Abstract
SPS1 encodes a sporulation-specific protein with homology to the Ste20/p21-activated kinase family. Deletion of SPS1 impinges on the formation of the spore wall, which surrounds each of the haploid nuclei generated by the meiotic divisions. Here, we demonstrate that the new internal membranes that surround the meiotic nuclei appear normal in the absence of Sps1p. Analyses of spore wall layers by immunohistochemistry suggest that the inner layers are not efficiently deposited. The defect in spore wall morphogenesis is most likely a consequence of mislocalization of enzymes required for the synthesis of the spore wall layers as both Chs3p, the major chitin synthase in yeast, and Gsc2/Fks2p, a glucan synthase transcriptionally upregulated during sporulation, fail to reach the prospore membrane in the sps1 mutant. Furthermore, localization of Chs3p to the prospore membrane is not dependent on Shc1p, a sporulation-specific homolog of Chs4p, which is required for recruitment of Chs3p to the bud neck in vegetative cells. Sps1p colocalized with Chs3p to peripheral and internal punctate structures and prospore membranes. We propose that Sps1p promotes sporulation, in part, by regulating the intracellular movement of proteins required for spore wall formation.
Gametogenesis is an essential process allowing eukaryotic organisms to reproduce sexually. It requires coordination of many diverse, yet interrelated molecular events: genetic exchange, haploidization, and cellular differentiation. The reduction in ploidy occurs through meiosis, in which haploid cells are derived from a diploid progenitor via one round of DNA replication followed by two successive rounds of chromosome segregation. The meiotic cell cycle is coupled to a program of cellular differentiation, which ultimately packages the haploid nuclei into gametes. In the yeast Saccharomyces cerevisiae, meiosis is coupled to the process of spore formation, resulting in four haploid nuclei each encapsulated in a membrane (the prospore membrane) and spore wall. The sporulation program commences when diploid MATa/MATα cells are starved for nitrogen in the presence of a nonfermentable carbon source (reviewed in reference 13). These nutritional signals ultimately lead to transcriptional induction of a large number of genes, which are classified according to their temporal expression pattern (5, 22). Early genes are involved in chromosome alignment and recombination, while middle genes regulate chromosome segregation and prospore membrane formation. Mid-late and late genes are responsible for synthesis and deposition of spore wall components.
Prospore membrane formation initiates at the spindle pole body (SPB), the equivalent of the mammalian centrosome, originating from an outer meiotic plaque comprised of the meiosis-specific proteins Mpc70p/Spo21p, Mpc54p, Spo74p, and Ady4p (1, 11, 18). A complex of proteins, including Don1p, is located at the leading edge of the growing prospore membrane (11). The SPB appears to be a hub for the fusion of secretory vesicles originating from the Golgi apparatus to form the prospore membrane (17). As the prospore membrane engulfs haploid nuclei, it pinches off, resulting in a double membrane. The late-acting SEC genes, essential for post-Golgi trafficking, play a critical role in growth of the prospore membrane (17), as do Spo14p, a phospholipase D (26), and a meiosis-specific Sec9p/SNAP-25 homolog, Spo20p (17). The septin complex is also located at the leading edge of the prospore membrane (6) and is believed to facilitate the deposition of spore wall components, which are deposited within the lumen of the double membrane in a specific temporal order (33). Deposition of spore wall components may also originate at the SPB, as certain temperature-sensitive alleles of MPS1, required for duplication and assembly of SPB components in both mitosis and meiosis (3), display a phenotype similar to that of mutants defective in spore wall formation (32).
The spore wall contains four distinct layers: the first layer closest to the membrane consists of mannans and mannoproteins (33). The second layer is comprised of β-1,3-glucan. Both glucan and mannan are components of the vegetative cell wall. There are two glucan synthase genes in yeast; one, GSC2/FKS2 (hereafter referred to as GSC2), has been demonstrated to play a role in sporulation (16). The third layer contains chitosan, a meiosis-specific derivative of chitin. The CHS3 gene product is responsible for the majority of chitin synthesis and is required for formation of the chitosan spore layer (20, 37). Chitin is deacetylated by the sporulation-specific CDA1 and CDA2 gene products to form chitosan (4). The fourth layer of the spore wall contains dityrosine, which is visualized by electron microscopy (EM) as a darkly staining layer and which fluoresces under short-wave UV light. Dityrosine is synthesized from two tyrosine molecules by reactions involving the DIT1 and DIT2 gene products (2) and is translocated through the prospore membrane by a membrane-associated transporter, DTR1 (7). The third and fourth layers of the spore wall confer resistance to environmental stress. Once plated on rich medium, the spore germinates, sheds the spore wall, and reenters the mitotic cell cycle.
SPS1 is a sporulation-specific gene expressed during the meiotic divisions, classifying it as a middle sporulation gene (8). sps1 mutants have no vegetative phenotype but fail to form birefringent spores. sps1 cells induced in sporulation lack the chitosan and dityrosine layers of the spore wall as detected by EM, sensitizing the cells to environmental perturbations (8). Deletion of SPS1 also results in an altered transcriptional profile of mid-late and late genes (8). The SPS1 gene is predicted to encode a serine/threonine protein kinase with a molecular mass of approximately 55 kDa. Sps1p shares 42% identity in its kinase domain to Ste20p, a member of the p21-activated kinase family involved in the yeast pheromone response.
In this study, we demonstrate that prospore membrane formation appears unaltered in the absence of Sps1p; however, the inner layers of the spore wall are inefficiently deposited within the lumen of the double membrane. Chs3p and Gsc2p, key enzymes responsible for the synthesis of spore wall layers, fail to reach the prospore membrane in sps1 mutants. Furthermore, Sps1p colocalizes with Chs3p during sporulation, suggesting that Sps1p regulates the intracellular location of these enzymes for spore wall synthesis.
