Abstract
Cell surface hydrophobicity (CSH) of Candida species enhances virulence by promoting adhesion to host tissues. Biochemical analysis of yeast cell walls has demonstrated that the most significant differences between hydrophobic and hydrophilic yeasts are found in the acid-labile fraction of Candida albicans phosphomannoprotein, suggesting that this fraction is important in the regulation of the CSH phenotype. The acid-labile fraction of C. albicans is unique among fungi, in that it is composed of an extended polymer of β-1,2-mannose linked to the acid-stable region of the N-glycan by a phosphodiester bond. C. albicans serotype A and B strains both contain a β-1,2-mannose acid-labile moiety, but only serotype A strains contain additional β-1,2-mannose in the acid-stable region. A knockout of the C. albicans homolog of the Saccharomyces cerevisiae MNN4 gene was generated in two serotype B C. albicans patient isolates by using homologous gene replacement techniques, with the anticipation that they would be deficient in the acid-labile fraction and, therefore, demonstrate perturbed CSH. The resulting mnn4Δ-deficient derivative has no detectable phosphate-linked β-1,2-mannose in its cell wall, and hydrophobicity is increased significantly under conditions that promote the hydrophilic phenotype. The mnn4Δ mutant also demonstrates an unanticipated perturbation in the acid-stable mannan fraction. The present study reports the first genetic knockout constructed in a serotype B C. albicans strain and represents an important step for dissecting the regulation of CSH.
The modification of proteins by carbohydrate is conserved throughout eukaryotic species. The stepwise addition of carbohydrate to nascent proteins occurs shortly after translocation of the polypeptide into the endoplasmic reticulum, and posttranslational glycosylation is limited to a subset of total cellular proteins that transit through the secretory pathway. Core carbohydrate addition to specific amino acid residues is essentially identical in all eukaryotes. However, the maturation of glycans is variable between species and can vary between cell types in a single organism. The addition of glycan to a protein plays various functional roles for the protein and increases the apparent molecular mass of the protein. The dissection of carbohydrate maturation pathways has been accomplished by the parallel use of biochemical analysis of glycosylation in many cell types and the identification of microbial mutants defective in carbohydrate addition.
The cell walls of fungi are composed of highly branched polymers of glycan with embedded, highly glycosylated proteins and serve an essential role in maintaining the structural integrity of the cell (46). The addition of glycans to cell wall glycoproteins in the budding yeast Saccharomyces cerevisiae has been elucidated by the identification of a panel of viable loss-of-function mutants at various points in the pathway (see, e.g., references 3-7, 20, and 45), and the basic enzymology of carbohydrate addition is essentially conserved between species. Glycoproteins isolated from these mutants are smaller in mass than those observed from the wild-type parent strain, due to the decreased amount of carbohydrate. Glycosylation in all eukaryotes can occur via the addition of carbohydrate to either asparagine residues (N-linked) or to serine or threonine residues (O-linked). Eukaryotic N-linked glycosylation core carbohydrate addition occurs by the transfer of a branched 9-mannose-3-glucose structure to select asparagine residues on the polypeptide backbone by means of the oligosaccharyltransferase complex shortly after the protein enters the lumen of the endoplasmic reticulum (reviewed in reference 8). This core group is then modified and extended in yeast by the addition of a long chain of α-1,6-mannose by proteins coded by OCH1, MNN9, MNN10, VAN1, and others (reviewed in reference 13). This outer chain is subsequently modified by the addition of α-1,2-linked mannose side branches, via the action of MNN2, MNN5, and KTR1 through KTR3 (13). These side branches are terminated by an α-1,3-mannose unit, attached by Mnn1p. Mannosylphosphate is added to a subset of the side branches by the action of the MNN4 and MNN6 genes, which has the functional effect of contributing a net negative charge to the cell wall. Phenotypically, the ability of the cell wall to bind the cationic dye Alcian blue can differentiate between strains containing mannosylphosphate and those that do not contain the structure (6, 17). The mannosylphosphate is further modified by the addition of a variable number of mannose residues. These resides are termed the acid-labile region, due to the mild acid sensitivity of the phosphodiester bond linking them to the remainder of the glycoprotein.
The relationship between the MNN4 and MNN6 genes is complex and not yet resolved. The initial genetic description of the S. cerevisiae mnn4-1 allele (6) indicated that this mutant operated in a dominant-negative manner relative to the wild-type locus in heterozygous strains, and this conclusion was further confirmed by complete deletion of the MNN4 gene (41). A closely related gene sequence (YJR061w) does not complement the mnn4Δ defect, and deletion of that gene does not result in the loss of the ability of the strain to bind the Alcian blue (32). Sequence analysis of the MNN4 gene product did not indicate an obvious role of the protein in mannosylphosphate transfer but suggested a role in regulating the activity of MNN6 (42). The MNN6 gene product in S. cerevisiae shares significant structural similarity with the KRE2/MNT1 family of mannosyltransferases, and deletion of that gene is recessive (56). Jigami and Odani (32) have proposed on the basis of this relationship that Mnn4p acts as a positive regulator of Mnn6p mannosylphosphate transfer in a dose-dependent manner with an unidentified negative regulator of transferase activity. In vitro demonstration of this model has not been accomplished. Functionally, null mutants of the mnn4 and mnn6 genes present essentially the same phenotype, resulting in complete loss of detectable cell wall phosphate detected by Alcian blue binding.
The cell wall of the commensal pathogen Candida albicans is biochemically similar to that of S. cerevisiae, and sequencing of the C. albicans genome has readily identified gene homologs in many of the carbohydrate maturation pathways that have previously been described in S. cerevisiae. A modification unique to C. albicans is the stepwise addition of β-1,2-mannose polymers to the mannosylphosphate that is added in S. cerevisiae by the MNN4 and MNN6 genes. The biosynthetic and genetic basis for β-1,2-mannose addition has not been described, although it is presumed to be the result of two or more enzymes. This conclusion is based on the observations of Suzuki and coworkers, who have identified enzymatic activities responsible for in vitro β-mannosyltransferase activities that vary in their substrate specificity (54).
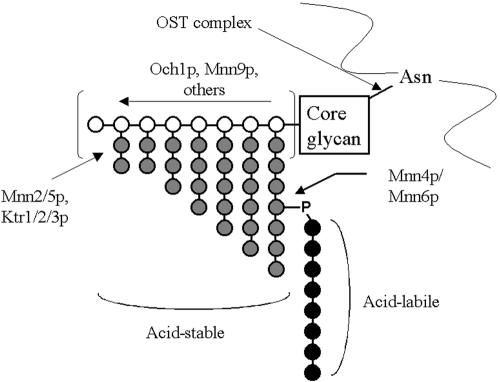
Clinical isolates of C. albicans can be broadly differentiated into two serotypes, originally described by Hasenclever and Mitchell (23), based on their immunoreactivity with specific agglutinating sera (Candida Check; Iatron Laboratories). The individual differences between serotypes can be traced to differences in their cell wall glycoproteins. Specifically, β-1,2-mannose is found in both the acid-stable and acid-labile region of serotype A strains, whereas it is only in the acid-labile region of serotype B strains (Fig. 1). Suzuki et al. have speculated (54) that the lack of β-1,2-mannose in the acid-stable region in serotype B strains is the result of a mutational event affecting one of the enzymes responsible for β-1,2-mannose addition.
FIG. 1.
N-linked carbohydrate structure of serotype B C. albicans mannoproteins. White circles represent α-1,6-linked mannose, gray circles represent α-1,2-linked mannose, and black circles represent β-1,2-linked mannose attached to α-1,2-mannan chains via a phosphodiester bond that defines the acid-labile mannan region. The extended α-1,6-linked mannose backbone can be very extensive, and α-1,2-mannose side chains can be of variable length along the backbone. Mannosylphosphate addition can also occur at multiple sites along the α-1,2-mannose side chains, resulting in significant heterogeneity of the mature glycan. Serotype A C. albicans strains contain additional β-1,2-mannose extending the α-1,2-mannose residues in the acid-stable region which is not phosphate linked.
Recently, a homolog of the C. albicans MNN4 gene has been described (29), and elimination of that gene in the standard strain (strain CAI4 [16]) for knockout generation results in a derivative that is negative for cell wall mannosylphosphate addition and does not have an acid-labile mannan fraction. Strain CAI4 is derived from a serotype A isolate and, as a result, still contains β-1,2-mannose in its acid-stable region. The C. albicans MNN4 gene is one of a family of related gene products in this fungal species that can be identified by computer analysis of the C. albicans genome, and the contribution of the rest of the members of the family to mannosylphosphate addition is not known.
