-specific expression (29, 41). Then B16 cells were transiently transfected with pTRP1 and pTRP2 and exposed to forskolin or α-MSH for 48 h. As a control, we transfected a reporter plasmid containing the tyrosinase promoter (pTyro; −2236 to +59) that we had previously found to be responsive to cAMP-elevating agents (5). As expected, forskolin and α-MSH, respectively, induced 30- and 20-fold increases in the luciferase activity in cells transfected with pTyro. Further, we observed a 20-fold stimulation of the luciferase activity by forskolin treatment in cells transfected with pTRP1 and pTRP2, demonstrating that TRP1 and TRP2 promoters are responsive to cAMP elevation (Fig. 2). The effects evoked by α-MSH on TRP1 and TRP2 promoters were slightly less significant (15- and 10-fold stimulation, respectively). Additionally, in cells transfected with pTyro, pTRP1, and pTRP2, the luciferase activities were dramatically (80-, 42-, and 37-fold, respectively) increased by cotransfection with an expression vector encoding the catalytic subunit of PKA. The effects of forskolin, α-MSH, and PKA were severely impaired by transfection with an expression plasmid encoding PKI, emphasizing the role of PKA in the effects of cAMP-elevating agents on these promoter activities. The effect of PKI on the cAMP pathway was specific, since transfection with PKI-encoding vector did not affect the response to phorbol ester of a TRE-LUC reporter plasmid in B16 cells (not shown). The results presented above demonstrate that tyrosinase, TRP1, and TRP2 promoters contain cis-acting elements involved in the regulation of their transcriptional activities by cAMP through PKA activation.
FIG. 2.
cAMP-elevating agents and PKA stimulate tyrosinase, TRP1, and TRP2 promoter activities in B16 cells: reversal of these effects by PKI. B16 cells were transfected with 0.3 μg of luciferase reporter plasmid pTyro, pTRP1, or pTRP2 and 0.05 μg of pCMVβGAL. In control (CONT), α-MSH, and forskolin conditions, 0.2 μg of empty pCDNA3 or 0.1 μg of empty pCDNA3 plus 0.1 μg of pCDNA3 encoding PKI was cotransfected with reporter plasmids. Cells were treated for 48 h with 20 μM forskolin or 1 μM α-MSH. To study the effects of PKA expression, B16 cells were transfected with 0.3 μg of luciferase reporter plasmid and 0.1 μg of pCDNA3 encoding PKA plus 0.1 μg of empty pCDNA3 or 0.1 μg of pCDNA3 encoding PKA plus 0.1 μg of pCDNA3 encoding PKI. Luciferase activity was normalized to β-galactosidase activity, and the results were expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± standard errors of five experiments performed in triplicate.
Localization of cis-acting elements in TRP1 promoter responsible for cAMP response.
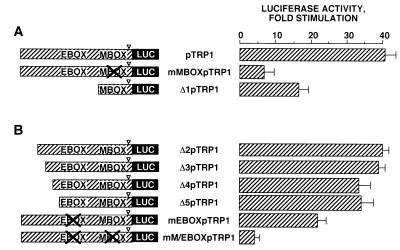
In an attempt to localize the cis-acting elements responsible for cAMP response of the TRP1 promoter, we constructed reporter plasmids containing deletions and mutations in the 5′-flanking region of the TRP1 promoter. Our previous results (5) have demonstrated that the M box upstream of the TATA box confers on the tyrosinase promoter its responsiveness to cAMP. Comparison of the tyrosinase, TRP1, and TRP2 promoter sequences shows that the M box (GTCATGTGCT) is highly conserved in these promoters. Hence, we constructed two reporter plasmids; in one, the M box was mutated in the pTRP1 construct (mMBOXpTRP1); the second construction had a deletion just upstream of the M box (Δ1pTRP1). Then we studied the effects of PKA expression on the transcriptional activity of mMBOXpTRP1 and Δ1pTRP1. We observed 7- and 16-fold increases in luciferase activity by PKA in cells transfected with mMBOXpTRP1 and Δ1pTRP1, respectively (Fig. 3A). The effects of PKA were markedly reduced compared to pTRP1 (42-fold stimulation), indicating that the regulation of TRP1 gene expression by PKA in B16 melanoma cells involves the M box just upstream of the TATA box. However, other regulatory elements located between bp −1247 and −83 in the promoter also participate in the cAMP response.
FIG. 3.
Regulation of TRP1 transcriptional activity by PKA is mediated by the M box (−44 to −33) and the E box (−238 to −233). B16 cells were transfected with 0.3 μg of pTRP1, mMBOXpTRP1, and Δ1pTRP1 (A) or Δ2pTRP1, Δ3pTRP1, Δ4pTRP1, Δ5pTRP1, mEBOXpTRP1, and mM/EBOXpTRP1 (B), 0.2 μg of pCDNA3, empty or encoding PKA, and 0.05 μg of pCMVβGAL. After 48 h, luciferase activity was assayed as described in the text and normalized to β-galactosidase activity. ▿, TATA-box position. Results are expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± standard errors of five experiments performed in triplicate.
