ABSTRACT
Ruminant livestock, including cattle, sheep, goats, and camels, possess a distinctive digestive system with complex microbiota communities critical for feed conversion and secondary metabolite production, including greenhouse gases. Yet, there is limited knowledge regarding the diversity of rumen microbes and metabolites benefiting livestock physiology, productivity, climate impact, and defense mechanisms across ruminant species. In this study, we utilized metataxonomics and metabolomics data from four evolutionarily distinct livestock species, which had fed on diverse plant materials like grass, shrubs, and acacia trees, to uncover the unique signature microbes and secondary metabolites. We established the presence of a distinctive anaerobic fungus called Oontomyces in camels, while cattle exhibited a higher prevalence of unique microbes like Psychrobacter, Anaeromyces, Cyllamyces, and Orpinomyces. Goats hosted Cleistothelebolus, and Liebetanzomyces was unique to sheep. Furthermore, we identified a set of conserved core microbes, including Prevotella, Rickenellaceae, Cladosporium, and Pecoramyces, present in all the ruminants, irrespective of host genetics and dietary composition. This underscores their indispensable role in maintaining crucial physiological functions. Regarding secondary metabolites, camel’s rumen is rich in organic acids, goat’s rumen is rich in alcohols and hydrocarbons, sheep’s rumen is rich in indoles, and cattle’s rumen is rich in sesquiterpenes. Additionally, linalool propionate and terpinolene were uniquely found in sheep rumen, while valencene was exclusive to cattle. This may suggest the existence of species-specific microbes and metabolites that require host rumen-microbes’ environment balance. These results have implications for manipulating the rumen environment to target specific microbes and secondary metabolite networks, thereby enhancing livestock productivity, resilience, reducing susceptibility to vectors, and environmentally preferred livestock husbandry.
IMPORTANCE
Rumen fermentation, which depends on feed components and rumen microbes, plays a crucial role in feed conversion and the production of various metabolites important for the physiological functions, health, and environmental smartness of ruminant livestock, in addition to providing food for humans. However, given the complexity and variation of the rumen ecosystem and feed of these various livestock species, combined with inter-individual differences between gut microbial communities, how they influence the rumen secondary metabolites remains elusive. Using metagenomics and metabolomics approaches, we show that each livestock species has a signature microbe(s) and secondary metabolites. These findings may contribute toward understanding the rumen ecosystem, microbiome and metabolite networks, which may provide a gateway to manipulating rumen ecosystem pathways toward making livestock production efficient, sustainable, and environmentally friendly.
KEYWORDS: ruminants, metabolomics, rumen, fermentation, microbiota, metataxonomic, metabolites
INTRODUCTION
Livestock are an important part of the ecosystem, especially because they are a major driver in most rural landscapes, diversifying belowground microbes, soil health, function, fertility, and crop productivity. Globally, it is estimated that more than 1.2 billion people are making a living in the livestock sector across the various value chains (1, 2).
Ruminant livestock provide humans with foods, such as milk and meat from non-human-edible plant material, even in arid and semi-arid ecologies, where crop production is not possible due to erratic rainfall and frequent drought, thus the only means to sustainably use such vast land is through sustainable livestock husbandry. Livestock in arid and semi-arid areas are vulnerable to climate shocks, which can be determined by the duration, frequency, and severity of the shocks, as well as the location of the stocks and related assets (3–5). Additionally, livestock’s vulnerability to climate change also depends on their sensitivity, which is determined by the breed, feeding system, efficiency, and resilience (5, 6). For instance, the one-humped camel (Camelus dromedarius) is the most efficient and resilient animal well adapted to arid and semi-arid ecologies with limited resources, this is recently evidenced as pastoralists shifted from cattle to camel keeping even at higher altitudes (7–10). This can be taken as a climate change adaptation strategy and has the potential to improve livestock climate resilience if the underlying mechanism is understood. However, the underlying mechanisms responsible for the observed variations in resilience between different livestock are not clear.
The rumen, a large fermentation chamber in ruminant livestock, harbors diverse and complex microbial communities that play crucial roles in the digestion and fermentation of feedstuff (11, 12) and the production of diverse metabolites including greenhouse gases (13–16). The relationship between some members of the microbiome and rumen function is well known (13, 17). The role of diet on microbial diversity has been investigated (16, 18, 19). Whereas host genetics have been studied in determining rumen microbes (20–22), most of the studies have been done on a single species and biased towards cattle, and no comparative studies have been reported between diverse ruminant animals that vary both in feeding regime and resilience, which is the focus of this study.
We hypothesize that livestock vary in their rumen microbes and secondary metabolites that have useful traits for livestock resilience and efficiency. The rumen environment hosts the most complex diverse microbial communities consisting of bacteria, fungi, protozoa, etc. Therefore, understanding the diversity and the pivotal role of the rumen microbes and secondary metabolites in digesting fibrous feed, providing nutrients to the host animal, defense, and determining livestock host-environment interaction is key for sustainable animal husbandry. Pertinent global issues of interest include climate resilience, the fight against climate change, and vector-borne diseases through rumen environment manipulation to make livestock part of the solution. Here, using four ruminant livestock that vary in feeding regime, drought resilience, and disease prevalence, we show that each livestock species created a mutual association with signature microbes and secondary metabolites that provide useful ecological traits.
RESULTS
Distribution of bacterial and fungal populations in the rumen
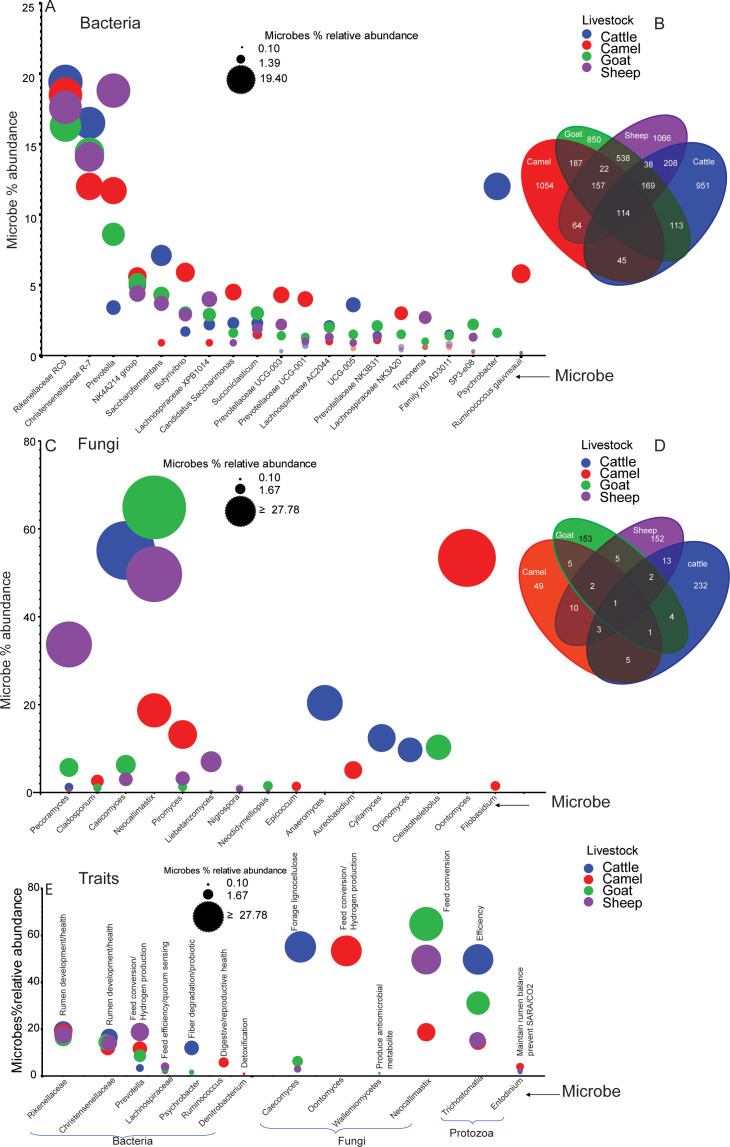
To correlate the secondary metabolites with rumen microbes, we performed genomic analysis of the two main rumen domains, bacteria and fungi. The taxonomic analysis of bacterial and fungal populations in the rumens of cattle, sheep, goats, and camels revealed a variation in the dominance of core groups of rumen microbes among the four ruminants (Fig. 1). A total of 1,052 species-level, bacterial amplicon sequence variants (ASVs) were uniquely identified in camels, 949 in cattle, 1,065 in sheep, and 847 in goats, respectively (Fig. 1B). Whereas 113 bacterial ASVs were shared by all the four ruminants, 187 ASVs were shared by both camels and goats, while 208 ASVs were common in cattle and sheep (Fig. 1B). Additionally, among the analyzed bacterial ASVs, 36,363, 47,659, 59,338, and 48,806 bacterial ASVs were unclassified in cattle, camels, goats, and sheep, respectively.
Fig 1.
Dominant bacteria and fungi across the four livestock species. (A) Bubble plots showing the qualitative and quantitative differences of unique and shared bacteria. (B) Venn diagrams showing the total number of the identified unique and shared bacterial microbes. (C) Bubble plots showing the qualitative and quantitative differences of unique and shared fungi communities among the four livestock. (D) Venn diagrams show the total number of the identified unique and shared fungi. (E) Association analysis between selected microbes and host traits. Trait analysis was done based on references 13, 23–38. Plots generated using microbes’ relative abundance data with at least 1% relative abundance in one of the four livestock species.