MATERIALS AND METHODS
Yeast genetic manipulations.
Routine growth and manipulation of S. cerevisiae strains were performed as described previously (24). Yeast strains used in this study are listed in Table 1.
TABLE 1.
Yeast strainsa
| Strain | Genotype | Source or reference |
|---|---|---|
| Y3690 | MATaHIS4 leu2-kho::LYS2ura3-1trp1::hisGlys2MATα his4 leu2::hisGho::LYS2 ura3-1 trp1::hisG lys2 | This study |
| Y592 | Y3690 but homozygous sps1::TRP1 | This study |
| Y3710 | Y592 + 2μm GFP-SPO14 LEU2 (pME1096) | This study |
| Y4998 | Y3690 + 2μm GFP-SPO14 LEU2 (pME1096) | This study |
| Y4145 | Y3690 + CEN4 SPR28-GFP URA3 (pSB19) | This study |
| Y4146 | Y592 + CEN4 SPR28-GFP URA3 (pSB19) | This study |
| YKS53-1 | MATaLEU2ho::hisGura3lys2DON1-eGFP-kanMXMATα leu2::hisG ho::hisG ura3 lys2 DON1-eGFP-kanMX | 11 |
| Y3947 | YKS53-1 but homozygous sps1::URA3 | This study |
| Y3059 | YKS53-1 but homozygous spo14::URA3 | This study |
| AN120 | MATaleu2ARG4RME1hoΔ::LYS2ura3trp1::hisGlys2 MATα leu2 arg4-NspI rme1Δ::LEU2 hoΔ::LYS2 ura3 trp1::hisG lys2 | 17 |
| Y4535 | AN120 but homozygous CHS3-GFP | This study |
| Y4537 | AN120 but homozygous GSC2-GFP | This study |
| Y4584 | Y4537 but homozygous sps1::TRP1 | This study |
| Y4620 | Y4535 but homozygous sps1::TRP1 | This study |
| Y4745 | AN120 but homozygous DTR1-GFP | This study |
| Y4741 | Y4745 but homozygous sps1::TRP1 | This study |
| Y4748 | AN120 but homozygous dtr1::kanMX | This study |
| Y4749 | Y4748 but homozygous sps1::TRP1 | This study |
| Y4743 | AN120 but homozygous SHC1-GFP | This study |
| Y4746 | Y4743 but homozygous sps1::TRP1 | This study |
| Y4716 | Y4535 but homozygous shc1::kanMX | This study |
| Y5050 | AN120 but homozygous GFP-SPS1 | This study |
| Y5088 | AN120 but heterozygous GFP-SPS1 and CHS3-dsRed | This study |
All strains are of SK1 background.
The lithium acetate procedure was used for DNA-mediated transformation of yeast cells (9). Gene replacements and disruptions (see below) were performed by the one-step method (25). dtr1::kanMX and shc1::kanMX, which replace the entire open reading frame with the marker kanMX4 (39), were made by PCR amplification of regions ∼300 bp upstream and downstream of the disrupted open reading frames from previously constructed knockout strains purchased from Research Genetics. Green fluorescent protein (GFP) and dsRed chromosomal fusions were constructed by using Longtine et al. (14) and Sheff and Thorn (31) reagents, respectively. All integrants were confirmed by PCR and the appropriate synthetic oligonucleotide primers.
Plasmids.
GFP-SPO14 was expressed from a high-copy-number (2μm) LEU2 plasmid (pME1096) and has been previously described (26). pKR466 (spo14::URA3, which removes 4,014 bp of the 5,052-bp open reading frame) was used to generate the spo14 chromosomal deletion/disruption and has been described previously (23). GFP-SPR28 was expressed from a low-copy-number (CEN) URA3 plasmid (pSB19) (38). p18-2 (sps1::URA3, which inserts URA3 at the 3′ end of SPS1) (21) and YIpΔST (sps1::TRP1, which inserts TRP1 at the 5′ end of SPS1) (8) were obtained from J. Segall (University of Toronto).
To construct the GFP-SPS1 fusion, the SPS1 promoter (500 bp upstream of the ATG) was amplified with a 5′ oligonucleotide primer containing a BglII site and a 3′ oligonucleotide primer containing a PacI site. The resulting PCR product was digested with BglII and PacI and inserted in place of the GAL1 promoter in the corresponding sites in pFA6a-kanMX6-PGAL1-GFP (14) to generate pME2330. This product was used as a template to amplify the fusion fragment for substitution at the SPS1 locus.
Analysis of GFP fusion proteins.
Strains expressing GFP-SPO14 (26), DON1-GFP (11), SPR28-GFP (38), DTR1-GFP, SHC1-GFP, CHS3-GFP, GSC2-GFP, GFP-SPS1, and CHS3-dsRED (Table 1) were grown and sporulated as described previously (27). At various times after induction of sporulation, living cells were examined by FITC (fluorescein isothiocyanate) or rhodamine optics on a Zeiss Axioskop 2 fluorescence microscope. To monitor progression in sporulation and ensure cells were at the appropriate stage, parallel cultures were fixed and stained with 4,6-diamidinophenylindole (DAPI).
Immunofluorescence of spore wall layers.
