Abstract
Cilia and flagella are central to many biological processes in a diverse range of organisms. The kinetoplastid protozoa are very appealing models for the study of flagellar function, particularly in the light of the availability of extensive trypanosomatid genome information. In addition to the highly conserved 9 + 2 axoneme, the kinetoplastid flagellum contains a characteristic paraflagellar rod structure (PFR). The PFR is necessary for full motility and provides support for metabolic regulators that may influence flagellar beating. However, there is an intriguing puzzle: one clade of endosymbiont-containing kinetoplastids apparently lack a PFR yet are as motile as species that possess a PFR and are able to attach to the invertebrate host epithelia. We investigated how these organisms are able to locomote despite the apparent lack of PFR. Here we have identified a PFR1 gene in the endosymbiont-bearing trypanosome Crithidia deanei. This gene is expressed in C. deanei and is able to partially complement a pfr1 null mutation in Leishmania mexicana cells, demonstrating that the encoded protein is functional. Careful reexamination of C. deanei flagellar ultrastructure revealed a greatly reduced PFR missed by many previous analyses. This affirms the PFR as a canonical organelle of kinetoplastids. Moreover, although PFR proteins have been conserved in evolution, primary sequence differences contribute to particular PFR morphotypes characteristic of different kinetoplastid species.
Cilia and flagella are central to many biological processes in a diverse range of organisms. In the order Kinetoplastida—the group of flagellates that include trypanosomatid parasites and bodonids—the flagellum is the classical organelle of motility (42). In parasitic species, the kinetoplastid flagellum has also evolved to be an organelle of attachment to the invertebrate vector, playing a critical role in parasite transmission to the vertebrate host (42). Moreover, recent work shows that the kinetoplastid flagellum is also involved in cell division; positioning of the new flagellum, one of the earliest event in cell duplication, defines the axis of polarity in the dividing cell and the position of the internal organelles (31). Finally, flagellar wave reversal in a Ca2+-dependent manner as an avoidance response has been studied in trypanosomatids (39), suggesting that the flagellum in these species may also be a specialized sensory organelle.
The main component of both cilia and flagella is the axoneme. This microtubule-based organelle was most probably present in the ancestor of all modern eukaryotes and is highly conserved in several deeply diverged lines. In many organisms, the axoneme is augmented by extra-axonemal structures—for example, the fibrous sheath in mammalian spermatozoa and the R-fiber of dinoflagellates. In the Kinetoplastida, a characteristic structure known as the paraflagellar rod (PFR) runs alongside the axoneme to form the flagellum. This PFR is an elegant and stable lattice-like arrangement of protein filaments which is composed of two related major proteins, PFR1 and PFR2 (19), and several minor ones (30, 44).
The PFR appears to be necessary for correct flagellar function in kinetoplastids. In Trypanosoma brucei, ablation of PFR2 protein expression by RNA interference disrupts PFR construction and results in cell paralysis (2, 4, 5). Deletion of the PFR1 and/or PFR2 genes from Leishmania mexicana also prevents the formation of a native PFR structure and produces cells with lower swimming velocities and severe flagellar waveform perturbations (29, 37). Part of these phenotypes may result from the fact that the PFR provides a support for the incorporation of at least two metabolic regulators into the flagellum (34). Interestingly, the electron-dense plaques that form when trypanosomatids attach to invertebrate epithelia contain filaments that appear similar to those of the PFR and appear to originate in the PFR itself (6, 10, 41), leading to the hypothesis that the PFR may be the critical organelle mediating attachment to vector cell surfaces.
However, given these functions, it is intriguing that the possession of the PFR structure does not appear to be universal within kinetoplastids. The members of one particular group of trypanosomatid species (exemplified by Crithidia deanei, C. oncopelti, C. desouzai, Blastocrithidia culicis, and Herpetomonas roitmani) have been described as lacking a PFR (17). These species also share other ultrastructural features (17), the most obvious of which is possession of an enslaved endosymbiont bacterium (13), most probably the result of a single acquisition in the common ancestor of the above species. Surprisingly, these endosymbiont-harboring species are able to attach to the invertebrate host epithelia (15) and are actively motile (21, 39). The cell movement and the flagellar beating features have been extensively studied, and they show the same major characteristics as other trypanosomatids (22-24, 39). A detailed description of C. oncopelti flagellar beat amplitude and frequency has been compared with that of C. fasciculata (which possesses a PFR), and no obvious differences could be seen (3; C. Gadelha and K. Gull, unpublished data).
There is an interesting conundrum here: the kinetoplastid PFR is implicated in both motility and attachment, yet the endosymbiont-bearing trypanosomes are motile and capable of attachment despite the apparent lack of a PFR. Here, we address this dichotomy by asking what has happened to the genes for the major PFR proteins in the endosymbiont-bearing Crithidia deanei. We show that this organism possesses a gene encoding PFR1 and that this protein is expressed. Despite some sequence differences, this gene is able to partially rescue a PFR1 deletion mutation in Leishmania mexicana. Furthermore, careful reexamination of the C. deanei flagellar ultrastructure revealed a greatly reduced PFR that had been missed by many previous analyses, hence reaffirming the PFR as a canonical organelle of kinetoplastids. Moreover, the data show that small differences in primary sequence between highly conserved PFR proteins may impose many of the differences in morphology characteristic of different kinetoplastid species.
MATERIALS AND METHODS
Cells.
Promastigotes of Leishmania mexicana wild type (WHO strain MNYC/BZ/62/M379) and PFR1 deletion mutants (Δpfr1::NEO Δpfr1::HYG, referred to here simply as Δpfr1) (29) were cultured at 28°C in Medium 199 with Earle's Salts and l-glutamine (Gibco) supplemented with 40 mM HEPES, 5% (vol/vol) fetal bovine serum (Gibco), and 5 μg of hemin per ml. L. mexicana complemented cell lines Δpfr1::NEO Δpfr1::HYG [pNUS-GFPcB] (referred to here as Δpfr1[GFP]), Δpfr1::NEO Δpfr1::HYG [pNUS-LmxPFR1cB] (Δpfr1[Lmx]), and Δpfr1::NEO Δpfr1::HYG [pNUS-CdePFR1cB] (Δpfr1[Cde]) were cultured as above, with the addition of 20 μg of phleomycin (Sigma) per ml. Crithidia deanei, C. oncopelti, and C. fasciculata coanomastigotes and Herpetomonas megaseliae promastigotes were grown at 28°C in brain heart infusion medium supplemented with 5% (vol/vol) fetal calf serum. Procyclic Trypanosoma brucei 427 cells were cultured at 28°C in SDM 79 medium supplemented with 10% (vol/vol) fetal calf serum.
Gene cloning.