Bacteria, being the main members of the rumen microbiome, were widely dominant across the four livestock groups analyzed, comprising most of the species richness, with some bacterial genera being livestock specific (Fig. 1A and B). Further analysis of the identified bacterial ASVs revealed the 20 most abundant bacterial genera present among the four livestock species (Fig. 1A and C). In all four ruminants, the Rickenellaceae RC9, Christensenellaceae R-7 group, NK4A214 group, and Succiniclasticum group are conserved both in their presence and abundance (Fig. 1A). The genus Ruminococcus gauvreauii was abundantly present in camels and in small amount in goats. Prevotella, and Prevotellaceae a hydrogen-producing bacterial genus, was dominant in camels, however, less abundant in cattle, goats, and sheep (Fig. 1A). The Psychrobacter genus was found uniquely in cattle and goats but absent in sheep and camels (Fig. 1A; Table S1). All the remaining bacterial genera were conserved in all four livestock species but with varying abundance. Compared to camels and sheep, cattle and goats had more bacterial diversity due to an additional genus, Psychrobacter (Fig. 1A; Table S1).
A comprehensive analysis was conducted on the ASVs of fungi in four livestock species, namely cattle, camels, goats, and sheep. The results showed a total of 232 ASVs in cattle, 49 ASVs in camels, 153 ASVs in goats, and 152 ASVs in sheep at the genus level (Fig. 1C and D). Furthermore, 1,639, 1,450, 6,666, and 1,022 fungal ASVs were unclassified in cattle, camels, goats, and sheep, respectively. Among the identified fungal ASVs, a diverse population of 17 highly prevalent fungal genera was found (Fig. 1C). The analysis further showed that only one fungal ASV was common in all four livestock species, while five were common in camels and goats, whereas cattle and sheep shared 13 ASVs (Fig. 1D). Goats had the highest representation of fungal genera with camels having the least representation among the four ruminants (Fig. 1D). The anaerobic fungal genus Caecomyces was abundantly present in cattle and in small amount in goats and sheep but missing in camels. The aerobic fungus genus Oontomyces was exclusively found in camels in high abundance. On the other hand, Neocallimastix was the most abundant in both goats and sheep, present in small amounts in camels but missing in cattle. Pecoramyces was found only in sheep and goats; in the past, it was much more abundant but missing from camels and cattle (Fig. 1C; Table S2). Liebetanzomyces were only found in sheep. Furthermore, Anaeromyces, Orpinomyces, and Cyllamyces were unique to cattle, whereas Piromyces were the major groups in camels (Fig. 1C; Table S2). Nigrospora was absent in cattle but present in the other three livestock in small amounts. Caecomyces, which was dominant in cattle, was absent in camels, though present in both goats and sheep. Cleistothelebolus was distinct to goats and may be considered a signature fungal community in goat rumen (Fig. 1C; Table S2). Only Cladosporium and Pecoramyces were found to be conserved among the four livestock, thus suggestive of their roles for conserved function. Furthermore, camels harbor different protozoans as compared to other livestock (data not shown).
Alpha and beta diversity
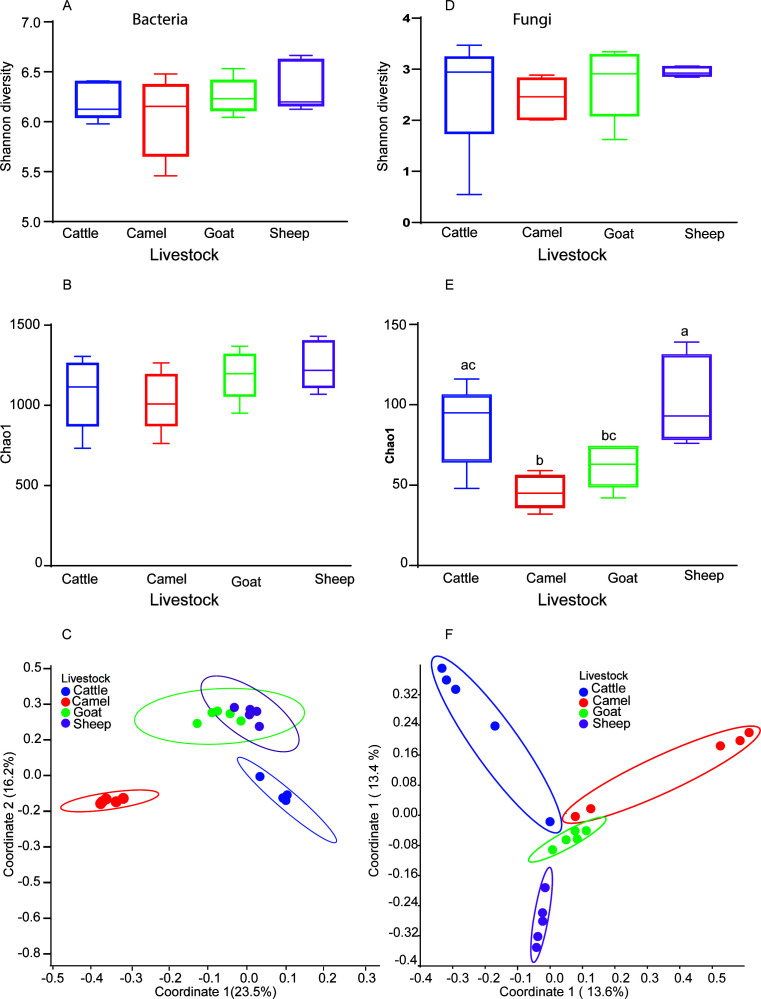
The four ruminants showed greater diversity in bacterial microbes as revealed by the high Shannon alpha diversity index, which takes into consideration both abundance and richness; however, there was no significant difference between the four livestock species, F = 1.18, P = 0.35 and Chao1 value F = 1.59, P = 0.23 (Fig. 2A and B). Beta diversity, variation of bacterial communities between the four livestock species phylogenetically, was assessed by calculating the principal coordinate analysis (PCoA) of different rumen bacterial domains using unweighted UniFrac distance dissimilarity and permutational multivariate analysis of variance (PERMANOVA) permutations 9,999 (P < 0.05) based on the Bray-Curtis dissimilarity distance matrix using all ASVs as an input. The investigation indicated that cattle exhibit a high degree of similarity among themselves, whereas camels also demonstrate a comparable level of similarity within themselves. Nevertheless, the goats and sheep exhibited a close grouping, indicating a lack of distinction between the two species (Fig. 2C).
Fig 2.
Diversity of bacteria and fungi across the four livestock species. (A) Shannon diversity index for bacteria. (B) Chao1 index for bacteria. (C) Beta diversity PCoA ellipse clusters showing the distribution of bacteria. (D) Shannon diversity index for fungi. (E) Chao1 richness index for fungi. (F) Beta diversity PCoA ellipse clusters showing the distribution of fungi.
Furthermore, the Shannon diversity index (Fig. 2D) did not show any notable variations in fungal microorganisms across the four livestock species. Nevertheless, the Chao1 index, which specifically quantifies microbial richness, indicated that camels had lower richness compared to cattle and sheep (F = 7.63, P = 0.002, analysis of variance [ANOVA] followed by Tukey’s multiple comparisons test) (Fig. 2E). The beta diversity analysis showed that there is individual variation between cattle and camel populations but still tend to cluster separately (P < 0.05, PERMANOVA) (Fig. 2F).
Dietary composition assessment in livestock rumen
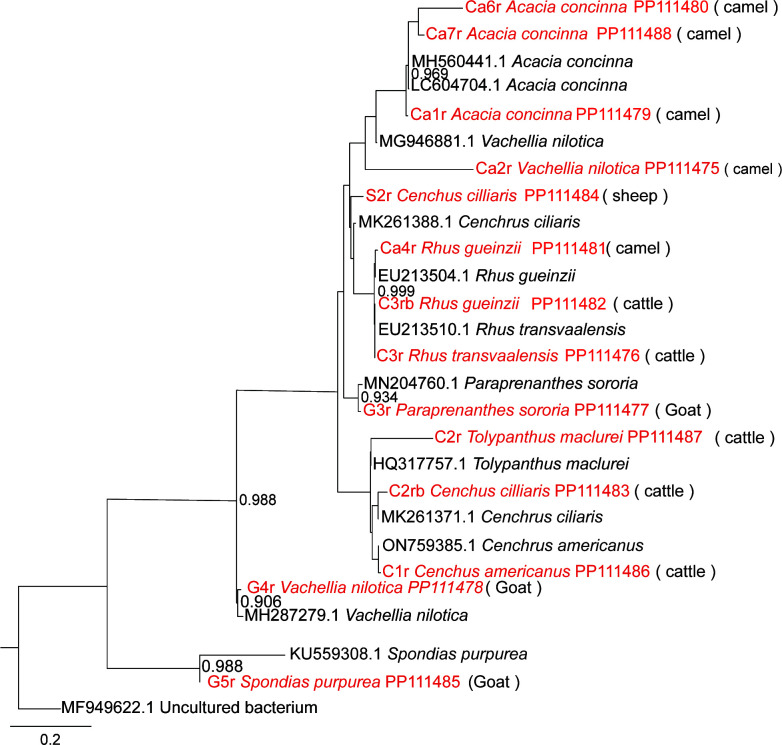
Besides the host’s individual genetic makeup, the composition of the host’s diet influences the types and amounts of substrates available to the microbial community, which, in turn, influence the production of secondary metabolites (20). Therefore, we evaluated the dietary composition in the rumens of the four livestock species. We established that the four ruminants fed on diverse plant materials. This may be attributed to the fact that feeding in pastoralist setup is not controlled or restricted, and hence livestock have access to a wide range of plant materials. For instance, we found that in addition to grasses (Poaceae), Cenchus cilliaris and Cenchus americanus, which had been consumed by cattle and sheep, cattle had consumed other plant species such as Rhus gueinzii and Rhus transvaalensii despite being predominantly grazers (Fig. 3). Unlike cattle and sheep, camels and goats are specially adapted to feed on leaves, fruits of high-growing woody plants, soft shoots, and shrubs, such as Acacia concinna, Paraprenanthes sororia, Vachellia nilotica, and Searsia tripartita (Fig. 3), which are predominantly found within arid and semi-arid areas. Therefore, it points to the diversity in dietary composition among the ruminants, which influences both the metabolite compound and microbial population composition among the ruminants.
Fig 3.