Our protocol is from the work of Tachikawa et al. (33). Cells were transferred to sporulation medium, and aliquots were removed at 5, 7, 9, and 11 h postinduction and fixed in 0.1 M K2HPO4-KH2PO4 and 3.7% formaldehyde. Cell pellets were resuspended in SHA buffer (1 M sorbitol, 0.1 M HEPES [pH 7.5], 5 mM NaN3) and spheroplasted with 27 μg of zymolyase 100T/ml in the presence of 10 mM dithiothreitol for 45 min at 30°C. Spheroplasts were permeabilized in SHA buffer plus 0.1% Triton X-100. Permeabilized cells were adhered to poly-l-lysine-coated slides and fixed in ice-cold methanol for 6 min and in acetone for 0.5 min. Slides were blocked for 10 min in WT buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1% nonfat milk, 0.1% Tween 20, 0.5 mg of bovine serum albumin/ml, 5 mM NaN3) and rinsed twice in WT buffer. Mouse monoclonal anti-β-1,3-glucan antibodies (1:3,000) (Aaron Neiman, State University of New York [SUNY], Stony Brook) were incubated with the cells for 2 h under humid conditions. Cells were washed 12 times in WT buffer and incubated with goat anti-mouse rhodamine-conjugated secondary antibodies (Molecular Probes, Eugene, Oreg.) at a 1:200 dilution and FITC-concanavalin A (FITC-ConA; 0.5 mg/ml) (Neta Dean, SUNY, Stony Brook) for 2 h in the dark. Following extensive washes with WT buffer, slides were mounted with ProLong antifade reagent (Molecular Probes) containing DAPI to monitor meiotic progression. Signals were detected with a Zeiss Axioskop microscope.
Calcofluor white staining was performed by resuspending fixed cells in SHA buffer plus 0.1% Triton X-100 containing 0.1 mg of calcofluor white/ml. The cells were washed twice in SHA buffer, harvested, and adhered to poly-l-lysine-coated slides. Slides were mounted with ProLong antifade reagent without DAPI. Fluorescence was visualized as described above under the DAPI channel.
Dityrosine assays.
For dityrosine assays, cells were incubated in sporulation medium for 24 h and collected by centrifugation. Cell pellets were resuspended in 1 ml of sterile H2O and then broken by vortexing with glass beads on ice. The whole-cell extracts were deproteinated with chloroform-methanol (1:1, vol/vol) by vortexing. The upper phase was collected by centrifugation at 1,600 × g for 5 min, and NH4OH was added to 10% (vol/vol). Samples were analyzed in a Spex Industries (Edison, N.J.) 212 Fluorolog spectrofluorometer operating in the ratio mode, and dityrosine was detected by an excitation scan from 270 to 370 nm with emission at 420 nm.
FM4-64 staining and latrunculin A (latA) treatment.
Sporulating cultures of the wild type and sps1 mutants where ∼50% of the cells had reached the meiotic divisions were incubated on ice with 1 μg of FM4-64 (Molecular Probes)/ml for 10 min. The cells were washed and resuspended in sporulation medium and examined by fluorescence microscopy at the indicated times.
Twenty micrograms of latA (Calbiochem, San Diego, Calif.) per milliliter was added for 15 min, and then the cells were washed and examined by fluorescence microscopy. Endocytosis was assessed in latA-treated cells by staining with FM4-64 (data not shown).
RESULTS
Prospore membrane formation and septin organization appear normal in sps1 mutants.
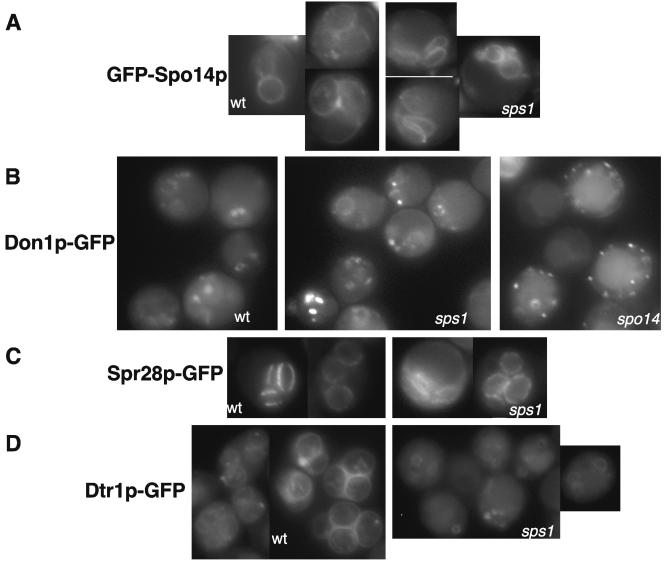
It was previously shown that sps1 mutants form aberrant spore structures (8). To more precisely determine the sporulation defect of sps1 mutants, we examined several cytological markers of spore morphogenesis. An early event in spore formation is the synthesis of the prospore membrane, which encapsulates the haploid nuclei that arise from the meiotic divisions and provides the scaffold for subsequent spore wall deposition. To determine whether sps1 mutants synthesize prospore membranes, we examined the localization of GFP-Spo14p, a phospholipase D, which is essential for and marks the prospore membrane (26) in live cells induced in sporulation. GFP fluorescence appeared normal in the sps1 mutant: GFP-Spo14p localized to the SPBs during the meiotic divisions and then moved with the growing prospore membrane, indicative of two bilobed structures corresponding to prospore membranes capturing haploid nuclei. Eventually, GFP-Spo14p fluorescence was visualized around each spore as observed in the wild type (Fig. 1A).
FIG. 1.
Prospore membrane formation appears normal in sps1 mutants. (A) GFP-Spo14p staining in the wild type (wt) and sps1 mutants; (B) Don1p-GFP staining in the wt and sps1 and spo14Δ mutants. (C) Spr28p-GFP staining in the wt and sps1 mutants; (D) Dtr1p-GFP staining in the wt and sps1 mutants. Living cells were visualized at the time of the meiotic divisions.
We also examined the localization of Don1p-GFP, which marks vesicular structures at the time of the meiotic divisions and then localizes to the leading edge of the growing prospore membrane, forming a range of doughnut- and bar-shaped structures (11). During a time course in sporulation, Don1p-GFP localization in the sps1 mutant was similar to that of the wild type (Fig. 1B). This is in contrast to Don1p-GFP localization in mutants that fail to synthesize prospore membranes, such as spo14. In this case, Don1p-GFP fluorescence accumulates in punctate dots within the cell (Fig. 1B). Thus, Sps1p does not appear to be required for prospore membrane formation and most likely regulates a later step in spore formation.
