ABSTRACT
Rotavirus infection is a leading cause of gastroenteritis in children worldwide; the genome of this virus is composed of 11 segments of dsRNA packed in a triple-layered protein capsid. Here, we investigated the role of nucleolin, a protein with diverse RNA-binding domains, in rotavirus infection. Knocking down the expression of nucleolin in MA104 cells by RNA interference resulted in a remarkable 6.3-fold increase in the production of infectious rhesus rotavirus (RRV) progeny, accompanied by an elevated synthesis of viral mRNA and genome copies. Further analysis unveiled an interaction between rotavirus segment 10 (S10) and nucleolin, potentially mediated by G-quadruplex domains on the viral genome. To determine whether the nucleolin–RNA interaction regulates RRV replication, MA104 cells were transfected with AGRO100, a compound that forms G4 structures and selectively inhibits nucleolin–RNA interactions by blocking the RNA-binding domains. Under these conditions, viral production increased by 1.5-fold, indicating the inhibitory role of nucleolin on the yield of infectious viral particles. Furthermore, G4 sequences were identified in all 11 RRV dsRNA segments, and transfection of oligonucleotides representing G4 sequences in RRV S10 induced a significant increase in viral production. These findings show that rotavirus replication is negatively regulated by nucleolin through the direct interaction with the viral RNAs by sequences forming G4 structures.
IMPORTANCE
Viruses rely on cellular proteins to carry out their replicative cycle. In the case of rotavirus, the involvement of cellular RNA-binding proteins during the replicative cycle is a poorly studied field. In this work, we demonstrate for the first time the interaction between nucleolin and viral RNA of rotavirus RRV. Nucleolin is a cellular protein that has a role in the metabolism of ribosomal rRNA and ribosome biogenesis, which seems to have regulatory effects on the quantity of viral particles and viral RNA copies of rotavirus RRV. Our study adds a new component to the current model of rotavirus replication, where cellular proteins can have a negative regulation on rotavirus replication.
KEYWORDS: rotavirus, nucleolin, virus–host cell interactions, RNA-binding protein
INTRODUCTION
Rotavirus-induced gastroenteritis poses a significant global health burden, particularly affecting children under 5 years old. Although the availability of vaccines, such as Rotateq and Rotarix, has contributed to a reduction in the incidence of severe cases of diarrhea, this virus continues to cause substantial morbidity and mortality worldwide (1). Rotaviruses are members of the Sedoreoviridae family (2), nonenveloped viruses characterized by a triple-layered protein capsid. Their genome is composed of 11 segments of double-stranded RNA (dsRNA) that encode six structural viral proteins (VP1, VP2, VP3, VP4, VP6, and VP7) and six nonstructural proteins (NSP1, NSP2, NSP3, NSP4, NSP5, and NSP6) (3).
During infection, the rotavirus employs intricate strategies to translate viral proteins and replicate its genome by modulating the cellular environment and hijacking host proteins (3). While substantial knowledge exists regarding the functions of viral proteins, our understanding of the cellular proteins involved in the rotavirus replication cycle remains limited. Several proteins with RNA-binding domains (RBDs) have been found to change their cellular localization upon rotavirus infection; silencing the expression of human antigen R protein (HuR), heterogeneous nuclear ribonucleoprotein (hnRNP) L, I, and D, ATP5B, and argonaute2 (Ago2) increases the production of viral progeny (4–6), suggesting that they have a role during the replication cycle of rotavirus.
Nucleolin (Ncl), a eukaryotic, mostly nuclear phosphoprotein, possesses a central region with an RBD comprising four RNA recognition motifs that enable interactions with RNA molecules (7). This protein plays diverse roles in ribosome assembly, rDNA transcription, and the modification and processing of pre-rRNA (7). Nucleolin has been reported to participate in cell binding and entry of several RNA viruses, thereby influencing their initial infection process (8–10). In addition, it has been implicated in the viral replication and translation of caliciviruses (11, 12) and the assembly of dengue virus particles (13).
In this work, we report that the number of viral RNA (vRNA) copies and the viral progeny increase when the expression of nucleolin was knocked down in MA104 cells. Immunoprecipitation assays reveal that nucleolin interacts with the RNA of rotavirus more likely through its binding to G-quadruplex RNA structures. Accordingly, using synthetic sequences predicted to form G-quadruplex structures and that mimic sequences in the rotavirus gene segment 10, we were able to confirm that these regions block the binding of nucleolin causing an increase in the production of infectious viral particles.