We constructed additional deletions and mutations to identify these elements involved in the cAMP response of the TRP1 promoter. The responsiveness of constructs Δ2pTRP1 and Δ3pTRP1 to PKA was similar to that of the wild type, pTRP1 (Fig. 3B). Δ4pTRP1 and Δ5pTRP1 showed only a slightly (35-fold) decreased response not significantly different from that of pTRP1. On the other hand, when we mutated the E box (mEBOXpTRP1) in pTRP1, luciferase activity was stimulated only 24-fold by PKA. Double mutation of both M and E boxes in pTRP1 (mM/EBOXpTRP1) led to a nearly complete loss of the cAMP responsiveness of the TRP1 promoter (threefold). Taken together, these results indicate that the M box (−44 to −33) and the E box (−238 to −233) play a key role in the cAMP responsiveness of the TRP1 promoter.
Characterization of regulatory elements involved in the cAMP responsiveness of the TRP2 promoter.
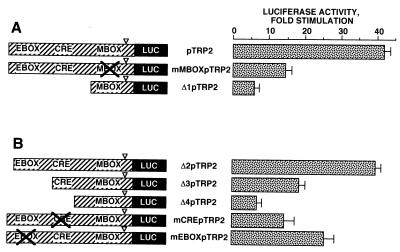
A similar approach was used to identify the cis-acting elements involved in the transcriptional regulation of TRP2 promoter activity by PKA. A first series of constructs (Fig. 4A) showed that mutation of the M box upstream of the TATA box (mMBOXpTRP2) and deletion of the 5′-flanking region upstream of the M box (Δ1pTRP2) greatly impaired the responsiveness of the TRP2 promoter to PKA (14- and 6-fold stimulation, respectively). These results indicate that the M box is involved in the transcriptional regulation of the TRP2 promoter activity by PKA and that important cis-regulatory elements conferring cAMP responsiveness on the TRP2 promoter are located between bp −585 and −144.
FIG. 4.
Regulation of TRP2 transcriptional activity by PKA involved the M box (−129 to −135), the CRE-like motif (−239 to −232), and the E box (−346 to −340). B16 cells were transfected with 0.3 μg of pTRP2, mMBOXpTRP2, and Δ1pTRP2 (A) or Δ2pTRP2, Δ3pTRP2, Δ4pTRP2, mCREpTRP2, and mEBOXpTRP2 (B), 0.2 μg of pCDNA3, empty or encoding PKA, and 0.05 μg of pCMVβGAL. After 48 h, luciferase activity was assayed as described in the text and normalized to β-galactosidase activity. ▿, TATA-box position. Results are expressed as fold stimulation of the basal luciferase activity from unstimulated cells. Data are means ± standard errors of five experiments performed in triplicate.
To further characterize these elements, we constructed a second series of deletions and mutations (Fig. 4B). The responsiveness of Δ2pTRP2 to PKA was similar to that observed with the initial reporter plasmid pTRP2 (37-fold), while after transfection with Δ3pTRP2, we observed only a 17-fold stimulation of the luciferase activity by PKA. Further deletion (Δ4pTRP2) led to a dramatic (sixfold) decrease in the cAMP sensitivity of the promoter. Hence, our data suggest that important regulatory elements are located between bp −475 and −273 and between bp −273 and −220. The mutation of the E box (mEBOXpTRP2) clearly impaired the effects of PKA on the TRP2 promoter (25-fold stimulation). Interestingly, we found a sequence (TGAGGTCA) very close to a classical CRE (TGACGTCA) located between bp −239 and −232 in the promoter. When mCREpTRP2, containing mutations in the CRE-like motif (−239 to −232), was transfected, stimulation of the luciferase activity was markedly (14-fold) reduced in response to PKA. Taken together, the results show that the M box (−129 to −135), the E box (−346 to −340), and the CRE-like motif (−239 to −232) play a pivotal role in the regulation of the transcriptional activity of the TRP2 promoter by cAMP.
Stimulation of tyrosinase, TRP1, and TRP2 promoter activities by PKA in normal human melanocytes.
All of the results presented above pertain to the transformed B16 melanoma cell line. Thus, we wished to verify that tyrosinase, TRP1, and TRP2 promoters respond to cAMP in normal, i.e., nontransformed, cells. To accomplish this, normal human melanocytes from skin phototype II or III were transfected with pTyro, pTRP1, and pTRP2 with or without the expression plasmid encoding PKA. The results from three different experiments performed on three different melanocyte cultures are shown in Table 1. Expression of PKA increased the luciferase activity in normal human melanocytes transfected with pTyro, pTRP1, and pTRP2. Similar effects were observed with cAMP-elevating agents such as forskolin or α-MSH (not shown). These results indicate that the transcriptional activities of tyrosinase, TRP1, and TRP2 promoters are stimulated by the cAMP pathway in normal human melanocytes.
TABLE 1.
