ABSTRACT
Unspliced HIV-1 RNAs function as messenger RNAs for Gag or Gag-Pol polyproteins and progeny genomes packaged into virus particles. Recently, it has been reported that fate of the RNAs might be primarily determined, depending on transcriptional initiation sites among three consecutive deoxyguanosine residues (GGG tract) downstream of TATA-box in the 5′ long terminal repeat (LTR). Although HIV-1 RNA transcription starts mostly from the first deoxyguanosine of the GGG tract and often from the second or third deoxyguanosine, RNAs beginning with one guanosine (G1-form RNAs), whose transcription initiates from the third deoxyguanosine, were predominant in HIV-1 particles. Despite selective packaging of G1-form RNAs into virus particles, its biological impact during viral replication remains to be determined. In this study, we revealed that G1-form RNAs are primarily selected as a template for provirus DNA rather than other RNAs. In competitions between HIV-1 and lentiviral vector transcripts in virus-producing cells, approximately 80% of infectious particles were found to generate provirus using HIV-1 transcripts, while lentiviral vector transcripts were conversely selected when we used HIV-1 mutants in which the third deoxyguanosine in the GGG tract was replaced with deoxythymidine or deoxycytidine (GGT or GGC mutants, respectively). In the other analyses of proviral sequences after infection with an HIV-1 mutant in which the GGG tract in 3′ LTR was replaced with TTT, most proviral sequences of the GGG-tract region in 5′ LTR were found to be TTG, which is reasonably generated using the G1-form transcripts. Our results indicate that the G1-form RNAs serve as a dominant genome to establish provirus DNA.
IMPORTANCE
Since the promoter for transcribing HIV-1 RNA is unique, all viral elements including genomic RNA and viral proteins have to be generated by the unique transcripts through ingenious mechanisms including RNA splicing and frameshifting during protein translation. Previous studies suggested a new mechanism for diversification of HIV-1 RNA functions by heterogeneous transcriptional initiation site usage; HIV-1 RNAs whose transcription initiates from a certain nucleotide were predominant in virus particles. In this study, we established two methods to analyze heterogenous transcriptional initiation site usage by HIV-1 during viral infection and showed that RNAs beginning with one guanosine (G1-form RNAs), whose transcription initiates from the third deoxyguanosine of the GGG tract in 5′ LTR, were primarily selected as viral genome in infectious particles and thus are used as a template to generate provirus for continuous replication. This study provides insights into the mechanism for diversification of unspliced RNA functions and requisites of lentivirus infectivity.
KEYWORDS: HIV-1, genome, LTR, RNA, transcriptional initiation site, infectivity, diversification
INTRODUCTION
To infect new cells, HIV-1 has to assemble infectious virus particles that contain viral proteins and two copies of its genomic RNA (1–3). In infected cells, HIV-1 particle assembly initiates with the transcription of viral RNA from integrated provirus DNA to produce the genomic RNA and the full range of mRNAs encoding the viral proteins. Since the promoter for transcribing HIV-1 RNAs is unique, some RNA transcripts undergo RNA splicing to become mRNAs encoding viral envelope protein, regulatory proteins, and accessory proteins (4). The others are not spliced and function as mRNAs for Gag proteins and Gag-Pol polyproteins via frameshifting as well as viral genomic RNA of progeny virus. Genomic RNAs are incorporated into virus particles, depending on cis-acting RNA “packaging signals” in the 5′-leader of unspliced HIV-1 RNA (5, 6), and most or nearly all HIV-1 particles were found to contain dimerized genomic RNAs (7). Thus, genomic RNA selection had been originally thought to be determined by dimerization-dependent secondary and tertiary structures of a single unspliced HIV-1 transcript (8, 9), switchable structures between dimerized RNAs in which packaging signals are accessible and not-dimerized RNAs in which packaging signals are masked (10–13).
In 2015, Masuda et al. reported heterogenous transcriptional start site usage by HIV-1 and two functionally distinct pools of resultant RNAs (14). There are three forms of HIV-1 RNA whose transcription initiates from each of three consecutive deoxyguanosine residues (GGG tract) downstream of TATA-box in the 5′ long terminal repeat (LTR), and RNAs whose transcription initiates from the first G (G3-form HIV-1 RNA) were predominantly expressed in cells (14). However, RNAs whose transcription initiates from the third G (RNAs beginning with a single guanosine: G1-form HIV-1 RNA) occupied almost 70% of genomic RNA in virus particles. Interestingly, 5′-capped G1-form 5′-leader RNAs adopted a dimeric structure, but most of 5′-capped G2- or 5′-capped G3-form leader RNAs existed as a monomer under physiological-like ionic condition (15). Furthermore, the G2- and G3-form RNAs were found to be enriched on polyribosomes in cells instead of being packaged into virus particles (15). It is also reported that abortive forms of minus-strand strong-stop cDNA (−sscDNA) were more abundantly generated during reverse transcription from the G3-form RNA than from the G1-form RNA in the same study (14), suggesting that transcriptional start site heterogeneity affects not only genomic RNA packaging but also reverse-transcription and possibly other processes during HIV-1 replication.
Nuclear magnetic resonance (NMR) analysis revealed that the G1-form RNAs adopted a dimeric multi-hairpin structure in which the 5′-cap structure was hidden, but the G2- or G3-form RNAs adopt an alternate structure in which splice donor residues and the 5′-cap structure were accessible (16, 17). Further NMR analysis of the joint region between the TAR and polyA stems in 5′-leader RNA revealed that they form a coaxial stem in the case of the G1-form RNA while the two stems are structurally independent in the case of the G2- and G3-form RNAs (18). Collectively, the number of G at the 5′-terminus of HIV-1 RNA, depending on transcriptional initiation sites, is suggested to dictate conformational regulation and finally fate of HIV-1 RNA.
In the present study, we show that the G1-form RNA is not only physically predominant in virus particles but also selectively used as a template RNA for generating provirus DNA by reverse transcription in the context of single-round HIV-1 infection.