MATERIALS AND METHODS
Cell culture and viruses
The African green monkey kidney epithelial cell line MA104 (ATCC) was cultivated in Dulbecco’s modified Eagle’s medium (DMEM) (Thermo Scientific HyClone, Logan, UT) and supplemented with 5% fetal bovine serum (Biowest, Kansas, MO) at 37°C in a 5% CO2 atmosphere. Rhesus rotavirus strain RRV and the human strain Wa were obtained from H.B. Greenberg (Stanford University, Stanford, CA), the bovine rotavirus UK was donated by D.R. Snodgrass (Moredun Research Institute, Edinburgh, United Kingdom), and Simian rotavirus strain SA11 was obtained from M. Estes (Baylor College of Medicine, Houston, TX). The virus was propagated in MA104 cells, and the resultant virus was titrated and stored at −70°C.
Antibodies
Polyclonal antibodies targeting purified rotavirus RRV triple-layered particles (TLPs) and vimentin were generated in rabbits, following previously described protocols (14). Rabbit polyclonal sera against NSP3 were prepared as described previously (15). Monoclonal antibodies specific to nucleolin and PPI were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase-conjugated goat anti-rabbit polyclonal antibody was sourced from KPL (Gaithersburg, MD), while horseradish peroxidase-conjugated goat anti-mouse antibody was acquired from Millipore Merck KGaA (Darmstadt, Germany). Streptavidin magnetic beads were procured from New England Biolabs (Massachusetts, US).
Cell infection and virus titration
Prior to infection, the different strains of rotavirus were activated by incubating with trypsin (10 µg/mL, Gibco, Life Technologies, Carlsbad, CA) for 30 minutes at 37°C. Monolayers of MA104 cells were grown in a T-75 flask and infected with a multiplicity of infection (MOI) of 1 for 15 hours. The infected cells were lysed by three freeze–thaw cycles. The viral lysate’s infection titers were determined using a focus-forming assay. Briefly, confluent cells in 96-well plates were incubated with two-fold serial dilutions of the viral lysate (previously activated with trypsin as above) for 60 minutes at 37°C. After the adsorption period, the virus inoculum was removed, the cells were washed once, and fresh minimum essential medium (MEM) was added. The infection was allowed to proceed for 15 hours at 37°C. Rotavirus-infected cells were detected using a rabbit hyperimmune serum to rotavirus in an immunoperoxidase focus-forming assay, following previously described protocols (16).
siRNA transfection
Small interfering RNAs (siRNAs) targeting nucleolin (SMARTpool siGENOME Human Ncl, M-003854-01-0005) and “Non-Targeting” (SMARTpool siGENOME Non-Targeting, D-001210-04-20) control siRNAs were obtained from GE Healthcare Dharmacon (Lafayette, CO). The siRNA transfection was performed in MA104 cells grown in 48-well plates using a reverse transfection method, as previously described (16). Briefly, the transfection mix [15 µL Oligofectamine/mL (Invitrogen, Eugene, OR), mixed with 2.5 µM of the appropriate siRNA] was added to the well, and 20,000 cells/well were subsequently added and incubated for 16 hours at 37°, the transfection mix was then replaced by DMEM, and the cells were further incubated for up to 72 hours at 37°C.
Immunoblot analysis
For immunoblot analysis, cells were lysed using Laemmli sample buffer (50 mM Tris-HCl pH 6.8, 2% sodium dodecyl sulfate (SDS), 0.1% bromophenol blue, 10% glycerol, 5% β-mercaptoethanol) and denatured by boiling at 95°C for 5 minutes. The protein lysates were subjected to electrophoresis on 10% SDS-polyacrylamide gels, followed by transfer of proteins to an Immobilon membrane (Millipore) using N-cycloexyl-3-aminopropanesulfonic acid (CAPS) buffer (CAPS 10 mM, 10% methanol). The membrane was then blocked for 1 hour at room temperature with 5% skim milk in phosphate buffered saline (PBS)–0.1% Tween. Next, the membrane was incubated with primary antibodies diluted 1:3,000 in a solution of 1% skim milk in PBS–0.1% Tween, for 1 hour at room temperature. The binding of the primary antibodies was detected using horseradish peroxidase-conjugated anti-rabbit immunoglobulins (Thermo Scientific, Waltham, MA) at a 1:3,000 dilution or with peroxidase-conjugated anti-mouse immunoglobulins (Thermo Scientific, Waltham, MA) at a 1:3,000 dilution. Protein bands were developed using the Western Lightning system (PerkinElmer, MA). Densitometry analysis of the western blot images was performed using ImageJ software (17).