Phylogenetic tree showing plant diet composition in the rumens of the four livestock. The plant species identified in this study from each livestock and their corressponding accession numbers are indicated in red, while their ex-type strains are shown in black. Phylogenetic analysis based on maximum likelihood with a bootstrap of 1,000 pseudoreplicates, with MF949622.1 uncultured bacterium as the outgroup taxa. Maximum likelihood bootstrap values equal to or greater than 65% are shown on the nodes, while the scale bar indicates 0.2 changes.
Ruminal metabolite composition in livestock
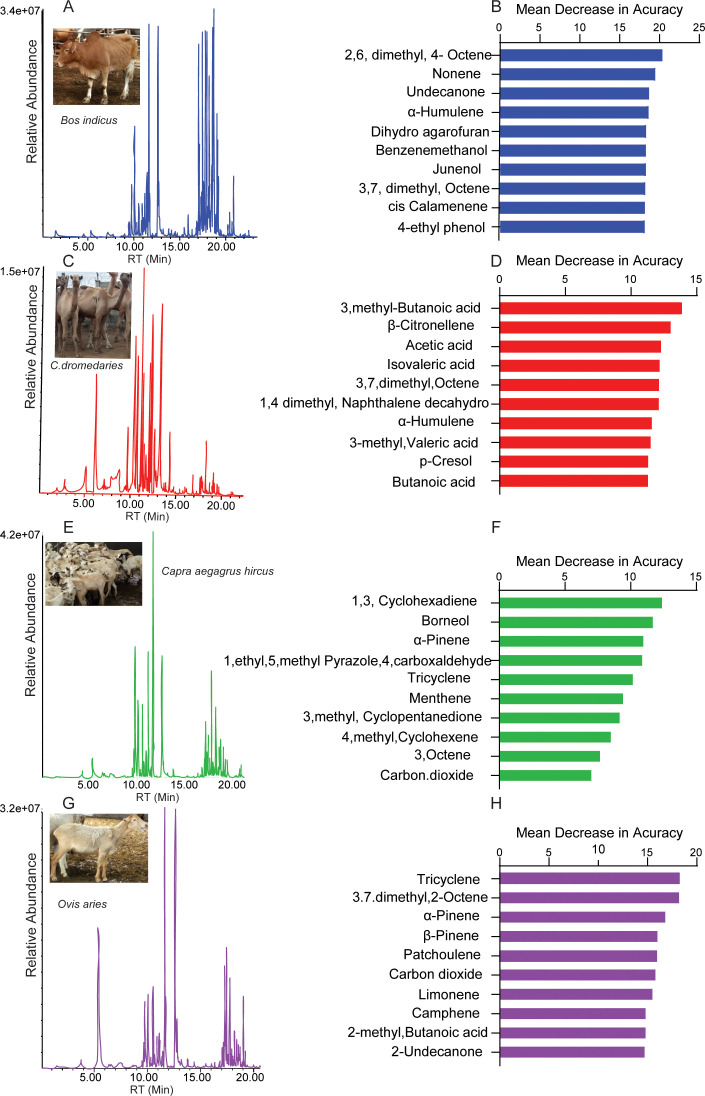
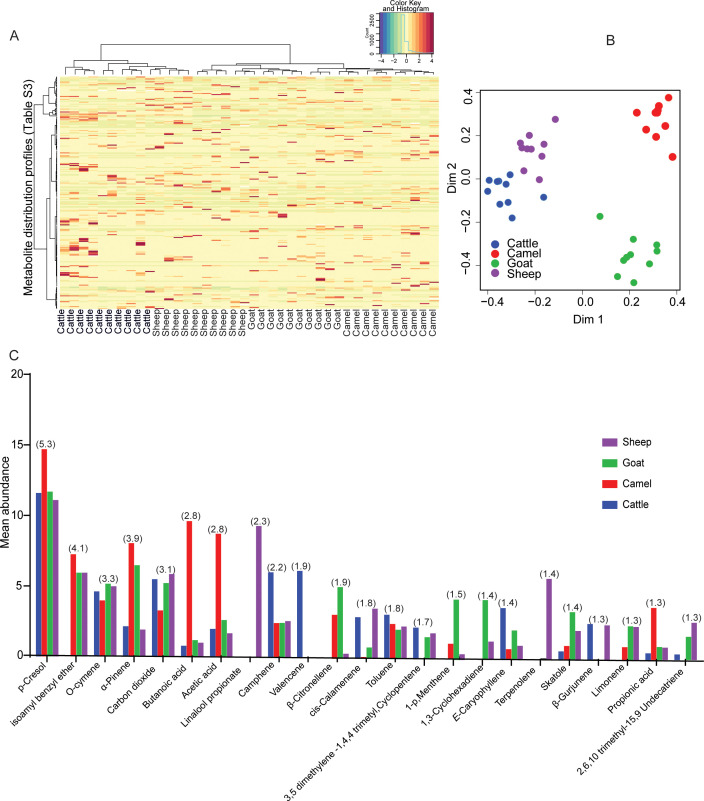
In the present study, a total of 162 metabolite compounds (Table S3) were identified in the bovine rumen content of four livestock; cattle, sheep, goats, and camels by gas chromatography-mass spectrometry (GC-MS). The detected compounds represented various chemical classes, including alcohols, ketones, phenols, volatile fatty acids, terpenes, esters, and hydrocarbons. Although most major classes of secondary metabolites have ubiquitous distributions among the four livestock, each livestock species has its own signature secondary metabolites (Fig. 4A). A random forest classification was conducted to reveal the top 10 predictive compounds for individual ruminant species (Fig. 4B). 2, 6 dimethyl 4-octene, 3-methylbutanoic acid, 1, 3-cyclohexene, and tricyclene being the most predictive secondary metabolite compounds of cattle, camels, goats, and sheep rumen, respectively. In camel, 5 out of the 10 predictive compounds are acids, signifying the diversity of acid in camel rumen. However, we did not observe the dominance of any specific chemical class in the other livestock; diverse classes of compounds contribute to predictive signature odors. The diversity and contrast in metabolite composition among the ruminants were revealed by the clustering and segregation of the respective species based on their metabolite composition by multidimensional scaling (MDS) and matrix plot (Fig. 5A and B). While some species, such as cattle and sheep, which are grazers clustered in proximity, camels and goats, which are browsers, were distinctly clustered apart from the other ruminants and from each other (Fig. 5B), thus, demonstrating a similarity in metabolite composition among grazers (cattle and sheep) but not clear with browsers (camels and goats). Overall, the four livestock dissimilarity based on their rumen secondary metabolites was 72.5%. Twenty-three compounds, based on quantitative and qualitative differences contributed to more than 50% of the variation (Fig. 5C). Specific metabolite compounds were found to be unique to specific livestock and some were shared between two or more ruminant species. For instance, compounds like isoamyl benzyl ether and beta-citronellene were absent in cattle, while linalool propionate and Valencene were distinctively detected in sheep and cattle, respectively. Similarly, cis-calamine and 1, 5, 9-undecatriene were absent among camels, while β-gurjunene was absent in both camels and goats and limonene was missing in cattle. Additionally, 1-p menthene was not found in cattle. On the other hand, 1, 3-cyclohexadiene is found solely in goat and sheep rumens.
Fig 4.
Rumen secondary metabolite profiles for various ruminant livestock. (A, C, E, and G) GC-MS chromatogram of metabolite compounds in the ruminal fluid of various livestock, cattle, camels, sheep, and goats, respectively. ( B, D, F, H) Histograms showing the classification of the top 10 important compounds from different livestock rumens based on their mean decrease in accuracy (MDA) of the random forest analysis. Metabolites with the highest MDA value as shown on the histogram are the most important and consequently most predictive for each species. Animals’ pictures are original.
Fig 5.
Diversity of rumen secondary metabolites across the four livestock species. (A) Heatmap-coded matrix showing the relative percent contribution of individual compounds to the total composition of each livestock species (for details, please see Table S3). (B) MDS plot showing the segregation of ruminants based on metabolite composition. (C) Histogram showing the classification of the top 23 metabolite compounds contributed to 50% dissimilarity between the four livestock based on similarity percentage analysis; the number in parenthesis is the percent contribution of a given compound for the dissimilarity.