As part of our ongoing studies of cell surface hydrophobicity (CSH) as a virulence contributor of C. albicans, we described variations in the acid-labile region of serotype B clinical isolates and their correlation with CSH (36, 37). Specifically, acid-labile mannan lengths in phenotypically hydrophobic cells are generally longer than in hydrophilic cells. Acid-stable mannan is comparable in structure in hydrophobic and hydrophilic cell populations, suggesting the hypothesis that regulation of a single modification is able to affect cell wall ultrastructure and surface hydrophobic characteristics (35). Because serotype A strains still contain readily detectable amounts of β-1,2-mannose in their acid-stable regions, we have constructed a deletion derivative of the MNN4 gene in two serotype B strains by using a standard homologous recombination approach for gene targeting in order to test the hypothesis that the β-1,2-mannose structure directly affects CSH. The consequences of this knockout on adhesion and pathogenesis were also assessed.
MATERIALS AND METHODS
Strains and growth.
Strains used in this study are summarized in Table 1. The mnn4Δ/mnn4Δ derivative of CAI4, designated CDH5, was kindly provided by N. Gow and R. Hobson (Aberdeen University). The mnn9Δ/mnn9Δ derivative of CAI4, designated SS-19, was obtained from C. Specht (Boston University). The mnn9 deletion derivative contains a significant N-glycan defect and only adds the core glycan to proteins; it does not add mannosylphosphate. CAI4 and serotype B strains LGH1095 (ATCC MYA-2719 and A9 [57]) were used for the generation of knockout derivatives and were grown in phosphate-buffered 0.67% yeast nitrogen base (Difco)-2% glucose (YNB2G) medium at either 25 or 37°C to early stationary phase as described (28). Strains with a ura− phenotype were grown in medium containing 1% yeast extract, 2% peptone, and 2% dextrose. All strains were maintained as frozen stocks (−80°C) in 100 mM sucrose-2 mM ZnSO4 (25).
TABLE 1.
C. albicans derivatives of CAI4 used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| LGH1095 | Wild-type, serotype B | 28 |
| CA-DSH4 | LGH1095 derivative, mnn4Δ/mnn4Δ | This study |
| CA-DSH6 | CA-DSH4 derivative, mnn4Δ/mnn4Δ::MNN4::RP10::caSAT1 | This study |
| A9 | Wild-type, serotype B | 57 |
| CA-JMH1 | A9 derivative, mnn4Δ/mnn4Δ | This study |
| CAI4 | ura3Δ::λimm434/ura3Δ::λimm434 | 16 |
| CDH5 | uraΔ::λimm434/ura3Δ::λimm434 mnn4Δ::hisG-URA3-hisG/mnn4Δ::hisG | 29 |
| CA-DSH5 | ura3Δ::λimm434/ura3Δ::λimm434 mnn4-3Δ::hisG/mnn4-3Δ::hisG | This study |
| SS19-4 | ura3Δ::λimm434/ura3Δ::λimm434 mnn9Δ::hisG/mnn9Δ::hisG | 52 |
Knockout strategy.
Potential homologs of the S. cerevisiae MNN4 gene were identified in the unannotated C. albicans genome database by using the FASTA algorithm. Elimination of the MNN4-3 gene in strain CAI4 to generate strain CA-DSH5 was accomplished by using the modified URA-blaster strategy (58) with PCR primers flanking the putative MNN4 gene open reading frame (ORF). This gene sequence was the first gene identified during our screen for MNN4 homologs in the initial publicly accessible iteration of the C. albicans genomic database.
To knockout the MNN4 homolog in the URA+ serotype B strains, we used the positive selection scheme used by Wirsching et al. (59) for targeted gene disruption in C. albicans. PCR fragments of 600 to 800 bp in size flanking the ORF of the MNN4 gene were generated by using the MNN4 sequence to design primers (mnn4F1, GGCCCCTCGTTTATAATAGCAACAAC; mnn4F2, GCTCGAGGAACCAAAGAATAATAACCCTG; mnn4F3, GCTCGAGGCTAAATAGTCAATGTTGTAAT; mnn4R1, CCGCGGGATAATGGGTTTATGGAAATTG; mnn4R2, CCGCGGGCAATTGACTTTTGGTAAATC; and mnn4R3, GGAGCTCGCTTAACACCTCATTTTAATCC). Two MNN4 knockout cassettes were cloned to eliminate either allele of the MNN4 locus. Primers mnn4F1/mnn4F2 and mnn4R2/mnn4R3 were used to amplify MNN4 segments at the 5′ and 3′ ends of the ORF, respectively, for one knockout cassette, and primer sets mnn4F1/mnn4F3 and mnn4R1/mnn4R3 were used to amplify segments for a second cassette. Fragments were cloned into plasmid pSFI1 to flank an expression cassette conferring mycophenolic acid resistance and the FLP recombinase gene under the control of the C. albicans inducible secreted aspartyl proteinase promoter. The expression cassette is in turn flanked by tandem repeats that are able to recombine and loop out the cassette after integration and induction by growth in medium containing bovine serum albumin (BSA) as the sole nitrogen source.
Strains LGH1095 and A9 were transformed by the standard LiAc-mediated method (58) or by electroporation (44) and plated on YNB2G agar plates containing 10 μg of mycophenolic acid (Sigma) per ml for incubation at 30°C until colonies appeared. Candidate colonies were restreaked to fresh plates, and potential heterozygotes were assessed by Southern blotting for proper insertion of the transforming cassette at the MNN4 locus. Mycophenolic acid (MPA)-resistant heterozygotes were reverted to MPA sensitivity by incubation in yeast carbon base-BSA liquid medium at 30°C for 3 days. Cells from growth in yeast carbon base-BSA medium were plated to single colonies and scored for MPA sensitivity or resistance. MPA-sensitive clones were retransformed as before to knock out the other locus by selection on MPA-containing plates. Potential homozygotic transformants were screened phenotypically by growing clones in 96-well plates and were assayed for Alcian blue binding in microtiter plates, based on the observation of multiple investigators (6, 17, 29) that elimination of the MNN4 gene results in greatly diminished levels of dye binding.
For subsequent reintegration of the intact MNN4 gene into the deletion mutant, a reintegration vector was constructed by replacing the URA3 gene of vector CIP10 (39) with the nourseothricin resistance cassette from vector pSFS2 (44) to construct vector pNATCIP. A full-length MNN4 genomic clone was generated by PCR by using the primers CAMNN4-7 (GAACAAGAGCTCTCTTCTTTTTCTTTTATAAC; [29]) and CAMNN4-Pst (GCTATGGTAACTGCAGTTGAAACCGG) and subcloned into pNATCIP, which allowed targeting of the reintegration vector either to the RP10 locus advocated by Murad et al. (39) and used for reintegration of the C. albicans MNN4 gene by Hobson et al. (29) by digestion with StuI or to the native MNN4 locus by digestion with BglII. Transformation of the deletion derivative to generate reintegrants was accomplished by electroporation according to the method of Reuss et al. (44) by using gel-purified linearized plasmid DNA, allowing a 3-h recovery period postelectroporation prior to plating on agar plates containing 1% yeast extract, 2% peptone, and 2% dextrose supplemented with 100 μg of nourseothricin (Werner BioAgents; Jena, Germany).
Phenotypic analysis of the mnn4Δ/mnn4Δ mutant.
Growth rate and germination efficiency were measured as previously described (50). Whole glycoproteins were analyzed following enzymatic release from intact cells by digestion with Zymolyase 100T (ICN) in the presence of 1 M sorbitol as an osmotic stabilizer as described (49). Protein samples were then subjected to Western blot analysis by using monoclonal antibody (MAb) B6.1, recognizing a β-1,2-mannotriose structure (9), and MAb B6 (21), which recognizes an incompletely characterized epitope on the acid-stable region (both antibodies courtesy of J. Cutler, Research Institute for Children, New Orleans, La.).