Real-time RT-PCR
Confluent MA104 cells were seeded in 48-well plates and infected with RRV at a MOI of 3. The cells were lysed at different times post-infection using the TRIzol (Thermo Scientific HyClone, Logan, UT) reagent, and the RNA was extracted using the manufacturer’s instructions. The primers used for the amplification of gene 10 of rotavirus were previously described as oligos forward: TCCTGGAATGGCGTATTTTC and reverse: GAGCAATCTTCATGGTTGGAA (18). Reverse transcription-polymerase chain reaction (RT-PCR) was employed to specifically determine the levels of each RNA strand, following the previously described method (18). The quantitative analysis of data was performed using the ABI Prism 7000 analysis software program. After PCR amplification, the fluorescence values of all samples between the logarithmic phases of the amplification curves were used to set a cutoff line using the ABI Prism software. The logarithm of the concentration of each sample was plotted against the cycle number, where the amplification curve of the sample reached the cutoff line (CT). The amount of positive or negative strand RNA from experimental samples was determined by extrapolating the CT value into the corresponding standard curve. The standard curve was prepared using a plasmid of DNA with the NSP4 gene sequence of rotavirus RRV present at 101–1011 copy numbers. Experimental samples were collected at 0, 4, 8, and 12 hours post-infection (hpi).
Immunoprecipitation assays
Confluent MA104 cells were grown in six-well plates and infected with RRV at an MOI of 5. At 8 hpi, the cells were washed with PBS-ethylenediaminetetraacetic acid (EDTA) (0.2 mM) and harvested by scraping. The resulting cell pellet was centrifuged for 10 minutes at 1,000 RPM at 4°C and subsequently washed twice with 2 mL of PBS at 4°C, followed by centrifugation for 10 minutes at 1,000 RPM. The resulting pellet was then resuspended in 1 mL of RIPA buffer (50 mM Tris-HCl at pH 7.5, 1% Triton, 0.5% sodium deoxycholate, 0.005% SDS, 1 mM EDTA, and 150 mM NaCl), containing protease inhibitor (Roche, Basilea) and RNase inhibitors (NEB, Massachusetts) and subjected to three rounds of sonication (20 seconds each). The resulting cell lysate was centrifuged for 10 minutes at 14,000 RPM, and the supernatants were subsequently clarified with 50 µL of magnetic beads coupled to protein G (Dynabeads Protein G-LS10003D, Thermo Scientific) at 4°C for 1 hour with agitation. Fifteen microliters of magnetic bead Dynabeads was pre-treated with 10 mg/mL of tRNA, and antibodies were added to the pre-cleared lysate and incubated overnight at 4°C with agitation. After this time, the beads were washed four times with RIPA buffer. The beads were split into two separate tubes: the first tube was used to identify the immunoprecipitated proteins through western blot, while the second tube was used for RNA extraction using TRIzol, and the RRV segments were detected by qPCR.
The primers used for the amplification of three genes of rotavirus were previously described as oligo forward CAGACCCGGGTACCTATTAAAGCTATACA and reverse CAGACCCGGGCCGCGGTCACATCTAAGCG for segment 1, oligo forward CAGACCCGGGTACCTTTTTAAACGAAGTCT and reverse CAGACCCGGGCCGCGGTCACATCCTCTCACT for segment 4, and oligo forward CAGACCCGGGTACCTTTTAAAAGTTCTGTT and reverse CAGACCCGGGCCGCGGTCACATCATACAATTC for segment 10 (18).
Analysis of putative G-quadruplex (G4) domains
The QGRS mapper web server (19) was utilized to search for putative G-quadruplex domains. The positive polarity RNA strand sequences of the 11 segments of rotavirus RRV were examined. The following parameters were employed: maximum domain length of 45 nucleotides, mini groups of guanines together of 2, and possible loop sizes of up to 36 nucleotides (19). The search was performed using the QGRS mapper web server, available at https://bioinformatics.ramapo.edu/QGRS/analyze.php.