We used all publicly available PFR1 and PFR2 sequences to identify conserved regions that could be used to design degenerate oligonucleotides capable of amplifying PFR genes from a wide range of Euglenozoa. Kinetoplastid and euglenoid sequences—namely, sequences from Crithidia fasciculata (AY568294 and AY568293), Leishmania mexicana (AY198411 and U45884), Euglena gracilis (AF044217), Lepocinclis ovata (AF296721 and AF263944), Khawkinea quartana (AF296722, AF263943, and AF263945), Gyropaigne lefevrei (AF263946), Phacus smulkowskianus (AF188117 and AF296720), and Distigma curvatum (AF188118), and sequence from the genome-sequencing projects for Leishmania major (www.sanger.ac.uk/Projects/L_major), L. infantum (www.sanger.ac.uk/Projects/L_infantum), L. braziliensis (www.sanger.ac.uk), Trypanosoma brucei (www.sanger.ac.uk/Projects/T_brucei), T. congolense (www.sanger.ac.uk/Projects/T_congolense), T. vivax (www.sanger.ac.uk/Projects/T_vivax), T. b. gambiense (www.sanger.ac.uk/Projects/T_b_gambiense), and T. cruzi (www.tigr.org/tdb/e2k1/tca1)—were aligned. Oligonucleotides 5′-ACGACGCSATCCAGAAGGC and 5′-CTTSGCGTTSGGGTCGAA span a 189-amino-acid (567-bp) fragment (residues 284 to 473 of T. brucei PFR1). These were used in PCR with C. deanei, C. oncopelti, C. fasciculata, H. megaseliae, or T. brucei genomic DNA templates (prepared as described in reference 43). Amplicons from each species were cloned, and several clones were sequenced. C. fasciculata, H. megaseliae, and T. brucei templates generated amplicons encoding fragments of both PFR1 and PFR2 proteins, whereas those from C. deanei and C. oncopelti gave only PFR1 sequence. For amplification of tandemly repeated PFR genes, oligonucleotides 5′-AGCGGCTGGAGGAGAT and 5′-GGTCGCAGTTGTACAC were used. We also used 5′-GTTCCTSGACGTGTG and 5′-CCATGTTGCCGGACTCAAC to specifically amplify PFR2 sequences. Both of these combinations resulted in PFR fragments from C. fasciculata, H. megaseliae, and T. brucei templates but not from C. deanei and C. oncopelti.
The C. deanei central region was used to generate complete PFR1 mRNA sequence—amplifying from this central region toward both ends by reverse transcription-PCR (RT-PCR). RNA was produced using the High-Pure RNA isolation kit (Roche). First-strand cDNA synthesis used either random hexadeoxynucleotides or oligo(dT)15 primers and the Omniscript reverse transcriptase kit (Qiagen). The N terminus was amplified using 5′-AGTTTCTGTACTWTATTG and 5′-GAATTCTAATTGAATATGTGT (crithidial mini-exon sequence), and the C terminus was amplified using 5′-ACGACGCSATCCAGAAGGC and oligo(dT)20.
Sequence analysis.
Multiple alignments of protein sequences were made using the ClustalW algorithm and manually adjusted as necessary. Phylograms were inferred from these alignments, using both maximum-parsimony (MP) and neighbor-joining (NJ) methods as implemented by the software PAUP*4.0-β10 (Sinauer Associates Inc.). MP trees were built by full heuristic searches with tree-bisection-reconnection (TBR) swapping, and start trees were generated by simple stepwise addition. Gaps were interpreted as missing data. NJ tree inference employed a mean distance measure and an objective of minimum evolution. A total of 100 bootstrap replicates were made for both methods.
Southern blot analysis.
DNA transfer to membranes was performed as described elsewhere (43). Fluorescein-labeled probes were generated by random priming (Gene Images kit; Amersham Life Science) from the following unlabeled DNA: CdePFR1, TbrPFR1, TbrPFR2, LmxPFR1, LmxPFR2, CfaPFR1, and CfaPFR2. For cross-species hybridizations, medium-stringency conditions were used: hybridization was performed overnight at 56°C in 0.1% (wt/vol) sodium dodecyl sulfate (SDS), 5% (wt/vol) dextran sulfate-5% (vol/vol) blocking solution (Amersham)-5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and blots were washed to a stringency of 0.1% SDS-0.3× SSC at 60°C. Hybridizations with species-specific probes were preformed under high-stringency conditions (hybridized at 60°C overnight as above and then washed to stringency of 0.1% SDS-0.1× SSC at 62°C). Hybridized probe was detected with an anti-fluorescein alkaline phosphatase-conjugated antibody followed by addition of the chemiluminescent substrate CDP-star (Amersham Life Science).
Plasmid construction and transfection of Leishmania.
The pNUS-GFPcB vector, kindly supplied by E. Tetaud and A. Fairlamb (40), allows the coexpression of an introduced gene (between glutathionylspermidine synthetase 5′ and phosphoglycerate kinase 3′ sequences from C. fasciculata) and a phleomycin-resistance marker (BLE). pNUS-GFPcB was digested with XhoI, filled in with T4 DNA polymerase, and digested with NdeI to remove the green fluorescent protein fragment. Genomic PCR was used to amplify the LmxPFR1 open reading frame (ORF) with 5′-CATATGGATGATGACCCCTGAAGATG and 5′-GATATCAATGCACATACCCTCCAGCT and to amplify the CdePFR1 ORF with 5′-CATATGAAAAAGGAAAGAATATGTCT and 5′-GATATCCCTTCATTCTTYTTTCACTT. These amplicons were ligated into the EcoRV site of pBluescriptSK(+), released by NdeI and EcoRV digestion, and ligated into pNUS-GFPcB prepared as described above, yielding plasmids pNUS-LmxPFR1cB and pNUS-CdePFR1cB, respectively. The identity and direction of the introduced fragments was checked by sequencing.
L. mexicana Δpfr1 cells were transfected with episomal plasmids by electroporation. Cells were harvested, washed twice in Cytomix (120 mM KCl2, 0.5 mM CaCl2, 10 mM K2HPO4, 10 mM KH2PO4, 25 mM HEPES, 2 mM EDTA, 5 mM MgCl2 [pH 7.6]), and resuspended to a final concentration of 5 × 107 cells ml−1. A total of 2.5 × 107 cells were placed in a 4-mm cuvette, and 10 μg of plasmid were added prior to electroporation twice at 1.7 kV with three × 100-μs pulses (Electro Square Porator; BTX). Following transfection, the cells were allowed to recover in normal growth medium for 16 h, after which time transformants were selected with 20 μg of phleomycin per ml. Episome-containing cell lines were maintained as populations in the presence of phleomycin.
Antibodies.
Monoclonal antibodies (MAb) used in this study were F4, which recognizes both PFR1 and PFR2 in Leishmania spp. (kindly supplied by Diane McMahon-Pratt [26]); L8C4, which recognizes PFR2 in T. brucei; and L13D6, which recognizes both PFR1 and PFR2 in T. brucei (27) but only PFR1 in Leishmania spp. (data not shown).
Immunofluorescence.