Expression of PKA increases luciferase activity in normal human melanocytes transfected with pTyro, pTRP1, and pTRP2a
| Vector | Fold stimulation
|
||
|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |
| pTyro | 3.8 | 5 | 6.6 |
| pTRP1 | 9.2 | 6.9 | 10 |
| pTRP2 | 4.6 | 7.2 | 8.2 |
Three different cultures of normal human melanocytes were transfected by electroporation with 20 μg of reporter plasmid, 10 μg of pCDNA3, or 10 μg of pCDNA3 encoding the catalytic subunit of PKA and 3 μg of pCMVβGAL; 48 h later, luciferase activity was measured and corrected by β-galactosidase activity. The results represent the fold stimulation of luciferase activity in cells transfected with PKA expression vector compared to cells transfected with empty pCDNA3. The effects of PKA on the promoterless construct PGL2B represent means of 1.3 ± 0.5 for three independent experiments.
Microphthalmia transactivates TRP1 and TRP2 promoters.
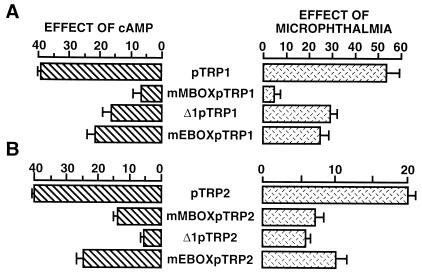
To thoroughly study the role of microphthalmia in the cAMP responsiveness of TRP1 and TRP2 promoters, we investigated the effects of microphthalmia on the different TRP1 and TRP2 constructs and compared these effects with their cAMP responses. TRP1 and TRP2 reporter constructs were cotransfected with an expression plasmid encoding microphthalmia. Microphthalmia induced a 40-fold stimulation of the TRP1 promoter (Fig. 5A). In three other constructs, mMBOXpTRP1, Δ1pTRP1, and mEBOXpTRP1, the effects of microphthalmia and those of cAMP were markedly decreased. The TRP2 promoter was also transactivated (20-fold) by microphthalmia (Fig. 5B). For the mMBOXpTRP2, Δ1pTRP2, and mEBOXpTRP2 constructs, which showed decreased cAMP responsiveness, the activation by microphthalmia was clearly reduced. It appears that the effects of cAMP on TRP1 and TRP2 promoter activities correlate with the ability of microphthalmia to transactivate the promoters.
FIG. 5.
The effects of microphthalmia on TRP1 and TRP2 promoter constructs correlate with their cAMP responsiveness. Histograms on the left represent the cAMP responsiveness of the different TRP1 (A) and TRP2 (B) constructs as determined in Fig. 3 and 4. To study the effects of microphthalmia on these different constructs, B16 cells were cotransfected with 0.3 μg of the reporter plasmid and 0.05 μg of pCMVβGAL plus 0.04 μg of empty pCDNA3 or pCDNA3 encoding microphthalmia. Reporter plasmids were pTRP1, mMBOXpTRP1, Δ1pTRP1, and mEBOXpTRP1 (A) and pTRP2, mMBOXpTRP2, Δ1pTRP2, and mEBOXpTRP2 (B). After 48 h, luciferase activity was assayed as previously described and normalized to β-galactosidase activity. Results are expressed as fold stimulation of the luciferase activity from cells transfected with empty pCDNA3. Data are means ± standard error of five experiments performed in triplicate.
cAMP increases the binding of microphthalmia to TRP1 and TRP2 M boxes.
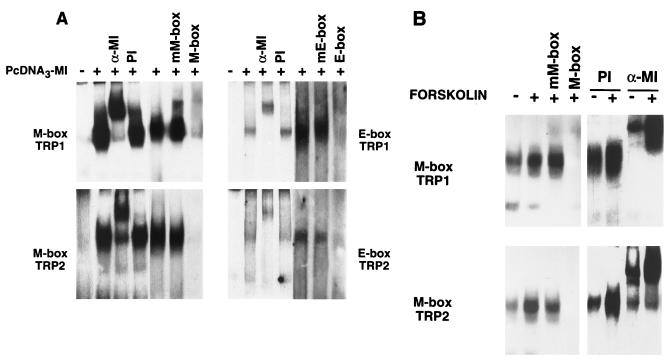
M and E boxes involved in the cAMP responsiveness of TRP1 and TRP2 promoters contain a core sequence, CANNTG, that binds helix-loop-helix transcription factors. Thus, we analyzed the binding activity of microphthalmia produced by in vitro transcription-translation to M and E boxes from TRP1 and TRP2 promoters (Fig. 6A). We observed strongly labeled complexes with TRP1 and TRP2 M boxes that were shifted by antimicrophthalmia antibody but not by preimmune serum. Further, these complexes were displaced by homologous unlabeled probe but not by oligonucleotides containing mutations corresponding to those introduced in mMBOXpTRP1 and mMBOXpTRP2. Complexes were also observed with the TRP1 E box (−238 to −233) and with the TRP2 E box (−346 to −340), but these complexes were markedly less labeled than the M-box complexes. E-box complexes were specifically shifted by antimicrophthalmia antibody and displaced by the corresponding unlabeled oligonucleotides but not by oligonucleotides bearing mutations in the E-box core motif. No DNA binding activity was observed when the transcription-translation reaction was performed in the absence of microphthalmia cDNA. These results indicate that microphthalmia has a high affinity for TRP1 and TRP2 M boxes but binds weakly to TRP1 and TRP2 E boxes. Next, using B16 cell nuclear extracts, we studied the effect of cAMP on microphthalmia binding to TRP1 and TRP2 M boxes (Fig. 6B). We observed that the TRP1 and TRP2 M-box binding activities were increased in nuclear extracts from cAMP-treated cells. The complexes were completely inhibited by addition of an excess of the unlabeled probe but not by the oligonucleotide bearing mutations in the core M-box motif. These complexes were totally and specifically displaced by antimicrophthalmia antibody, demonstrating that cAMP increased the binding of microphthalmia to TRP1 and TRP2 M boxes.