RESULTS
Infectious HIV-1 particles were successfully produced in the absence of G1-form HIV-1 transcripts
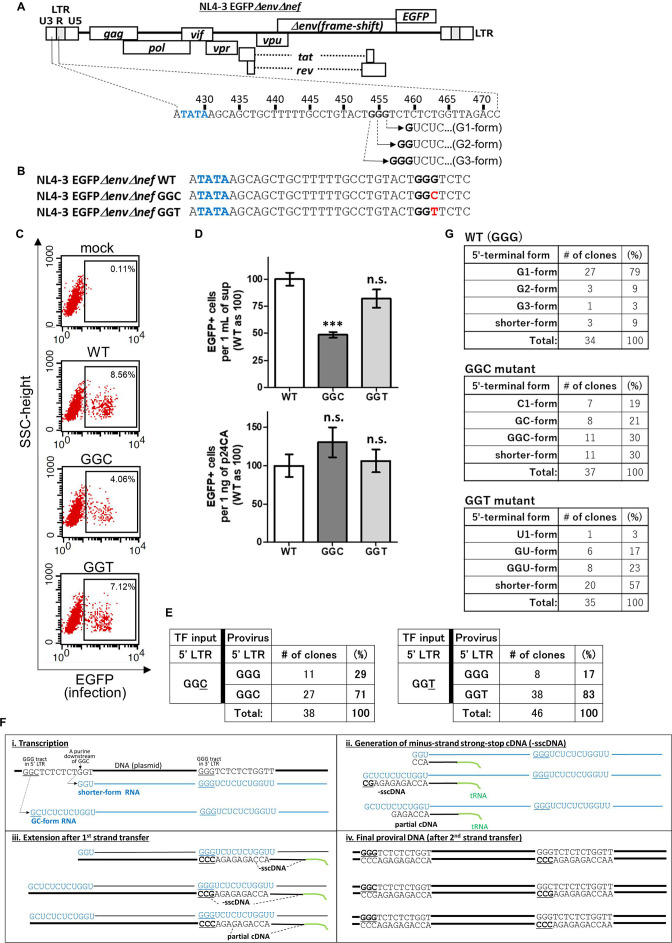
Although three deoxyguanosines (G) in a conserved GGG tract in the U3/R junction of 5′ LTR of HIV-1 (the GGG tract is located from 454 to 456 in pNL4-3) are known as the transcription initiation sites of HIV-1 RNA, RNAs whose transcription initiates from the third G of the GGG tract (RNAs beginning with single guanosine: G1-form HIV-1 RNA, Fig. 1A) are predominantly incorporated into virus particles (14). First of all, we addressed a question whether the G1-form HIV-1 transcripts are essential to produce infectious HIV-1 particles. To assess whether infectious HIV-1 particles could be produced in the absence of the G1-form HIV-1 transcripts, we generated two mutants of HIV clone DNA pNL4-3EGFPΔenvΔnef (19) by replacing the third G at 456 with deoxycytidine (C, GGC mutant) or deoxythymidine (T, GGT mutant) (Fig. 1B). These mutants are expected not to express the G1-form because transcription usually initiates from a purine such as deoxyadenosine (A) and G but rarely from a pyrimidine such as deoxythymidine (T) or deoxycytidine (C) (20). Even though transcription initiates from the nucleotide they will be U1- or C1-form RNAs (RNAs beginning with single uridine or cytidine, respectively) but not the G1-form RNAs. Using these mutants, we prepared vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped virus as reported previously (21) and assessed whether the prepared viruses are infectious by infecting human T-cell line, MT-4 cells. Forty-eight hours after exposure of MT-4 cells with the supernatant of 293T cells transfected with each HIV clone DNA, expression of the marker protein EGFP by MT-4 cells was evaluated by flow cytometry. Some cells exposed with the GGC or GGT mutant virus were found to express EGFP (Fig. 1C), suggesting that infectious particles could be produced in the absence of the G1-form HIV-1 transcripts. By counting EGFP-positive (EGFP+) MT-4 cells gated in Fig. 1C, we evaluated number of EGFP+ cells produced with 1 mL of supernatant or with 1 ng of p24 (capsid protein of HIV-1). We found that EGFP+ cells by the GGC mutant virus are significantly less to the level of approximately 50% of wild-type (WT) control, but the GGT mutation was tolerable (Fig. 1D, upper graph). On the other hand, EGFP+ cells produced with 1 ng of p24 (capsid protein of HIV-1) were not significantly different among WT and the mutants (Fig. 1D, lower graph). Based on the results, it was assumed that HIV-1 RNAs other than the G1-form can be also used as viral genome in infectious particles.
Fig 1.
Infectious particles can be produced in the absence of the G1-form HIV-1 transcripts. (A) A schematic represents initiation sites of HIV-1 RNA transcription. The GGG tract is located at the U3/R junction in 5′ LTR of HIV-1 clone DNA (pNL4-3EGFPΔenvΔnef). Sequences of 5′-termini of HIV-1 RNA whose transcription initiates from the first, the second, and the third deoxyguanosine are also shown (G3-, G2-, and G1-form, respectively). TATA-box and the GGG tract are highlighted by blue and black bold letters, respectively. (B) Nucleotide sequences of the GGC and GGT mutants of pNL4-3EGFPΔenvΔnef are shown. The mutated nucleotides are highlighted by red bold letters. (C) MT-4 cells were exposed with supernatant containing VSV-G-pseudotyped NL4-3EGFPΔenvΔnef wild-type (WT), GGC, or GGT mutants and subjected to flow cytometric analysis at 48 h after infection to assess whether these mutant plasmids can produce infectious particles. From three independent experiments, one representative dot plot is shown. EGFP-positive cells are gated in black squares. (D) Numbers of EGFP-positive cells produced by the VSV-G-pseudotyped NL4-3EGFPΔenvΔnef WT, GGC, or GGT mutant viruses in 1 mL of supernatant (upper) or 1 ng of p24 antigen (lower) were evaluated. The numbers for WT were arbitrarily set as 100, and error bars are ±standard error of the mean from three independent experiments. Asterisk code for statistical significance compared to WT: ***P < 0.001; n.s., not significant, P > 0.05. (E) Sequencing analyses of provirus after infection with the VSV-G-pseudotyped NL4-3EGFPΔenvΔnef GGC mutant (n = 38 clones) or GGT mutant (n = 46 clones) mutants are shown (left and right panels, respectively). TF input: a plasmid for transfection to express HIV-1 transcripts (pNL4-3EGFPΔenvΔnef-based mutant plasmid). (F) A schematic represents processes of HIV-1 replication from transcription to provirus DNA production. Proviral sequences could be different, depending on transcriptional initiation site in the case of the GGC mutant of NL4-3EGFPΔenvΔnef. The cases of a shorter-form (upper) and the GC-form (lower) RNA are shown. (G) Results of 5′ rapid amplification of cDNA end analyses using RNA purified from virus particles of the VSV-G-pseudotyped NL4-3EGFPΔenvΔnef WT virus (n = 34 clones), GGC mutant virus (n = 37 clones), and GGT mutant virus (n = 35 clones) are shown (first, second, and 3rd panels, respectively).
To verify this point, we analyzed sequences of provirus in newly infected cells. DNA purified from MT-4 cells 48 h post-infection with the GGC or GGT mutants of VSV-G-pseudotyped virus was used as template of nested PCR for amplifying fragment including 5′ LTR U3/R junction of provirus. Then amplified DNA was cloned in a plasmid, pCR4, and proviral sequences were analyzed. As shown in Fig. 1E, in 71% of analyzed clones, proviral sequences of mutated region are GGC (Fig. 1E, left panel) which can be generated from RNAs whose transcription initiates from the first or the second G of the GGC mutant (GGC- or GC-form, respectively; a schematic of reverse transcription using the GC-form RNA as template is shown in Fig. 1F). Interestingly, in 29% of analyzed clones, proviral sequences of mutated region are GGG (Fig. 1E, left panel) which cannot be generated from the GGC- or GC-form. They might be possibly generated either from RNAs whose transcription initiates from nucleotides downstream of the GGC sequence (shorter-form RNAs: an example with RNA whose transcription initiates from G at 464 is shown in Fig. 1F, first RNA) or by premature strand transfer (22) that partial cDNA (Fig. 1F, panel ii, derived from the third RNA) was used as a primer for reverse transcription after first-strand transfer. To assess whether there is the GGC-, GC- or shorter-form HIV-1 RNAs are in virus particles, we carried out 5′ rapid amplification of cDNA end (5′ RACE) analyses using RNA purified from VSV-G-pseudotyped virus to analyze sequences of 5′-terminal edge of HIV-1 genomic RNA. First of all, we found that the G1-form HIV-1 RNA occupied 79% of genomic RNA in virus particles of WT virus (Fig. 1G, first panels), suggesting that we successfully confirmed a previous observation by our own experiment (14). In the virus particle of NL4-3EGFPΔenvΔnef virus GGC mutant, we found the GGC-, GC- and shorter-form RNAs (Fig. 1G, second panels), by which proviral sequences GGC or GGG could be reasonably generated (as shown in Fig. 1F, first or second RNA). In the experiments using the GGT mutant, GGG and GGT were found as proviral sequences of mutated region (Fig. 1E, right panels) and we found the shorter-form RNAs in addition to the U1-, GU- and GGU-form RNAs in virus particle of the GGT mutant virus (Fig. 1G, third panels). From these results, in the absence of the G1-form RNA, HIV-1 transcripts other than the G1-form are concluded to be incorporated into virus particles and used as a template RNA to establish the HIV-1 provirus state, namely, genomic RNA in infectious virus particles.