After prospore membrane closure, spore wall components are deposited in the lumen between the double membranes (15). Septins are organized at the leading edge of the prospore membrane and are believed to facilitate spore wall deposition (6). Septin localization is altered in mutants where prospore membrane closure is not sensed or has failed (33). To determine whether sps1 mutations affect septin organization, Spr28p-GFP, a sporulation-specific septin (38), was examined in cells induced in sporulation. Septins form parallel bar structures and then eventually surround the prospores. Analogous localization patterns were observed in both wild-type and sps1 cells (Fig. 1C), suggesting that septins organize properly in the absence of Sps1p. Thus, prospore membrane closure and septin organization occur in the absence of Sps1p.
sps1 mutants do not lay down spore wall components.
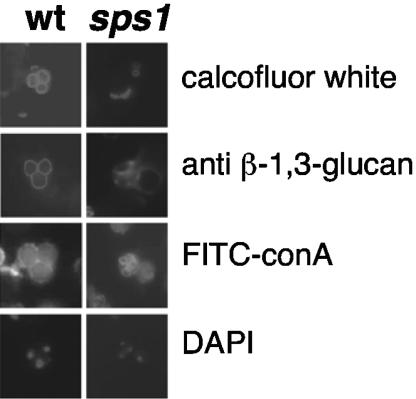
Initial observations by EM revealed that sps1 mutant spores do not contain the third and fourth layers of the spore wall (8). The first two layers, mannan and glucan, appear electron lucent and thus are not easily resolved by EM. To determine whether sps1 mutants contained the inner layers of the spore wall, we used an immunofluorescence approach developed by Tachikawa and coworkers (33).
The first layer of the spore wall is comprised of mannans; additionally, mannoproteins are incorporated into the prospore membrane (10). Thus, detection with the mannan-binding lectin ConA conjugated to FITC (FITC-ConA) will label both the prospore membrane and the mannan layer (33). Similar to the wild type and as expected from the analysis of the prospore membrane, sps1 mutants displayed distinct FITC-ConA-positive circles (Fig. 2). However, as this analysis does not distinguish between the prospore membrane and the mannan layer of the spore wall, it is unclear whether sps1 mutants lay down the first layer of the spore wall.
FIG. 2.
sps1 mutants do not display the full array of spore wall layers. The left panels show wild-type (wt) cells with characteristic staining patterns of calcofluor white (chitosan), anti-glucan antibodies (β-1,3-glucan), and FITC-ConA (mannan) around each individual spore. The fourth spore of the tetrad is out of the plane of focus. sps1 mutants do not display staining with calcofluor white or anti-glucan antibodies around each spore-like structure but do have distinct FITC-ConA staining.
The glucan layer was detected by using a monoclonal antibody against β-1,3-glucan and visualized with rhodamine-conjugated secondary antibodies. Compared to wild-type cells, which displayed anti-β-1,3-glucan staining around individual prospores, sps1 mutants did not exhibit staining around prospore-like structures, indicating that the prospores lacked the second layer of the spore wall (Fig. 2).
The third layer of the spore wall, comprised of chitosan, was detected by using the chitin-binding dye calcofluor white. Wild-type cells showed calcofluor white staining around each individual spore (Fig. 2). Consistent with EM analysis (8), sps1 mutants did not display this pattern, as no asci were observed with the characteristic staining. Instead, we observed small round bud scars (Fig. 2). Taken together with the EM analysis, these results indicate that the chitosan layer of the spore wall is not laid down properly in sps1 mutants.
Interestingly, a very small fraction of sps1 cells (<1%) displayed prospore-like structures that were positive for glucan staining but not chitosan. This suggests that sps1 mutants produce prospores of varying maturity as reported previously (8). Nonetheless, our results indicate that once prospore membranes form, neither the glucan nor chitosan layer is deposited in the majority of sps1 mutant cells.
Dityrosine is not incorporated into the sps1 prospores.
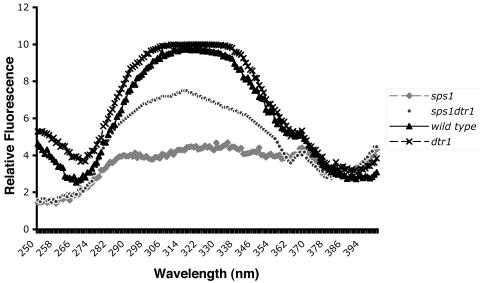
sps1 mutant spores do not contain detectable levels of dityrosine when visualized by short-wave UV light or EM but display increased levels of DIT1 mRNA (8). To monitor total dityrosine, we measured fluorescence at 315 nm in whole-cell extracts and found that sps1 mutants contain severely reduced levels of dityrosine compared to the wild type (Fig. 3). To distinguish between a failure to synthesize dityrosine and the inability to incorporate dityrosine into the spore wall, we constructed an sps1 dtr1 double mutant. Dtr1p is a membrane protein that is responsible for translocation of dityrosine through the prospore membrane. Thus, inactivation of DTR1 results in intracellular accumulation of dityrosine (7). In sps1 mutants, Dtr1p-GFP localized to prospore membranes (Fig. 1D). Furthermore, dityrosine levels were increased in whole-cell extracts from sps1 dtr1 mutants (Fig. 3). Thus, dityrosine is synthesized and transported out of the cell but fails to be incorporated into the spore wall in the sps1 mutant.
FIG. 3.
Dityrosine is synthesized but not incorporated into the spore wall in sps1 mutants. Extracts were prepared and fluorescence was measured as described in Materials and Methods. The intensity of emission was measured at 420 nm. The peak at 315 nm corresponds to dityrosine.
sps1 mutants display defects in the localization of Gsc2p and Chs3p to the prospore membrane.