L. mexicana promastigotes were settled onto poly-l-lysine-coated slides and fixed in methanol at −20°C for 10 min. The cells were labeled with L13D6 in phosphate-buffered saline (PBS) (140 mM NaCl, 3 mM KCl, 10 mM sodium phosphate, 2 mM potassium phosphate [pH 7.2]) containing 0.5% (wt/vol) skim milk and visualised with tetramethylrhodamine-5-isothiocyanate (TRITC)-conjugated anti-mouse immunoglobulins (Sigma) in PBS containing 1% (wt/vol) bovine serum albumin. The cells were embedded in Vectashield with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories). Cytoskeletons were prepared from live cells settled on slides by extraction with 1% (vol/vol) NP-40 in PEME buffer (100 mM PIPES, 1 mM MgSO4, 0.1 mM EDTA, 2 mM EGTA [pH 6.9]), followed by fixation as above. C. deanei cells and cytoskeletons were prepared as above, with the exception of being fixed in 1% paraformaldehyde in PBS for 5 min.
1D SDS-PAGE and Western blot analysis.
For whole-cell protein samples, cells were harvested, washed in PEME buffer with 5 μM E-64d (Sigma), pelleted and immediately resuspended in boiling Laemmli buffer (2% SDS, 10% [vol/vol] glycerol, 400 mM β-mercaptoethanol, 50 mM Tris-HCl [modified pH 7.2]). For detergent fractionation, cells were harvested and washed as above and resuspended in PEME buffer containing 1% (vol/vol) NP-40, 200 μg of DNase I per ml, and protease inhibitors (5 μM E-64d, 10 mM 1,10-phenanthroline, 50 μM leupeptin, 7.5 μM pepstatin A, 500 μM phenylmethylsulfonyl fluoride, 100 μM tosyl-lysyl-chloromethyl ketone [TLCK], 100 μM tolylsulfonyl phenylalanyl chloromethyl ketone [TPCK], 1 mM EDTA [all from Sigma]). The cells were incubated for 5 min on ice and centrifuged at 3,400 × g for 15 min at 4°C. Pelleted material was washed once in PEME buffer, and protein in the supernatant was acetone precipitated. Both samples were resuspended in boiling Laemmli buffer and boiled for 5 min. SDS-polyacrylamide gel electrophoresis (PAGE) and immersion transfer to nitrocellulose membrane were performed using standard techniques described elsewhere (36). For immunoblotting, the membranes were blocked with 3% (wt/vol) skim milk in TTBS (20 mM Tris-HCl, 500 mM NaCl, 0.05% Tween 20 [pH 7.5]), labeled with anti-PFR monoclonal antibody with 0.5% skim milk, and developed with horseradish peroxidase-conjugated anti-mouse immunoglobulins (Sigma).
MS.
Briefly, SDS-PAGE gels were fixed for 30 min in 10% methanol-5% acetic acid and stained with SYPRO ruby (Sigma). Bands were excised over 280-nm UV light and macerated. Gel fragments were washed in 50% acetonitrile-50 mM ammonium bicarbonate (pH 8.5) solution, dehydrated in 100% acetonitrile, and air dried. Proteins were then digested for 16 h with ∼20 μg of trypsin (mass spectrometry [MS] grade; Promega) per ml in 25 mM ammonium bicarbonate at 37°C. Electrospray ionization (ESI)-MS/MS was performed on a Micromass Q-ToF micro instrument (Waters), and masses were analyzed by the MASCOT search engine (Matrix Science).
Electron microscopy.
Thin-section transmission electron microscopy (TEM) for Leishmania samples was performed by harvesting cells and fixing them in 2.5% glutaraldehyde-2% paraformaldehyde-100 mM sodium phosphate (pH 6.5) for 2 h at 4°C. Samples were postfixed in 1% osmium tetroxide in 100 mM sodium phosphate buffer (pH 6.5) for 1 h at 4°C, washed several times in double-distilled water, en bloc stained with 0.5% aqueous uranyl acetate for 16 h at 4°C, dehydrated through acetone and propylene oxide, and embedded in TAAB 812 embedding resin. C. deanei cells were washed in PBS, fixed in 2.5% glutaraldehyde-100 mM sodium phosphate (pH 7.0) for 1 h at room temperature, postfixed in 0.5% osmium tetroxide in 100 mM sodium phosphate buffer (pH 7.0) for 20 min at 4°C, dehydrated through ethanol, and embedded in Spurr (TAAB). Whole-mount cytoskeletons were prepared by settling cells onto carbon-coated, charged Formvar grids, extracting lipids with 1% (vol/vol) NP-40 in PEME buffer, fixing the cells in 2.5% glutaraldehyde in PEME buffer, and negatively staining them with 0.7% gold-thioglucose in water.
Motility analysis.
L. mexicana cell lines were grown to a density of 5 × 106 cells ml−1 and equilibrated to 22°C for 1 h. These cells (in medium) were placed onto slides and covered with a glass coverslip raised above the slide surface with 50-μm-thick tape (ca. two cell lengths). To prevent cell adherence, both slide and coverslip were precoated with poly-l-glutamate. The edges of the coverslip were then sealed to avoid capillary flow of liquid. Time-lapse sequences of images captured every 1 s for 1 min were made at 22°C and low magnification (10×) using phase-contrast illumination. For each cell line, velocity measurements (assuming movement along the z-axis to be negligible) were made by tracking ∼100 cells over all 60 frames.
Nucleotide sequence accession numbers.
Sequence data reported here are available from GenBank under accession numbers AY785777, AY785778, AY785779, and AY785780.
RESULTS
C. deanei possesses a gene encoding PFR1.
To determine whether an endosymbiont-containing organism (apparently not possessing a PFR) possesses any putative PFR genes, we designed degenerate oligonucleotides which were able to amplify both PFR1 and PFR2 genes from any trypanosomatid species. PCR from genomic DNA of C. deanei amplified a PFR gene fragment. To complete the sequence of this gene fragment and to test for transcription, we performed RT-PCR on C. deanei RNA, using either crithidial mini-exon or poly(T) primers along with PFR-specific primers. This revealed a full-length, fully processed mRNA that clearly encoded a homologue of a major PFR protein (Fig. 1A). We used the same strategy to isolate a PFR gene from the endosymbiont-bearing species C. oncopelti and genes from the nonendosymbiont-bearing trypanosomes C. fasciculata and H. megaseliae and, as a control, the previously described genes of T. brucei.
FIG. 1.
C. deanei possesses a gene encoding PFR1. (A) Alignment of the major PFR proteins from C. deanei, L. major, and T. brucei. Black shading indicates identical residues, and grey shading indicates similar residues. (B) Phylogenetic analysis of PFR sequences of Kinetoplastida. The MP unrooted phylogram shown was inferred from protein alignments. Topology support from 100 bootstrap replicates using either MP or NJ methods are shown next to nodes (MP/NJ). Partial sequences are shown in grey.
Figure 1A shows an alignment of the putative PFR protein from C. deanei with PFR1 and PFR2 from T. brucei and L. major. The encoded protein is 591 amino acids in length and has a predicted pI of 5.28—consistent with other known major PFR proteins. Phylogenetic analyses showed unambiguously that the proteins encoded by the endosymbiont-bearing species (C. deanei and C. oncopelti) are PFR1 orthologues (Fig. 1B). In contrast, both PFR1 and PFR2 sequences were isolated from C. fasciculata, H. megaseliae, and T. brucei.