The GGG tract in U3/R junction of 5′ LTR is dispensable but important for efficient production of infectious virus particles
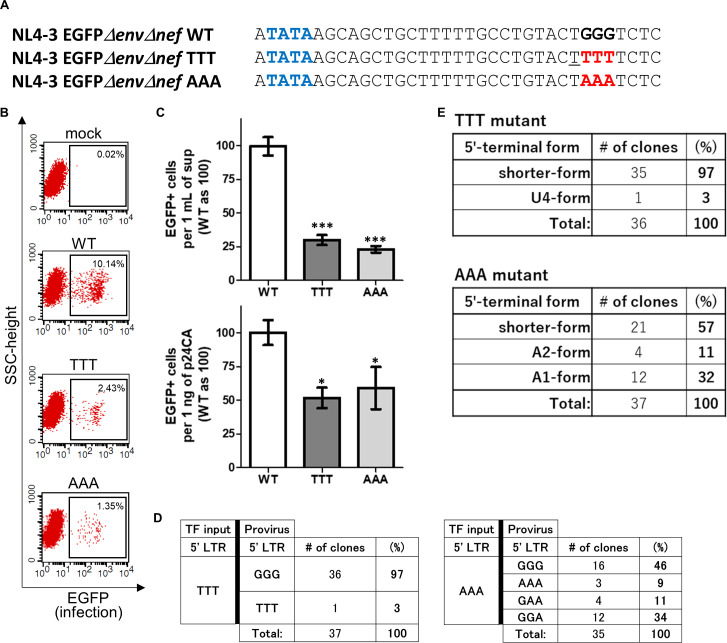
Next, we generated pNL4-3EGFPΔenvΔnef mutants by replacing the entire GGG tract with TTT or AAA (Fig. 2A) to assess whether the GGG tract itself is essential for HIV-1 to generate infectious virus particles. As shown in Fig. 2B, infectious VSV-G-pseudotyped HIV-1 particles were successfully produced by these mutant plasmids, suggesting that the GGG tract in U3/R junction of 5′ LTR is not an absolute requisite for successful production of infectious particles. As done for Fig. 1D, we also evaluated the number of EGFP+ cells produced by WT, TTT, or AAA mutant viruses with 1 mL of supernatant or 1 ng of p24 and found that a significantly smaller number of EGFP+ cells were produced by each mutant virus than the WT in both results per supernatant and per p24 antigen (Fig. 2C; upper or lower graph, respectively). To understand what HIV-1 RNA transcripts were used as genome in infectious VSV-G-pseudotyped particles, we analyzed sequences of provirus as done for Fig. 1E. In the case of the TTT mutant, proviral sequences of the mutated region are GGG (97%) and TTT (3%) (Fig. 2D, left panels). It suggests that the shorter- or longer-form RNAs whose transcription initiates from a nucleotide downstream or upstream of TTT were probably used as genome in infectious particles for generating proviral sequences GGG or TTT, respectively. In fact, most RNAs in the virus particle of the NL4-3EGFPΔenvΔnef TTT mutant virus were found to be the shorter-form RNAs, and only one clone of the longer-form RNA (U4-form RNA) was found by 5′ RACE analyses using RNA purified from VSV-G-pseudotyped virus (Fig. 2E, upper panels).
Fig 2.
The GGG tract in the U3/R junction of 5′ LTR is not an absolute requisite for production of infectious HIV-1 particles. (A) Nucleotide sequences of the TTT and AAA mutants of NL4-3EGFPΔenvΔnef are shown. The mutated nucleotides are highlighted by red bold letters. (B) Flow cytometric analyses were carried out using the TTT and AAA mutants as done in Fig. 1C. (C) Numbers of GFP-positive cells produced by the VSV-G-pseudotyped NL4-3EGFPΔenvΔnef WT, TTT, or AAA mutant viruses in 1 mL of supernatant (upper) or 1 ng of p24 antigen (lower) were evaluated. Graphs are presented as described in Fig. 1D. Asterisk code for statistical significance compared to WT: *P = 0.001 < P < 0.05, ***P < 0.001. (D) Sequencing analyses of proviral sequences after infection with the VSV-G-pseudotyped NL4-3EGFPΔenvΔnef TTT (n = 37 clones) and AAA (n = 35 clones) mutants are shown (left and right panels, respectively). TF input: a plasmid for transfection to express HIV-1 transcripts (pNL4-3EGFPΔenvΔnef-based mutant plasmid). (E) Results of 5′ RACE analyses using RNA purified from virus particles of the VSV-G-pseudotyped NL4-3EGFPΔenvΔnef TTT mutant virus (n = 36 clones) and AAA mutant virus (n = 37 clones) are shown (upper and lower panels, respectively).
Conversely, multiple clones of proviral sequences such as GGG, AAA, GAA, and GGA were found from cells infected with the AAA mutant virus (Fig. 2D, right panels). In the 5′ RACE analyses using RNA purified from VSV-G-pseudotyped AAA mutant virus, the A1-, A2-, and shorter-form RNAs were found in virus particles (Fig. 2E, lower panels). When A2-form RNAs whose transcription initiates from the second A of AAA mutation were used as template for reverse transcription, the terminal of −sscDNA is likely to be 5′-(continued)-GAGATT-3′. On the other hand, sequences of template RNA (U3/R junction of 3′ LTR) is 5′-(continued)-GGGUCUC-(continued)-3′; hence, sequences between regions underlined such as GAGA and UCUC will be matched, but mismatch will occur after the first-strand transfer between GG in template and TT on the edge of the −sscDNA shown as bold letters. However, HIV-1 reverse transcriptase was reported to overcome misinserted nucleotides and elongate from mispairs between template and 3′-terminal DNA both in cell-free system and during viral replication in cells (23, 24). Thus, proviral sequences GAA and GGA can be reasonably produced from the A2- and A1-form HIV-1 RNAs, respectively. Here, we established a method to understand the 5′ edge of genomic RNA used for HIV-1 infection by analyzing proviral sequences using mutant viruses of the GGG tract in LTR. Notably, we concluded that the reason why the GGG tract in 5′ LTR (from 454 to 456 in the case of pNL4-3EGFPΔenvΔnef) is conserved among most HIV-1 strains (14, 25) is not because it is an absolute requisite to produce infectious HIV-1 particles.
Selective usage of the G1-form HIV-1 transcripts as genome in infectious virus particles rather than the other forms
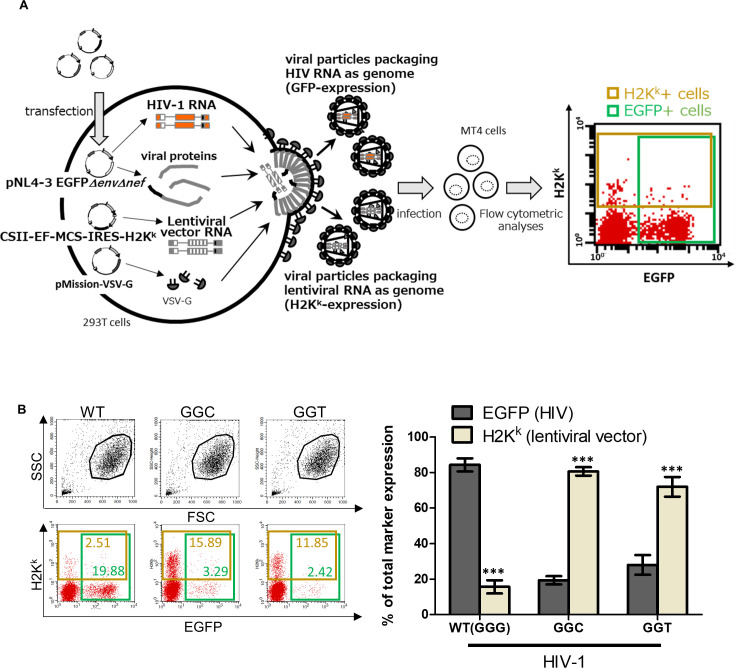
Next, we focused on difference in function between the G1-form transcripts and other forms and addressed selective usage of the G1-form over other forms of HIV-1 transcripts by using competition assay. Based on the existence of a GGG tract downstream of TATA-box in a plasmid coding genomic RNA of lentiviral vector, pCSII-EF-MCS-IRES-H2Kk (26), we carried out a competition between HIV-1 transcripts and lentiviral vector transcripts to assess which transcripts were used as viral genome in infectious particles (a schematic of the experiment is shown as Fig. 3A). We transfected 293T cells with pNL4-3EGFPΔenvΔnef, pCSII-EF-MCS-IRES-H2Kk, and pMission-VSV-G expressing envelope protein of VSV-G-pseudotyped virus, harvested supernatant 48 h post-transfection, exposed MT-4 cells with the harvested supernatant, and assessed which marker protein MT-4 cells express 48 h after the exposure. Since EGFP or mouse H2Kk antigen is encoded by genome of HIV-1 (NL4-3EGFPΔenvΔnef) or lentiviral vector, EGFP+ or H2Kk+ cells (cells in a green box or a dark yellow box in Fig. 3A) were defined as cells in which provirus derived from HIV genome or lentiviral vector genome integrated, respectively. As shown in dot plots of Fig. 3B, we calculated percentage of each EGFP+ or H2Kk+ out of total marker expression (number of EGFP+ cells + number of H2Kk+ cells; double-positive cells were counted as EGFP+ cells and also as H2Kk+ cells) to evaluate which transcripts play a role as genome in infectious particles. To avoid the multiple infection of cells with virus carrying the same marker gene, we collected results in which positive cells of each marker is less than 30%. In the first competition between RNAs derived from pNL4-3EGFPΔenvΔnef (WT) and pCSII-EF-MCS-IRES-H2Kk, EGFP or H2Kk occupied more than 80% or less than 20% of total marker expression, respectively (Fig. 3B, left bars labeled as WT in the bar graph), suggesting that more than 80% of infectious particles used HIV-1 transcripts as template for generating provirus. It might be reasonable because viral proteins are expressed from pNL4-3EGFPΔenvΔnef. Notably, we observed a completely opposite trend in a competition using the GGC or GGT mutants of NL4-3EGFPΔenvΔnef instead of WT (Fig. 3B; center or right bars in the bar graph, respectively). In the competition, H2Kk occupied more than 70% of total marker expression, suggesting that lentiviral vector transcripts are preferentially used as genomic RNA in infectious virus particles in the absence of the G1-form HIV-1 transcripts.