Adhesion of yeasts to extracellular matrix proteins was measured on coated tissue culture plastic in a static binding assay (38, 50) and on coated glass capillaries in a dynamic shear binding assay (18). The static adhesion assay was performed by allowing yeast cells to adhere to fibronectin-coated tissue culture dishes for 15 min at 37°C, and nonadherent cells were gently washed off. Dishes were then overlaid with corn meal agar and incubated overnight at 30°C for subsequent quantification of CFU. Sterile glass capillaries were prepared for the shear force assay by coating with fibronectin overnight as described elsewhere (38), and the capillaries were then washed five times with assay buffer (Dulbecco's phosphate buffer with Mg2+ and Ca2+, pH 7.4). The capillary was placed in line into a recirculating buffer system and exposed to a constant shear force of 2 dynes/cm2 (equivalent to venous capillary shear force), and yeast cells were introduced via a sampling port. Adhesion was measured microscopically as yeast foci attached per unit area of capillary after 8 min of incubation.
Assays.
CSH was measured by the hydrophobic microsphere assay developed in our laboratory (24, 27). Microsphere attachment is assessed by bright field microscopy at a magnification of ×400. The percentage of cells with three or more attached spheres is recorded as the percent hydrophobicity. Hexose content was determined by the phenol-sulfuric acid method of Dubois et al. (14). Sample absorbances were compared to a mannose standard curve. Protein concentration was determined by the bicinchoninic acid assay (Pierce Chemical) (51). BSA was used as the standard protein to generate a standard curve. Phosphate concentration was determined by using the method of Ames and Dubin (1) with K2HPO4 to make a standard curve and d-glucose-6-phosphate as a control. Oligosaccharide concentration was determined by measuring the concentration of reducing ends as reported by Dygert et al. (15).
Alcian blue binding of C. albicans derivatives was assessed as described previously (41) and according to the method of Wang et al. (56). The cationic dye Alcian blue binds to phosphate in the yeast cell wall. A culture of early stationary phase yeast was pelleted and suspended in 0.05% Alcian blue dye dissolved in 0.02 N HCl and incubated for 15 min at room temperature. The cells were then pelleted and assessed visually for dye binding. To quantify dye binding, constant numbers of cells were resuspended in dye as above, and dye remaining in the supernatant after binding was measured spectrophotometrically and compared to a dye standard curve. Residual levels of Alcian blue dye binding in mutants is due to incompletely dissolved dye.
Systemic challenge.
Six-week-old female AJ mice were challenged via injection into the lateral tail vein with 105 to 106 live yeast cells suspended in Hanks' buffered saline and monitored three times daily for development of disease. For coinfection, a suspension of equal numbers (2 × 105 to 5 × 106 total CFU) of C. albicans strain LGH1095 and its mnn4Δ/mnn4Δ derivative were injected into mice and monitored for disease progression. Kidneys were recovered from animals following euthanasia and homogenized in sterile water (55), and infective CFU were isolated following plating of the homogenate onto YNB2G agar plates. The phenotype of individual colonies was determined by overnight growth in liquid culture in 96-well plates, followed by determination of Alcian blue binding as described above. All animal work was carried out in American Association of Laboratory Animal Care-approved facilities by using protocols approved for use by the University of Virginia Institutional Animal Care and Use Committee.
Mannoprotein preparation.
Glycans (glucan and mannoprotein) were extracted from the cell wall by using a protocol based on that of Peat et al. (43) and described previously (35, 37). The washed cell pellet was immediately suspended in 50 ml of distilled H2O, and the cell suspension was autoclaved at 121°C for 90 min. Mannoprotein was separated from glucan by using a sequential ethanol precipitation strategy based on that of Lloyd (34, 36). Fractionation of mannan into acid-labile and acid-stabile fractions was carried out by mild acid hydrolysis as described previously (35) and labeled with 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS) for fluorophore-assisted carbohydrate electrophoresis (FACE) analysis (37). Acid-stable fractions were further hydrolyzed in acid as previously described (35) to preferentially digest the α-1,6-glycosidic backbone, leaving the side branches intact for subsequent FACE analysis. Dextran (α-1,6-glucose) (catalog no. 31391; Fluka) was subjected to acid hydrolysis (60), fluorescently labeled, and used as an electrophoretic mobility marker.
FACE.
FACE was performed essentially as described by Jackson (31) and modified by Goins and Cutler (19). Prior to use, gel plates were soaked in 10% (wt/vol) methanolic KOH, washed with detergent (7X; ICN), rinsed with deionized water, and finally rinsed with 95% ethanol. Electrophoresis was carried out in a Hoeffer SE600 gel electrophoresis apparatus (Amersham). Run parameters were 600 V for 4 to 6 h or until the ANTS front reached 1 cm from the bottom of the gel. The gel was cooled with a recirculating water bath set to 4°C. Gels were imaged with a Kodak EDAS 290 system containing a DC290 digital camera and UV transilluminator (302 nm). Image contrast and brightness were optimized by using Photoshop version 7.0 (Adobe).
RESULTS
Elimination and reintegration of MNN4.
The present study describes the first knockout of a gene involved in N-linked glycosylation in a serotype B isolate of C. albicans. The MNN4 gene is essential for the biosynthesis of the acid-labile fraction of cell wall mannoproteins in yeast, and elimination of this gene product in both S. cerevisiae and C. albicans has the effect of blocking the addition of mannosylphosphate to α-1,2-mannose side branches on N-glycans (Fig. 1). This mannosylphosphate is further modified by an enzyme presumably unique to Candida species that transfers mannose to extend β-1,2-linked mannose polymers of various length. Although C. albicans knockouts are generally made in the CAI4 strain, this strain was derived from a serotype A clinical isolate, which has been demonstrated to add β-1,2-mannose to the acid-stable mannan fraction and to show less hydrophilicity than serotype B strains when grown at 37°C (37). As a result, although an mnn4Δ/mnn4Δ knockout derived from CAI4 does not bind the dye Alcian blue (29), it retained significant immunoreactivity with an antibody of clinical relevance (21) that recognizes a β-1,2-manntriose.
Elimination of the MNN4 gene in two serotype B C. albicans strains resulted in derivatives that are virtually negative for binding the cationic dye Alcian blue (Fig. 2) and generates white cell pellets in comparison to the dark blue pellets seen for the parent strains. This confirms and extends the observation made for a knockout derivative of the same gene in the standard strain background used for C. albicans genetic studies (29). Heterozygotic strains were observed to generate an intermediate phenotype for Alcian blue binding. The exponential phase growth rate of the LGH1095 mnn4Δ/mnn4Δ derivative was comparable to the parent strain in YNB2G medium at 25°C. The A9 mnn4Δ/mnn4Δ demonstrated a slight (∼20%) reduction in exponential phase growth rate, but stationary phase cell densities (26 h) were comparable. A moderate growth defect (as assessed by colony size) was observed by plating the mnn4Δ/mnn4Δ strain on Sabauroud's glucose plates at 24 h, but comparable colony sizes were observed at later time points. Both strain backgrounds showed similar yeast-to-hyphal transition of the derivatives upon reinoculation at low cell density into defined media (Table 2). Dye binding in the LGH1095 and A9 mnn4Δ/mnn4Δ derivatives was diminished to levels observed for the serotype A mnn4Δ/mnn4Δ strain in comparison to the parent strains (Table 3). Reintegration of the MNN4 gene in the LGH1095 mnn4Δ/mnn4Δ derivative to the MNN4 locus by digestion of the targeting cassette with BglII appeared to be efficient, as greater than 90% of the transformants recovered Alcian blue dye binding. In contrast, only about 10% of colonies recovered from transformants with StuI-digested DNA targeting the RP10 locus demonstrated recovered Alcian blue binding. This result is consistent with the observation of Hobson et al. (29) that multiple copies of the reintegrated MNN4 gene are necessary to rescue the mutant phenotype when the RP10 locus is used to target reintegration.