C. deanei PFR1 is a single copy gene.
To date, the major PFR protein genes have been found to be present as tandemly duplicated arrays in all the kinetoplastids for which information is available (Table 1). This is thought to be a consequence of a need for high levels of mRNA from these genes. The newly identified C. deanei PFR gene (CdePFR1) was used as a probe in Southern hybridization to restriction endonuclease-digested C. deanei genomic DNA in an assay of gene copy number. This showed that C. deanei PFR1 is a single-copy gene (data not shown), in comparison to the multiple copies present in endosymbiont-lacking species. This conclusion was reinforced by the results of a strategy to amplify tandemly repeated PFR genes, which produced amplicons from the species with multiple copies of PFR1, but not from C. deanei or C. oncopelti (data not shown).
TABLE 1.
Distribution of PFR1 copy number across the Kinetoplastida
| Species (strain) | PFR1 copy no.a | Reference |
|---|---|---|
| T. brucei (927) | 5 | www.sanger.ac.uk/Projects/T_brucei |
| T. brucei (427) | 4 | 11 |
| L. major | 3 | www.sanger.ac.uk/Projects/L_major |
| L. mexicana | 4 | 29 |
| H. megaseliae | ≥2 | This study |
| C. fasciculata | ≥2 | This study |
| C. oncopelti | 1 | This study |
| C. deanei | 1 | This study |
Copy number per haploid genome.
Complementation experiments.
Phylogenetic inference using a maximum-parsimony method indicates that CdePFR1 protein has accumulated a greater number of changes since the last common ancestor than have PFR1 proteins from endosymbiont-lacking species (as shown by the branch lengths in Fig. 1B). We addressed whether CdePFR1 is a functional PFR protein (in spite of sequence divergence) by using a complementation approach. Maga et al. (29) have previously described the L. mexicana double-gene-knockout line Δpfr1::NEO Δpfr1::HYG—referred to here simply as Δpfr1— which does not assemble a native PFR structure and shows severely reduced swimming motility. We complemented this cell line with an episomal copy of either C. deanei or L. mexicana PFR1, to produce cell lines Δpfr1[Cde] and Δpfr1[Lmx], respectively.
We used the MAb L13D6 (which recognizes both PFR1 and PFR2 in T. brucei but only PFR1 in Leishmania spp.) to check for PFR1 expression and correct localization in complemented cell lines. In these immunofluorescence experiments, a bright signal was observed along the entire flagellum of wild-type and both complemented cell lines, Δpfr1[Cde] and Δpfr1[Lmx] (Fig. 2A). This signal was also seen in cells that had been detergent extracted to produce cytoskeletons, demonstrating a tight association with the axoneme (data not shown). These results show the correct targeting and incorporation of CdePFR1 into the flagellar cytoskeleton of L. mexicana.
FIG. 2.
Cells complemented with C. deanei PFR1 build a PFR structure of reduced thickness that incorporates endogenous PFR2. Wild-type L. mexicana samples are shown alongside PFR1 deletion mutants (Δpfr1) and mutants complemented with L. mexicana PFR1 (Δpfr1[Lmx]) and C. deanei PFR1 (Δpfr1[Cde]). (A) Phase-contrast microscopy and corresponding immunofluorescence images. DNA is labeled with DAP1 (blue), and the PFR is identified by the L13D6, anti-PFR1 MAb (red). Scale bars, 5 μm. (B) Western blot analysis of cell lines. Protein samples from 2 × 107 cells were separated by SDS-PAGE and analyzed with MAb F-4, which recognizes both-PFR1 and PFR2 in Leishmania spp. Whole-cell protein (wc) was separated alongside cell equivalents of detergent-soluble (sol) and insoluble (ins) fractions. (C) Thin-section TEM of flagella from the cell lines. The positions of the axoneme (Ax) and three domains of the PFR—proximal (p), intermediate (i), and distal (d)—are indicated on the wild-type. Scale bar, 200 nm. (D) Negatively stained whole-mount cytoskeletons of complemented cell lines. Axoneme (Ax) and PFR structures are indicated. Scale bar, 200 nm.
Given that CdePFR1 protein assembles into the Leishmania PFR structure in complemented cells and remains associated on detergent extraction, we then asked if it was able to recruit other PFR proteins to the structure. SDS-PAGE and Western blot analysis using MAb F4 (which recognizes both PFR1 and PFR2 in Leishmania spp.) were performed on whole-cell lysates and detergent-soluble and -insoluble fractions from L. mexicana cells lines. In the absence of PFR1 in the deletion mutant Δpfr1, PFR2 accumulates mostly in the detergent-soluble fraction (Fig. 2B). However, when PFR1 is expressed in the complemented strains, PFR2 is recruited to the cytoskeleton fractions, although some remains detergent soluble. This is true of both Δpfr1[Lmx] and Δpfr1[Cde] complemented lines (Fig. 2B). Therefore, not only is CdePFR1 incorporated into the leishmanial Δpfr1 cytoskeleton but also it enables the recruitment of endogenous LmxPFR2 to the flagellum. These results are consistent with the model of PFR organization (29), in which PFR1 and PFR2 are present throughout the structure although only PFR1 is involved in attachment to the axoneme.
TEM of thin sections and whole-mount cytoskeletons was used to identify the level of structural organization of the assembled PFR within the flagellum of complemented cell lines. Seen in cross-section, the PFR of wild-type cells consists of three ultrastructural domains, named the proximal, intermediate, and distal domains in order of increasing distance from the central axis of the axoneme (Fig. 2C). The proximal domain is connected via bridges to axonemal doublets 4 to 7, maintaining the tight association of axoneme and PFR. In the deletion mutant Δpfr1, the main PFR structure was absent and only the connecting bridges to the axoneme were seen (Fig. 2C and D). This phenotype is rescued in the Δpfr1[Lmx] strain, where a wild-type PFR was observed in the flagellum. In Δpfr1[Lmx] cells, all three domains of the PFR were restored. Interestingly, although the deletion mutant complemented with the C. deanei PFR1, Δpfr1[Cde], constructed a major portion of the PFR, the structure lacked much of the detailed intermediate and distal organisation seen in wild-type Leishmania and Δpfr1[Lmx] cells (n > 100; Fig. 2C and D).