FIG. 6.
Microphthalmia binds to TRP1 and TRP2 M and E boxes. (A) TRP1 M box (−44 to −33), TRP1 E box (−238 to −233), TRP2 M box (−129 to −135), and TRP2 E box (−346 to −340) were used as probes. Gel shift assays were performed in an in vitro transcription-translation reaction using empty pCDNA3 or pCDNA3 encoding microphthalmia (pCDNA3-MI). For competition experiments, unlabeled homologous and mutated oligonucleotides were added in 50-fold excess. (B) B16 nuclear extracts from control cells (−) or cells treated with forskolin for 6 h (+) were incubated with labeled TRP1 M box and TRP2 M box. Where indicated, reactions were carried out in the presence of preimmune serum (PI) or a specific antibody directed against the C terminus of the microphthalmia (α-MI). For competition experiments, unlabeled homologous and mutated oligonucleotides were added in 50-fold excess. Autoradiograms were exposed for 15 h at −80°C, except for competition experiments with TRP1 or TRP2 E box, which were exposed for 48 h.
cAMP increases expression of microphthalmia.
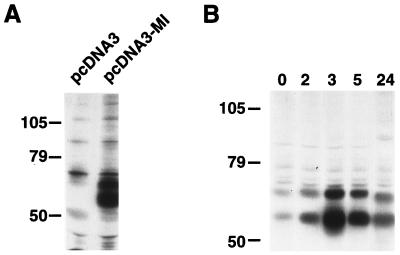
To investigate the mechanism by which cAMP increases the binding of microphthalmia, we studied the effect of forskolin on the expression of microphthalmia in B16 cells. First, NIH 3T3 cells nontransfected or transfected with an expression vector encoding microphthalmia were labeled with [35S]methionine-cysteine mix. In nontransfected cells, immunoprecipitation with antibody to the C terminus of microphthalmia revealed several weak bands that were also present with preimmune serum (not shown). In NIH 3T3 cells transfected with microphthalmia-encoding plasmid, we observed two strongly labeled bands around 70 and 60 kDa corresponding to differentially processed forms of microphthalmia (Fig. 7A). In B16 cells, we observed in basal conditions two major labeled bands at 70 and 60 kDa corresponding to microphthalmia. Incubation with forskolin led to a strong increase in the labeling of these two bands. The maximal expression of microphthalmia was observed after 3 h of forskolin treatment and then decreased after 5 h to return near the basal level after 24 h (Fig. 7B). Thus, we conclude that the effect of cAMP on the binding of microphthalmia to the M boxes is due to an increase in microphthalmia level in B16 cells.
FIG. 7.
cAMP increases microphthalmia expression in B16 mouse melanoma cells. B16 melanoma cells and NIH 3T3 cells nontransfected or transfected with microphthalmia were labeled with [35S]methionine-cysteine mix for 30 h. (A) NIH 3T3 cells were solubilized and immunoprecipitated with antimicrophthalmia antibody. (B) B16 melanoma cells were exposed to 20 μM forskolin for the indicated time, and then solubilized proteins were immunoprecipitated with antimicrophthalmia antibody. The immune complexes were analyzed by gel electrophoresis and autoradiography. Molecular masses, indicated on the left, are expressed in kilodaltons.
DISCUSSION
During the last few years, controversial results concerning the regulation of tyrosinase, TRP1, and TRP2 mRNA levels have been published. For instance, Abdel-Malek et al. (2) have shown that α-MSH-induced melanogenesis is accompanied by an increased amount of tyrosinase, TRP1, and TRP2 proteins without any changes in mRNA levels. On the other hand, Kuzumaki et al. (26) demonstrated that cAMP-elevating agents increase both tyrosinase and TRP1 expression at mRNA levels. In the same way, TRP1 and TRP2 mRNA levels, in mice with different coat color mutations, were shown to tightly correlate with eumelanin synthesis (23). In the course of investigating the regulation of TRP1 and TRP2 expression during cAMP-induced melanogenesis, we clearly showed in this report that cAMP increases tyrosinase, TRP1, and TRP2 at both protein and mRNA levels. Our results demonstrate a coordinated regulation of tyrosinase, TRP1, and TRP2 expression at mRNA levels during cAMP-induced melanogenesis. Next, we showed that cAMP-elevating agents such as forskolin and α-MSH or expression of the catalytic subunit of PKA stimulate the transcriptional activity of the TRP1 and TRP2 promoters. The role of PKA in the melanogenic pathway was confirmed by transfection with an expression plasmid encoding PKI, which dramatically reduces the effect of forskolin, α-MSH, and PKA. These results clearly demonstrate that α-MSH effects on melanogenesis are mediated through the activation of the cAMP pathway and PKA.