Metabolite composition by chemical functional groups
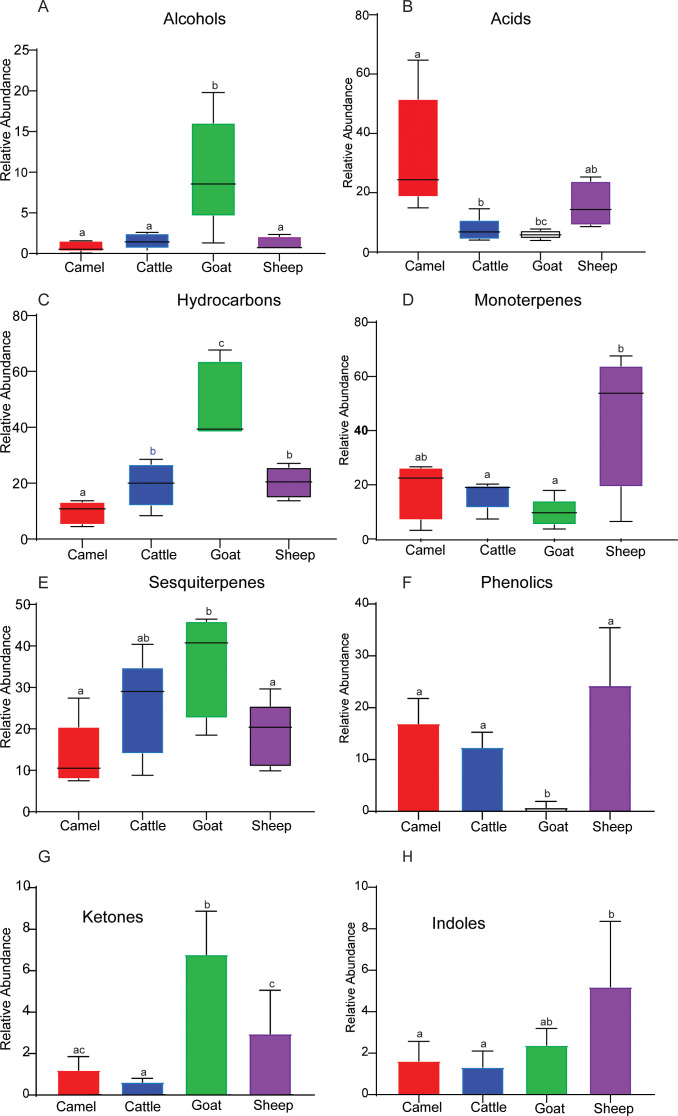
We then evaluated the chemical identities and variation in the distribution of volatile organic compounds across the four ruminant species, with compounds categorized based on their functional group classification, including phenols, alcohols, indoles, monoterpenes, sesquiterpenes, acids, hydrocarbons, and ketones. We found significant differences in the relative abundance of certain chemical classes, such as alcohols, hydrocarbons, monoterpenes, acids, and sesquiterpenes, among the four livestock groups, with cattle, sheep, camels, and goats displaying varying relative abundance of these compounds (Fig. 6A through G, P < 0.05). Cattle, sheep, and camels had significantly lower alcohol and sesquiterpene concentrations compared to goats (Fig. 6A and E, ANOVA, P < 0.05). Similarly, camels were established to contain the highest acid concentration compared to cattle, sheep, and goats (ANOVA, P = 0.004, Fig. 6B), which resulted in an acidic rumen environment in camels that had been established when the rumen pH was determined for the four ruminants. Camel ruminal pH was significantly lower (pH 6.3–6.5) compared to cattle, goats, and sheep, which had a relatively neutral pH ranging from 7.0 to 7.4 (ANOVA, P < 0.05). Interestingly, when we examined the carbon dioxide emissions, the three livestock species (cattle, sheep, and goats) produced almost twice as much carbon dioxide as camels. This intriguing array of differences in chemical composition and carbon dioxide emissions among livestock highlights the diversity of these animals in our environment. On the other hand, phenols, ketones, and hydrocarbons were more abundant in goats compared to other livestock, and sheep had more indoles compared to other livestock (ANOVA, P < 0.05, Fig. 6H).
Fig 6.
Dominant secondary metabolites by functional groups. (A–H) Box plots showing variations of chemical families of identified metabolite compounds among different ruminants. Bar graphs followed by different letters are statistically different in their abundance based on the analysis of variance (P ≤ 0.05).
DISCUSSION
Microbiome and diet composition
In this comparative study, we reveal an intricate network of rumen microorganisms that coexist, interact, and vie for resources, ultimately maintaining a delicate equilibrium of diverse end products, including secondary metabolites. These products not only fuel microbial growth without autotoxicity but also yield beneficial outcomes for the host. A better understanding of the impact of the rumen microbiome on host well-being and performance could open the door to innovative approaches and strategies for enhancing desired traits in livestock using nature-inspired methods.
Three bacterial genera (Rikenellaceae RC9 gut group, Prevotella, NK4A214 group, and Christensenellaceae R-7 group) were highly conserved both in their presence and abundance among the four livestock species regardless of genetics. This may suggest that they are core rumen bacteria, essential for highly conserved common traits or functions for these animals (Fig. 1E). However, there were unique bacteria in each host. For instance, the detection of Anaeromyces, Cyllamyces, and Orpinomyces in cattle only may be suggestive of the existence of species-specific microbes that require host rumen-microbes’ environment balance. Similarly, two fungi genera Cladosporium and Pecoramyces are conserved among the four livestock, although varied in abundance. This demonstrates that they are essential for a conserved function, which may be dependent on their relative abundance in each species.
Several fungi genera were unique. Some were present only in one animal species, while some others were shared between either two or three animals but not by all four. Even those that were shared varied in their abundance, which may be integral to their environment and potentially compatible with the rumen environment and host requirements. The significant variation in fungi microbes between the four ruminant livestock may be explained to a significant extent by host genetics (13, 39–41). This unique microbe-host framework variation in microbial composition between hosts may affect microbially mediated ecosystem processes. Additionally, this variation may as well be dependent on host phylogenetic relatedness and trait-based patterns of ecologies (39).
Each of the bacterial and fungal communities established in the present study plays a specific metabolic role in the rumen (42–44). For instance, bacterial species like Ruminococcus, Lachnospiraceae, Christensenellaceae, and Prevotella are associated with hydrogen production during rumen fermentation (22, 44). There are some microbes that were ubiquitous in all four ruminant species, demonstrating their wide rumen environment adaptation. For instance, camel rumen has an acidic pH compared to other livestock.
The next step is to explicitly link the observed microbial and secondary metabolite diversity and network with the basic evolutionary principle, which is biological fitness. We have shown the variation between diverse microbes among the four livestock that varies in feeding behavior, drought resilience, and disease susceptibility (45). For instance, camels and small ruminants, to some extent, are the most resilient to frequent drought among the analyzed livestock compared to cattle (7–10). This could be due to the abundant presence of unique anaerobic fungi, Oontomyces, originally identified from Indian camel, (46) and bacteria (Prevotella) in camels that have demonstrated high capability of diet conversion (23, 24, 39, 47). Additionally, the fungus Neocallimastix, which is present in camels, sheep, and goats but absent in cattle, has been shown to be effective in the bioconversion of poor diet such as lignocellulose into useful products (25, 48), which may have contributed to their resilience. These microbes combined with other physiological mechanisms such as suppression of cholesterol biosynthesis in the kidneys of camels and retention and reabsorption of water (49) may be responsible for camels’ drought resilience. Thus, camel’s evolutionary success to dry climate is partly may be due to the ability to engage in mutualistic interactions with useful microbes that provide novel ecological adaptation traits. Furthermore, such knowledge will give us the opportunity to manipulate the rumen environment to make livestock less susceptible to vectors, efficient in converting their diet to animal protein, and to make livestock environmentally friendly. For instance, in one study, the addition of a fungal inoculant to the diet of dairy cows was found to increase the production of propionate and decrease the production of acetate (50), which is a precursor of greenhouse gas production. Furthermore, microbiome work in humans and rodents has revealed that microbes play essential roles in host health and function (51, 52). Similarly, in our previous work, these various livestock exhibited various susceptibilities to various pathogens (53), which may depend on their mutualistic association with useful microbes.
A recent study to establish the role of the rumen microbiome in dairy cow productivity and greenhouse gas emissions revealed that a heritable subset of the core rumen microbiome influenced the efficiency of feed utilization and the environmental impact of dairy farming (13). This demonstrated that the core microbiome had a significant explanatory role in relation to dietary components within a controlled experimental setting. In our experiment, it was difficult to dissect the role of diet for the microbes and chemo-diversity as the animals were from a free grazing and browsing setup and fed on diverse diets. If we assume that diet may structure rumen microbes, we would have expected similarity both in microbes and secondary metabolites between browsers (camels and goats) and between grazers (cattle and sheep). However, we did not establish a clear link between diet and microbes. For instance, only one bacterial genus, Psychrobacter, was absent in camels and sheep. If diet shapes the rumen microbes, browsers (camels and goats) should share more similar microbes than camels have in common with cattle and sheep, and vice versa. On the other hand, if microbes dictate diet, cattle and sheep share more similar microbes than what camels and cattle share. But sheep and goats shared more bacteria than either of them shared between camel and cattle. This may also be because there are no strict browsers and grazers under the free grazing setting, as they can easily shift between various diets depending on feed availability.
The various plants consumed by the various livestock are characterized by high fiber content, rich in secondary metabolites, and bioactive compounds including tannins, flavonoids, alkaloids, and terpenoids, hence may have potential health benefits for ruminants (54–56). The utilization of shrubs and woody plants in livestock diets has been shown to increase rumen metabolite richness compared to diets based on traditional forage sources (57–59). Studies showed that feeding goats on Acacia saligna, a shrub species, led to increased diversity and richness of rumen metabolites compared to a control diet based on alfalfa hay (60–63). The composition of the plant diet can have significant impacts on the production of metabolites in the rumen. For instance, Grasses (Poaceae) contain fermentable cellulose, hemicellulose, lignin, and protein, which are broken down by rumen microbes into various metabolites, including acetate and propionate (17, 64), which are ingredients in greenhouse gas formation and energy source. Hence, the variability in plant diets can have a significant impact on rumen metabolite production and composition in livestock.
Secondary metabolite composition and diversity
Rumen fermentation is a complex process that results in the production of various metabolites (13, 50). We established a wide range of secondary metabolite compounds in the rumens of the four livestock species. This highlights the interplay between host genetics, diet, and microbes, most of which are associated with various biochemical activities in livestock rumen. The detection of metabolite compound classes, such as volatile fatty acids, aromatic hydrocarbons, terpenes, hydrocarbons, phenols, and alcohols, displays the diversity and complexity of metabolic synthetic pathways in livestock rumens, leading to the production of several diverse metabolites (63). The detection of plant-derived metabolite compounds, such as camphene, α-pinene, and β-caryophyllene, including fecal predictive indolic and phenolic compounds like p-cresol (a byproduct of protein breakdown in the animal gut) and skatole, which had previously been reported in various animals metabolic by-products like animal feces, has a role in livestock-vectors interaction (65–68). This demonstrates that metabolites are conserved as they pass through various digestion processes. But we also observed less complexity in some metabolites. For example, phenols in the rumen are less complex compared to livestock urine (53), which shows that metabolites may gain complexity after they leave the rumen.
Even though the examined metabolite composition varied among the ruminants, minimal intraspecific variation was realized among individual species herd (Fig. S1). Despite the intraspecific diversity among the four ruminants, identical metabolite compound classes were detected, suggesting shared biosynthetic pathways during rumen metabolism. This phenomenon further points to the possibility of some metabolite compounds having a conserved function regardless of the host genotype. This has also been shown by some studies, which also documented that rumen secondary metabolites may not be affected by host-specific microbes, host genotype, or livestock population dynamics (13, 16, 26, 69).