Fig 3.
Lentiviral vector transcripts rather than HIV-1 transcripts are primarily used as template for generating provirus in the absence of the G1-form HIV-1 transcripts. (A) A schematic represents a competition between HIV-1 transcripts and lentiviral vector transcripts. MT-4 cells were exposed with supernatant of 293T cells transfected with an HIV-1 proviral plasmid (pNL4-3EGFPΔenvΔnef), a lentiviral vector transfer plasmid (pCSII-EF-MCS-IRES-H2Kk), and the pMission-VSV-G. EGFP- or mouse antigen H2Kk-positive cells in green or dark yellow boxes represent cells infected with VSV-G-pseudotyped virus particles packaging HIV-1 or lentiviral vector genome, respectively. (B) Results of the competition between RNAs transcribed from pNL4-3EGFPΔenvΔnef WT, GGC, or GGT mutant and those from lentiviral vector pCSII-EF-MCS-IRES-H2Kk are shown. From three independent experiments, one representative dot plot is shown. Living cells are gated in black lines (the upper dot plots), and EGFP- or mouse antigen H2Kk-positive cells are gated in green or dark yellow boxes, respectively (the lower dot plots). In the bar graph, the Y-axis represents the percentage of each EGFP-positive (dark gray) or H2Kk-positive (bright gray) per total marker expression (EGFP positive + H2Kk positive). WT (GGG) or the GGC and GGT mutants of pNL4-3EGFPΔenvΔnef are used (left, center, and right bars, respectively). Error bars are ±standard eror of the mean of three independent experiments. Asterisk code for statistical significance compared to EGFP+: ***P < 0.001.
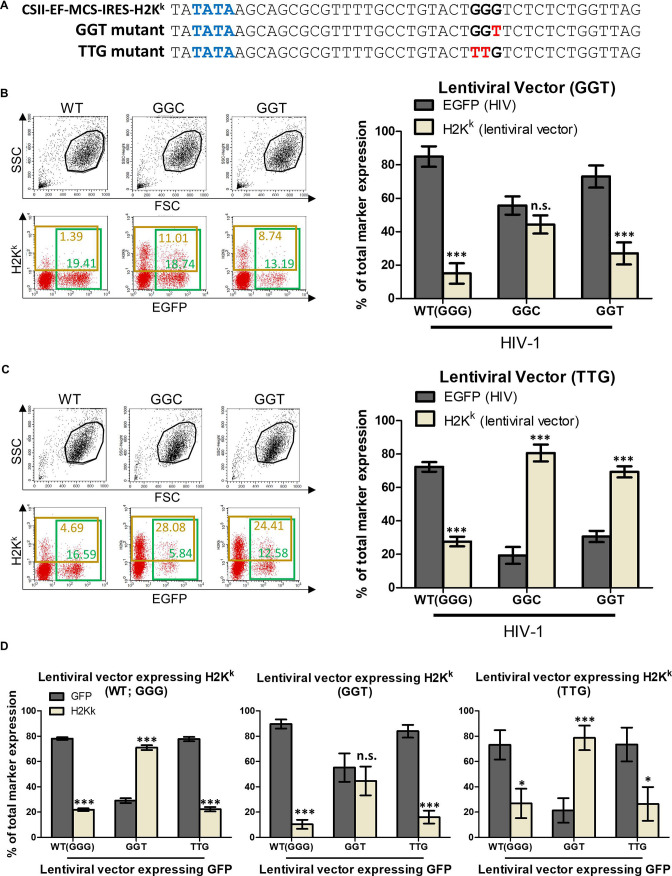
Next, we addressed a question whether the aforementioned opposite trend might not be observed in the absence of the G1-form of both HIV-1 RNAs and lentiviral vector RNAs. To assess the possibility, we generated a GGT mutant of pCSII-EF-MCS-IRES-H2Kk (Fig. 4A) and carried out the competition assay using it (Fig. 4B). In the competition using the pCSII-EF-MCS-IRES-H2Kk GGT mutant versus pNL4-3EGFPΔenvΔnef GGC or GGT mutants, lentiviral vector transcripts were not dominantly selected as genome in infectious virus (Fig. 4B; center or right bars in the bar graph, respectively), suggesting that the third G in the GGG tract of pCSII-EF-MCS-IRES-H2Kk would be necessary for the opposite trend that lentiviral vector transcripts are dominantly selected as genome in infectious particles. To assess whether the third G in the GGG tract of pCSII-EF-MCS-IRES-H2Kk suffices to recapitulate the initially found genome usage inversion shown in Fig. 3B, we generated a TTG mutant of pCSII-EF-MCS-IRES-H2Kk (Fig. 4A) and found that it is sufficient (Fig. 4C, center and right bars in the bar graph). Thus, we concluded that the G1-form transcripts are primarily selected as genome in infectious virus particles rather than the other forms in the case of not only HIV-1 RNA but also lentiviral vector RNA.
Fig 4.
The G1-form viral transcripts may be primarily used as template for generating provirus DNA rather than the other form transcripts. (A) A schematic represents sequences including TATA-box and a GGG tract in 5′ LTR of a transfer plasmid expressing genomic RNA of lentiviral vector (pCSII-EF-MCS-IRES-H2Kk). Nucleotide sequences of the GGT and TTG mutants are also shown. TATA-box, the GGG tract, and the mutated nucleotides are highlighted by blue, black, and red bold letters, respectively. (B and C) Results of the competition between RNAs transcribed from pNL4-3EGFPΔenvΔnef WT, GGC, or GGT mutant and those from the GGT (B) or the TTG (C) mutants of lentiviral vector transfer plasmid pCSII-EF-MCS-IRES-H2Kk are shown. Graphs are presented as described in Fig. 3B. Asterisk code for statistical significance compared to EGFP (HIV): ***P < 0.001; n.s., not significant, P > 0.05. (D) Results of the competition between RNAs transcribed from two lentiviral vectors. pCSII-EF-H2Kk WT, GGT or TTG mutant (left, center, and right graphs, respectively) and pCSII-EF-GFP WT, GGT, or TTG mutant (first, second, and third bars in each graph, respectively) were used. Y-axis represents the percentage of each GFP-positive (dark gray bars) or H2Kk-positive (bright gray bars) per total marker expression (GFP positive + H2Kk positive). Error bars are ±standard eror of the mean of three independent experiments. Asterisk code for statistical significance compared to GFP-positive: *P = 0.001 < P < 0.05, ***P < 0.001; n.s., not significant, P > 0.05.