G-quadruplex formation and transfection
The G-quadruplex-forming sequences used in these assays were AGRO100: 5′-GGTGGTGGTGGTTGTGGTGGTGGTGG-3′, CRO26: 5′-CCTCCTCCTCCTTCTCCTCCTCCTCC-3′, G4-1: 5′-GGAGGATCCTGGAATGG-3′, G4-2: 5′-GGTTGAGCTGCCGTCGTCTGTCTGCGGAAGCGGCGG-3′, and G4-3: 5′-GGACGTTAATGGAAGGAACGG-3′.
G-quadruplex folding was carried out as described previously (20). Briefly, each G4 oligonucleotide was suspended in folding buffer (10 mM Tris-HCl, pH 7.5, 100 mM KCl, and 0.1 mM EDTA), and the formation of secondary structures was achieved by heating the ssDNA to 95°C for 5 minutes and cooling it down to 4°C in steps of 2°C per minute. The folded G4 sequences were transfected into MA104 cells in 48-well plates as described previously for the siRNA transfections.
Statistical analysis
The statistical significance of the experiments was evaluated using the Mann–Whitney U test with a significance level of 95%. The GraphPad Prism 5 program was employed for the statistical analysis. The asterisks displayed in the graphs represent different degrees of statistical significance: *P < 0.05, **P < 0.01, and ***P < 0.001.
RESULTS
Nucleolin negatively regulates rotavirus replication
To advance the characterization of the role of RNA-binding cellular proteins on rotavirus replication, we evaluated the role of nucleolin, an RNA-binding protein known to interact with several RNA viruses, in the replication cycle of rotaviruses. For this, the effect of silencing the expression of nucleolin by RNA interference (RNAi) on the yield of infectious rotavirus progeny during a single replicative cycle was evaluated. In these assays, MA104 cells seeded in 48-well plates were transfected with a pool of siRNAs specifically targeting nucleolin, and 72 hours post-transfection, the cells were infected with four different strains of rotavirus: rhesus rotavirus (RRV), the simian rotavirus SA11 (SA11), the bovine rotavirus (UK), and the Wa rotavirus strain (Wa) at an MOI of 3. At 15 hpi, the infected cells were harvested, and the viral titer of the virus produced under these conditions was determined by a focus-forming assay (16) (Fig. 1A).
Fig 1.
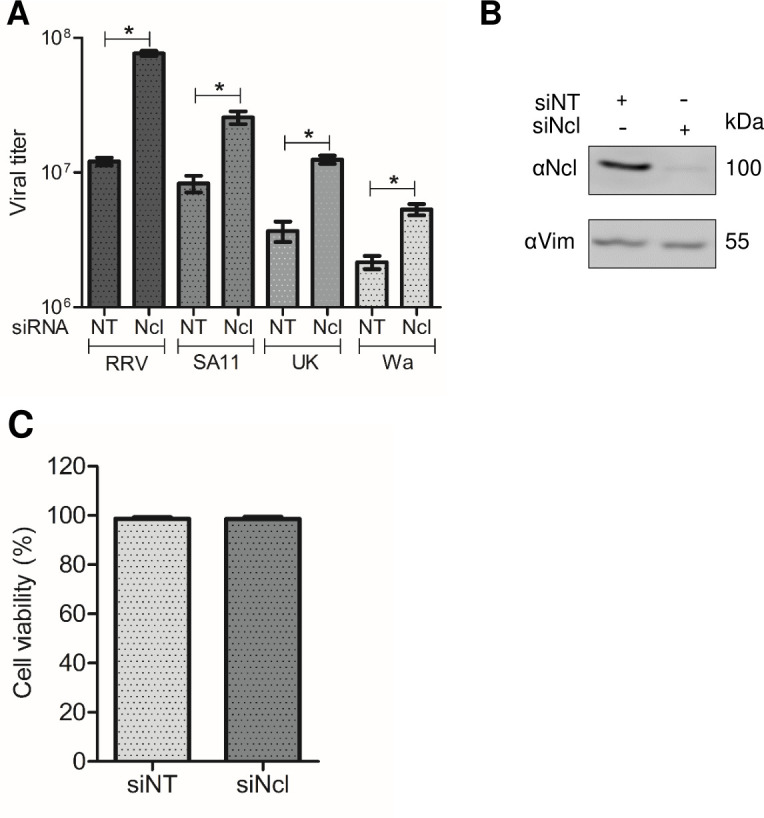
Knockdown of nucleolin expression promotes an increase in RRV, SA11, UK, and Wa strains of rotavirus titers. (A) MA104 cells grown in 48-well plates were transfected with either nontargeting (NT) siRNA or nucleolin siRNA. At 72 hours post-transfection (hpt), cells were infected with RRV, SA11, UK, and Wa rotavirus at an MOI of 3, harvested at 15 hpi. Data shown represent the arithmetic mean ± standard deviation of three independent experiments. (B) Cells transfected with the indicated siRNAs were harvested at 72 hpt, and the proteins were detected by immunoblot analysis, using antibodies to nucleolin (Ncl) and vimentin (Vim) as a loading control. Densitometry analysis of the Ncl and Vim bands in the western blot was performed using ImageJ software. (C) Cell viability was assessed using the LDH release assay following the manufacturer’s specifications. The arithmetic means ± standard deviation of three independent experiments are shown.