Wild-type L. mexicana swims actively in culture. On the other hand, L. mexicana PFR null mutants—Δpfr1 and Δpfr2— display severe reduction in flagellar and cellular motility (28). Having seen that the C. deanei PFR1 could enable the elaboration of a substantial PFR in the Leishmania null mutant, Δpfr1, we next asked if this endowed the cells with increased mobility. We used time-lapse microscopy to monitor the cellular motility of the L. mexicana cell lines produced here. Figure 3 shows the distribution of individual cell speeds and also the mean speeds for each cell line. These data show that expression of episomally encoded LmxPFR1 in Δpfr1[Lmx] essentially rescues the defective motility phenotype (two-sample unpaired t test; Δpfr1[Lmx] versus Δpfr1 mean speed; P = 0.002), although the full wild-type mean speed could not be reached (Δpfr1[Lmx] versus wild type; P = 0.003). Expression of CdePFR1 also significantly increased cellular motility (Δpfr1[Cde] versus Δpfr1; P = 0.006). Interestingly, the mean cell speeds of Δpfr1[Lmx] and Δpfr1[Cde] were not significantly different (P = 0.11), despite Δpfr1[Cde] cells clearly not being able to reach the same maximal speeds as Δpfr1[Lmx] cells (as shown by the distribution in Fig. 3A).
FIG. 3.
C. deanei PFR1 partially rescues the L. mexicana Δpfr1 motility defect. (A). Distribution of cell speeds for wild-type (wt), Δpfr1, Δpfr1[Lmx], and Δpfr1[Cde] cell lines. Speeds shown are mean for each cell (of n total) over a 60-s period. (B) Mean speeds of cell lines. Standard errors are represented by bars.
The results shown here indicate that the endosymbiont-bearing kinetoplastid C. deanei—previously described as lacking a PFR—encodes a functional, expressed PFR1 protein that possesses all necessary information to be transported and incorporated into the flagellar cytoskeleton of the PFR-containing species L. mexicana. CdePFR1 interacts with the axoneme-connecting bridges and recruits endogenous LmxPFR2 to assemble a PFR structure, albeit reduced in thickness and intermediate and distal organization compared to that of Leishmania itself. In addition, this structure is able to partially rescue the motility defect of L. mexicana PFR1 deletion mutants.
The crithidial PFR.
The functional expression of CdePFR1 in an exogenous system and the knowledge that its RNA was present in C. deanei cells clearly raised the issue of whether the CdePFR1 protein was actually expressed in C. deanei. A single band of CdePFR1 was detected by Western blot analysis using the MAb L13D6 against whole-cell and detergent-insoluble protein preparations (Fig. 4A). To exclude the possibility of cross-reactivity with an epitope-containing unrelated protein, the 75-kDa band detected in C. deanei cytoskeletons was sequenced by ESI-MS/MS. The peptide sequences obtained were all consistent with the predicted protein of the CdePFR1 ORF (Fig. 1A).
FIG. 4.
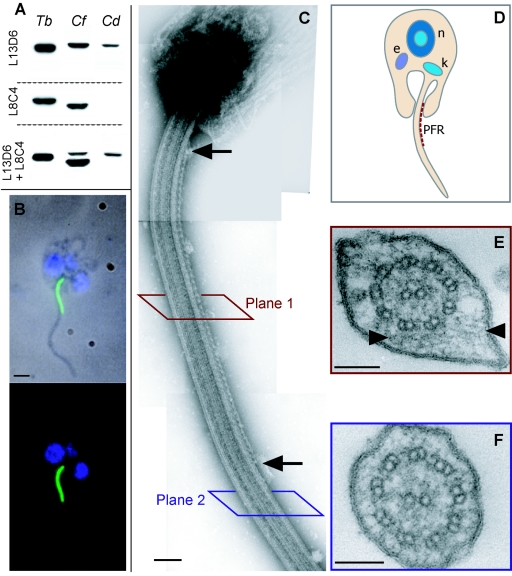
The PFR of C. deanei. (A) Western blot analysis of major PFR proteins. Protein samples from 0.5 × 107, 1 × 107, and 2 × 107 cytoskeletons of T. brucei, C. fasciculata, and C. deanei, respectively, were separated by SDS-PAGE and analysed with MAb L13D6 (reacts with PFR1 in Crithidia spp. and with PFR1 and PFR2 in T. brucei) or MAb L8C4 (reacts with PFR2 only). A Western blot using both MAbs together is also shown. Note that PFR1 and PFR2 of T. brucei are not resolved on these blots. (B) Phase-contrast microscopy and corresponding immunofluorescence image for a C. deanei coanomastigote. DNA is labeled with DAPI (blue), and the PFR is identified by MAb L13D6 (green). Scale bars, 1 μm. (C) Ultrastructure of C. deanei PFR visualized by TEM of negatively stained whole-mount cytoskeletons. Arrows show the limits of the PFR structure along the flagellum. Scale bar, 200 nm. (D) Schematic representation of C. deanei showing the position of the PFR (also the kinetoplast [k], nucleus [n] and endosymbiont [e]). (E and F) TEM on thin sections reveal a putative PFR structure (arrowheads) in some sections (E), but much of the flagellum is free of PFR (F). Scale bars, 100 nm.
The same L13D6 antibody was used in immunofluorescence analysis to label C. deanei. A strong signal was detected from inside the flagellar pocket up to the first third of the flagellar length in all cells observed (Fig. 4B). Notably, this flagellar signal was sensitive to the long fixation times (>5 min) and higher concentrations of cross-linking fixatives (>2% paraformaldehyde) that are often used for immunofluorescence. For that reason, we used fixation conditions that did not preserve cell morphology as well as other methods (see Materials and Methods) but allowed the detection of a PFR signal in C. deanei.
We then carefully reexamined the ultrastructure of the flagellum in C. deanei. Analysis of negatively-stained whole-mount cytoskeletons was most informative (Fig. 4C). These images revealed that this species does indeed posses a PFR. It is, however, much reduced in width and length compared to that of endosymbiont-lacking kinetoplastids and can be visualised in these negatively stained preparations extending along one side of the flagellum as a thin punctuate line. Importantly, the length and position of the C. deanei PFR are consistent with the length of the paraflagellar signal seen by immunofluorescence (Fig. 4B). This much reduced form explains why many previous thin-section studies concluded that the PFR was absent. Thin cross sections of the flagellum along most of its length (e.g., plane 2 in Fig. 4C) should produce images of flagella lacking PFR, as in Fig. 4F (representative of 33 of 41 cross sections). Only when we searched more thoroughly did we obtain flagellar cross sections that exhibited a small extra-axonemal structure that we interpret as being the reduced crithidial PFR (8 of 41 cross sections). This structure is much less obvious than the unambiguous structure seen by negative staining but is always between microtubule doublets 4 and 7 and is attached to the axoneme by putative connecting bridges (Fig. 4E).
PFR2 appears to be absent from endosymbiont-bearing species.