We also confirmed these results by competition assay using two lentiviral vectors (CSII-EF-GFP and CSII-EF-H2Kk), which were generated using the same backbone plasmid and different marker genes. We generated WT (GGG), GGT, and TTG mutants of both lentiviral vectors and carried out a competition assay using them. When we used WT of CSII-EF-H2Kk with CSII-EF-GFP WT (GGG), GGT mutant, or TTG mutant (Fig. 4D, left graph), H2Kk occupied less than 25%, more than 70%, or less than 25% of the total marker expression, respectively, suggesting that H2Kk was predominant only when the G1-form RNA of CSII-EF-GFP was absent (Fig. 4D, second bars in left graph). Conversely, H2Kk occupied only 45% when the GGT mutants of both CSII-EF-H2Kk and CSII-EF-GFP were used (Fig. 4D, second bars in center graph) probably because the G1-form RNAs of CSII-EF-H2Kk are also absent as well as the G1-form RNAs of CSII-EF-GFP. In addition, we observed that H2Kk occupied more than 75% in a competition using the GGT mutant of CSII-EF-GFP and the TTG mutant of CSII-EF-H2Kk (Fig. 4D, second bars in right graph). Thus, results by the assay using two lentiviral vectors confirmed the conclusion that the G1-form transcripts are primarily selected as template for generating provirus for successful infection if they are present, which is also suggested by results of Fig. 3B and 4B and C. However, it is still possible that the results in Fig. 3 and 4 were biased by recombination between two different competitor transcripts in virus-producing cells or in virus particles.
Proviral sequences which can be generated with the G1-form HIV-1 transcripts as template were dominantly found
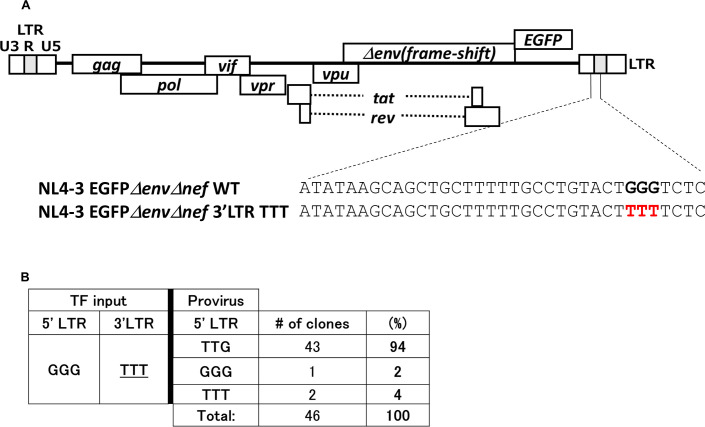
To confirm the conclusions that the G1-form viral RNAs are dominantly selected as genome of infectious virus, we finally generated a pNL4-3EGFPΔenvΔnef mutant plasmid in which a GGG tract in the U3/R junction of 3′ LTR was replaced with TTT (Fig. 5A). To evaluate what HIV-1 RNA transcripts were used as genome in infectious particles, we analyzed proviral sequences as done for Fig. 1E and 2D. From analyses of 46 clones of proviral sequences, as proviral sequences of the region for the GGG tract in 5′ LTR, we found that TTG occupied 94% of proviral sequences (43 clones, Fig. 5B), which can be generated from the G1-form HIV-1 transcripts with the activity of HIV-1 reverse transcriptase to elongate from mispairs between template RNA and 3′ terminal edge of primer DNA during reverse transcription (Fig. 6, upper panel). GGG or TTT were also found to occupy 2% and 4% of proviral sequences (one clone and two clones, respectively; Fig. 5B). This result confirms that the G1-form HIV-1 RNAs whose transcription initiates from the third deoxyguanosine of the GGG tract in the U3/R junction of 5′ LTR are predominantly selected as genome in infectious virion.
Fig 5.
Proviral sequences which can be generated with the G1-form HIV-1 transcripts as template were dominantly found. (A) A schematic represents the GGG tract located at the U3/R junction in 3’LTR of HIV-1 clone DNA (pNL4-3EGFPΔenvΔnef). Nucleotide sequences of the 3′ LTR TTT mutant of NL4-3EGFPΔenvΔnef are shown. The mutated nucleotides are highlighted by red bold letters. (B) Sequencing analyses of proviral sequences after infection with the VSV-G-pseudotyped NL4-3EGFPΔenvΔnef 3’LTR TTT mutant are shown (n = 46 clones). TF input: a plasmid for transfection to express HIV-1 transcripts (pNL4-3EGFPΔenvΔnef-based mutant plasmid).
Fig 6.
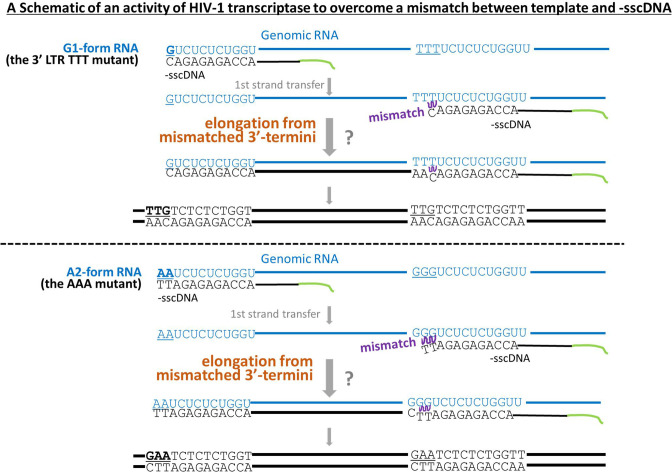
Presumed processes to generate provirus DNA from a genomic RNA of the 3′ LTR TTT or AAA mutant of HIV-1. A schematic represents provirus DNA production by overcoming mismatched 3′ termini between template RNA and minus-strand strong-stop cDNA (−sscDNA) is shown. The cases of the G1-form HIV-1 RNA of the 3′ LTR TTT mutant (upper panel) and an HIV-1 RNA beginning with two adenosines (an A2-form HIV-1 RNA of the AAA mutant, lower panel) are shown. RNAs and DNAs are written in blue and black, respectively.