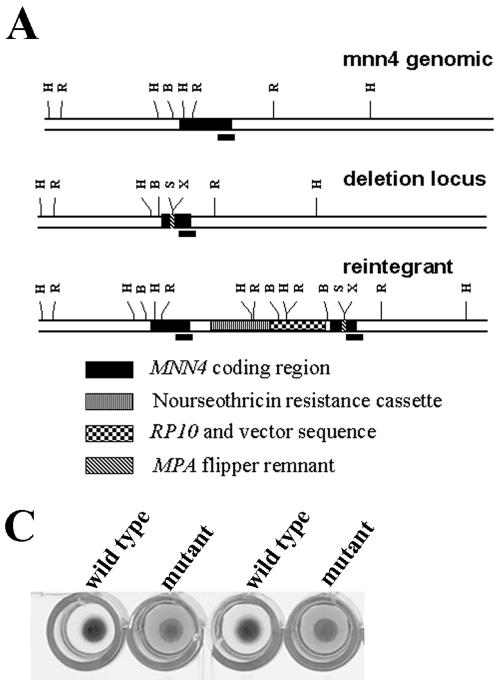
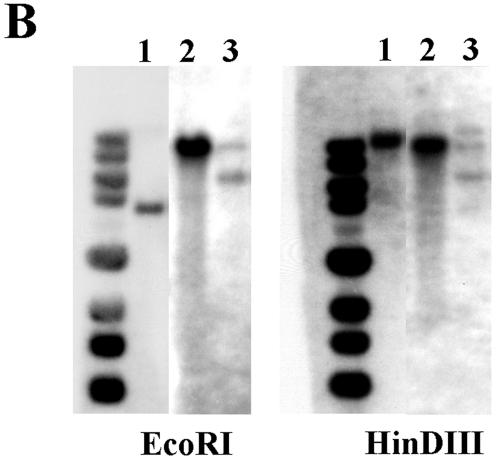
FIG. 2.
(A) Restriction map of MNN4 locus. The genomic region containing the C. albicans MNN4 gene (ORF, nucleotides 27312 to 30302 of contig19-10162; GenBank accession no. AACQ01000084) is mapped schematically for the wild-type, MPA-sensitive deletion, and the nourseothricin-resistant (reintegrant) loci. Hash marks indicate HinDIII (H), EcoRI (R), BglII (B), SacII (S), and XhoI (X) sites in the sequence, and the black bar indicates the probe fragment from the 3′ end of the MNN4 gene sequence used for Southern blotting. (B) Southern analysis of LGH1095 and derivatives. Genomic DNA prepared from strains LGH1095 (lanes 1), CA-DSH4 (lanes 2), and CA-DSH6 (lanes 3) was digested by restriction enzymes EcoRI or HinDIII, separated by agarose electrophoresis, and transferred to nylon for Southern blotting. Blots were hybridized in formamide-containing hybridization buffer overnight at 45°C with 32P-labeled probe corresponding to the 3′ end of the MNN4 open reading frame, washed at high stringency at 62°C, and then exposed to film. (C) Alcian blue binding by LGH1095 and mnn4 derivative. Colonies of either strain LGH1095 or the isogenic mnn4-deficient derivative (CA-DSH4) were grown overnight in YNB2G medium in 96-well microtiter plates. Following overnight growth, cells were pelleted and resuspended in a solution of 0.05% Alcian blue dye in 0.02 N HCl for 15 min at room temperature. The microtiter plate was again centrifuged and was inspected visually. Wild-type cell clones are able to bind all of the dye in solution and generate blue cell pellets. Mutant cell clones do not bind the dye, generate creamy white cell pellets, and leave unbound dye in the well that can be quantified by spectrophotometry. Comparable results are observed with strain CA-JMH1.
TABLE 2.
Phenotypic characterization of mnn4Δ mutantsa
| Parameter | Strain (culture temperature)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| LGH1095 (25°C) | A9 (25°C) | LGH1095 (37°C) | A9 (37°C) | DSH4 (25°C) | JMH1 (25°C) | DSH4 (37°C) | JMH1 (37°C) | |
| Exponential phase doubling time (min) | ≈50 | ≈100 | ND | ≈60 | ≈50 | ≈120 | ND | ≈100 |
| % Germination at 1 hb | >50 | 50 | >50 | 50 | ||||
| % Germ tubes | 58 | 45 | 65 | 55 | ||||
| % Pseudohyphae | 24 | 2 | 20 | 4 | ||||
| % Yeast formc | 18 | 53 | 15 | 41 | ||||
| Hydrophobicity (% phobic cells) | 98.5 ± 0.7 | 95.3 ± 2.2 | 1.7 ± 0.8 | 4.2 ± 1.1 | 98.9 ± 0.6 | 88.7 ± 4.5 | 31.3 ± 9.8 | 11.2 ± 1.5 |
| Adhesion (mean no. of foci per field) | 58.3 ± 3.2 | 50.8 ± 10.5 | 2.5 ± 0.5 | 3.3 ± 0.9 | 65.0 ± 0.6 | 33.4 ± 7.7 | 19.9 ± 10.9 | 6.0 ± 2.6 |
All assays were performed as described in Materials and Methods. For germination determination, cell morphology was assessed at 3 h post/inoculation (80 min for strain A9 and its derivative) into germination media as either having true germ tube morphology (no constriction at the hyphal neck), pseudohyphal morphology (constriction at hyphal neck), or yeast form. Similar ratios of each cell type were seen for both strain backgrounds. ND, not determined.
Cells grown at 37°C do not form true hyphae; consequently, germination data are presented only for cells grown at 25°C transferred to germination media.
100% yeast at time zero for cells grown at 25°C.
TABLE 3.
Relative Alcian blue binding by glycosylation-deficient C. albicans derivatives grown at 37°C
| Value | Alcian blue binding (μg/OD unit) by strain (n):a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1095 (4) | DSH4 (4) | DSH6 (2) | A9 (4) | JMH1 (4) | CAI4 (3) | CDH5 (4) | DSH5 (2) | SS19-4 (3) | |
| Avg | 42.5 | 6.0 | 38.3 | 49.2 | 7.4 | 27.5 | 7.3 | 12.9 | 5.9 |
| SD | 4.38 | 6.21 | NA | 5.70 | 4.91 | 6.70 | 7.56 | NA | 2.73 |
Data are presented as micrograms of Alcian blue bound per optical density (OD) unit of early stationary phase (26-h culture) cells. n, number of independent experiments used to generate values for each strain; NA, not available.
Although there is an increasing number of other C. albicans genes that appear to be redundant or cooperative members of multiple paralog gene families, elimination of a single ORF led to a derivative that is phenotypically similar to the homologous genetic knockout in S. cerevisiae. The function of other members of the MNN4 gene paralog family is unknown; however, knockout of one of these genes during the initial phase of this study (orf19.5557; this represents the second most similar ORF to the S. cerevisiae MNN4 gene sequence by FASTA analysis with an E value of 10−7 and contains significant conservation of domain order with the budding yeast protein) in the strain CAI4 background had the result of partially lowering total Alcian blue binding (Table 3, strain CA-DSH5). This result was not unexpected, however, as a recent survey of 622 nonessential S. cerevisiae deletion strains found that approximately 10% showed alterations in Alcian blue binding (11), concluding that many diverse cellular pathways can ultimately contribute to the amount of cell wall mannosylphosphate.
Hydrophobicity and adhesion of the mnn4Δ/mnn4Δ knockout.
Our phenotypic analysis of the two serotype B mnn4Δ/mnn4Δ deletion strains had determined that the CSH of the mutants was indistinguishable from the parent strain in cells grown under conditions that promote the hydrophobic phenotype. When grown under conditions that promote the hydrophilic phenotype at 37°C, wild-type strain cell populations are composed of less than 5% hydrophobic cells, whereas hydrophobicity in the mnn4Δ/mnn4Δ knockout derivatives reproducibly increased to 25 to 60% (Tables 2 and 4). This result was different from what has been observed in the serotype A mnn4Δ/mnn4Δ deletion derivative CDH5, which demonstrated CSH levels comparable to the parent strain CAI4 under all growth conditions (data not shown).
TABLE 4.