Studies have highlighted a direct relationship between bacterial and fungal populace with rumen metabolome (70–72). These microorganisms work together in a symbiotic relationship with the host to break down complex plant polysaccharides and fiber into simple sugars, which can then be fermented into volatile fatty acids, microbial proteins, and other metabolites that can be absorbed by the host animal (16, 73). Thus, the various secondary metabolites identified may provide various functions to the host. Ruminants, such as cattle, sheep, and goats, utilize hydrocarbons as an energy source, largely contained in plant carbohydrates like glucose and sucrose, by fermenting them in their rumen into volatile fatty acids, which are then absorbed and utilized for energy (74, 75). In this study, we established notable differences in hydrocarbons, terpenes, ketones, and indoles relative abundance among the ruminants. Such variations clarify relevant aspects, such as diet composition, breed, and environment, since the detection and concentration of most ruminal metabolite compounds are influenced by these factors (75–78). The diversity and importance of different compound classes of rumen metabolome in livestock were further demonstrated by the detection of terpenes, which have been linked to improved nutrient utilization and digestive health (79). In addition to terpenes, chemical compound classes like acids, phenols, indoles, ketones, and alcohols also varied significantly among the ruminants (Fig. 6).
The acid profile significantly differs between the four livestock species, with camels having the highest. Acids are involved in the hydrolysis of complex carbohydrates, such as cellulose, lignin, and hemicellulose, into simpler sugars that can be further metabolized by rumen microbes (80). This may be ascribed to the fact that acids are energy sources for the host animal and can be used as precursors for energy production during special conditions. For instance, fatty acids, such as acetate, are used by the host animal as a precursor for fatty acid synthesis in adipose tissues, which can then be utilized as an energy source during times of high energy demand, such as during lactation or periods of feed restriction (80); thus, the diverse acids produced in camel rumen may have contributed to camel rumen acidic pH and resilience even during extended droughts with limited feed availability in arid and semi-arid ecologies. Phenols and indoles are aromatic compounds that are derived from lignin, which is present in the cell wall of plants in addition to being produced during the fermentation of plant material in the rumen. Phenols and ketones exhibit antimicrobial properties that can help to maintain a healthy microbial balance in the rumen (81). Additionally, the antioxidant properties of both ketones and phenols can help reduce oxidative stress in the rumen and improve animal health (82, 83). Alcohols provide energy for rumen microbes in addition to being a carbon source for the synthesis of microbial protein (84, 85). On the other hand, ketones have been shown to be an alternative energy source for ruminants in addition to preventing ketosis (86). Furthermore, elucidation of maternal, genetic, and environmental factors, such as rumen environment (for instance, pH, nutrient, etc.), may provide novel insights into possible mechanisms for manipulating the rumen microbial and secondary metabolites composition to enhance long-term host health, performance, and climate resilience.
Conclusion
This study sheds light on the intricate web of interactions within the rumen ecosystem, highlighting the coexistence, interaction, and competition among various microorganisms. This interplay results in a delicate balance of end products, including secondary metabolites that fuel microbial growth and offer benefits to the host animals. The identification of highly conserved microbial genera across different livestock species suggests the existence of core rumen microbes essential for common traits or functions. Simultaneously, the presence of unique bacteria and fungi in specific hosts underscores the role of species-specific microbes in maintaining the balance of the rumen environment. Furthermore, this study also highlights the intricate relationship between diet, host genetics, and microbial composition in the rumen, which contributes to the diversity of secondary metabolites. These metabolites play critical roles in livestock rumen metabolism. While certain compound classes are conserved among different ruminants, their abundance can vary significantly, which can, in turn, confer unique ecological traits to the host organisms.
Our results demonstrate that rumen fermentation at the interface of host genetics, microbes, and diets has a significant implication in the production of complex secondary metabolites. Hence, it has the potential to revolutionize the livestock sector. By linking the rumen microbiome to host health and performance, we can explore novel strategies and treatments inspired by nature. For instance, the microbial communities in camels and goats may contribute to their resilience in arid environments, offering valuable insights into drought-resistant livestock breeding. Furthermore, the manipulation of the rumen environment could lead to livestock that are less susceptible to disease vectors and more efficient at converting their diet into animal protein while reducing greenhouse gas production. Overall, this study underscores the importance of the rumen microbiome and its associated secondary metabolites in shaping the health, performance, resilience, and environmental impact of livestock. By further exploring the evolutionary and ecological aspects of these interactions, we can unlock new opportunities for sustainable and environmentally friendly livestock husbandry.
MATERIALS AND METHODS
Collection of rumen content
Bovine rumen contents were collected from 10 different freshly slaughtered Boran cattle (Bos indicus), goats (Capra aegagrus hircus), sheep (Ovis aries), and camels (Camelus dromedaries) from their respective slaughterhouses in Nairobi (−1.18623, 36.90744) and Machakos (−1.46500, 36.98166) Counties in Kenya, Africa. The samples (500 mL each) were kept in sterile airtight freeze-resistant 1 L odor collection glass jars (Sigma Scientific, USA) and transported in a cooler box to the laboratory for metabolite compound collection and analysis.
Genomic DNA extraction
To extract genomic DNA from the rumen contents of cattle, sheep, camels, and goats, 200 µL of the sample was mixed with an equal volume of buffered phenol and 20 µL of 20% SDS in a 2 mL centrifuge tube (Eppendorf, Germany). After adding 0.5 g of 2 mm zirconia beads (BioSpec Inc., USA), the mixture was shaken thrice in a mini tissue lyser (Qiagen, Hilden, Germany) at a frequency of 30 Hz for 90 seconds. The lysate was then centrifuged at 14,000 rpm on a 5417R centrifuge (Eppendorf, Germany) for 10 minutes, and the supernatant was transferred to a 1.5 mL clean tube (Eppendorf, Germany). Afterward, 200 µL of buffered phenol was added to the supernatant, the mixture was briefly vortexed, and then centrifuged at 14,000 rpm at 4°C for 15 minutes. The DNA was then precipitated by adding 500 µL absolute ethanol to the supernatant in a clean 1.5 mL centrifuge tube and centrifuged at 14,000 rpm at 4°C for 5 minutes. The supernatant was discarded, and the DNA pellet was washed with 500 µL of 70% ethanol and then centrifuged for 5 minutes. Finally, the pellet was suspended in 100 µL of preheated elution buffer G (ISOLATE II Genomic DNA kit, Bioline Meridian). The DNA quality and quantity were checked by a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Aliquots of 50 µL of the obtained DNA extracts were sent to Macrogen Inc. (Netherlands) for Illumina next-generation sequencing targeting 16S rRNA and ITS1 for bacteria and fungi, respectively. The remaining amounts (50 µL) were utilized for PCR for plant diet identification.
PCR amplification for diet composition screening
PCR amplification targeting two chloroplast markers, consisting of coding (rbcL gene) and non-coding gene spacer region (trnH-psbA) primers (Table S4), was done according to reference (87). The obtained amplicons were then sent to Macrogen Inc (Netherlands) for sequencing. Using Geneious software, obtained sequences were cleaned, edited, and aligned, resulting in a congruent sequence made up of contigs from both the forward and reverse sequences. The plant species were then identified by aligning the processed sequences against the GenBank database using the NCBI BLAST1 search engine. Subsequent phylogenetic analyses were done using the MEGA software version 11 (88).
Metabolite extraction and analysis
Metabolite compounds from cattle, camel, sheep, and goat rumen contents were extracted using the headspace, solid phase microextraction (HS-SPME) technique as detailed in reference (89). Stableflex 24Ga, manual holder SPME fibers (65 µm, polydimethyl siloxane/divinylbezene, Supelco, Bellefonte, PA, USA) were used to trap the volatile metabolite compounds and later analyzed by gas chromatography (HP-7890A, Agilent Technologies, USA) coupled with mass spectrometry (5975C, Agilent Technologies, USA). The chromatograms were subsequently examined using the Agilent MSD Productivity ChemStation software designed for GC and GC/MS systems (Agilent Technologies, USA). The integration process for the respective compound peaks employed a probability-based matching algorithm, with an initial peak width set at 0.034 and an initial threshold of 15.7. To identify individual compounds, a computer-aided approach was used, comparing their retention times and corresponding mass spectral data to the MSD library (specifically, NIST 2005, NIST 05a, and Adams MS HP, all from the USA). For a compound to be considered correctly identified, its spectra needed to exhibit a minimum probability match factor exceeding 80%. Metabolite compounds that were present in at least 7 out of the 10 ruminal fluid samples analyzed for each livestock species were considered as positively detected in the rumen (89).
Data analysis
Multivariate statistical analyses were conducted based on the nature of the obtained data using R studio statistical software version 4.2.1 (90), PAST software Version 4.03 (91), and GraphPad Prism version 9. Similarity percentages (SIMPER) and one-way ANOSIM with Bray-Curtis dissimilarity index were used to compare the profiles and establish the dissimilarity contribution of identified metabolite compounds based on their peak areas across the four livestock species. The metabolite compounds were then classified using the R software package “Random Forest,” version 4.2.1. The random forest analysis was executed by running 1,000 iterations (ntree) with 10 compounds randomly selected at each split (mtry = √q, where q is the total number of compounds). Based on the function “importance (),” we generated the mean decrease in accuracy (MDA), which provides an important score for each metabolite compound. For each livestock, the metabolite with the highest MDA value was considered the most important. An MDS plot and a classical cluster dendrogram were used to visualize the output of analyzed metabolite compound profiles in each livestock. We then used Pearson’s correlation to establish how metabolite compounds compared among individual ruminants’ herd populations. The detected metabolite compounds from across the four ruminants were then pooled based on their chemical identities, after checking for normality using the Shapiro-Wilk test (P > 0.05). A pairwise comparison of the mean relative abundance of respective metabolite compounds in each chemical entity was analyzed by ANOVA among the four ruminants. Statistical significance was declared at P < 0.05.