DISCUSSION
To assess the importance of transcriptional initiation site heterogeneity in viral infection, single-round viral infection by using VSV-G-pseudotyped virus was employed in this study. As shown in Fig. 1C, infectious virus can be produced in the absence of the G1-form HIV-1 RNA. This result indicates that the G1-form HIV-1 RNA is not an absolute requisite for infectious HIV-1. In other words, not only HIV-1 particles incorporating the G1-form RNA as viral genome but also those carrying other HIV-1 RNAs are infectious. In fact, after infection with the GGC mutant virus, 71% of proviral sequences at the region of the GGG tract were GGC (Fig. 1E, left panels), which can be generated from RNAs whose transcription initiates from the first G (G454, GGC-form) or the second G (G455, GC form) as shown in Fig. 1F. On the other hand, 29% of proviral sequences were GGG (Fig. 1E), which is unlikely to be generated from GGC- or GC-form RNAs. Since HIV-1 RNAs whose transcription is reported to be initiated from the GGG tract of the wild-type HIV-1 (14), we expected that most transcription starts probably at first G (G454) and second G (G455) in the GGC sequences but possibly does also at a nucleotide upstream or downstream of GGC sequences (sequences are shown in Fig. 1A and B). In fact, the shorter-form RNAs (30%) were also found in addition to the C1-, GC-, and GGC-form RNAs (19%, 21%, and 30%, respectively; total of 70%) in virus particle of NL4-3EGFPΔenvΔnef GGC mutant virus by 5′ RACE analyses using RNA purified from VSV-G-pseudotyped virus (Fig. 1G, second panels). Based on the idea, proviral sequences GGG derived after infection with the GGC mutant virus (29%, Fig. 1E; the left panels) is likely to be generated from genomic RNAs whose transcription initiates from a nucleotide downstream of GGC sequences (shorter-form RNAs, Fig. 1F). After infection with the TTT mutant virus, GGG (97%) and TTT (3%) were found as proviral sequences of mutated region (Fig. 2D, left panels). Interestingly, it might be also reasonable because 97% of genomic RNAs in viral particles of the TTT mutant virus were found to be the shorter-form (Fig. 2E, upper panel), by which GGG could be generated as proviral sequences. In addition, in the 5′ RACE assay, we also found a U4-form RNA (3%) whose transcription initiates probably from deoxythymidine (T) just one nucleotide upstream of the TTT mutation (T highlighted by underline in Fig. 2A). Thus, proviral sequences TTT (3%) found in Fig. 2D are likely to be generated by using the U4-form RNA as template for reverse transcription. Many varieties of proviral sequences such as GGG, AAA, GAA, and GGA were found after infection with the AAA mutant virus (Fig. 2D, right panels). Proviral sequence GGG or AAA would be derived by using the shorter- or longer-form RNAs whose transcription initiates from a nucleotide downstream or upstream of AAA as template, respectively. We could not exactly determine from what genomic RNAs proviral sequences GAA or GGA can be derived, but it is possible that they were produced by reverse transcription using the shorter-form RNAs with unexpected random mutation(s). However, if mutations were inserted randomly resultant proviral sequences should vary and sequences other than GAA or GGA would have been found. As described previously, HIV-1 reverse transcriptase was reported to overcome the mismatch between template and primer and extend the mismatched 3′ termini by incorporation and polymerization of the next complementary nucleotide both in cell-free system and during viral replication in cells (23, 24). From a previous study, HIV-1 reverse transcriptase has an ability to extend over mismatches of as many as three bases during viral replication (23). Thus, from the aspect of the feature of HIV-1 transcriptase, proviral sequences such as AAA, GAA, and GGA can be reasonably generated with RNAs whose transcription initiates from each A of AAA (the A3-, A2-, and A1-form RNAs; the A2-form are shown in the lower panel of Fig. 6). In fact, we found the A1-, A2-, and shorter-form RNAs in virus particles of the AAA mutant virus in our 5′ RACE analyses (Fig. 2E, lower panels). Although in the assay we did not find any A3- or longer-form RNA which could generate proviral sequences AAA, we showed a possibility that HIV-1 transcription could initiate a nucleotide upstream of the GGG tract and the resultant longer-form RNA can be incorporated into virus particles (the U4-form RNA of the TTT mutant virus; Fig. 2E, upper panels) and used as template to generate provirus DNA (proviral sequence TTT; Fig. 2D, left panels). In addition, we are also interested in the fact that the A2-, A1-, and shorter-form RNAs were incorporated into virus particles (Fig. 2E, lower panels) and used as template RNA for reverse transcription for generating provirus DNA in the case of the AAA mutant virus (Fig. 2D, right panels). This suggests that there is no/little strict preference for packaging of the progeny genomes and diversification of HIV-1 RNA functions by heterogeneous transcriptional initiation sites usage might not work well in the case of the AAA mutant virus. It is far different from the situation of wild-type HIV-1 with the intact GGG tract that function of unspliced full-length RNAs is regulated according to a disciplined order depending on number(s) of guanosine in the 5′-terminus of RNAs (14). This result might suggest the reason why the GGG tract but not an AAA tract had been conserved as transcriptional initiation sites in 5′ LTR of HIV-1 genome.
We employed the competition assay between HIV-1 transcripts and lentiviral vector transcripts to assess which transcript is dominantly selected as genome in infectious particles (Fig. 3A). In the assay, we used 1 µg of the HIV-1 plasmid (pNL4-3EGFPΔenvΔnef, which expresses the genome of EGFP-expressing HIV-1 and viral proteins) and 0.2 µg of the lentiviral transfer plasmid (pCSII-EF-MCS-IRES-H2Kk, which expresses genome of the mouse antigen H2Kk-expressing lentiviral vector) for transfection. The ratio of these plasmids is critical for the assay, especially for the competitions using the GGT or TTG mutants of pCSII-EF-MCS-IRES-H2Kk (Fig. 4B or 4C, respectively). The competition experiments strongly indicated preferential usage of the G1-form HIV-1 transcripts or even the G1-form of lentiviral vector transcripts as genomic RNA of infectious particles (Fig. 3B, 4B and C). By previous studies (14), it was already reported that the G1-form RNAs are physically predominant in virus particles, but here we newly show that they would be selectively used as template of reverse transcription to establish successful infection if they are present.
Since differences between genomes of NL4-3EGFPΔenvΔnef and lentiviral vector might possibly affect the results of the assay, we also carried out the competition assay using two similar lentiviral vectors carrying different marker genes (CSII-EF-GFP and CSII-EF-H2Kk) to minimize differences between two competitor RNAs (Fig. 4D). For the assay, we used the GGT and TTG mutants as well as WT (GGG) of CSII-EF-GFP and CSII-EF-H2Kk. From results of the assay, we also concluded that the G1-form viral RNAs are predominantly used as template for reverse transcription to generate proviral DNA if the G1-form viral RNAs are present.
In the competitions shown in Fig. 3 and 4, we could not exclude the possibility of recombination between HIV transcripts and lentiviral vector transcripts in virus-producing cells or in virus particles even though location of open reading flame for EGFP and H2Kk are completely different in genomic RNAs of HIV-1 and lentiviral vector. Thus, the conclusions had to be confirmed by another experimental method (Fig. 5B). In fact, most proviral sequences were found to be TTG (94%), and others such as GGG (2%) and TTT (4%) were also found among clones we tested after infection with the 3′ LTR TTT mutant virus (Fig. 5A and B). Proviral sequence TTG could be derived from the G1-form HIV-1 transcripts as template with an ingenious feature of HIV-1 reverse transcriptase that it overcomes the mismatch between uridine (U) in genomic RNA template and deoxycytidine in −sscDNA (a schematic is shown in the upper panel of Fig. 6). It is again mentioned that our conclusions are stated with results not only of the competition assay but also of the proviral sequence analyses.
Masuda et al. also demonstrated that abortive forms of −sscDNA were more abundantly generated from the G3-form RNA than the G1-form RNA (14). In fact, after infection with the 3′ LTR TTT mutant, 94% of proviral sequences tested (43 out of 46 clones) were found to be TTG (Fig. 5B), whereas frequency of the G1-form RNA in virus particles was reported not to be over 90% but 79% in this study (Fig. 1G, first panels), approximately 70% (14), almost 80% (25), and just less than 80% (27) when 293T cells were transfected with a pNL4-3-based HIV-1 plasmid carrying the intact GGG tract in 5′ LTR. This difference might be caused by superiority of the G1-form RNA in viral processes after RNA packaging into virus particles, probably the process of reverse transcription. By viewing from the aspect of viral evolution, not only RNA packaging or reverse-transcription but also all processes during HIV-1 replication should be under selective pressures. The importance of transcriptional initiation site heterogeneity in HIV-1 replication might be further understood by following investigation.
Interestingly, a recent study showed that heterogeneity of transcriptional initiation sites and selective packaging of certain forms of unspliced viral RNA are conserved features of primate immunodeficiency viruses (25). In fact, the GGG tract in 5′ LTR and preferentially the packaged G1-form RNAs were found to be conserved among transmitted founder viruses of HIV-1 group M subtypes B and C and an SIV isolated from chimpanzees or gorillas (25), suggesting that these features probably improve viral replication or viral fitness in host primates. In fact, the third G in the GGG tract of 5′ LTR or the GGG tract itself was shown not to be essential for producing infectious particles (Fig. 1C and 2B), but infectious particles of WT virus were more efficiently produced than those of mutants which cannot express the G1-form RNA (the GGC and GGT mutant in Fig. 1D and TTT and AAA mutants in Fig. 2C), probably suggesting its importance for efficient production of infectious virus particles.
MATERIALS AND METHODS
Cells and transfection
HEK293T cells were maintained at 37°C with 5% CO2 in Dulbecco’s Modified Eagle Medium (Gibco, Waltham, MA) supplemented with 10% heat-inactivated fetal bovine serum (GE Healthcare, Logan, UT, USA) and penicillin-streptomycin (Fujifilm Wako, Osaka, Japan). MT-4 cells were maintained at 37°C with 5% CO2 in RPMI-1640 Medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (GE Healthcare) and penicillin-streptomycin (Fujifilm Wako). HEK293T cells were transfected using polyethylenimine (PEI, PolyScience, Niles, IL, USA).