Alongside our identification of PFR1 sequences from C. deanei and C. oncopelti, we also undertook an extensive search for PFR2 in these organisms. A large number of techniques were employed (a more detailed account of each can be found in the relevant section of Materials and Methods). PCR from genomic DNA, using degenerate PFR primers, resulted in amplicons of both PFR1 and PFR2 fragments from C. fasciculata, H. megaseliae, and T. brucei templates (number of independent clones PFR1:PFR2-Cf 4:3, Hm 7:5, Tb 3:3), but produced only PFR1 fragments from C. deanei and C. oncopelti (Cd 11:0, Co 6:0). Similar results were achieved in RT-PCR experiments using different primer sets (Cf 0:14, Tb 4:2, Cd 24:0, Co 5:0). Moreover, two sets of PFR2-specific primers produced amplicons when used with C. fasciculata, H. megaseliae, and T. brucei gDNA templates, but not with C. deanei or C. oncopelti. Southern blot analyses using T. brucei, L. mexicana, and C. fasciculata PFR2 coding regions as probes could easily identify PFR2 sequences in gDNA from each of C. fasciculata, H. megaseliae, and T. brucei (i.e., even when LmxPFR2 was used to probe for TbrPFR2) but did not unambiguously identify a sequence in C. deanei or C. oncopelti gDNA (data not shown).
Searches for PFR2 protein showed similar results to those for the PFR2 gene. A MAb specific for PFR2 protein (L8C4; Fig. 4A) produced no strong reaction to protein samples from C. deanei and C. oncopelti separated by SDS-PAGE. An antibody reacting with both PFR1 and PFR2 proteins in most species (MAb F4 [26]), produced a strong reaction to only one band in one-dimensional SDS-PAGE with protein samples from C. deanei and C. oncopelti. Furthermore, protein sequencing from one-dimensional SDS-PAGE and also two-dimensional electrophoresis demonstrated that both major PFR proteins could be found in C. fasciculata and T. brucei protein preparations. In C. deanei protein samples, the strongly reacting band contained PFR1 protein, as expected, whereas taking weakly reacting bands revealed only abundant, unrelated proteins (Hsp70 and Hsp80). Each of the above techniques has limitations. However, taken together, it is very striking that we have found no evidence for the presence of PFR2 protein nor encoding DNA or RNA in kinetoplastid species that harbor the endosymbiont.
DISCUSSION
Evolutionary distribution of the PFR.
The PFR is an extra-axonemal structure that appears to be restricted to the sister orders Kinetoplastida and Euglenida. Dinoflagellates were once included in the PFR-containing group because of the presence of an extra-axonemal structure within the flagellum. However, these structures do not resemble the paracrystalline PFR of kinetoplastids and euglenoids. In addition, these two groups of excavate protozoa are phylogenetically distinct from the dinoflagellates (phylum Alveolata), from which they diverged very early in eukaryote evolution (8). At present, no sequence is available for proteins of the dinoflagellate para-axial structure; however, it is most probable that this structure and the PFR are examples of convergent, rather than divergent, evolution, since it is known that extra-axonemal structures have evolved independently several times across eukaryotes.
Previous structural studies have reported a lack of PFR in kinetoplastids possessing an endosymbiotic bacterium (12, 17). During the early phases of the identification of PFR1 and PFR2 in T. brucei, one antibody used gave a weak Western blot signal to proteins of C. deanei but not C. oncopelti (38). The suggestion was made, therefore, that if this signal was from a bona fide PFR protein, it was not incorporated into a structural entity (38). There are, of course, many explanations for such discrepant observations. Here, we have resolved this issue by demonstrating that the endosymbiont-bearing C. deanei does indeed possess a gene encoding a major PFR protein (PFR1) and that this protein is recognized by antibodies raised to PFR proteins from other species. Moreover, this protein forms part of a flagellar structure in C. deanei that, although reduced, is undoubtedly recognizable as a PFR.
It is worth noting that the PFR1 phylogeny shown here agrees with those inferred from other molecular data (14, 20) in supporting the monophyly of the endosymbiont-bearing species (as would be expected for a rare event such as enslavement). Our phylogeny, in common with the rRNA data (20), also suggests that non-endosymbiont-bearing Crithidia species diverged from those possessing an endosymbiont soon after the divergence of the leishmanial and trypanosomal lines and that the nomenclature of the kinetoplastids often does not reflect evolutionary relatedness (for example, the genera Crithidia and Herpetomonas encompass both endosymbiont-bearing and endosymbiont-lacking species and are not monophyletic taxa).
PFR structural organization.
Visualized by thin-section TEM, the PFR consists of three morphologically distinct regions. These are named the proximal, intermediate, and distal domains, according to their position relative to the axoneme in cross section (Fig. 2C). There are some variations in form, size, and arrangement, but overall the PFRs of all kinetoplastids exhibit a very similar tripartite pattern of construction (1, 16, 18). The PFR has a permanent position relative to the flagellum axoneme, with the proximal region linked to axonemal doublets 4 through 7. Genetic ablation of the major PFR proteins of the L. mexicana flagellum has yielded a set of structural assembly rules (29). (i) The connecting bridges that link the PFR to the axoneme do not contain either PFR1 or PFR2, and they assemble in the absence of a native PFR structure. A possible component of these bridges in T. brucei is the protein I17, which has been detected between the PFR and the axoneme (25). (ii) PFR1 is expressed and targeted to the flagellum in the absence of PFR2, where it forms stable associations with axoneme-connecting bridges. (iii) PFR2 is targeted to the flagellum but cannot be assembled into the PFR in the absence of PFR1. (iv) PFR2 incorporation is required for the subsequent assembly of the intermediate and distal domains. (v) PFR1 and PFR2 are present in each of the three domains of the PFR.
Episomal complementation of L. mexicana PFR1 deletion mutants with C. deanei PFR1 allowed the localization of CdePFR1 along the length of the flagellum in a manner similar to that of LmxPFR1. In these cells, CdePFR1 was stably bound to the axoneme and was able to stably recruit endogenous LmxPFR2. However, L. mexicana Δpfr1[CdePFR1] cells were not able to build full PFR structures; they lacked the intermediate and distal domains. This indicates that, in Leishmania, PFR1 not only binds to the axonemal bridges as described above but also is necessary for the construction of the PFR intermediate domain. This second function is evidently not merely a product of PFR binding, because CdePFR1 retains this function. Moreover, the processes of binding to axonemal bridges and seeding (in the presence of PFR2) the construction of the intermediate domain are at least partially independent, since the CdePFR1 protein is able to substitute in the former, but not in the latter.
Wild-type C. deanei cells build a reduced PFR that lacks intermediate and distal domains. In these cells, the proximal domain is also much smaller than those of Leishmania or Trypanosoma species. Interestingly, although we could readily identify a homologue of PFR1 in C. deanei, we found no evidence of PFR2 when using a wide variety of techniques. Moreover, in terms of ultrastructure, the PFR of C. deanei is not like that formed in L. mexicana Δpfr1[CdePFR1] but is reminiscent of the residual structure formed in L. mexicana PFR2 deletion mutants. Hence, the cryptic nature of the PFR of endosymbiont-bearing kinetoplastids appears to be linked to the selective loss of PFR2 (but retention of PFR1).
The major proteins of the PFR are highly conserved within the kinetoplastids. Despite this, the exact ultrastructure of the PFRs varies from species to species. Our data demonstrate that (in addition to likely major gene losses such as in the endosymbiont-containing kinetoplastids) small variations in the PFR primary sequence can result in significant changes in the overall PFR structure.
PFR function.