Given the failure of sps1 mutants to synthesize either the glucan or chitosan layer of the spore wall, we examined the localization of two key enzymes responsible for synthesis of these components during sporulation. Gsc2p, a glucan synthase, forms an imperfect redundant pair with Gsc1p and is required for regulated glucan synthesis in response to environmental cues, including sporulation (16). Chs3p, chitin synthase III, synthesizes 90% of bulk chitin in vegetatively growing yeast and is also required for synthesis of the chitosan layer during sporulation (20, 37). We constructed functional GFP-tagged versions of these enzymes and examined their localization in living wild-type and sps1 cells.
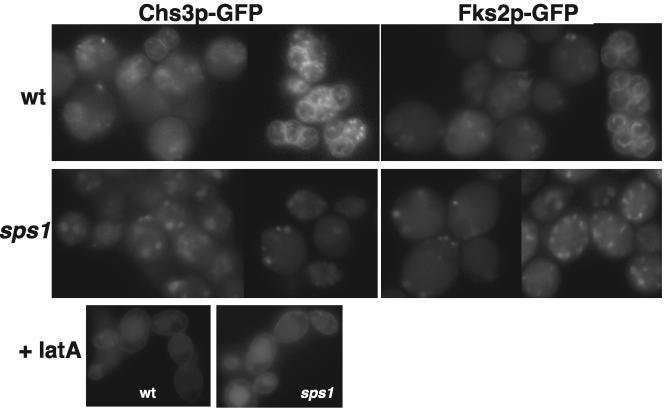
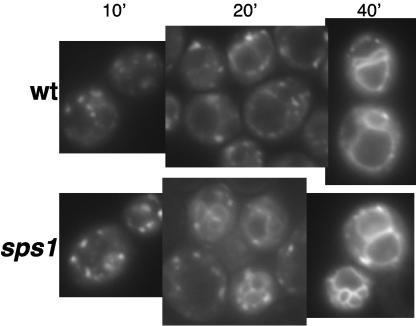
Cells induced in sporulation show plasma membrane (PM) and internal punctate staining of Gsc1p-GFP and Chs3p-GFP (data not shown). As the cells progress through the meiotic divisions, the majority of Gsc2p-GFP and Chs3p-GFP resided in intracellular structures in both wild-type and sps1 cells (Fig. 4). As spore morphogenesis continued, Gsc2p-GFP and Chs3p-GFP were visualized at the prospore membrane in wild-type cells (Fig. 4). However, neither Gsc2p-GFP nor Chs3p-GFP was found at the prospore membrane in sps1 cells (Fig. 4). In contrast, Chs3p-GFP reached the prospore membrane in smk1 mutants (data not shown), which arrest at a stage of sporulation similar to that in sps1 mutants (12). Thus, sps1 mutants appear to be specifically defective in localization of these enzymes to the prospore membrane.
FIG. 4.
Chs3p-GFP and Gsc2p-GFP fail to reach the prospore membrane in sps1 mutants. Chs3p-GFP localization is shown in living wild-type (wt) and sps1 cells and with the addition of latA (+latA). Gsc2p-GFP (Fks2p-GFP) localization is shown in living wt and sps1 cells.
Sporulating cells accumulate PM-associated Chs3p-GFP during endocytic arrest.
The failure of sps1 cells to localize Gsc2p-GFP and Chs3p-GFP to the prospore membrane could reflect either a failure to properly transport these molecules through the secretory pathway or a failure to redirect pools of these enzymes from the PM to the prospore membrane. To distinguish between these possibilities, we inhibited endocytosis to determine at which stage Chs3p trafficking is blocked. To that end, we first determined whether the absence of Sps1p results in endocytic defects and used the vital stain FM4-64 to monitor uptake during sporulation. Figure 5 shows that wild-type cells internalized FM4-64 from the PM to internal sites and then to the vacuolar membrane during sporulation. Similarly, sps1 mutants (Fig. 5) displayed distinct vacuolar membrane staining with the same kinetics as the wild type. Thus, sps1 mutants are proficient for endocytosis.
FIG. 5.
sps1 mutants are proficient for endocytosis. Wild-type (wt) cells and sps1 mutants stained with FM4-64 are shown. Numbers above the panels refer to time (in minutes) after FM4-64 was washed out.
We next transiently blocked endocytosis in the wild type and sps1 mutants by using the actin-depolymerizing agent latA. latA was added to cells undergoing the meiotic divisions and incubated for 15 min. The cells were then washed and immediately examined by fluorescence microscopy. The majority of wild-type cells showed PM and punctate cytosolic staining of Chs3p-GFP (Fig. 4), while the majority of the untreated cells displayed punctate cytosolic staining as well as distinct prospore membrane staining in some cells (Fig. 4). sps1 cells also displayed Chs3p-GFP PM and punctate cytosolic staining when treated with latA (Fig. 4); however, Chs3p remained primarily in the cytosol and failed to accumulate at the prospore membrane in the absence of latA (Fig. 4). Taken together, our results suggest that Chs3p reaches the PM in sps1 mutants and becomes internalized by endocytosis, but fails to localize to prospore membranes.
Shc1p-GFP is not properly localized in sps1 mutants, but its inactivation does not alter Chs3p-GFP localization.