We found an increase in the viral titer obtained using the four strains of rotavirus from cells in which nucleolin was silenced as compared to cells transfected with a control NT siRNA. The fold increase obtained for every strain was 6.3 for RRV, 4.8 for SA11, 3.3 for UK, and 2.4 for Wa (Fig. 1A). This observation suggests that nucleolin is a negative regulator of rotavirus infection of four different strains. The efficacy of the knockdown of nucleolin was found to be 91%, as evaluated by densitometry of western blots developed with an anti-nucleolin antibody, normalized to the loading control, using an anti-vimentin antibody (vimentin) (Fig. 1B). Under the conditions studied, no discernible changes in the cell viability, as determined by the LDH release assay, were detected in cells treated with the siRNA directed to nucleolin (Fig. 1C).
Nucleolin has no influence on rotavirus cell entry or viral protein synthesis
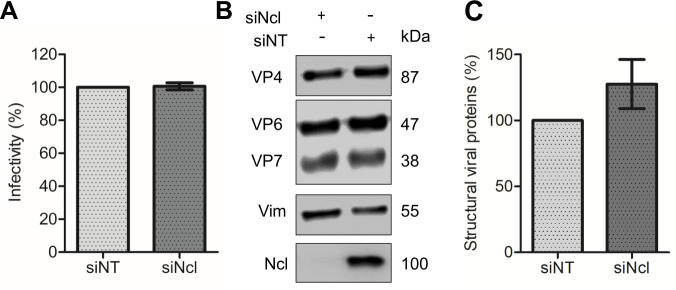
To define the step of the virus replication cycle affected by the knockdown of nucleolin, we evaluated the initial stages of virus replication. For this, MA104 cells that had been previously treated with either nucleolin or NT siRNAs were infected with RRV at an MOI of 0.02. The cells were fixed, and then, using a rabbit polyclonal antibody raised against purified rotavirus particles (anti-TLPs), the number of infected cells was quantified by a focus-forming assay. The viral titer measured in both nucleolin-silenced and NT control cells exhibited no statistically significant difference between the two groups (Fig. 2A), indicating that the reduction in nucleolin does not impact virus cell entry.
Fig 2.
Low nucleolin expression does not have an effect on rotavirus infectivity or viral protein synthesis. (A) MA104 cells grown in 96-well plates were transfected with either NT or Ncl siRNA, as described previously. Subsequently, cells were infected with RRV at an MOI of 0.02, and the virus titer was determined at 15 hpi by a foci-forming assay, as described under Materials and Methods. The number of infected cells transfected with the NT siRNA was considered as 100% infectivity. (B) MA104 cells were transfected with either NT or Ncl siRNAs, followed by infection with RRV at an MOI of 3. At 15 hours post-infection, cells were harvested, and proteins were detected through immunoblot analysis. The analysis utilized a polyclonal antibody recognizing rotavirus structural proteins (TLPs), an anti-Ncl antibody to validate interference, and a vimentin (Vim) antibody as a loading control. (C) Densitometry analysis of three rotavirus structural proteins (VP4, VP6, and VP7) and Vim obtained from the western blot shown in (B) was performed using ImageJ software. The arithmetic means ± standard deviation of three independent experiments are presented.