From the data presented here, we propose that the cryptic PFR built by C. deanei is sufficient to perform the functions common to all kinetoplastids, namely, motility and attachment to the invertebrate host epithelium. Expression of C. deanei PFR1 in L. mexicana Δpfr1 cells was able to rescue the motility defect of the null cells. However, this rescue was only partial: complemented cells moved faster than did Δpfr1 cells but did not achieve the rates of movement seen in the fastest wild-type cells. This may simply be the result of expressing a nonoptimal version of PFR1, but it is tempting to speculate that it could be linked to the lack of intermediate and distal PFR domains.
It has been shown recently that the PFR is not simply an architectural attribute but, rather, acts as a matrix into which enzymes such as adenylate kinases (34) and calmodulin (35) can be built. These proteins, and possibly many others, are thought to regulate the metabolic environment of the flagellum. Given the small diameter and reduced structural complexity of the C. deanei PFR, one may ask whether the PFR in species with an endosymbiont can provide an adequate platform for such regulatory enzymes. If this is the case, how can the crithidial flagellum perform the same functions as in any other kinetoplastid?
One obvious candidate to consider when looking for metabolic alterations is the endosymbiont. This organelle carries sufficient information to code for a complex set of proteins, and it is capable of independent protein synthesis (33). Moreover, it relieves the host cell from dependence on exogenous heme (9), ornithine metabolism enzymes (7), purines, and various amino acids (32). Therefore, it seems plausible that the endosymbiont could compensate the host with some enzymatic functions that would allow the loss of all but a rudimentary PFR. However, when making such suggestions, it must be remembered that both possession of the endosymbiont and lack of a full PFR are traits in the kinetoplastids restricted to a single clade. Since both are monophyletic, the two may result from entirely independent events.
Novel PFR components.
Until recently, little progress has been made in the molecular identification of minor PFR components. In this regard, comparative proteomic approaches hold much promise—a comparison of T. brucei cells with and without ablation of PFR2 by RNA interference has recently been employed to identify four novel PFR components including the adenylate kinases previously mentioned (34). The creation in this study of a complemented cell line (Δpfr1[Cde]) that builds a proximal domain but no intermediate or distal domains may be a valuable tool in a more detailed description of PFR composition.
Whilst demonstrating that the extra-axonemal PFR is not absent in endosymbiont-bearing kinetoplastids (as previously reported [12, 17]) and that this organelle is therefore most probably universal in the kinetoplastids and euglenoids, we have been able to provide further insights into the relationship of molecular diversity and PFR morphotypes in these fascinating protists.
Acknowledgments
We thank Diane McMahon-Pratt for the MAb F4, Emmanuel Tetaud and Alan H. Fairlamb for pNUS-GFPcB, David Woolley and Paulo Marcio de Faria e Silva for valuable discussion, Sue Vaughan for help with TEM micrographs, and Ben Thomas for ESI-MS/MS analyses.
This work is supported by grants from the Wellcome Trust, CNPq, and PRONEX-MCT, Brasil. K.G. is a Wellcome Trust Principal Research Fellow. C.G. is supported by CAPES, Brasil.
REFERENCES
- 1.Al-Shammary, R. J., N. M. Shoukrey, S. E. Al-Shewemi, E. A. Ibrahim, M. A. Al-Zahrani, and A. S. Al-Tuwaijri. 1995. Leishmania major: in vitro ultrastructural study of the paraxial rod of promastigotes. Int. J. Parasitol. 25:443-452. [DOI] [PubMed] [Google Scholar]
- 2.Bastin, P., T. Sherwin, and K. Gull. 1998. Paraflagellar rod is vital for trypanosome motility. Nature 391:548-548. [DOI] [PubMed] [Google Scholar]
- 3.Bastin, P., A. Bagherzadeh, K. R. Matthews, and K. Gull. 1996. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 77:235-239. [DOI] [PubMed] [Google Scholar]
- 4.Bastin, P., T. J. Pullen, T. Sherwin, and K. Gull. 1999. Protein transport and flagellum assembly dynamics revealed by analysis of the paralysed trypanosome mutant snl-1. J. Cell Sci. 112:3769-3777. [DOI] [PubMed] [Google Scholar]
- 5.Bastin, P., T. J. Pullen, F. F. Moreira-Leite, and K. Gull. 2000. Inside and outside of the trypanosome flagellum: a multifunctional organelle. Microbes Infect. 2:1865-1874. [DOI] [PubMed] [Google Scholar]
- 6.Brooker, B. E. 1971. Flagellar attachment and detachment of Crithidia fasciculata to the gut wall of Anopheles gambiae. Protoplasma 73:191-202. [DOI] [PubMed] [Google Scholar]
- 7.Camargo, E. P., and E. Freymuller. 1977. Endosymbiont as supplier of ornithine carbamoyl transferase in a trypanosomatid. Nature 270:52-53. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith, T., and E. E.-Y. Chao. 2003. Phylogeny of Choanozoa, Apusozoa, and other Protozoa and early eukaryote megaevolution. J. Mol. Evol. 56:540-563. [DOI] [PubMed] [Google Scholar]
- 9.Chang, K.-P., and W. Trager. 1974. Nutritional significance of symbiotic bacteria in two species of the hemoflagellates. Science 183:531-532. [DOI] [PubMed] [Google Scholar]
- 10.Current, W. L. 1980. Cryptobia sp. in the snail Triadopsis multilineata (Say): fine structure of attached flagellates and their mode of attachment to the spermatheca. J. Protozool. 27:278-287. [Google Scholar]
- 11.Deflorin, J., M. Rudolf, and T. Seebeck. 1994. The major components of the paraflagellar rod of Trypanosoma brucei are two similar, but distinct proteins which are encoded by two different gene loci. J. Biol. Chem. 269:28745-28751. [PubMed] [Google Scholar]
- 12.de Souza, W., and M. C. M. Motta. 1999. Endosymbiosis in protozoa of the Trypanosomatidae family. FEMS Microbiol. Lett. 173:1-8. [DOI] [PubMed] [Google Scholar]