Plasmids
pNL4-3EGFPΔenvΔnef (19) and pCSII-EF-MCS-IRES-H2Kk (26) were described previously. pCSII-EF-H2Kk and pCSII-EF-GFP were generated by inserting fragment for H2Kk ORF or EGFP ORF between XhoI and XbaI sites of pCSII-EF-MCS-IRES-H2Kk. The mutants of pNL4-3EGFPΔenvΔnef (GGC, GGT, TTT, AAA, and 3′ LTR TTT), pCSII-EF-MCS-IRES-H2Kk, pCSII-EF-H2Kk, and pCSII-EF-GFP (GGT and TTG) were constructed by overlap-extension PCR. All mutations were confirmed by sequencing analyses.
Evaluation of infected cells
Approximately 2 × 106 of 293T cells grown in a six-well plate were transfected with 3 µg of proviral plasmid pNL4-3EGFPΔenvΔnef WT or mutants and 1 µg of pMISSION-VSV-G (Sigma-Aldrich, St. Louis, MO, USA) to produce VSV-G pseudotyped HIV-1 vectors, and culture medium was changed once 24 h post-transfection. The supernatants containing VSV-G-pseudotyped HIV-1 vector were harvested 48 h post-transfection and filtered through 0.45-μm pore filters (Merck Millipore, Burlington, MA, USA). Approximately 2 × 106 of MT-4 cells were incubated with an optimized amount (achieving less than 30% EGFP-positive population) of viral supernatants. At 48 h after infection, cells were harvested and analyzed on FACSCalibur flow cytometer and analyzed using BD Cell Quest Pro software (BD Bioscience, San Diego, CA, USA). The number of infected cells produced with 1 mL of supernatant containing VSV-G-pseudotyped HIV-1 or 1 ng of p24 antigen quantified using RETROtek HIV-1 p24 Antigen ELISA kit (ZeptoMetrix Corp., Buffalo, NY, USA) was calculated.
Sequencing of provirus DNA
Approximately 2 × 106 of 293T cells grown in a six-well plate were transfected with 3 µg of proviral plasmid pNL4-3EGFPΔenvΔnef mutants and 1 µg of pMISSION-VSV-G; the culture medium was changed three times 24 h post-transfection, and supernatants containing VSV-G-pseudotyped HIV-1 vector were harvested 48 h post-transfection and filtered through 0.45-μm pore filters (Merck Millipore). The harvested supernatants were incubated with RNase-free Recombinant DNase I (Takara Bio, Shiga, Japan) at 37°C for 20 min. Approximately 2 × 106 of MT-4 cells were incubated with DNase I-treated supernatant in a 15-mL tube at 37°C for 60 min and washed with 10 mL of medium three times. Forty-eight h post-infection, EGFP expression was confirmed by microscopy, and genomic DNA was purified using DNeasy Blood & Tissue Kit (Qiagen Inc, Hilden, Germany). The first PCR was carried out using Prime STAR HS (Takara) with primers (seqFW238-NL: 5′-CGGAGGGAGAAGTATTAGTG-3′, seqRV998gag-NL: 5′-GTCTGAAGGGATGGTTGTAG-3′), which could amplify sequences in 5′ LTR but not those in 3′ LTR. The cycling condition included a denaturation step (94°C for 1 min), followed by 35 cycles of denaturation (94°C for 30 s), annealing (52°C for 30 s), and extension (72°C for 50 s). The first PCR product was subsequently subjected to nested PCR using primers (seqFW266-NL: 5′-GACAGCCTCCTAGCATTTC-3′, seqRV964gag-NL: 5′-GTCTACAGCCTTCTGATGTC-3′) with the same condition above except for annealing (60°C for 30 s). DNA fragments with appropriate size from the second PCR product were cloned into pCR4 using Zero Blunt TOPO PCR Cloning Kit (Invitrogen, Waltham, MA). Sequences of cloned fragments were analyzed by Eurofins Genomics (Tokyo, Japan). More than 33 clones were analyzed per mutant. To avoid possibilities that transfected plasmid DNA was amplified and analyzed, we used intact or mutants of pNL4-3EGFPΔenvΔnef because their sequences of 5′ LTR and 3′ LTR are different.
Competition assay between HIV-1 and lentiviral vector transcripts
Approximately 2 × 106 of 293T cells grown in a six-well plate were transfected with 1 µg of the HIV-1 plasmid (pNL4-3EGFPΔenvΔnef WT or mutants), 0.2 µg of the lentiviral transfer plasmid (pCSII-EF-MCS-IRES-H2Kk WT or mutants), and 1 µg of pMission-VSV-G (Sigma-Aldrich), and the culture medium was changed once 24 h post-transfection. At 48 h after transfection, supernatants containing VSV-G-pseudotyped viruses were harvested and filtered through 0.45-μm pore filters (Merck Millipore). Approximately 6 × 105 of MT-4 cells were incubated with an optimized amount (attaining less than 30% EGFP- and H2Kk-positive population) of viral supernatants. At 48 h after infection, cells were harvested, stained with Alexa Fluor 647-conjugated anti-H2Kk antibody (BioLegend, San Diego, CA, USA), and analyzed using FACSCalibur flow cytometer and BD Cell Quest Pro software (BD Bioscience). The percentage of each EGFP+ or H2Kk+ per total marker expression (EGFP+ +H2Kk+) was calculated.
Competition assay between two lentiviral vector transcripts
Approximately 2 × 106 of 293T cells grown in a six-well plate were transfected with 1 µg of the pCSII-EF-GFP WT or mutants, 0.75 µg of the pCSII-EF-H2Kk WT or mutants, 1 µg of pCAG-HIVgp (kindly provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED, Japan), and 0.5 µg of pCMV-VSV-G-RSV-Rev (kindly provided by the RIKEN BRC through the National BioResource Project of the MEXT/AMED), and the culture medium was changed once 24 h post-transfection. Approximately 2 × 106 of MT-4 cells were incubated with an optimized amount of viral supernatants (attaining less than 30% EGFP- and H2Kk-positive populations). The following procedures were done as done for the competition assay between HIV-1 and lentiviral vector transcripts.
5′ RACE assay
Approximately 7 × 106 of 293T cells grown in 10-cm plate were transfected with 15 µg of the HIV-1 plasmid (pNL4-3EGFPΔenvΔnef mutants) and 6 µg of pMission-VSV-G (Sigma-Aldrich), and the culture medium was changed three times 24 h post-transfection. At 48 h after transfection, supernatants containing VSV-G-pseudotyped virus were harvested, filtered through 0.45-μm pore filters (Merck Millipore), and concentrated using PEG-IT (System Biosciences, Mountain View, CA, USA). Viral RNAs in virus particles were purified using Isogen (Fujifilm Wako), and purified RNAs were subjected to SMARTer RACE 5′/3′ Kit (Takara Bio USA Inc., San Jose, CA, USA). As shown in Rawson et al. (25), RT primer 5′-GGTGGCTCCTTCTGATAATG-3′ and reverse PCR primer: 5′-GATTACGCCAAGCTTTCGTTCTAGCTCCCTGCTTG-3′ were used. Sequences of cloned fragments were analyzed by Eurofins Genomics. More than 33 clones were analyzed per mutant or WT.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 6. Data are presented as means with error bars indicating standard eror of the mean from three independent experiments. Student’s t-test was used for comparison between EGFP expression and H2Kk expression in the competitions (Fig. 3B, 4B through D). One-way analysis of variance with the Dunnett’s multiple comparison test was used for comparison of infectivity (Fig. 1D and 2C) (asterisk code for statistical significance: *P = 0.001 < P < 0.05; ***P < 0.001; not significant, P > 0.05).
ACKNOWLEDGMENTS
T.Y. thanks Dr. Yoshio Koyanagi for kindly providing pCSII-EF-MCS-IRES-H2Kk and Dr. Yuko Yoshida for fruitful discussion.