The regulation of tyrosinase, TRP1, and TRP2 promoters by PKA was observed in other human or mouse melanoma cell lines such as G361 or S91 (data not shown). Furthermore, we also observed a stimulation of the melanogenic promoter activities by PKA in normal human melanocytes, emphasizing the physiological relevance of the cAMP effects. It should be noted that human melanocytes express high basal levels of melanogenic proteins (13), probably because culture conditions of normal melanocytes are unable to prevent a spontaneous differentiation of the cells. Thus, consequent to the high basal melanogenesis, the effect of PKA on tyrosinase, TRP1, and TRP2 promoters appears markedly less pronounced in human melanocytes than in mouse melanoma cells. In other cell types, such as NIH 3T3 mouse fibroblasts, cAMP does not change the activity of the melanogenic promoters (not shown). These results indicate that a cell-specific mechanism is involved in the cAMP response of the melanogenic promoters. We then attempted to localize and to identify the cis-regulatory elements conferring on TRP1 and TRP2 promoters their cAMP responsiveness. We demonstrated that the M box and the E box upstream of the TATA box play a key role in the cAMP response of TRP1 and TRP2 promoters. Recently, we also reported that the cAMP response of the tyrosinase promoter involved a M box and an E box surrounding the TATA box (5). Thus, the mechanisms of regulation of tyrosinase, TRP1, and TRP2 promoters by cAMP appear to rely on similar regulatory elements. However, it should be noted that in the tyrosinase promoter, the cAMP-sensitive E box located at the initiator site binds tightly to microphthalmia and is absolutely necessary for the cAMP response. In TRP1 and TRP2 promoters, the M boxes bind avidly to microphthalmia, but the E boxes, involved in the cAMP response, interact weakly with microphthalmia. Thus, microphthalmia appears to require the core motif CATGTG to provide a strong interaction with DNA, since this motif is found in all of the M boxes and in the tyrosinase E box, while TRP1 and TRP2 E boxes have CAAGTG and CAATTG, respectively, as core sequences. We cannot exclude the possibility that in intact cells and in the context of the intact promoter, microphthalmia tightly binds to TRP1 and TRP2 E boxes. However, mutations of TRP1 and TRP2 E boxes moderately affect cAMP sensitivity and the microphthalmia effect on the promoters, suggesting that in intact cells, microphthalmia does not interact strongly with these motifs. In the TRP2 promoter, a CRE-like sequence also participates in the cAMP response, indicating that transcription factors of the CREB family are involved in the stimulation of the TRP2 promoter activity by cAMP. However, this CRE motif has to cooperate with the M box to give full cAMP sensitivity to the TRP2 promoter. Noteworthy, the same cis-acting elements (M and E boxes) were thought to mediate the tissue-specific expression (4) and the cAMP responses of the tyrosinase, TRP1, and TRP2 genes. Thus, it should be considered that the cAMP pathway participates in the regulation of the tissue-specific expression of the melanocyte-specific genes.
Although microphthalmia and M boxes play a pivotal role in the regulation of tyrosinase, TRP1, and TRP2 promoter activities by cAMP, it appears that each promoter responds to cAMP through specific mechanisms because of the relative position of the regulatory elements or the intrinsic nature of the elements cooperating with the M box. This is particularly true for the TRP2 promoter, which contains a CRE-like element involved in cAMP sensitivity; in tyrosinase and TRP1 promoters, no CRE participates in the cAMP response. These observations reveal, besides a common regulatory mechanism, the existence of specific processes involved in the control of tyrosinase, TRP1, and TRP2 gene expression that could allow a differential expression pattern of the melanogenic enzymes under particular physiological and pathological conditions.
We have previously demonstrated that cAMP increases microphthalmia binding to the M box and to the E box. Since microphthalmia has a strong stimulating effect on the tyrosinase promoter, we have proposed that microphthalmia, through the binding to M and E boxes, mediates the effects of cAMP on tyrosinase gene expression (5). In the present report, we clearly showed a strong stimulation of TRP1 and TRP2 promoter activities by microphthalmia. Consistently, microphthalmia or its human homolog MITF (microphthalmia-associated transcription factor) was shown to transactivate the TRP1 promoter (39, 40). However, the TRP2 promoter has been shown to be unresponsive to MITF (39), while we clearly showed in this report that microphthalmia stimulates the TRP2 promoter activity. This discrepancy could be explained either by specific behaviors of human versus mouse microphthalmia or by the different cellular contexts.