Compositional analysis of C. albicans mannan
| Strain | nb | Composition (% dry weight) of mannan sample from:a
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Hydrophilic cells
|
Hydrophobic cells
|
||||||||
| CSH | Protein | Hexose | PO4 | CSH | Protein | Hexose | PO4 | ||
| A9 | 3/3 | 4.0 (1.0) | 5.13 (0.47) | 79.9 (13.7) | 3.17 (0.11) | 93.6 (3.1) | 6.0 (0.87) | 74.3 (3.6) | 3.9 (0.1) |
| JMH1 | 3/1 | 11.0 (1.7) | 3.4 (0.87) | 82.5 (11.0) | 3.3 (3.2) | 92.5 | 4.2 | 71.3 | 5.1 |
| LGH1095 | 3/3 | 1.4 (1.7) | 5.6 (1.1) | 80.9 (2.6) | 3.7 (0.1) | 99.2 (0.7) | 4.93 (0.93) | 77.6 (5.8) | 4.7 (0.5) |
| DSH4 | 3/2 | 61 | 4.23 (0.32) | 92.8 (3.3) | 2.3 (0.95) | 99.3 | 4.25 | 57.5 | 10.3 |
Hydrophilic cells were grown as described in the text at 37°C to early stationary phase (26 h), and hydrophobic cells were grown at 25°C. Data are the means (SD) of determinations of total protein, hexose, and phosphate in mannan samples (dried to constant mass).
n, no. of independent mannan preparations from hydrophilic cells/no. of independent mannan preparations from hydrophobic cells.
Adhesion of the mutant derivative grown at 37°C as assessed in a static binding assay to fibronectin-coated plates was slightly elevated relative to strain LGH1095. Cells of either type grown at 25°C to promote the hydrophobic phenotype bound equally well. A much more pronounced difference was observed when there was adhesion of yeasts to coated capillaries under physiologic shear forces (Table 2). The knockout derivative grown at 37°C bound significantly more frequently to fibronectin-coated capillaries than the parent strain, correlating with the observed differences in hydrophobicity. The increased difference between the mutant and parent strains in the shear assay demonstrates the benefits of the assay for assessing relative changes in adhesion.
Virulence of the mnn4Δ/mnn4Δ knockout.
We have also examined the relative virulence of the knockout strain by systemic infection via the lateral tail vein in AJ mice. Both the mutant and parent strains were virulent at 105 or 106 CFU/dose, and the disease course progresses with similar kinetics in both cases (data not shown). We have additionally used the mnn4Δ/mnn4Δ derivative together with strain LGH1095 in coinfection experiments and recovered CFU from kidneys of animals at the experimental endpoint. Colonies (24 per mouse; 5 mice per infective dose; 3 different doses of between 2 × 10 to 5 × 106 CFU per dose) were then randomly chosen and plated in 96-well plates, grown overnight, and phenotypically scored as either the knockout or parent. Equal ratios of mutant to parent cells were observed directly after infection and up to 3 days postinfection, indicating that neither strain has a short-term infective advantage relative to the other. The serotype A mnn4Δ/mnn4Δ mutant was also found to be equally virulent in mice in comparison to its cognate parent strain and did not demonstrate a deficit in macrophage interaction (29).
Altered immunoreactivity of carbohydrate epitopes in the mnn4Δ/mnn4Δ mutant.
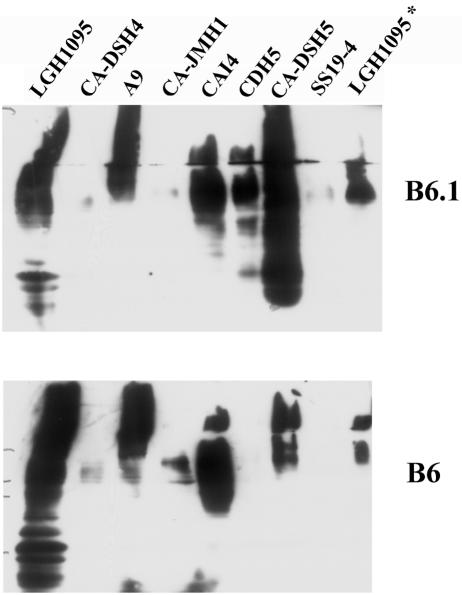
The most significant phenotypic effects of the mnn4Δ/mnn4Δ mutant are on the mature structure of cell surface glycoproteins. Both the serotype A and serotype B mnn4Δ/mnn4Δ derivatives lack an acid-labile mannan fraction on N-linked wall glycoproteins. The serotype B mutants were unreactive with an antibody that recognizes the β-1,2-mannotriose structure that is added distally to the mannosylphosphate, which is missing due to the disruption of MNN4 (Fig. 3, top panel). However, the serotype A mnn4Δ/mnn4Δ strain CDH5 retained significant immunoreactivity with MAb B6.1. Deletion derivatives constructed in both serotypes showed an unanticipated altered immunoreactivity with an antibody that is reactive with an incompletely characterized epitope on the acid-stable mannan region (Fig. 3, bottom panel).
FIG. 3.
Immunoreactivity of glycosylation-deficient C. albicans derivatives with anticarbohydrate antibodies. Cell wall proteins from stationary phase cultures of C. albicans glycosylation mutants were extracted by digestion with Zymolyase 100T (a β-1,3-glucanase; ICN Biochemicals) and separated by on 12% polyacrylamide gels for transfer to nitrocellulose. Western blots were probed either with MAb B6.1, which recognizes a β-1,2-mannotriose, or MAb B6, which recognizes an incompletely characterized epitope in the acid-stable mannan fraction. Lanes were loaded with equal amounts of protein, which resulted in a consistent pattern across the blot as visualized by staining with Ponceau S. The sample on the right (LGH1095*) represents total cell lysate generated by glass bead breakage of a cell pellet (49).
FACE analysis.
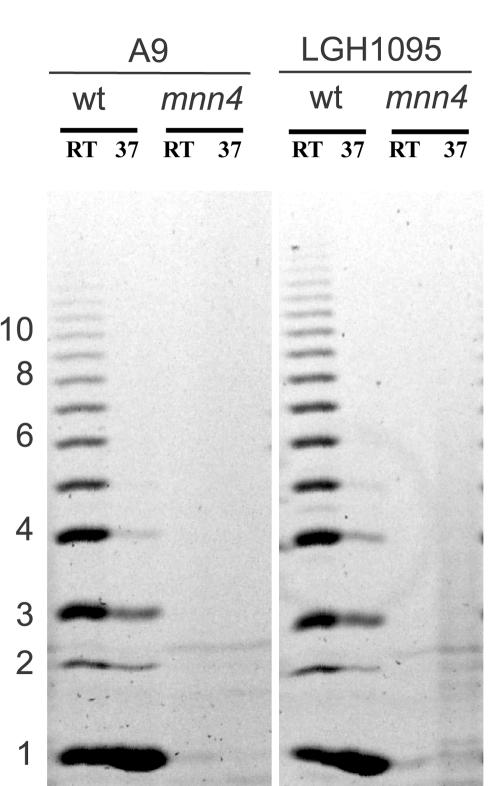
Absence of the acid-labile fraction in the serotype B mnn4Δ/mnn4Δ mutants was further demonstrated by FACE analysis (Fig. 4). Mannoproteins extracted from wild-type, hydrophobic cells produced greater amounts of acid-labile oligosaccharides. The concentration of oligosaccharide was 5- to 10-fold higher in these samples than in any other sample. An unexpected result was that a small amount of oligosaccharide was detected in the acid-labile fraction of the mnn4-disrupted strains. Because of the sensitivity of the assay, the detected oligosaccharide could represent environmental contaminants. The material detected by the oligosaccharide assay can be seen as a faint signal in some of the mnn4 lanes. The weakness of this signal relative to the wild-type lanes suggests that this is not due to bona fide acid-labile mannan. Furthermore, none of the bands in the mnn4 lanes comigrates with bands in the wild-type samples. This indicates that none of the mnn4 bands are β-1,2-oligomannosides because electrophoretic mobility is affected by glycosidic linkage anomericity (α versus β) and position and the component monosaccharides (J. Masuoka and K. C. Hazen, unpublished results.) More likely, it is due to a small amount of acid hydrolysis of the underlying glucan that takes place even in 10 mM HCl when heated in boiling water for 60 min.
FIG. 4.
Acid-labile mannan analysis of C. albicans derivatives by purification and FACE. Bulk mannan was prepared from wild-type (LGH1095 and A9) and mnn4Δ/mnn4Δ cells grown at either 25°C (RT) (hydrophobic cells, lane 1) or 37°C (hydrophilic cells, lane 2) and extracted to obtain the acid-labile fraction as described in the text. Equal moles of reducing end sugars were labeled with ANTS, electrophoretically separated, and visualized by UV-fluorescence. wt, wild type.