- 13.Du, Y., G. McLaughlin, and K.-P. Chang. 1994. 16S ribosomal DNA sequence identities of β-proteobacterial endosymbiont in three Crithidia species. J. Bacteriol. 176:3081-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du, Y., D. A. Maslov, and K.-P. Chang. 1994. Monophyletic origin of β-division proteobacterial endosymbionts and their coevolution with insect trypanosomatid protozoa Blastocrithidia culicis and Crithidia spp. Proc. Natl. Acad. Sci. USA 91:8437-8441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fampa, P., M. S. Correa-da-Silva, D. C. Lima, S. M. P. Oliveira, M. C. M. Motta, and E. M. B. Saraiva. 2003. Interaction of insect trypanosomatids with mosquitoes, sand fly and the respective insect cell lines. Int. J. Parasitol. 33:1019-1026. [DOI] [PubMed] [Google Scholar]
- 16.Farina, M., M. Attias, T. Souto-Padron, and W. de Souza. 1986. Further studies on the organization of the paraxial rod of trypanosomatids. J. Protozool. 33:552-557. [Google Scholar]
- 17.Freymuller, E., and E. P. Camargo. 1981. Ultrastructural differences between species of trypanosomatids with and without endosymbiont. J. Protozool. 28:175-182. [DOI] [PubMed] [Google Scholar]
- 18.Fuge, H. 1969. Electron microscopic studies of the intraflagellar structures of trypanosomes. J. Protozool. 16:160-166. [DOI] [PubMed] [Google Scholar]
- 19.Gadelha, C., J. H. LeBowitz, J. Manning, T. Seebeck, and K. Gull. 2004. Relationships between the major kinetoplastid paraflagellar rod proteins: a consolidating nomenclature. Mol. Biochem. Parasitol. 136:113-115. [DOI] [PubMed] [Google Scholar]
- 20.Hollar, L., J. Lukes, and D. A. Maslov. 1998. Monophyly of endosymbiont containing trypanosomatids: phylogeny versus taxonomy. J. Eukaryot. Microbiol. 45:293-297. [DOI] [PubMed] [Google Scholar]
- 21.Holwill, M. E. 1965. The motion of Strigomonas oncopelti. J. Exp. Biol. 42:125-137. [DOI] [PubMed] [Google Scholar]
- 22.Holwill, M. E., and J. L. McGregor. 1974. Micromanipulation of the flagellum of Crithidia oncopelti. I. Mechanical effects. J. Exp. Biol. 60:437-444. [DOI] [PubMed] [Google Scholar]
- 23.Holwill, M. E., and J. L. McGregor. 1975. Control of flagellar wave movement in Crithidia oncopelti. Nature 255:157-158. [DOI] [PubMed] [Google Scholar]
- 24.Holwill, M. E., and J. L. McGregor. 1976. Effects of calcium on flagellar movement in the trypanosome Crithidia oncopelti. J. Exp. Biol. 65:229-242. [DOI] [PubMed] [Google Scholar]
- 25.Imboden, M., N. Muller, A. Hemphill, R. Mattioli, and T. Seebeck. 1995. Repetitive proteins from the flagellar cytoskeleton of African trypanosomes are diagnostically useful antigens. Parasitology 110:249-258. [DOI] [PubMed] [Google Scholar]
- 26.Ismach, R., C. M. Cianci, J. P. Caulfield, P. J. Langer, A. Hein, and D. McMahon-Pratt. 1989. Flagellar membrane and paraxial rod proteins of Leishmania: characterization employing monoclonal antibodies. J. Protozool. 36:617-624. [DOI] [PubMed] [Google Scholar]
- 27.Kohl, L., T. Sherwin, and K. Gull. 1999. Assembly of the paraflagellar rod and the flagellum attachment zone complex during the Trypanosoma brucei cell cycle. J. Eukaryot. Microbiol. 46:105-109. [DOI] [PubMed] [Google Scholar]
- 28.Maga, J. A., and J. H. LeBowitz. 1999. Unravelling the kinetoplastid paraflagellar rod. Trends Cell Biol. 9:409-413. [DOI] [PubMed] [Google Scholar]
- 29.Maga, J. A., T. Sherwin, S. Francis, K. Gull, and J. H. LeBowitz. 1999. Genetic dissection of the Leishmania paraflagellar rod, a unique flagellar cytoskeleton structure. J. Cell Sci. 112:2753-2763. [DOI] [PubMed] [Google Scholar]
- 30.Moreira-Leite, F. F., W. de Souza, and N. L. da Cunha-e-Silva. 1999. Purification of the paraflagellar rod of the trypanosomatid Herpetomonas megaseliae and identification of some of its minor components. Mol. Biochem. Parasitol. 104:131-140. [DOI] [PubMed] [Google Scholar]
- 31.Moreira-Leite, F. F., T. Sherwin, L. Kohl, and K. Gull. 2001. A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science 294:587-591. [DOI] [PubMed] [Google Scholar]
- 32.Mundim, M. H., I. Roitman, M. A. Hyermans, and E. W. Kitajima. 1974. Simple nutrition of Crithidia deanei, a reduvid trypanosomatid with an endosymbiont. J. Protozool. 21:518-521. [DOI] [PubMed] [Google Scholar]
- 33.Novak, E., E. F. Haapalainem, S. Silva, and J. F. Silveira. 1988. Protein synthesis in isolated symbiont from the flagellate protozoon Crithidia deanei. J. Protozool. 35:375-378. [Google Scholar]
- 34.Pullen, T. J., M. L. Ginger, S. J. Gaskell, and K. Gull. 2004. Protein targeting of an unusual, evolutionary conserved adenylate kinase to a eukaryotic flagellum. Mol. Biol. Cell 15:3257-3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ridgley, E., P. Webster, C. Patton, and L. Ruben. 2000. Calmodulin-binding properties of the paraflagellar rod complex from Trypanosoma brucei. Mol. Biochem. Parasitol. 109:195-201. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 37.Santrich, C., L. Moore, T. Sherwin, P. Bastin, C. Brokaw, K. Gull, and J. H. LeBowitz. 1997. A motility function for the paraflagellar rod of Leishmania parasites revealed by PFR-2 gene knockouts. Mol. Biochem. Parasitol. 90:95-109. [DOI] [PubMed] [Google Scholar]
- 38.Schlaeppi, K., J. Deflorin, and T. Seebeck. 1989. The major component of the paraflagellar rod of Trypanosoma brucei is a helical protein that is encoded by two identical, tandemly linked genes. J. Cell Biol. 109:1695-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugrue, P., M. R. Hirons, J. U. Adam, and M. E. Holwill. 1988. Flagellar wave reversal in the kinetoplastid flagellate Crithidia oncopelti. Biol. Cell 63:127-1331. [DOI] [PubMed] [Google Scholar]
- 40.Tetaud, E., I. Lecuix, T. Sheldrake, T. Baltz, and A. H. Fairlamb. 2002. A new expression vector for Crithidia fasciculata and Leishmania. Mol. Biochem. Parasitol. 120:195-204. [DOI] [PubMed] [Google Scholar]
- 41.Tetley, L., and K. Vickerman. 1985. Differentiation in Trypanosoma brucei: host-parasite cell junctions and their persistence during acquisition of the variable antigen coat. J. Cell Sci. 74:1-19. [DOI] [PubMed] [Google Scholar]
- 42.Vickerman, K. 1994. The evolutionary expansion of the trypanosomatid flagellates. Int. J. Parasitol. 24:1317-1331. [DOI] [PubMed] [Google Scholar]
- 43.Wickstead, B., K. Ersfeld, and K. Gull. 2004. The small chromosomes of Trypanosoma brucei involved in antigenic variation are constructed around repetitive palindromes. Genome Res. 14:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodward, R., M. J. Carden, and K. Gull. 1994. Molecular characterization of a novel, repetitive protein of the paraflagellar rod in Trypanosoma brucei. Mol. Biochem. Parasitol. 67:31-39. [DOI] [PubMed] [Google Scholar]