This work was supported by a grant (23fk0410041h9903) to T.Y., G.K., and T. Masuda by the Japan Agency for Medical Research and Development Research Project on AIDS/HIV and a grant (1224) to T.Y. from the Takeda Science Foundation.
We also recognize support from Dr. Masako Nishizawa, Dr. Shoji Yamaoka, Dr. Koji Sakai, Dr. Shigeyoshi Harada, Dr. Sayuri Seki, Dr. Kosuke Miyauchi, Mr. Yuki Honda and Ms. Shioko Kojima.
T.Y. conceived the study and designed the experiments. T.Y. and Y.K. performed the experiments. T.Y., Y.K., H.Y., G.K., K.H., T. Matano, and T. Masuda analyzed the data. T.Y. and T. Masuda contributed to reagents and materials. T.Y. wrote the paper. All authors reviewed the manuscript.
Contributor Information
Takeshi Yoshida, Email: takeshi-yoshida@umin.ac.jp.
Viviana Simon, Icahn School of Medicine at Mount Sinai, New York, New York, USA.
REFERENCES
- 1. Freed EO. 2015. HIV-1 assembly, release and maturation. Nat Rev Microbiol 13:484–496. doi: 10.1038/nrmicro3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bieniasz P, Telesnitsky A. 2018. Multiple, switchable protein:RNA interactions regulate human immunodeficiency virus type 1 assembly. Annu Rev Virol 5:165–183. doi: 10.1146/annurev-virology-092917-043448 [DOI] [PubMed] [Google Scholar]
- 3. Olson ED, Musier-Forsyth K. 2019. Retroviral Gag protein-RNA interactions: implications for specific genomic RNA packaging and virion assembly. Semin Cell Dev Biol 86:129–139. doi: 10.1016/j.semcdb.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beemon KL. 2022. Retroviral RNA processing. Viruses 14:1113. doi: 10.3390/v14051113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rein A. 2019. RNA packaging in HIV. Trends Microbiol 27:715–723. doi: 10.1016/j.tim.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuzembayeva M, Dilley K, Sardo L, Hu W-S. 2014. Life of psi: how full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 454–455:362–370. doi: 10.1016/j.virol.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen J, Nikolaitchik O, Singh J, Wright A, Bencsics CE, Coffin JM, Ni N, Lockett S, Pathak VK, Hu WS. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci U S A 106:13535–13540. doi: 10.1073/pnas.0906822106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lu K, Heng X, Summers MF. 2011. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol 410:609–633. doi: 10.1016/j.jmb.2011.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D’Souza V, Summers MF. 2005. How retroviruses select their genomes. Nat Rev Microbiol 3:643–655. doi: 10.1038/nrmicro1210 [DOI] [PubMed] [Google Scholar]
- 10. Lu K, Heng X, Garyu L, Monti S, Garcia EL, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Divakaruni SS, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. 2011. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 334:242–245. doi: 10.1126/science.1210460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russell RS, Liang C, Wainberg MA. 2004. Is HIV-1 RNA dimerization a prerequisite for packaging? yes, no, probably? Retrovirology 1:23. doi: 10.1186/1742-4690-1-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pereira-Montecinos C, Toro-Ascuy D, Ananías-Sáez C, Gaete-Argel A, Rojas-Fuentes C, Riquelme-Barrios S, Rojas-Araya B, García-de-Gracia F, Aguilera-Cortés P, Chnaiderman J, Acevedo ML, Valiente-Echeverría F, Soto-Rifo R. 2022. Epitranscriptomic regulation of HIV-1 full-length RNA packaging. Nucleic Acids Res 50:2302–2318. doi: 10.1093/nar/gkac062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. 2004. Dimerization of retroviral RNA genomes: an inseparable pair. Nat Rev Microbiol 2:461–472. doi: 10.1038/nrmicro903 [DOI] [PubMed] [Google Scholar]
- 14. Masuda T, Sato Y, Huang Y-L, Koi S, Takahata T, Hasegawa A, Kawai G, Kannagi M. 2015. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep 5:17680. doi: 10.1038/srep17680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kharytonchyk S, Monti S, Smaldino PJ, Van V, Bolden NC, Brown JD, Russo E, Swanson C, Shuey A, Telesnitsky A, Summers MF. 2016. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc Natl Acad Sci U S A 113:13378–13383. doi: 10.1073/pnas.1616627113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brown JD, Kharytonchyk S, Chaudry I, Iyer AS, Carter H, Becker G, Desai Y, Glang L, Choi SH, Singh K, Lopresti MW, Orellana M, Rodriguez T, Oboh U, Hijji J, Ghinger FG, Stewart K, Francis D, Edwards B, Chen P, Case DA, Telesnitsky A, Summers MF. 2020. Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 368:413–417. doi: 10.1126/science.aaz7959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ding P, Kharytonchyk S, Kuo N, Cannistraci E, Flores H, Chaudhary R, Sarkar M, Dong X, Telesnitsky A, Summers MF. 2021. 5′-cap sequestration is an essential determinant of HIV-1 genome packaging. Proc Natl Acad Sci U S A 118:e2112475118. doi: 10.1073/pnas.2112475118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Obayashi CM, Shinohara Y, Masuda T, Kawai G. 2021. Influence of the 5′-terminal sequences on the 5′-UTR structure of HIV-1 genomic RNA. Sci Rep 11:10920. doi: 10.1038/s41598-021-90427-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yao W, Yoshida T, Hashimoto S, Takeuchi H, Strebel K, Yamaoka S. 2020. Vpu of a simian immunodeficiency virus isolated from greater spot-nosed monkey antagonizes human BST-2 via two AxxxxxxxW motifs. J Virol 94:e01669-19. doi: 10.1128/JVI.01669-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shatkin AJ. 1987. mRNA caps – old and newer hats. Bioessays 7:275–277. doi: 10.1002/bies.950070611 [DOI] [PubMed] [Google Scholar]
- 21. Yao W, Strebel K, Yamaoka S, Yoshida T. 2022. Simian immunodeficiency virus SIVgsn-99CM71 Vpu employs different amino acids to antagonize human and greater spot-nosed monkey BST-2. J Virol 96:e0152721. doi: 10.1128/JVI.01527-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Klaver B, Berkhout B. 1994. Premature strand transfer by the HIV-1 reverse transcriptase during strong-stop DNA synthesis. Nucleic Acids Res 22:137–144. doi: 10.1093/nar/22.2.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu H, Goodman MF. 1992. Comparison of HIV-1 and avian myeloblastosis virus reverse transcriptase fidelity on RNA and DNA templates. J Biol Chem 267:10888–10896. doi: 10.1016/S0021-9258(19)50101-6 [DOI] [PubMed] [Google Scholar]
- 24. Perrino FW, Preston BD, Sandell LL, Loeb LA. 1989. Extension of mismatched 3′ termini of DNA is a major determinant of the infidelity of human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci U S A 86:8343–8347. doi: 10.1073/pnas.86.21.8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rawson JMO, Nikolaitchik OA, Shakya S, Keele BF, Pathak VK, Hu WS. 2022. Transcription start site heterogeneity and preferential packaging of specific full-length RNA species are conserved features of primate lentiviruses. Microbiol Spectr 10:e0105322. doi: 10.1128/spectrum.01053-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoshida T, Kawano Y, Sato K, Ando Y, Aoki J, Miura Y, Komano J, Tanaka Y, Koyanagi Y. 2008. A CD63 mutant inhibits T-cell tropic human immunodeficiency virus type 1 entry by disrupting CXCR4 trafficking to the plasma membrane. Traffic 9:540–558. doi: 10.1111/j.1600-0854.2007.00700.x [DOI] [PubMed] [Google Scholar]
- 27. Nikolaitchik OA, Liu S, Kitzrow JP, Liu Y, Rawson JMO, Shakya S, Cheng Z, Pathak VK, Hu WS, Musier-Forsyth K. 2021. Selective packaging of HIV-1 RNA genome is guided by the stability of 5′ untranslated region polyA stem. Proc Natl Acad Sci U S A 118:e2114494118. doi: 10.1073/pnas.2114494118 [DOI] [PMC free article] [PubMed] [Google Scholar]