Interestingly, the effect of microphthalmia on different TRP1 and TRP2 promoter constructs tightly correlates with their cAMP responsiveness. Further, we showed that cAMP increases the binding of microphthalmia to TRP1 and TRP2 M boxes. These data strongly suggest that the effect of cAMP on TRP1 and TRP2 gene expression is mediated through an increased interaction of microphthalmia with M boxes, thereby leading to a transactivation of TRP1 and TRP2 promoters. At least two hypothesis could explain the stimulation of microphthalmia binding on the M box after cAMP treatment. cAMP could increase either microphthalmia expression or affinity of microphthalmia for its target sequences. Previous immunofluorescence studies with antimicrophthalmia antibody did not show a stimulation of microphthalmia expression after 24 h with forskolin. Although this result does not support the first hypothesis, we have reassessed the effect of cAMP on microphthalmia expression. Metabolic labeling followed by immunoprecipitation with specific antibody to microphthalmia demonstrates that cAMP increases microphthalmia expression. Microphthalmia appears as two bands around 60 and 70 kDa. The lower mobility of the higher band results from its phosphorylation on serine 73 by mitogen-activated protein kinases (16). The maximal expression of microphthalmia was obtained after 3 h of forskolin treatment, and consistent with our immunofluorescence studies, the level of microphthalmia returned near the basal level after 24 h with forskolin. The presence of a CRE in the microphthalmia promoter (10) suggests that PKA regulates microphthalmia expression through the classical pathway involving transcription factors of the CREB family. However, this hypothesis remains to be demonstrated. Interestingly, Rungta et al. (33) showed that α-MSH treatment significantly increases the tyrosinase mRNA levels within 16 h, and we previously observed an effect of forskolin on tyrosinase promoter after 6 h. Further, careful examination of the report of Abdel-Malek et al. (2) shows that the TRP1 mRNA amount was increased after 6 h with α-MSH. Thus, if we compare the kinetics of microphthalmia induction with those of tyrosinase and TRP1, the upregulation of the melanogenic enzymes is observed clearly after the maximal expression of microphthalmia. These observations are consistent with our former hypothesis suggesting that microphthalmia plays a key role in the stimulation of melanogenic gene expression.
Considering the physiological aspect of our findings, it should be mentioned that in humans, melanogenesis is stimulated by UVB, which upregulates the production of α-MSH by epidermal keratinocytes. Further, subcutaneous injection of α-MSH has been shown to stimulate local pigmentation (28). Thus, we can hypothesize that α-MSH, through the binding to its receptor coupled to the G protein αs and adenylate cyclase, increases the cAMP content in melanocytes. Then cAMP, through the activation of PKA, leads to an augmentation of microphthalmia expression. Consequently, the amount of microphthalmia bound to M or E boxes increases resulting in a stimulation of the melanogenic promoter activities. Taken together, our results disclosed the cascade of molecular events involved in the regulation of the melanogenic genes that could be of paramount importance in the control of skin pigmentation.
ACKNOWLEDGMENTS
We thank V. Hearing (Bethesda, Md.) for providing antityrosinase (PEP7) and anti-TRP2 (PEP8) antibodies and P. G. Parsons (Brisbane, Australia) for anti-TRP1 antibody B8G3. We also thank P. Sassone-Corsi and E. Lalli (Illkirch, France) for providing the expression vector encoding the catalytic subunit of PKA and R. Maurer (Portland, Oreg.) for the expression vector encoding PKI. We are grateful to A. Grima and C. Minghelli for illustration work and to J. C. Scimeca and C. Sable for critical reading of the manuscript.
This work was supported by Association pour la Recherche sur le Cancer (grant 6760) and Ligue Nationale Contre le Cancer.