To assess the impact of nucleolin on the synthesis of viral structural proteins that form the intermediate and outer capsid, we analyzed the amount of viral proteins by characterizing the abundance of three structural viral proteins (VP4, VP6, and VP7) by densitometry in lysates of cells in which nucleolin was knocked down. MA104 cells were transfected with nucleolin siRNA and subsequently infected at 72 hpt with rotavirus RRV at an MOI of 3. Cells were harvested at 15 hpi, and the infected cell lysates were analyzed by western blot SDS-PAGE using an anti-TLP polyclonal antibody. No statistically significant difference was observed between the control NT siRNA-treated cells and the nucleolin-silenced cells (Fig. 2B and C). These results confirm that the amount of rotavirus structural proteins remains unaffected in the context of reduced expression of nucleolin.
Rotavirus genome replication is enhanced in cells in which the expression of nucleolin was silenced
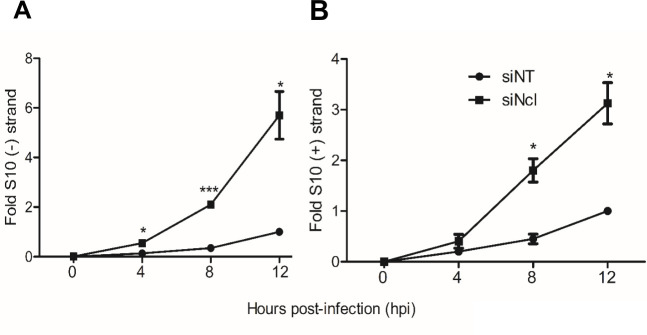
The synthesis of negative-sense RNA during rotavirus infection is a measure of genome replication since this is a mandatory step for the synthesis of the genomic double-stranded RNA present in the infectious viral progeny. To quantify the synthesis of genomic RNA in RRV-infected cells that had been previously treated with nucleolin siRNAs, the specific abundance of the negative strand of rotavirus S10 was amplified by RT-qPCR as an indicator. For this, MA104 cells knocked down for nucleolin were infected with RRV at an MOI of 3, and total RNA was extracted from samples collected at 0, 4, 8, and 12 hpi. Data are expressed as fold increase, and the amount of RNA produced at 12 hpi in cells transfected with the NT siRNA was taken as a unit of fold change, as previously reported (15).
To quantitate the amount of S10 negative and positive strands, we used a standard curve for the qPCR analysis, using a plasmid construct containing the cDNA of RRV S10 gene segment (see Materials and Methods). The synthesis of viral negative strand under conditions where the expression of nucleolin was silenced showed an increase starting at 4 hpi, remaining sustained until 12 hpi, and showing a substantial 5.8-fold increase compared with the RNA synthesis observed in control cells treated with the NT siRNA (Fig. 3A).
Fig 3.
The synthesis of the negative and positive strands of viral RNA is enhanced in nucleolin-silenced cells. MA104 cells grown in 48-well plates were transfected with either NT or Ncl siRNA as previously described and subsequently infected with RRV at an MOI of 3. Samples were collected at the indicated hpi, and total RNA was extracted using TRIzol. The levels of both strands of the RRV S10 were determined by real-time RT-PCR as described under Materials and Methods. Data are expressed as fold increases in which the RNA copies obtained from the cells transfected with the NT siRNA and infected at 12 hpi were used to normalize. (A) S10 negative strand levels and (B) S10 positive strand levels. The arithmetic means ± standard deviation of three independent experiments are presented.
We also determined the abundance of the S10 positive strand, which represents the sum of the double-stranded RNA genome synthesis and the mRNA synthesized from the transcription process. Similar to what we observed with the S10 negative strand, an increase in the abundance of the S10 RNA positive strand was observed at 8 hpi compared to control silenced cells, with a significant 3.5-fold enhancement at 12 hpi (Fig. 3B). These findings support the notion that under conditions in which nucleolin expression is silenced, there is an increased viral RNA synthesis that contributes to the observed increase in viral progeny production.