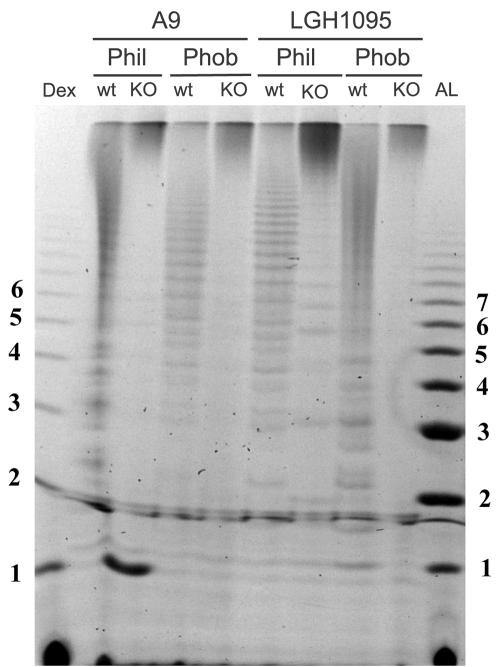
A perturbation of the acid-stable region is also apparent by FACE analysis where acetolysis of the acid-stable mannan was followed by electrophoretic separation of the released branches. Compared to the wild-type cells, acetolysates from the mnn4Δ/mnn4Δ strains showed a greatly simplified electrophoretic profile (Fig. 5). The number of bands was reduced, and their relative mobilities were changed in most cases. In addition, the majority of the fluorescent signal appeared at the top of the gel. One interpretation of this result is that the structure of the acid-stable mannan (e.g., the α-1,6-mannose backbone which is cleaved during acetolysis) in mnn4Δ/mnn4Δ cells has been altered so that it is more resistant to acetolysis than the corresponding fraction from wild-type cells. The significant loss of MAb B6 immunoreactivity seen in Fig. 3B could therefore be due to obscuring of the epitope as a result of this change in structure, without any other significant disruption of the mature mannan biosynthesis.
FIG. 5.
Acid-stable mannan analysis of C. albicans derivatives by purification and FACE. Bulk mannan was prepared as before from wild-type (LGH1095 and A9) and mnn4Δ/mnn4Δ cells grown at either 25°C (hydrophobic cells) or 37°C (hydrophilic cells). The α-1,6-mannose backbone of purified acid-stable mannan was cleaved by acetolysis as described in the text, and equal moles of cleaved reducing ends were labeled with ANTS and electrophoretically separated as described in the legend of Fig. 4. The left-hand lane is a labeled ladder of hydrolyzed dextran (α-1,6-glucose) as an approximate sizing standard, and the right-hand lane is a acid-labile ladder from phobic cells prepared as described in the legend of Fig. 4. wt, wild type; Dex, dextran; AL, acid-labile ladder; Phil, hydrophilic; Phob, hydrophobic; KO, knockout.
Compositional analysis of mannan prepared from hydrophobic (grown at 25°C) or hydrophilic (grown at 37°C) cells was performed to ascertain whether bulk mannan from wild-type or mnn4Δ/mnn4Δ mutant cells was comparable in makeup (Table 4). Relative levels of protein to hexose in mannoproteins were very similar between the wild-type strains and cognate mnn4Δ/mnn4Δ mutants, indicating that a glycan structure comparable in size is formed. Similar levels of phosphate were measured in both wild-type and mnn4 mutant cells. As Ballou et al. (6) observed in S. cerevisiae, disruption of MNN4 eliminates Alcian blue binding but does not eliminate phosphate from the cell wall (6) (Table 4).
DISCUSSION
We have used gene replacement by means of homologous recombination to independently knockout the MNN4 gene in two serotype B C. albicans strains. Standard molecular techniques in C. albicans typically use selection of a prototrophic marker for identifying transformants. We have used a positive selection scheme (59) to generate a deletion derivative that differs from the parent strain only at the gene of interest. The selection scheme with mycophenolic acid resistance acquisition is typically leakier than the standard prototrophic marker selection, resulting in the need to screen larger numbers of transformants for insertions of interest; however, the single gene difference between the two strains ultimately facilitates phenotypic comparisons between deletion derivatives and the parent strain. Of particular significance is the fact that the recently observed position effects on gene expression (33, 53) of the URA3 selectable marker are not relevant with these mutants, making virulence studies easier to interpret. Multiple investigators have reported that reintroduction of the URA3 selection marker at different chromosomal sites independent of other mutational events has profound effects on the ability of the yeast derivative to survive within the host, due to variability in the levels of expression of the URA3 gene (2, 10, 33).
The present study was undertaken to further examine the relationship between CSH and mannoprotein makeup. Previous work had demonstrated that cell walls appear different ultrastructurally between hydrophobic and hydrophilic cell populations (26). However, gross protein profiles and overall glycosylation appear similar between the two phenotypes, and the major glycosylation difference that has been found is an increase in the length of the acid-labile mannan fraction in hydrophobic cells (36, 37). This observation has led us to postulate that the length of this structure in wall mannoproteins affects the folding of cell wall fibrils, potentially by interacting with other proteins or carbohydrates (37). One can extend this reasoning to expect that elimination of the acid-labile fraction in the serotype B background might affect CSH, which we anticipated would result in conversion to the hydrophilic phenotype. We have found that the mnn4Δ/mnn4Δ mutant does have a significant effect on CSH; however, the mutant strains demonstrate an increased hydrophobic phenotype in comparison to the parents. The reason for this effect is unknown but does continue to support a role of the β-1,2-oligomannose group in the regulation of hydrophobicity, in that subtle differences in glycosylation can have a remarkable effect on CSH.
Readily detectable levels of phosphate in purified mannan from wild-type and mnn4 mutant derivatives confirmed earlier observations in S. cerevisiae. Ballou et al. (6), and before them Friis and Ottolenghi (17), speculated that quantity and location of the phosphate groups contributed to Alcian blue binding, suggesting that in the mnn4 mutant mannosylphosphorylation might occur in a region of the glycan inaccessible to dye binding. Both groups observed that mannosylphosphorylation was linked to only a single gene and correlated with the addition of a single specific group to maturing glycoproteins. But both groups also observed cell wall phosphate in the background of an mnn4 or mnn4-like mutant and speculated that this additional phosphate was deeper in the wall and inaccessible to dye. It was unclear from the previous analyses whether these additional phosphate groups are also attached to mannoprotein or are on another cell wall component. Several possible scenarios arise to fit these results. First, mannosylphosphorylation continues to occur in the mnn4 background, but the phosphate groups are added to glycan proximal to the polypeptide or are on glycoprotein components buried deeper in the cell wall. In either case, Ballou et al. (6) speculated that the phosphate groups might not be accessible enough to interact with the dye. Unlike Coomassie blue, Alcian blue does not form colloids and is relatively small, which likely allows it to penetrate the outer layers of the cell wall. Our results suggest that this interpretation is incorrect by assessing the phosphate content of purified mannoproteins. No acid-labile oligosaccharides were detected throughout the glycan in the mnn4Δ/mnn4Δ mutant, indicating that in the purified mannoproteins alteration of mannosylphosphorylation occurs on a global scale, not just at the distal ends of the glycans more likely to interact with the dye. Second, phosphate might exist in N-linked glycans aside from an acid-labile mannosylphosphate linkage, such as in the core portion of the N-glycan. Odani et al. (41) observed that the mannosylphosphate content of the core oligosaccharide in mnn4 mutants was greatly diminished relative to wild-type in S. cerevisiae, suggesting that this source of phosphate is also not significant in the C. albicans mnn4 mutant. Third, other sources of phosphate distinct from the acid-labile fraction mannosylphosphate that might contribute to cell wall-associated phosphate are potential plasma membrane contamination or the phospholipomannan fraction that has been recently described to mediate fungal cell adhesion (12).
An unanticipated perturbation of the acid-stable region is in apparent contradiction to the reported function of the MNN4 gene product as strictly a regulator of mannosylphosphate addition in S. cerevisiae (40, 42). No reported role of the MNN4 gene has been biochemically or genetically demonstrated elsewhere in the mature mannan structure. The standard model of glycoprotein biosynthesis holds that maturation of carbohydrate structure occurs in a sequential manner, due to the action of multiple gene products as the protein migrates through sites within the secretory pathway, and in yeast this is mainly due to the activity of numerous specific mannosyltransferases. Loss of a specific enzymatic activity would then be predicted not to have an effect on the structure that had been previously synthesized. Our results suggest that MNN4 function (and therefore mannosylphosphate or β-1,2-mannose addition) is important for proper glycan structure or function, and elimination of Mnn4p activity has unexpected repercussions on overall mannan structure. It is not known whether a perturbation in the acid-stable region is present in S. cerevisiae mnn4 mutants; however, mannan structure up to the point of β-1,2-mannose addition is similar between the two fungal species. It can be anticipated that if the presence of mannosylphosphate is necessary to form a proper glycan structure, then the S. cerevisiae mnn4 mutant would demonstrate a similar defect. If it is the lack of acid-labile associated β-1,2-oligomannose in mnn4Δ/mnn4Δ mutants that causes the acid-stable perturbation, this defect would be restricted to Candida species. Analysis of the cognate S. cerevisiae mutants is currently being done.