REFERENCES
- 1.Abdel-Malek Z, Swope V B, Amornsiripanitch N, Nordlund J J. In vitro modulation of proliferation and melanization of S91 melanoma cells by prostaglandins. Cancer Res. 1987;47:3141–3146. [PubMed] [Google Scholar]
- 2.Abdel-Malek Z, Swope V B, Suzuki I, Akcali C, Harriger M D, Boyce S T, Urabe K, Hearing V. Mitogenic and melanogenic stimulation of normal human melanocytes by melanotropic peptides. Proc Natl Acad Sci USA. 1995;92:1789–1793. doi: 10.1073/pnas.92.5.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barber J I, Townsend D, Olds D P A, King R A. Dopachrome oxydoreductase: a new enzyme in the pigment pathway. J Invest Dermatol. 1984;83:145–149. doi: 10.1111/1523-1747.ep12263381. [DOI] [PubMed] [Google Scholar]
- 4.Bentley N J, Eisen T, Goding C R. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol Cell Biol. 1994;14:7996–8006. doi: 10.1128/mcb.14.12.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertolotto C, Bille K, Ortonne J P, Ballotti R. Regulation of tyrosinase gene expression by cAMP in B16 melanoma cells involves two CATGTG motifs surrounding the TATA box: implication of the microphthalmia gene product. J Cell Biol. 1996;134:747–755. doi: 10.1083/jcb.134.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Day R N, Walder J A, Maurer R A. A protein kinase inhibitor gene reduces both basal and multihormone-stimulated prolactin gene transcription. J Biol Chem. 1989;264:431–436. [PubMed] [Google Scholar]
- 7.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Englaro W, Rezzonico R, Durand-Clément M, Lallemand D, Ortonne J P, Ballotti R. Mitogen-activated protein kinase pathway and AP-1 are activated during cAMP-induced melanogenesis in B-16 melanoma cells. J Biol Chem. 1995;270:24315–24320. doi: 10.1074/jbc.270.41.24315. [DOI] [PubMed] [Google Scholar]
- 9.Fuller B B, Viskochil D H. The role of RNA and protein synthesis in mediating action of MSH on melanoma cell cultures. Life Sci. 1979;24:2405–2416. doi: 10.1016/0024-3205(79)90448-x. [DOI] [PubMed] [Google Scholar]
- 10.Fuse N, Yasumoto K I, Suzuki H, Takahashi K, Shibahara S. Identification of a melanocyte-type promoter of the microphthalmia-associated transcription factor gene. Biochem Biophys Res Commun. 1996;219:702–707. doi: 10.1006/bbrc.1996.0298. [DOI] [PubMed] [Google Scholar]
- 11.Ganss R, Schutz G, Beermann F. The mouse tyrosinase gene. J Biol Chem. 1994;269:29808–29816. [PubMed] [Google Scholar]
- 12.Gordon P R, Mansur C P, Gilchrest B A. Regulation of human melanocyte growth, dendricity, and melanization by keratinocyte derived factors. J Invest Dermatol. 1989;92:565–572. doi: 10.1111/1523-1747.ep12709595. [DOI] [PubMed] [Google Scholar]
- 13.Halaban R, Pomerantz S H, Marshall S, Lambert D T, Lerner A B. Regulation of tyrosinase in human melanocytes grown in culture. J Cell Biol. 1983;97:480–488. doi: 10.1083/jcb.97.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hearing V J. Mammalian monophenol monooxygenase (tyrosinase): purification, properties and reactions catalyzed. Methods Enzymol. 1987;142:154–165. doi: 10.1016/s0076-6879(87)42024-7. [DOI] [PubMed] [Google Scholar]
- 15.Hearing V J, Tsukamoto K. Enzymatic control of pigmentation in mammals. FASEB J. 1991;5:2902–2909. [PubMed] [Google Scholar]
- 16.Hemesath, T. J., E. R. Price, C. Takemoto, T. Badalian, and D. E. Fisher. MAPK links microphthalmia to c-kit signaling in melanocytes. Nature, in press. [DOI] [PubMed]
- 17.Hirobe T, Takeuchi T. Induction of melanogenesis in vitro in the epidermal melanoblasts of newborn mouse skin MSH. J Embryol Exp Morphol. 1977;37:79–80. [PubMed] [Google Scholar]
- 18.Hunt G, Todd C, Cresswell J E, Thody A J. α-MSH and its analog Nle4DPhe7 α-MSH affect morphology, tyrosinase activity and melanogenesis in cultured human melanocytes. J Cell Sci. 1994;107:205–211. doi: 10.1242/jcs.107.1.205. [DOI] [PubMed] [Google Scholar]
- 19.Jackson J I, Cambers D M, Tsukamoto K, Copeland N, Gilbert D J, Jenkins N A, Hearing V J. A second tyrosinase-related protein, TRP-2, maps to and mutated at the mouse slaty locus. EMBO J. 1992;11:527–535. doi: 10.1002/j.1460-2075.1992.tb05083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiménez M, Kameyama K, Maloy W L, Tomita Y, Hearing V J. Mammalian tyrosinase: biosynthesis, processing, and modulation by melanocyte-stimulating hormone. Proc Natl Acad Sci USA. 1988;85:3830–3834. doi: 10.1073/pnas.85.11.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiménez-Cervantes C, Solano F, Kobayashi T, Urabe K, Hearing V J, Lozano J A, García-Borrón J C. A new enzymatic function in the melanogenic pathway: the 5,6-dihydroxyindole-2-carboxylic acid oxidase activity of tyrosinase-related protein-1 (TRP1) J Biol Chem. 1994;269:29198–29205. [PubMed] [Google Scholar]