Bioinformatics analysis
Initially, the data obtained from Illumina sequencing was assessed using nf-core- ampliseq (v2.4.0) workflow and nextflow (v22.10.0), with predefined parameters of trunclenf = 180 and trunclenr = 120. The workflow proceeded as follows: first, the quality of the reads was checked, using FASTQC (version 0.11.9). Cutadapt (v4.1) was then employed to trim reads and eliminate adapter sequences, following the method developed by Martin (92). Preprocessing was performed using the DADA2 tool (v1.26.0) for filtering and trimming, dereplication, sample inference, merging of paired-end reads, removal of chimeras, and taxonomic classification of the ASVs, as outlined in reference (93). Furthermore, DADA2 performed the classification of the ASVs’ taxa based on their taxonomic categorization (Silva database v138 was used on 16S rRNA, and unite database v8.3 was used on ITS1 rRNA).
Abundance visualization
To visualize the ASV count table and ASV taxonomy table generated by the DADA2 algorithm within the nf-core ampliseq workflow, R statistical software (version 4.2.1) was used for further analysis. The ASV count table, ASV taxonomy table, and metadata were put into a single phyloseq object using the Phyloseq package (version 1.40.0) in R. A subset_taxa() function was then employed to eliminate undesired taxa before converting it to a data frame for further manipulation using the phyloseq_to_df() function. Subsequent data frame manipulation was conducted by tidyverse package (version 1.3.2). Finally, ggplot2 (version 3.4.0) and Cairo (version 1.6.0) were used to produce the visual plots.
Alpha and beta diversity
The reads were rarefied to uniform sequencing depths using microbiome package (version 1.18.0), and rarefaction curves were drawn with vegan (version 2.6.2) using function rarecurves(). MicrobiotaProcess (v1.8.2) and ggplot2 (v3.3.6) were used to visualize alpha and beta diversity. Three alpha diversity metrics, the Shannon, Chao1, and Evenness indices, were used. We used the Shapiro-Wilk normality test followed by Kruskal-Wallis to determine the statistical significance of the alpha diversity metrics.
The PCoA of beta diversity was determined on weighted UniFrac distance matrix with plot_ordination() function, and PERMANOVA permutations 999 (P < 0.05) were performed based on the Bray-Curtis dissimilarity distance matrix.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge the financial support for this research by the following organizations and agencies: Max Planck Institute icipe partner group to M.N.G. and the Swedish International Development Cooperation Agency (Sida), the Swiss Agency for Development and Cooperation (SDC), the Australian Centre for International Agricultural Research (ACIAR), the Norwegian Agency for Development Cooperation (Norad), the Federal Democratic Republic of Ethiopia, and the Government of the Republic of Kenya. The views expressed herein do not necessarily reflect the official opinion of the donors.
We express our gratitude to Dr. Segenet Kelemu for the insightful discussions; Mr. John Ngiela and Mr. Joseck Otiwi for their valuable support during sample collection; and Mr. Onesmus Wanyama, Mr. John Mark Makwatta, and Mr. James Kabii for their guidance on chemical and molecular biology instrumentation.
V.O.O. designed the study, collected and analyzed the data, and wrote the manuscript, M.N.G. conceptualized and designed the study, analyzed the data, wrote the manuscript, and mobilized the resources. C.K., S.M., and V.N.O. contributed to the bioinformatics data analysis part of the work. G.O.B. and J.M.O. supervised, reviewed, and edited the manuscript.
Contributor Information
Merid N. Getahun, Email: mgetahun@icipe.org.
James R. Brown, Drexel University, Berwyn, Pennsylvania, USA
DATA AVAILABILITY
The data sets generated and/or analyzed during this study are all included in the paper and as supplemental materials. Additionally, all the analysis R scripts and packages used in this study are available at https://github.com/SamuelMwasya/Metataxonomics-rumen-microbes-visualization.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.01228-23.
Supplemental figure and tables.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. FAO, IFAD, UNICEF, WFP and WHO . 2020. The state of food security and nutrition in the world 2020: transforming food systems for affordable healthy diets. Rome, FAO. [Google Scholar]
- 2. Graham MW, Butterbach-Bahl K, du Toit CJL, Korir D, Leitner S, Merbold L, Mwape A, Ndung’u PW, Pelster DE, Rufino MC, van der Weerden T, Wilkes A, Arndt C. 2022. Research progress on greenhouse gas emissions from livestock in sub-Saharan Africa falls short of national inventory ambitions. Front Soil Sci 2:927452. doi: 10.3389/fsoil.2022.927452 [DOI] [Google Scholar]
- 3. FAO . 2023. Livestock production and climate change. Climate-Smart Agriculture Sourcebook. [Google Scholar]
- 4. Godde CM, Mason-D’Croz D, Mayberry DE, Thornton PK, Herrero M. 2021. Impacts of climate change on the livestock food supply chain; a review of the evidence. Glob Food Sec 28:100488. doi: 10.1016/j.gfs.2020.100488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salem HB, Rekik M, Lassoued N, Darghouth MA. 2011. Global warming and livestock in dry areas: expected impacts, adaptation and mitigation, p 341–366. In Climate change—socioeconomic effects. Intech, Rijeka. [Google Scholar]
- 6. Rust JM. 2019. The impact of climate change on extensive and intensive livestock production systems. Anim Front 9:20–25. doi: 10.1093/af/vfy028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Toulmin C. 2009. Securing land and property rights in sub-Saharan Africa: the role of local institutions. Land Use Policy 26:10–19. doi: 10.1016/j.landusepol.2008.07.006 [DOI] [Google Scholar]
- 8. Boru D, Schwartz M, Kam M, Degen AA. 2014. Cattle reduction and livestock diversification among borana pastoralists in Southern Ethiopia. Nomadic Peoples 18:115–145. doi: 10.3197/np.2014.180108 [DOI] [Google Scholar]
- 9. Kagunyu AW, Wanjohi J. 2014. Camel rearing replacing cattle production among the borana community in Isiolo county of northern Kenya, as climate variability bites. Pastoralism 4:13. doi: 10.1186/s13570-014-0013-6 [DOI] [Google Scholar]
- 10. Watson EE, Kochore HH, Dabasso BH. 2016. Camels and climate resilience: adaptation in Northern Kenya. Hum Ecol Interdiscip J 44:701–713. doi: 10.1007/s10745-016-9858-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. García-Yuste S. 2020. Sustainable and environmentally friendly dairy farms. Springer International Publishing, Cham. [Google Scholar]
- 12. Tapio I, Snelling TJ, Strozzi F, Wallace RJ. 2017. The ruminal microbiome associated with methane emissions from ruminant livestock. J Anim Sci Biotechnol 8:7. doi: 10.1186/s40104-017-0141-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wallace RJ, Sasson G, Garnsworthy PC, Tapio I, Gregson E, Bani P, Huhtanen P, Bayat AR, Strozzi F, Biscarini F, et al. 2019. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci Adv 5:eaav8391. doi: 10.1126/sciadv.aav8391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Medjekal S, Ghadbane M. 2021. Sheep digestive physiology and constituents of feeds. In Sheep farming-an approach to feed, growth and health. IntechOpen. [Google Scholar]
- 15. Zeng Y, Zeng D, Ni X, Zhu H, Jian P, Zhou Y, Xu S, Lin Y, Li Y, Yin Z, Pan K, Jing B. 2017. Microbial community compositions in the gastrointestinal tract of Chinese Mongolian sheep using illumina miseq sequencing revealed high microbial diversity. AMB Express 7:75. doi: 10.1186/s13568-017-0378-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang YK, Zhang XX, Li FD, Li C, Li GZ, Zhang DY, Song QZ, Li XL, Zhao Y, Wang WM. 2021. Characterization of the rumen microbiota and its relationship with residual feed intake in sheep. Animal 15:100161. doi: 10.1016/j.animal.2020.100161 [DOI] [PubMed] [Google Scholar]
- 17. Morgavi DP, Rathahao-Paris E, Popova M, Boccard J, Nielsen KF, Boudra H. 2015. Rumen microbial communities influence metabolic phenotypes in lambs. Front Microbiol 6:1060. doi: 10.3389/fmicb.2015.01060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jang SY, Kim EK, Park JH, Oh MR, Tang YJ, Ding YL, Seong HJ, Kim WH, Yun YS, Moon SH. 2017. Effects of physically effective neutral detergent fiber content on dry matter intake, digestibility, and chewing activity in Korean native goats (Capra hircus coreanae) fed with total mixed ration. Asian-Australas J Anim Sci 30:1405–1409. doi: 10.5713/ajas.16.0868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jami E, Israel A, Kotser A, Mizrahi I. 2013. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J 7:1069–1079. doi: 10.1038/ismej.2013.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hayes BJ, Donoghue KA, Reich CM, Mason BA, Bird-Gardiner T, Herd RM, Arthur PF. 2016. Genomic heritabilities and genomic estimated breeding values for methane traits in angus cattle. J Anim Sci 94:902–908. doi: 10.2527/jas.2015-0078 [DOI] [PubMed] [Google Scholar]
- 21. Roehe R, Dewhurst RJ, Duthie C-A, Rooke JA, McKain N, Ross DW, Hyslop JJ, Waterhouse A, Freeman TC, Watson M, Wallace RJ. 2016. Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet 12:e1005846. doi: 10.1371/journal.pgen.1005846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rooke JA, Wallace RJ, Duthie C-A, McKain N, de Souza SM, Hyslop JJ, Ross DW, Waterhouse T, Roehe R. 2014. Hydrogen and methane emissions from beef cattle and their rumen microbial community vary with diet, time after feeding and genotype. Br J Nutr 112:398–407. doi: 10.1017/S0007114514000932 [DOI] [PubMed] [Google Scholar]