The physiologic role of glycan mannosylphosphorylation in fungi has not been identified. Comparison of the S. cerevisiae Mnn4p sequence with the GenBank database readily identifies paralogous sequences in all fungal genomes deposited to date, arguing that the regulation of mannosylphosphate addition has been conserved during fungal divergence. Elimination of the MNN4 gene in S. cerevisiae and C. albicans demonstrated that mannosylphosphorylation is not essential for viability and in the latter species is not necessary for virulence in a hematogenous dissemination model (the present study) and for macrophage recognition (29). In light of these observations, several possible scenarios can be postulated about the role of Mnn4p-mediated mannosylphosphorylation in fungi and why it has been preserved among disparate fungal species. First, the phosphodiester bond is a labile linkage that might allow pathogenic fungi to rapidly alter antigenic characteristics or other surface properties. However, there is as yet no evidence that this mechanism is utilized for immune circumvention by fungi. Second, the carbohydrate-associated phosphate confers a net negative charge on the cell wall, which may mediate intercellular contact through ionic interactions or disrupt cell aggregation by electrostatic repulsion. Ionic interactions have been demonstrated in binding of a plant antifungal protein with yeast phosphomannan (30, 47). Third, mannosylphosphorylation might be necessary for proper glycan conformation, which in turn would allow proper structure and function of the cell wall elements. The preliminary data presented here are consistent with this possibility, but further experimentation is necessary to demonstrate a role for Mnn4p in affecting cell wall morphology in C. albicans and to extend it to other fungal species.
In summary, elimination of the MNN4 gene blocks the addition of β-1,2-mannose residues that extend from mannosylphosphate. The β-1,2-mannose polymer forms an epitope of diagnostic significance and of clinical and therapeutic relevance (12, 22, 48). Although there is not an immediately obvious effect on virulence in the mouse, absence of this carbohydrate modification leads to a significant change in CSH, which in turn affects adhesion to fibronectin. Loss of the acid-labile mannan fraction in a serotype B C. albicans strain does not result in significant changes in the ratio of protein to hexose, suggesting that the overall extent of glycosylation is relatively similar in the mnn4Δ/mnn4Δ strain background. An uncharacterized perturbation of the acid-stable region as assessed by FACE analysis of purified acid-stable mannan and immunodetection supports the hypothesis that loss of the acid-labile fraction results in an overall change in mannoprotein conformation and confirms its role in the regulation of cell surface hydrophobicity.
Acknowledgments
This work was supported with funds from the Public Health Service's National Institute of Allergy and Infectious Diseases grant R01 AI43997.
The authors thank Paula Veldhuis (Department of Pathology, University of Virginia) for performing the adhesion assays and statistical analysis, Neil Gow (University of Aberdeen) and Richard Hobson (University of Leeds) for helpful discussions and communicating strains and results prior to publication, and Jim Cutler (Research Institute for Children) for providing antibodies.
REFERENCES
- 1.Ames, B. N., and D. T. Dubin. 1960. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J. Biol. Chem. 235:769-775. [PubMed] [Google Scholar]
- 2.Bain, J. M., C. Stubberfield, and N. A. Gow. 2001. URA-status-dependent adhesion of Candida albicans mutants. FEMS Microbiol Lett. 204:323-328. [DOI] [PubMed] [Google Scholar]
- 3.Ballou, C. E. 1990. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 185:440-470. [DOI] [PubMed] [Google Scholar]
- 4.Ballou, C. E. 1976. Structure and biosynthesis of the mannan component of the yeast cell envelope. Adv. Microb. Physiol. 14:93-158. [DOI] [PubMed] [Google Scholar]
- 5.Ballou, C. E. 1982. Yeast cell wall and cell surface, p. 335-360. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 6.Ballou, C. E., K. A. Kern, and W. C. Raschke. 1973. Genetic control of yeast mannan structure. Complementation studies and properties of mannan mutants. J. Biol. Chem. 248:4667-4671. [PubMed] [Google Scholar]
- 7.Ballou, L., R. E. Cohen, and C. E. Ballou. 1980. Saccharomyces cerevisiae mutants that make mannoproteins with a truncated carbohydrate outer chain. J. Biol. Chem. 255:5986-5991. [PubMed] [Google Scholar]
- 8.Burda, P., and M. Aebi. 1999. The dolichol pathway of n-linked glycosylation. Biochim. Biophys. Acta 1426:239-257. [DOI] [PubMed] [Google Scholar]
- 9.Caesar-TonThat, T. C., and J. E. Cutler. 1997. A monoclonal antibody to Candida albicans enhances mouse neutrophil candidacidal activity. Infect. Immun. 65:5354-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, S., M. Nguyen, Z. Zhang, H. Jia, M. Handfeld, and C. Clancy. 2003. Evaluation of the roles of four Candida albicans genes in virulence by using gene disruption strains that express URA3 from the native locus. Infect. Immun. 71:6101-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conde, R., G. Pablo, R. Cueva, and G. Larriba. 2003. Screening for new yeast mutants affected in mannosylphosphorylation of cell wall mannoproteins. Yeast 20:1189-1211. [DOI] [PubMed] [Google Scholar]
- 12.Dalle, F., T. Jouault, P. A. Trinel, J. Esnault, J. M. Mallet, P. d′Athis, D. Poulain, and A. Bonnin. 2003. β-1,2- and α-1,2-Linked oligomannosides mediate adherence of Candida albicans blastospores to human enterocytes in vitro. Infect. Immun. 71:7061-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, N. 1999. Asparagine-linked glycosylation in the yeast golgi. Biochim. Biophys. Acta 1426:309-322. [DOI] [PubMed] [Google Scholar]
- 14.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 15.Dygert, S., L. H. Li, D. Florida, and J. A. Thoma. 1965. Determination of reducing sugar with improved precision. Anal. Biochem. 13:367-374. [DOI] [PubMed] [Google Scholar]
- 16.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friis, J., and P. Ottolenghi. 1970. The genetically determined binding of Alcian blue by a minor fraction of yeast cell walls. C. R. Trav. Lab. Carlsberg 37:327-341. [PubMed] [Google Scholar]
- 18.Glee, P. M., J. E. Cutler, E. E. Benson, R. F. Bargatze, and K. C. Hazen. 2001. Inhibition of hydrophobic protein-mediated Candida albicans attachment to endothelial cells during physiologic shear flow. Infect. Immun. 69:2815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goins, T. L., and J. E. Cutler. 2000. Relative abundance of oligosaccharides in Candida species as determined by fluorophore-assisted carbohydrate electrophoresis. J. Clin. Microbiol. 38:2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gopal, P. K., and C. E. Ballou. 1987. Regulation of the protein glycosylation pathway in yeast: structural control of n-linked oligosaccharide elongation. Proc. Natl. Acad. Sci. USA 84:8824-8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, Y., R. P. Morrison, and J. E. Cutler. 1998. A vaccine and monoclonal antibodies that enhance mouse resistance to Candida albicans vaginal infection. Infect. Immun. 66:5771-5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han, Y., M. H. Riesselman, and J. E. Cutler. 2000. Protection against candidiasis by an immunoglobulin G3 (IgG3) monoclonal antibody specific for the same mannotriose as an IgM protective antibody. Infect. Immun. 68:1649-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasenclever, H. F., and W. O. Mitchell. 1961. Antigenic studies of Candida. I. Observation of two antigenic groups in Candida albicans. J. Bacteriol. 82:570-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hazen, B. W., and K. C. Hazen. 1988. Modification and application of a simple, surface hydrophobicity detection method to immune cells. J. Immunol. Methods 107:157-163. [DOI] [PubMed] [Google Scholar]