- 22.Kameyama K, Jiménez M, Muller J, Ishida Y, Hearing V J. Regulation of mammalian melanogenesis by tyrosinase inhibition. Differentiation. 1989;42:28–36. doi: 10.1111/j.1432-0436.1989.tb00604.x. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi T, Viera W D, Potterf B, Sakai C, Imokawa G. Modulation of melanogenic protein expression during the switch from eu- to pheomelanogenesis. J Cell Sci. 1995;108:2301–2309. doi: 10.1242/jcs.108.6.2301. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi T, Urabe K, Winder A, Jiménez-Cervantes C, Imokawa G, Brewington T, Solano F, García-Borrón J C, Hearing V J. Tyrosinase related protein 1 (TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO J. 1994;13:5818–5825. doi: 10.1002/j.1460-2075.1994.tb06925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Körner A, Pawelek J. Mammalian tyrosinase catalyzes three reactions in the biosynthesis of melanin. Science. 1982;217:1163–1165. doi: 10.1126/science.6810464. [DOI] [PubMed] [Google Scholar]
- 26.Kuzumaki T, Matsuda A, Wakamatsu K, Ito S, Ishikawa K. Eumelanin biosynthesis is regulated by coordinate expression of tyrosinase and tyrosinase-related protein-1 genes. Exp Cell Res. 1993;207:33–40. doi: 10.1006/excr.1993.1159. [DOI] [PubMed] [Google Scholar]
- 27.Le Douarin N. The neural crest. Cambridge, England: Cambridge University Press; 1982. [Google Scholar]
- 28.Levine N, Sheftel S N, Eytan T, Dorr R, Hadley M E, Weinrach J C, Ertl G A, Toth K, McGee D L, Hurby V J. Induction of skin tanning by subcutaneous administration of a potent synthetic melanotropin. JAMA. 1991;226:2730–2736. [PubMed] [Google Scholar]
- 29.Lowings P, Yavuzer U, Goding R. Positive and negative elements regulate a melanocyte-specific promoter. Mol Cell Biol. 1992;12:3653–3662. doi: 10.1128/mcb.12.8.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molina C A, Foulkes N S, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 31.Prota G. Some new aspects of eumelanin chemistry. Prog Clin Biol Res. 1988;256:101–124. [PubMed] [Google Scholar]
- 32.Prota G. Melanins and melanogenesis. New York, N.Y: Academic Press; 1992. [Google Scholar]
- 33.Rungta D, Corn T D, Fuller B B. Regulation of tyrosinase mRNA in mouse melanoma cells by α-melanocyte-stimulating hormone. J Invest Dermatol. 1996;107:689–693. doi: 10.1111/1523-1747.ep12365578. [DOI] [PubMed] [Google Scholar]
- 34.Steel K P, Davidson D R, Jackson I J. TRP-2/DT, a new early melanoblast marker, shows that steel growth factor (c-kit ligand) is a survival factor. Development. 1992;115:1111–1119. doi: 10.1242/dev.115.4.1111. [DOI] [PubMed] [Google Scholar]
- 35.Swope V B, Abdel-Malek Z, Kassem L M, Nordlund J J. Interleukins 1 alpha and 6 and tumor necrosis factor-alpha are paracrine inhibitors of human melanocyte proliferation and melanogenesis. J Invest Dermatol. 1991;96:180–185. doi: 10.1111/1523-1747.ep12460991. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi H, Parsons P G. In vitro phenotypic alteration of human melanoma cells induced by differentiating agents. Pigment Cell Res. 1990;3:223–232. doi: 10.1111/j.1600-0749.1990.tb00294.x. [DOI] [PubMed] [Google Scholar]
- 37.Wong G, Pawelek J. Control of phenotypic expression of cultured melanoma cells by melanocyte stimulating hormone. Nature. 1973;241:213–215. doi: 10.1038/newbio241213a0. [DOI] [PubMed] [Google Scholar]
- 38.Wong G, Pawelek J. Melanocyte-stimulating hormone promotes activation of pre-existing tyrosinase molecules in Cloudman S91 melanoma cells. Nature. 1975;255:644–645. doi: 10.1038/255644a0. [DOI] [PubMed] [Google Scholar]
- 39.Yasumoto K I, Yokoyama K, Takahashi K, Tomita Y, Shibahara S. Functional analysis of microphthalmia-associated transcription factor in pigment cell-specific transcription of the human tyrosinase family genes. J Biol Chem. 1997;272:503–509. doi: 10.1074/jbc.272.1.503. [DOI] [PubMed] [Google Scholar]
- 40.Yavuzer U, Keenan E, Lowings P, Vachtenheim J, Currie G, Goding C R. The microphthalmia gene product interacts with the retinoblastoma protein in vitro and is a target for deregulation of melanocyte-specific transcription. Oncogene. 1995;10:123–134. [PubMed] [Google Scholar]
- 41.Yokoyama K, Yasumoto K I, Suzuki H, Shibahara S. Cloning of the human DOPAchrome tautomerase/tyrosinase-related protein 2 gene and identification of two regulatory regions required for its pigment cell-specific expression. J Biol Chem. 1994;269:27080–27087. [PubMed] [Google Scholar]
- 42.Yokoyama K, Suzuki H, Tomita Y, Shibahara S. Molecular cloning of and functional analysis of a cDNA coding for human DOPAchrome tautomerase/tyrosinase-related protein-2. Biochim Biophys Acta. 1994;1217:317–321. doi: 10.1016/0167-4781(94)90292-5. [DOI] [PubMed] [Google Scholar]