- 23. Dagar SS, Kumar S, Griffith GW, Edwards JE, Callaghan TM, Singh R, Nagpal AK, Puniya AK. 2015. A new anaerobic fungus (Oontomyces anksri gen.nov.,sp.nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol 119:731–737. doi: 10.1016/j.funbio.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 24. Brooke CG, Najafi N, Dykier KC, Hess M. 2019. Prevotella copri, a potential indicator for high feed efficiency in Western steers. Anim Sci J 90:696–701. doi: 10.1111/asj.13197 [DOI] [PubMed] [Google Scholar]
- 25. Dagar SS, Kumar S, Mudgil P, Puniya AK. 2018. Comparative evaluation of lignocellulolytic activities of filamentous cultures of monocentric and polycentric anaerobic fungi. Anaerobe 50:76–79. doi: 10.1016/j.anaerobe.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 26. Cunha CS, Veloso CM, Marcondes MI, Mantovani HC, Tomich TR, Pereira LGR, Ferreira MFL, Dill-McFarland KA, Suen G. 2017. Assessing the impact of rumen microbial communities on methane emissions and production traits in holstein cows in a tropical climate. Syst Appl Microbiol 40:492–499. doi: 10.1016/j.syapm.2017.07.008 [DOI] [PubMed] [Google Scholar]
- 27. Huang C, Ge F, Yao X, Guo X, Bao P, Ma X, Wu X, Chu M, Yan P, Liang C. 2021. Microbiome and metabolomics reveal the effects of different feeding systems on the growth and ruminal development of yaks. Front Microbiol 12:682989. doi: 10.3389/fmicb.2021.682989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alataş MS, Arık HD, Gülşen N, Kahraman O. 2022. Effects of entodinium caudatum monocultures in an acidotic environment on in vitro rumen fermentation. J Ani Feed Sci 31:379–389. doi: 10.22358/jafs/152643/2022 [DOI] [Google Scholar]
- 29. Wang Y, McAllister TA. 2002. Rumen microbes, enzymes and feed digestion-a review. Asian Australas J Anim Sci 15:1659–1676. doi: 10.5713/ajas.2002.1659 [DOI] [Google Scholar]
- 30. La Reau AJ, Suen G. 2018. The ruminococci: key symbionts of the gut ecosystem. J Microbiol 56:199–208. doi: 10.1007/s12275-018-8024-4 [DOI] [PubMed] [Google Scholar]
- 31. Xie Y, Sun H, Xue M, Liu J. 2022. Metagenomics reveals differences in microbial composition and metabolic functions in the rumen of dairy cows with different residual feed intake. Anim Microbio 4:19. doi: 10.1186/s42523-022-00170-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ricci S, Pacífico C, Castillo-Lopez E, Rivera-Chacon R, Schwartz-Zimmermann HE, Reisinger N, Berthiller F, Zebeli Q, Petri RM. 2022. Progressive microbial adaptation of the bovine rumen and hindgut in response to a step-wise increase in dietary starch and the influence of phytogenic supplementation. Front Microbiol 13:920427. doi: 10.3389/fmicb.2022.920427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Andrade BGN, Bressani FA, Cuadrat RRC, Cardoso TF, Malheiros JM, de Oliveira PSN, Petrini J, Mourão GB, Coutinho LL, Reecy JM, Koltes JE, Neto AZ, R de Medeiros S, Berndt A, Palhares JCP, Afli H, Regitano LCA. 2022. Stool and ruminal microbiome components associated with methane emission and feed efficiency in nelore beef cattle. Front Genet 13:812828. doi: 10.3389/fgene.2022.812828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brulc JM, Antonopoulos DA, Miller MEB, Wilson MK, Yannarell AC, Dinsdale EA, Edwards RE, Frank ED, Emerson JB, Wacklin P, Coutinho PM, Henrissat B, Nelson KE, White BA. 2009. Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA 106:1948–1953. doi: 10.1073/pnas.0806191105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wuertz S, Beça F, Kreuz E, Wanka KM, Azeredo R, Machado M, Costas B. 2023. Two probiotic candidates of the genus psychrobacter modulate immune response and disease resistance after experimental infection in turbot (Scophthalmus maximus, Linnaeus 1758). Fishes 8:144. doi: 10.3390/fishes8030144 [DOI] [Google Scholar]
- 36. Anderson RC, Majak W, Rassmussen MA, Callaway TR, Beier RC, Nisbet DJ, Allison MJ. 2005. Toxicity and metabolism of the conjugates of 3-nitropropanol and 3-nitropropionic acid in forages poisonous to livestock. J Agric Food Chem 53:2344–2350. doi: 10.1021/jf040392j [DOI] [PubMed] [Google Scholar]
- 37. Jančič S, Frisvad JC, Kocev D, Gostinčar C, Džeroski S, Gunde-Cimerman N. 2016. Production of secondary metabolites in extreme environments: food-and airborne Wallemia spp. produce toxic metabolites at hypersaline conditions. PLoS One 11:e0169116. doi: 10.1371/journal.pone.0169116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang Q, Difford G, Sahana G, Løvendahl P, Lassen J, Lund MS, Guldbrandtsen B, Janss L. 2020. Bayesian modeling reveals host genetics associated with rumen microbiota jointly influence methane emission in dairy cows. ISME J 14:2019–2033. doi: 10.1038/s41396-020-0663-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martiny JBH, Martiny AC, Brodie E, Chase AB, Rodríguez-Verdugo A, Treseder KK, Allison SD. 2023. Investigating the eco‐evolutionary response of microbiomes to environmental change. Ecol Lett 26 Suppl 1:S81–S90. doi: 10.1111/ele.14209 [DOI] [PubMed] [Google Scholar]
- 40. Henderson G, Cox F, Ganesh S, Jonker A, Young W, Janssen PH, Global Rumen Census Collaborators . 2015. Rumen microbial community composition varies withdiet and host, but a core microbiome is found across a widegeographical range. Sci Rep 5:14567. doi: 10.1038/srep14567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bertelsen MF, Brown-Kav A, Janssen PH. 2015. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci Rep 5:14567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petri RM, Schwaiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, McAllister TA. 2013. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 8:e83424. doi: 10.1371/journal.pone.0083424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wallace RJ, Rooke JA, McKain N, Duthie C-A, Hyslop JJ, Ross DW, Waterhouse A, Watson M, Roehe R. 2015. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:839. doi: 10.1186/s12864-015-2032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chiri E, Nauer PA, Lappan R, Jirapanjawat T, Waite DW, Handley KM, Hugenholtz P, Cook PLM, Arndt SK, Greening C. 2021. Termite gas emissions select for hydrogenotrophic microbial communities in termite mounds. Proc Natl Acad Sci USA 118:e2102625118. doi: 10.1073/pnas.2102625118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Denman SE, Martinez Fernandez G, Shinkai T, Mitsumori M, McSweeney CS. 2015. Metagenomic analysis of the rumen microbial community following inhibition of methane formation by a halogenated methane analog. Front Microbiol 6:1087. doi: 10.3389/fmicb.2015.01087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Getahun MN, Villinger J, Bargul JL, Muema JM, Orone A, Ngiela J, Ahuya PO, Saini RK, Torto B, Masiga DK. 2022. Molecular characterization of pathogenic African trypanosomes in biting flies and camels in surra-endemic areas outside the tsetse fly belt in Kenya. Int J Trop Insect Sci 42:3729–3745. doi: 10.1007/s42690-022-00896-2 [DOI] [Google Scholar]
- 47. Xue MY, Xie YY, Zhong Y, Ma XJ, Sun HZ, Liu JX. 2022. Integrated meta-omics reveals new ruminal microbial features associated with feed efficiency in dairy cattle. Microbio 10:32. doi: 10.1186/s40168-022-01228-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Doelman J, McKnight LL, Carson M, Nichols K, Waterman DF, Metcalf JA. 2019. Postruminal infusion of calcium gluconate increases milk fat production and alters fecal volatile fatty acid profile in lactating dairy cows. J Dairy Sci 102:1274–1280. doi: 10.3168/jds.2018-15148 [DOI] [PubMed] [Google Scholar]
- 49. Saye LMG, Navaratna TA, Chong JPJ, O’Malley MA, Theodorou MK, Reilly M. 2021. The anaerobic fungi: challenges and opportunities for industrial lignocellulosic biofuel production. Microorg 9:694. doi: 10.3390/microorganisms9040694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Aristizabal-Henao JJ, Lemas DJ, Griffin EK, Costa KA, Camacho C, Bowden JA. 2021. Metabolomic profiling of biological reference materials using a multiplatform high-resolution mass spectrometric approach. J Am Soc Mass Spectrom 32:2481–2489. doi: 10.1021/jasms.1c00194 [DOI] [PubMed] [Google Scholar]
- 51. Dagaew G, Wongtangtintharn S, Suntara C, Prachumchai R, Wanapat M, Cherdthong A. 2022. Feed utilization efficiency and ruminal metabolites in beef cattle fed with cassava pulp fermented yeast waste replacement soybean meal. Sci Rep 12:16090. doi: 10.1038/s41598-022-20471-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cho I, Blaser MJ. 2012. The human microbiome: at the interface of health and disease. Nat Rev Genet 13:260–270. doi: 10.1038/nrg3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lloyd-Price J, Abu-Ali G, Huttenhower C. 2016. The healthy human microbiome. Genome Med 8:51. doi: 10.1186/s13073-016-0307-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Getahun MN, Ngiela J, Makwatta JO, Ahuya P, Simon TK, Kamau SK, Torto B, Masiga D. 2022. Metabolites from trypanosome-infected cattle as sensitive biomarkers for animal trypanosomosis. Front Microbiol 13:922760. doi: 10.3389/fmicb.2022.922760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cardoso-Gutierrez E, Aranda-Aguirre E, Robles-Jimenez LE, Castelán-Ortega OA, Chay-Canul AJ, Foggi G, Angeles-Hernandez JC, Vargas-Bello-Pérez E, González-Ronquillo M. 2021. Effect of tannins from tropical plants on methane production from ruminants: a systematic review. Vet Anim Sci 14:100214. doi: 10.1016/j.vas.2021.100214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gemeda BS, Hassen A. 2015. Effect of tannin and species variation on in vitro digestibility, gas, and methane production of tropical browse plants. Asian-Australas J Anim Sci 28:188–199. doi: 10.5713/ajas.14.0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mohammed AS, Animut G, Urge M, Assefa G. 2020. Grazing behavior, dietary value and performance of sheep, goats, cattle and camels co-grazing range with mixed species of grazing and browsing plants. Vet Anim Sci 10:100154. doi: 10.1016/j.vas.2020.100154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Degen AA, Benjamin RW, Mishorr T, Kam M, Becker K, Makkar HPS, Schwartz HJ. 2000. Acacia saligna as a supplementary feed for grazing desert sheep and goats. J Agric Sci 135:77–84. doi: 10.1017/S0021859699007984 [DOI] [Google Scholar]