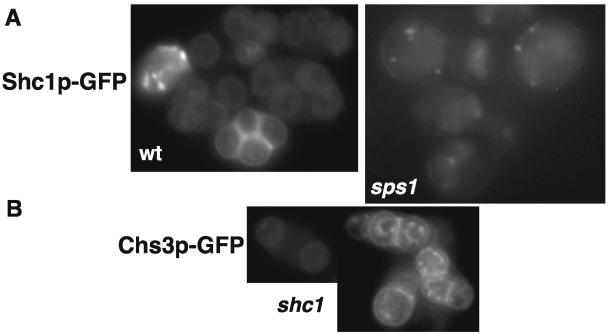
In vegetative cells, proper localization of Chs3p to the developing bud relies on Chs4p. During sporulation, Chs4p is replaced by the sporulation-specific homolog Shc1p (29). To determine the localization of Shc1p in sporulating cells, we constructed and examined Shc1p-GFP in the wild type and sps1 mutants. Shc1p-GFP localized to punctate dots within the cytoplasm and then to prospore membranes in the wild type (Fig. 6), a pattern similar to what is observed for Chs3p-GFP. Like Chs3p-GFP, Shc1p-GFP did not reach the prospore membrane in the majority of sps1 mutant cells examined (Fig. 6A). This result raises the possibility that the failure of Chs3p to reach the prospore membrane in sps1 mutants is due to a failure to properly localize Shc1p.
FIG. 6.
Shc1p-GFP is not properly localized in sps1 mutants but is not required for Chs3p-GFP localization to the prospore membrane. (A) Shc1p-GFP localization in wild-type (wt) and sps1 cells; (B) Chs3p-GFP localization in shc1Δ cells.
To determine whether Shc1p did indeed mediate the localization of Chs3p during sporulation, analogous to the role of Chs4p in vegetative cells, we examined the localization of Chs3p-GFP in shc1 mutants induced in sporulation. Chs3p-GFP displayed a pattern of localization indistinguishable from that of the wild type (Fig. 6B). Thus, while Shc1p functions like Chs4p to stimulate the enzymatic activity of Chs3p (29), it is not required for the localization of Chs3p to the prospore membrane.
Sps1p localizes to internal punctate structures and prospores.
To determine the intracellular localization of Sps1p, an SPS1 promoter-driven N-terminal GFP fusion was constructed and integrated onto the chromosome (see Materials and Methods). Unlike C-terminal fusions (data not shown), the full-length N-terminal fusion is fully functional.
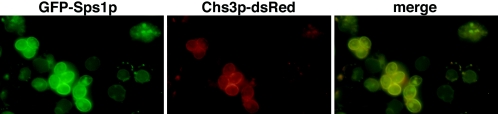
Very little fluorescence was observed in cells immediately after transfer to sporulation medium; however, as cells progressed through the meiotic divisions, a punctate staining pattern was observed (Fig. 7). In some cells, prospore membrane staining was also evident. The pattern of GFP-Sps1p staining was similar to that of Chs3p-GFP and Gsc2p-GFP staining. To determine whether Sps1p colocalized with these enzymes, we constructed a Chs3p-dsRed fusion and examined yeast strains harboring both GFP-Sps1p and Chs3p-dsRed in living cells. As shown in Fig. 7, the signals largely overlapped. Of 34 cells examined, 89.9% of Chs3p-dsRed signals colocalized with GFP-Sps1p, while 93.6% of GFP-Sps1p signals also contained Chs3p-dsRed. We also examined the colocalization of Chs3-dsRed with Gsc2p-GFP and found that 95.3% of the Chs3p-dsRed also contained GFP-Gsc2p and 68.5% of the Gsc2-GFP signal overlapped with Chs3p-dsRed. These results indicate that these proteins largely colocalize with each other in the cell.
FIG. 7.
GFP-Sps1p localizes to internal structures and prospores and displays colocalization with Chs3p-dsRed. Left (green), GFP-Sps1p localization in living cells induced in sporulation; center (red), Chs3p-dsRed localization in living cells induced in sporulation; right (yellow), merged image showing colocalization of GFP-Sps1p and Chs3p-dsRed.
DISCUSSION
Our results demonstrate that Sps1p, a member of the Ste20/p21-activated kinase family of kinases, regulates the movement of enzymes required for spore wall synthesis. Analysis of Chs3p and Gsc2p, key enzymes regulating chitin and glucan synthesis, respectively, reveals that these enzymes fail to reach the prospore membrane in the absence of Sps1p.
Secretory events involved in prospore membrane formation differ from those involved in spore wall maturation.
During sporulation, the secretory system plays a vital role in redirecting proteins that transit the Golgi apparatus to the prospore membrane (which will become the PM once the spore germinates) (17). Sps1p does not appear to regulate these trafficking events, as prospore membrane-associated GFP-Spo14p, Don1p-GFP, Spr28p-GFP, and Dtr1p-GFP are all detected on prospore membranes in sps1 mutants. In contrast, spo14Δ mutants, which fail to synthesize prospore membranes (26), show only punctate fluorescence of Don1p-GFP and no defined pattern for Spr28p-GFP (J. Connolly and J. Engebrecht, unpublished data). These Don1p-GFP punctate structures presumably correspond to post-Golgi vesicles, consistent with the hypothesis that Spo14p functions in vesicle fusion for prospore membrane formation (27). The difference in the phenotypes of spo14 and sps1 mutants with respect to localization of these membrane-associated proteins indicates that Sps1p functions downstream of this developmentally regulated secretory pathway.
Although prospore membranes are synthesized in sps1 mutants, they are not as uniform in shape as the membranes observed in the wild type (Fig. 1). We attribute the irregular shape of the sps1 membranes to the failure to elaborate the spore wall, which provides a rigid structure, and not to a defect in prospore membrane formation per se. While trafficking from the Golgi apparatus to the prospore membrane appears functional in the sps1 mutant, enzymes required for synthesis of the spore wall fail to reach the prospore membrane, suggesting that additional developmentally regulated pathways of protein movement are in operation during sporulation.
Sps1p regulates the movement of enzymes required for spore wall synthesis.