- 25.Hazen, K. C., L. D. Bourgeois, and J. F. Carpenter. 1988. Cryoprotection of antibody by organic solutes and organic solute/divalent cation mixtures. Arch. Biochem. Biophys. 267:363-371. [DOI] [PubMed] [Google Scholar]
- 26.Hazen, K. C., and B. W. Hazen. 1992. Hydrophobic surface protein masking by the opportunistic fungal pathogen Candida albicans. Infect. Immun. 60:1499-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hazen, K. C., and B. W. Hazen. 1987. A polystyrene microsphere assay for detecting cell surface hydrophobicity within Candida albicans populations. J. Microbiol. Methods 6:289-299. [Google Scholar]
- 28.Hazen, K. C., and B. W. Hazen. 1987. Temperature-modulated physiological characteristics of Candida albicans. Microbiol. Immunol. 31:497-508. [DOI] [PubMed] [Google Scholar]
- 29.Hobson, R. P., C. A. Munro, S. Bates, D. M. MacCallum, J. E. Cutler, S. E. M. Heinsbroek, G. D. Brown, F. C. Odds, and N. A. R. Gow. 2004. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279:39628-39635. [DOI] [PubMed] [Google Scholar]
- 30.Ibeas, J. I., H. Lee, B. Damsz, D. T. Prasad, J. M. Pardo, P. M. Hasegawa, R. A. Bressan, and M. L. Narasimhan. 2000. Fungal cell wall phosphomannans facilitate the toxic activity of a plant PR-5 protein. Plant J. 23:375-383. [DOI] [PubMed] [Google Scholar]
- 31.Jackson, P. 1990. The use of polyacrylamide-gel electrophoresis for the high-resolution separation of reducing saccharides labelled with the fluorophore 8-aminonaphthalene-1,3,6-trisulphonic acid. Biochem. J. 270:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jigami, Y., and T. Odani. 1999. Mannosylphosphate transfer to yeast mannan. Biochim. Biophys. Acta 1426:335-345. [DOI] [PubMed] [Google Scholar]
- 33.Lay, J., L. K. Henry, J. Clifford, Y. Koltin, C. E. Bulawa, and J. M. Becker. 1998. Altered expression of selectable marker URA3 in gene-disrupted Candida albicans strains complicates interpretation of virulence studies. Infect. Immun. 66:5301-5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd, K. O. 1970. Isolation, characterization, and partial structure of peptido galactomannans from the yeast form of Cladosporium werneckii. Biochemistry 9:3446-3453. [DOI] [PubMed] [Google Scholar]
- 35.Masuoka, J., and K. C. Hazen. 2004. Cell wall mannan and cell surface hydrophobicity in Candida albicans serotype A and B strains. Infect. Immun. 72:6230-6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Masuoka, J., and K. C. Hazen. 1997. Cell wall protein mannosylation determines Candida albicans cell surface hydrophobicity. Microbiology 143:3015-3021. [DOI] [PubMed] [Google Scholar]
- 37.Masuoka, J., and K. C. Hazen. 1999. Differences in the acid-labile component of Candida albicans mannan from hydrophobic and hydrophilic yeast cells. Glycobiology 9:1281-1286. [DOI] [PubMed] [Google Scholar]
- 38.Masuoka, J., G. Wu, P. M. Glee, and K. C. Hazen. 1999. Inhibition of Candida albicans attachment to extracellular matrix by antibodies which recognize hydrophobic cell wall proteins. FEMS Immunol. Med. Microbiol. 24:421-429. [DOI] [PubMed] [Google Scholar]
- 39.Murad, A. M., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. P. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 40.Nakayama, K., Y. Feng, A. Tanaka, and Y. Jigami. 1998. The involvement of MNN4 and MNN6 mutations in mannosylphosphorylation of o-linked oligosaccharide in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1425:255-262. [DOI] [PubMed] [Google Scholar]
- 41.Odani, T., Y. Shimma, A. Tanaka, and Y. Jigami. 1996. Cloning and analysis of the MNN4 gene required for phosphorylation of n-linked oligosaccharides in Saccharomyces cerevisiae. Glycobiology 6:805-810. [DOI] [PubMed] [Google Scholar]
- 42.Odani, T., Y. Shimma, W. X. H., and Y. Jigami. 1997. Mannosylphosphate transfer to cell wall mannan is regulated by the transcriptional level of the MNN4 gene in Saccharomyces cerevisiae. FEBS Lett. 420:186-190. [DOI] [PubMed] [Google Scholar]
- 43.Peat, S., W. J. Whelan, and T. E. Edwards. 1961. Polysaccharides of baker's yeast. Part IV. Mannan. J. Chem. Soc. 1961:29-34. [Google Scholar]
- 44.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 45.Rosenfeld, L., and C. E. Ballou. 1974. Genetic control of yeast mannan structure. J. Biol. Chem. 249:2319-2321. [PubMed] [Google Scholar]
- 46.Ruiz-Herrera, J. 1992. Fungal cell wall: structure, synthesis, and assembly. CRC Press, Inc., Ann Arbor, Mich.
- 47.Salzman, R. A., H. Koiwa, J. I. Ibeas, J. M. Pardo, P. M. Hasegawa, and R. A. Bressan. 2004. Inorganic cations mediate plant PR5 protein antifungal activity through fungal MNN1- and MNN4-regulated cell surface glycans. Mol. Plant Microbe Interact. 17:780-788. [DOI] [PubMed] [Google Scholar]
- 48.Sendid, B., J. L. Poirot, M. Tabouret, A. Bonnin, D. Caillot, D. Camus, and D. Poulain. 2002. Combined detection of mannanaemia and antimannan antibodies as a strategy for the diagnosis of systemic infection caused by pathogenic Candida species. J. Med. Microbiol. 51:433-442. [DOI] [PubMed] [Google Scholar]
- 49.Singleton, D. R., and K. C. Hazen. 2004. Differential surface localization and temperature-dependent expression of the Candida albicans CSH1 protein. Microbiology 150:285-292. [DOI] [PubMed] [Google Scholar]
- 50.Singleton, D. R., J. Masuoka, and K. C. Hazen. 2001. Cloning and analysis of a Candida albicans gene that affects cell surface hydrophobicity. J. Bacteriol. 183:3582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 52.Southard, S. B., C. A. Specht, C. Mishra, J. Chen-Weiner, and P. W. Robbins. 1999. Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins. J. Bacteriol. 181:7439-7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sundstrom, P., J. E. Cutler, and J. F. Staab. 2002. Reevaluation of the role of HWP1 in systemic candidiasis by use of Candida albicans strains with selectable marker URA3 targeted to the ENO1 locus. Infect. Immun. 70:3281-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Suzuki, A., Y. Takata, A. Oshie, A. Tezuka, N. Shibata, H. Kobayashi, Y. Okawa, and S. Suzuki. 1995. Detection of beta-1,2-mannosyltransferase in Candida albicans cells. FEBS Lett. 373:275-279. [DOI] [PubMed] [Google Scholar]
- 55.Walsh, T. J., C. McEntee, and D. M. Dixon. 1987. Tissue homogenization with sterile reinforced polyethylene bags for quantitative culture of Candida albicans. J. Clin. Microbiol. 25:931-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, X. H., K. Nakayama, Y. Shimma, A. Tanaka, and Y. Jigami. 1997. MNN6, a member of the KRE2/MNT1 family, is the gene for mannosylphosphate transfer in Saccharomyces cerevisiae. J. Biol. Chem. 272:18117-18124. [DOI] [PubMed] [Google Scholar]
- 57.Whelan, W. L., J. M. Delga, E. Wadsworth, T. J. Walsh, K. J. Kwon-Chung, R. Calderone, and P. N. Lipke. 1990. Isolation and characterization of cell surface mutants of Candida albicans. Infect. Immun. 58:1552-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]
- 59.Wirsching, S., S. Michel, and J. Morschhauser. 2000. Targeted gene disruption in Candida albicans wild-type strains: the role of the MDR1 gene in fluconazole resistance of clinical Candida albicans isolates. Mol. Microbiol. 36:856-865. [DOI] [PubMed] [Google Scholar]
- 60.Yamashita, K., T. Mizuochi, and A. Kobata. 1982. Analysis of oligosaccharides by gel filtration. Methods Enzymol. 83:105-126. [DOI] [PubMed] [Google Scholar]