- 59. Kewan KZ, Elkhouly AA, Negm AM, Javadi A. 2019. Feedstock values of some common fodder Halophytes in the Egyptian desert. In 22nd International Water Technology Conference, IWTC22; Ismailia: , p 12–13 [Google Scholar]
- 60. El-Waziry AM, Basmaeil SM, Al-Owaimer AN, Metwally HM, Ali MH, Al-Harbi MS. 2019. Effect of replacing alfalfa hay with acacia foliage on the growth performance, in vitro gas production and rumen fermentation in goats. Adv Anim Vet Sci 7:738–744. doi: 10.17582/journal.aavs/2019/7.9.738.744 [DOI] [Google Scholar]
- 61. Belanche A, Kingston-Smith AH, Griffith GW, Newbold CJ. 2019. A multiKingdom study reveals the plasticity of the rumen microbiota in response to a shift from non-grazing to grazing diets in sheep. Front Microbiol 10:122. doi: 10.3389/fmicb.2019.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Van Soest PJ, Robertson JB, Hall MB, Barry MC. 2020. Klason lignin is a nutritionally heterogeneous fraction unsuitable for the prediction of forage neutraldetergent fibre digestibility in ruminants. Br J Nutr 124:693–700. doi: 10.1017/S0007114520001713 [DOI] [PubMed] [Google Scholar]
- 63. Owens FN, Basalan M. 2016. Ruminal fermentation, p 63–102. In Millen DD, De Beni Arrigoni M, Lauritano Pacheco RD (ed), Rumenology. Springer International Publishing. [Google Scholar]
- 64. Mansourian S, Corcoran J, Enjin A, Löfstedt C, Dacke M, Stensmyr MC. 2016. Fecal-derived phenol induces egg-laying aversion in Drosophila. Curr Biol 26:2762–2769. doi: 10.1016/j.cub.2016.07.065 [DOI] [PubMed] [Google Scholar]
- 65. Ferreira LL, Sarria ALF, de Oliveira Filho JG, de Silva F de O, Powers SJ, Caulfield JC, Pickett JA, Birkett MA, Borges LMF. 2019. Identification of a non-host semiochemical from tick-resistant donkeys (Equus asinus) against amblyomma sculptum ticks. Ticks Tick Borne Diseases 10:621–627. doi: 10.1016/j.ttbdis.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Getahun MN, Ahuya P, Ngiela J, Orone A, Masiga D, Torto B. 2020. Shared volatile organic compounds between camel metabolic products elicits strong Stomoxys calcitrans attraction. Sci Rep 10:21454. doi: 10.1038/s41598-020-78495-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Spor A, Koren O, Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290. doi: 10.1038/nrmicro2540 [DOI] [PubMed] [Google Scholar]
- 68. Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, Fliegerova K, Griffith GW, Forster R, Tsang A, McAllister T, Elshahed MS. 2014. Anaerobic fungi (phylum neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol Ecol 90:1–17. doi: 10.1111/1574-6941.12383 [DOI] [PubMed] [Google Scholar]
- 69. Foroutan A, Fitzsimmons C, Mandal R, Piri-Moghadam H, Zheng J, Guo A, Li C, Guan LL, Wishart DS. 2020. The bovine metabolome. Metabolites 10:233. doi: 10.3390/metabo10060233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhu C, Li C, Wang Y, Laghi L. 2019. Characterization of yak common biofluids metabolome by means of proton nuclear magnetic resonance spectroscopy. Metabolites 9:41. doi: 10.3390/metabo9030041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Newbold CJ, Ramos-Morales E. 2020. Ruminal microbiome and microbial metabolome: effects of diet and ruminant host. Animal 14:s78–s86. doi: 10.1017/S1751731119003252 [DOI] [PubMed] [Google Scholar]
- 72. Wallace RJ, Snelling TJ, McCartney CA, Tapio I, Strozzi F. 2017. Application of meta-omics techniques to understand greenhouse gas emissions originating from ruminal metabolism. Genet Sel Evol 49:27. doi: 10.1186/s12711-017-0304-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Grossi G, Goglio P, Vitali A, Williams AG. 2019. Livestock and climate change: impact of livestock on climate and mitigation strategies. Anim Front 9:69–76. doi: 10.1093/af/vfy034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Khan N, Ali S, Zandi P, Mehmood A, Ullah S, Ikram M, Ismail I, Shahid MA, Babar MA. 2020. Role of sugars, amino acids and organic acids in improving plant abiotic stress tolerance. Pak J Bot 52. doi: 10.30848/PJB2020-2(24) [DOI] [Google Scholar]
- 75. Mokaya HO, Nkoba K, Ndunda RM, Vereecken NJ. 2022. Characterization of honeys produced by sympatric species of afrotropical stingless bees (hymenoptera, meliponini). Food Chem 366:130597. doi: 10.1016/j.foodchem.2021.130597 [DOI] [PubMed] [Google Scholar]
- 76. Clauss M, Kaiser T, Hummel J. 2008. The morphophysiological adaptations of browsing and grazing mammals, p 47–88. In Gordon IJ, Prins HHT (ed), The ecology of browsing and grazing. Vol. 195. Springer, Berlin Heidelberg. [Google Scholar]
- 77. Malheiros JM, Correia BSB, Ceribeli C, Cardoso DR, Colnago LA, Junior SB, Reecy JM, Mourão GB, Coutinho LL, Palhares JCP, Berndt A, de Almeida Regitano LC. 2021. Comparative untargeted metabolome analysis of ruminal fluid and feces of nelore steers (Bos indicus). Sci Rep 11:12752. doi: 10.1038/s41598-021-92179-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ward D, Schmitt MH, Shrader AM. 2020. Are there phylogenetic differences in salivary tannin‐binding proteins between browsers and grazers, and ruminants and hindgut fermenters. Ecol Evol 10:10426–10439. doi: 10.1002/ece3.6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Poulopoulou I, Hadjigeorgiou I. 2021. Evaluation of terpenes’ degradation rates by rumen fluid of adapted and non-adapted animals. Nat Prod Bioprospect 11:307–313. doi: 10.1007/s13659-020-00289-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tarasov D, Leitch M, Fatehi P. 2018. Lignin–carbohydrate complexes: properties, applications, analyses, and methods of extraction: a review. Biotechnol Biofuels 11:269. doi: 10.1186/s13068-018-1262-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Urrutia NL, Harvatine KJ. 2017. Acetate dose-dependently stimulates milk fat synthesis in lactating dairy cows. J Nutr 147:763–769. doi: 10.3945/jn.116.245001 [DOI] [PubMed] [Google Scholar]
- 82. de Paula EM, Samensari RB, Machado E, Pereira LM, Maia FJ, Yoshimura EH, Franzolin R, Faciola AP, Zeoula LM. 2016. Effects of phenolic compounds on ruminal protozoa population, ruminal fermentation, and digestion in water buffaloes. Livestock Sci 185:136–141. doi: 10.1016/j.livsci.2016.01.021 [DOI] [Google Scholar]
- 83. Mahfuz S, Shang Q, Piao X. 2021. Phenolic compounds as natural feed additives in poultry and swine diets: a review. J Anim Sci Biotechnol 12:48. doi: 10.1186/s40104-021-00565-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rossi R, Stella S, Ratti S, Maghin F, Tirloni E, Corino C. 2017. Effects of antioxidant mixtures in the diet of finishing pigs on the oxidative status and shelf life of longissimus dorsi muscle packaged under modified atmosphere1, 2. J Anim Sci 95:4986–4997. doi: 10.2527/jas2017.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Machado G, Santos F, Lourega R, Mattia J, Faria D, Eichler P, Auler A. 2020. Biopolymers from lignocellulosic biomass: feedstocks, production processes, and applications. Lig bioref tech:125–158. doi: 10.1002/9781119568858 [DOI] [Google Scholar]
- 86. Guliński P. 2021. Ketone bodies - causes and effects of their increased presence in cows’ body fluids: a review. Vet World 14:1492–1503. doi: 10.14202/vetworld.2021.1492-1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tawich SK, Bargul JL, Masiga D, Getahun MN. 2021. Supplementing blood diet with plant nectar enhances egg fertility in Stomoxys calcitrans. Front Physiol 12:646367. doi: 10.3389/fphys.2021.646367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. doi: 10.1093/molbev/msab120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Omondi VO, Bosire GO, Onyari JM, Getahun MN. 2022. A comparative investigation of volatile organic compounds of cattle rumen metabolites using HSSPME and porapak-Q odor trapping methods. Anal Chem Let 12:451–459. doi: 10.1080/22297928.2022.2100276 [DOI] [Google Scholar]
- 90. R Core Team . 2022. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: http://www.R-project.org [Google Scholar]
- 91. Hammer O, Harper DAT, Ryan PD. 2001. PAST: paleontological statistics software package for education and data analysis [Google Scholar]
- 92. Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 93. Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. doi: 10.1038/ismej.2017.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure and tables.
An accounting of the reviewer comments and feedback.
Data Availability Statement
The data sets generated and/or analyzed during this study are all included in the paper and as supplemental materials. Additionally, all the analysis R scripts and packages used in this study are available at https://github.com/SamuelMwasya/Metataxonomics-rumen-microbes-visualization.