The glucan synthase, Gsc2p, plays a minor role in glucan synthesis in vegetative cells but is essential for sporulation (16). During sporulation, Gsc2p localizes to prospore membranes for synthesis of the glucan spore wall layer. However, in the absence of Sps1p, Gsc2p fails to reach the prospore membrane and thus the glucan spore wall layer is absent. In addition to Gsc2p, sps1 mutants also fail to localize Chs3p and its regulator, Shc1p, to the prospore membrane. Chs3p is responsible for most of the chitin synthase activity in yeast (30) and is essential for synthesis of the chitosan layer of the spore wall (20, 37). Surprisingly, when the demand for chitin is increased, such as during spore wall synthesis, increases in CHS3 transcription are not observed (36). Chs3p is dynamic during both mitosis and meiosis and interacts with a number of protein partners in each cell compartment (28, 34, 40). Interaction with these proteins, in addition to a cleavage event mediated by Chs4p/Shc1p, results in a concomitant increase in chitin synthase activity (19), suggesting that regulation of Chs3p is achieved through spatial and temporal control. Just as it is important to localize chitin synthase activity to the bud neck in vegetative cells, the ability to synthesize the chitosan layer in the spore wall is dependent on the localization of the enzyme to the prospore membrane. To accomplish this, Chs3p is redirected to the prospore membrane. Sps1p localizes to internal structures that contain Chs3p, suggesting that Sps1p directly regulates the movement of this enzyme to the developing spore.
Factors governing chitin transport during vegetative growth, such as Chs6p and Chs7p, display spore wall defects, implying that at least some of the same machinery regulating the transport of chitin synthase is used during sporulation (29). On the other hand, Shc1p replaces Chs4p during sporulation, indicating that Chs3p must be specifically modified to carry out its role in sporulation. We found that, unlike Chs4p in vegetative cells, Shc1p is not required for the localization of Chs3p to the prospore membrane. This is presumably due to the fact that spore maturation occurs uniformly, while bud growth is highly polarized, and Chs4p is required to specifically target the enzyme to the site of bud growth. Chs3p and Shc1p localization is dependent on Sps1p, suggesting that Sps1p is an important regulator of chitin synthase activity during sporulation.
Sps1p functions after Gip1p-Glc7p-regulated prospore membrane closure.
sps1 mutants synthesize prospore membranes and organize septins, indicating that prospore membrane closure occurs, but fail to synthesize spore wall layers. Septin organization is regulated by the Gip1p-Glc7p phosphatase and is necessary for prospore membrane closure and subsequent spore wall maturation (33). In the gip1 mutant, septins are not organized and the spore wall layers are not synthesized. These results suggest that Sps1p functions downstream of the Gip1p-Glc7p phosphatase. The Gip1p-Glc7p phosphatase complex may antagonize Sps1p until septins are properly organized. Once this scaffold for spore wall synthesis is set up, Sps1p may be activated and regulate the movement of proteins important for spore wall synthesis.
Sps1p regulates the trafficking of Gsc2p, Chs3p, Shc1p, and perhaps other proteins essential for spore wall formation. This pathway of protein movement involves redirection of PM proteins to the prospore membrane. The localization of Sps1p to a large complex that contains Chs3p and Gsc2p suggests that Sps1p may directly target these enzymes, either through protein-protein interactions or by phosphorylation. During cell stress, Chs3p has been shown to be phosphorylated in a Pkc1p-dependent manner; this modification correlates with translocation of Chs3p from internal structures (chitosomes) to the PM (35). Thus, phosphorylation is one mechanism to drive movement of proteins in response to environmental changes (e.g., stress and starvation/sporulation). Alternatively, Sps1p may interact with or modify components of the intracellular trafficking pathway to allow spore wall enzymes to be redirected in the cell. Additionally, Sps1p regulates other aspects of sporulation. In particular, sps1 mutants display defects in the expression pattern of mid-late and late sporulation genes (8) and thus may play a role in the transcriptional program. Identification of the targets of Sps1p should elucidate the pathways regulated by this conserved protein.
Acknowledgments
This work was supported by grant GM66124 from the National Institutes of Health.
We thank Aimee Jaramillo-Lambert (UC, Davis) and Eric Sawey (SUNY, Stony Brook) for excellent technical assistance. We also thank James Trimmer (UC, Davis), Aaron Neiman (SUNY, Stony Brook), and Neta Dean (SUNY, Stony Brook) for helpful discussion and reagents. We gratefully acknowledge Roger Tsien (UC, San Diego, HHMI), who developed the tdimer2 (dsRed) reagent used in this study. Additionally, we acknowledge Jaime Connolly and Masayo Morishita for discussion and critical reading of the manuscript.
REFERENCES
- 1.Bajgier, B. K., M. Malzone, M. Nickas, and A. M. Neiman. 2001. SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol. Biol. Cell 12:1611-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briza, P., M. Eckerstorfer, and M. Breitenbach. 1994. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble LL-dityrosine-containing precursor of the yeast spore wall. Proc. Natl. Acad. Sci. USA 91:4524-4528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castillo, A. R., J. B. Meehl, G. Morgan, A. Schutz-Geschwender, and M. Winey. 2002. The yeast protein kinase Mps1p is required for assembly of the integral spindle pole body component Spc42p. J. Cell Biol. 156:453-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christodoulidou, A., V. Bouriotis, and G. Thireos. 1996. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. J. Biol. Chem. 271:31420-31425. [DOI] [PubMed] [Google Scholar]
- 5.Chu, S., J. DeRisi, M. Eisen, J. Mulholland, D. Botstein, P. O. Brown, and I. Herskowitz. 1998. The transcriptional program of sporulation in budding yeast. Science 282:699-705. [DOI] [PubMed] [Google Scholar]
- 6.Fares, H., L. Goetsch, and J. R. Pringle. 1996. Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J. Cell Biol. 132:399-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felder, T., E. Bogengruber, S. Tenreiro, A. Ellinger, I. Sa-Correia, and P. Briza. 2002. Dtrlp, a multidrug resistance transporter of the major facilitator superfamily, plays an essential role in spore wall maturation in Saccharomyces cerevisiae. Eukaryot. Cell 1:799-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friesen, H., R. Lunz, S. Doyle, and J. Segall. 1994. Mutation of the SPS1-encoded protein kinase of Saccharomyces cerevisiae leads to defects in transcription and morphology during spore formation. Genes Dev. 8:2162-2175. [DOI] [PubMed] [Google Scholar]
- 9.Ito, H., Y. Fukada, K. Murata, and A. Kimura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kane, S. M., and R. Roth. 1974. Carbohydrate metabolism during ascospore development in yeast. J. Bacteriol. 118:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knop, M., and K. Strasser. 2000. Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J. 19:3657-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krisak, L., R. Strich, R. S. Winters, J. P. Hall, M. J. Mallory, D. Kreitzer, R. S. Tuan, and E. Winter. 1994. SMK1, a developmentally regulated MAP kinase, is required for spore wall assembly in Saccharomyces cerevisiae. Genes Dev. 8:2151-2161. [DOI] [PubMed] [Google Scholar]
- 13.Kupiec, M., B. Byers, R. E. Esposito, and A. P. Mitchell. 1997. Meiosis and sporulation in Saccharomyces cerevisiae, p. 889-1036. In J. R. Pringle, J. R. Broach, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces. Cell cycle and cell biology, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [Google Scholar]
- 14.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 15.Lynn, R. R., and P. T. Magee. 1970. Development of the spore wall during ascospore formation in Saccharomyces cerevisiae. J. Cell Biol. 44:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mazur, P., N. Morin, W. Baginsky, M. el-Sherbeini, J. A. Clemas, J. B. Nielsen, and F. Foor. 1995. Differential expression and function of two homologous subunits of yeast 1,3-β-d-glucan synthase. Mol. Cell. Biol. 15:5671-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neiman, A. M. 1998. Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nickas, M., C. Schwartz, and A. Neiman. 2003. Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot. Cell 2:431-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono, N., T. Yabe, M. Sudoh, T. Nakajima, T. Yamada-Okabe, M. Arisawa, and H. Yamada-Okabe. 2000. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology 146:385-391. [DOI] [PubMed] [Google Scholar]
- 20.Pammer, M., P. Briza, A. Ellinger, T. Schuster, R. Stucka, H. Feldmann, and M. Breitenbach. 1992. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast 8:1089-1099. [DOI] [PubMed] [Google Scholar]
- 21.Percival-Smith, A., and J. Segall. 1986. Characterization and mutational analysis of a cluster of three genes expressed preferentially during sporulation of S. cerevisiae. Mol. Cell. Biol. 6:2443-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Primig, M., R. M. Williams, E. A. Winzeler, G. G. Tevzadze, A. R. Conway, S. Y. Hwang, R. W. Davis, and R. E. Esposito. 2000. The core meiotic transcriptome in budding yeasts. Nat. Genet. 26:415-423. [DOI] [PubMed] [Google Scholar]
- 23.Rose, K., S. A. Rudge, M. A. Frohman, A. J. Morris, and J. Engebrecht. 1995. Phospholipase D signaling is essential for meiosis. Proc. Natl. Acad. Sci. USA 92:12151-12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics, a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Rothstein, R. 1983. One step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 26.Rudge, S. A., A. J. Morris, and J. Engebrecht. 1998. Relocalization of phospholipase D activity mediates membrane formation during meiosis. J. Cell Biol. 140:2025-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudge, S. A., V. A. Sciorra, C. Zhou, M. Iwamoto, T. Strahl, A. J. Morris, J. Thorner, and J. Engebrecht. 2004. Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol. Biol. Cell 15:207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos, B., A. Duran, and M. H. Valdivieso. 1997. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:2485-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanz, M., J. A. Trilla, A. Duran, and C. Roncero. 2002. Control of chitin synthesis through Shc1p, a functional homologue of Chs4p specifically induced during sporulation. Mol. Microbiol. 43:1183-1195. [DOI] [PubMed] [Google Scholar]
- 30.Shaw, J. A., P. C. Mol, B. Bowers, S. J. Silverman, M. H. Valdivieso, A. Duran, and E. Cabib. 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114:111-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sheff, M. A., and K. S. Thorn. 2004. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21:661-670. [DOI] [PubMed] [Google Scholar]
- 32.Straight, P. D., T. H. Giddings, and M. Winey. 2000. Mps1p regulates meiotic spindle pole body duplication in addition to having novel roles during sporulation. Mol. Biol. Cell 11:3525-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tachikawa, H., A. Bloecher, K. Tatchell, and A. M. Neiman. 2001. Septin organization and spore wall formation. J. Cell Biol. 155:797-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trilla, J. A., A. Duran, and C. Roncero. 1999. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 145:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valdivia, R. H., and R. Schekman. 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 100:10287-10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valdivieso, M. H., L. Ferrario, M. Vai, A. Duran, and L. Popolo. 2000. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 182:4725-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdivieso, M. H., P. C. Mol, J. A. Shaw, E. Cabib, and A. Duran. 1991. CAL1, a gene required for activity of chitin synthase 3 in Saccharomyces cerevisiae. J. Cell Biol. 114:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Virgilio, C. D., D. J. DeMarini, and J. R. Pringle. 1996. SPR28, a sixth member of the septin gene family in Saccharomyces cerevisiae that is expressed specifically in sporulating cells. Microbiology 142:2897-2905. [DOI] [PubMed] [Google Scholar]
- 39.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 40.Ziman, M., J. S. Chuang, M. Tsung, S. Hamamoto, and R. Schekman. 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 9:1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]