Nucleolin interacts with rotavirus RNA segments 1, 4, and 10 during infection
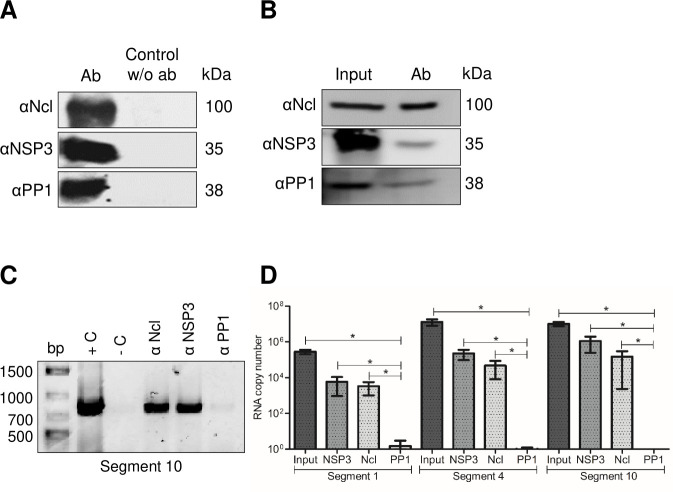
To demonstrate the potential interaction between nucleolin and rotavirus RNA, an immunoprecipitation (IP) assay was conducted. In this assay, a monoclonal antibody against nucleolin was utilized to immunoprecipitate infected or mock-infected cell lysates. The presence of rotavirus RNA in the immunoprecipitated complex was detected using RT-PCR to amplify the rotavirus RNA segments 1, 4, and 10. As a positive control, an immunoprecipitation using an antibody to the virus nonstructural protein NSP3 was included, since it is known that this protein specifically binds to all rotavirus RNA segments (21). Additionally, as a negative control, an antibody to protein phosphatase 1 (PP1), which has been previously shown to not interact with rotavirus RNA (5), was used.
The effectiveness of the IP assays was verified by western blot analysis of the precipitates using antibodies specific for nucleolin, NSP3, and PP1 (Fig. 4A). When the nucleolin monoclonal antibody was used to IP putative protein–RNA complexes in cell lysates, an RT-PCR amplicon of 751 bp, corresponding to S10 of rotavirus, was obtained (Fig. 4C) from infected cells. This amplicon was also detected when the polyclonal antibody to NSP3 was used in the IP. Importantly, no amplification bands were observed when using the PP1 antibody. To corroborate the binding of other RRV segments to nucleolin during viral infection, additional immunoprecipitation assays were conducted. Various pairs of primers were employed to amplify three distinct segments of rotavirus RRV, namely, segment 1 (S1), segment 4 (S4), and segment 10 (S10), as detailed in Materials and Methods. The verification of nucleolin, NSP3, and PP1 presence in the precipitates was executed using specific antibodies (Fig. 4B). Through RT-qPCR, we confirmed the presence of the three distinct rotavirus segments corresponding to S1, S4, and S10 (Fig. 4D). Altogether, these results provide evidence for a specific interaction between nucleolin and three different segments of RRV mRNA.
Fig 4.
Nucleolin binds to viral RNA in rotavirus-infected cells. Confluent MA104 cells grown in six-well plates were infected at an MOI of 5 and harvested at 8 hpi. Cells were lysed, and the cell lysates were used for immunoprecipitation assays using the indicated monoclonal or polyclonal antibodies to detect the proteins in the samples. The immunoprecipitated samples were resolved in an SDS-10% PAGE. The proteins were immunoprecipitated and identified through immunoblot analysis using specific antibodies: Ncl, NSP3, and PP1. (A) Visualization of proteins detected by IP with or without the addition of antibodies (w/o Ab). (B) Visualization of proteins in input and proteins detected in the IP samples for RNA detection. (C) The RNA present in every IP sample was extracted with TRIzol, and RRV segment 10 was amplified by end-point RT-PCR. The amplicon was resolved in 1% agarose gel. As a template, RNA from rotavirus was used as a positive control (+C) and water as a negative control (−C) of reaction. (D) The RNA corresponding to S1, S4, and S10 was identified using RT-qPCR, and the number of RNA copies relative to a standard curve is depicted. The arithmetic means ± standard deviation from three independent experiments are presented.
The presence of parallel G4 structures in rotavirus RRV genomic segments is identified in silico
The potential interaction between nucleolin and viral S10 RNA may be attributed to the presence of RNA-binding domains present in nucleolin. However, the precise regions of viral RNA that could potentially engage in these interactions have yet to be elucidated. Previous studies have highlighted that G-quadruplex domains (G4) present in the RNAs of various viruses can interact with nucleolin (22–24).
To determine the presence of putative G4 regions within the rotavirus genome, we employed the bioinformatic tool QGRS mapper, as previously described (19). We analyzed all 11 rotavirus RRV mRNAs. Table 1 shows the summary of the results obtained, which reveal the presence of G4 regions in every genomic RNA segment. A closer analysis of rotavirus sequences identified putative G4 domains in the three segments that we evaluated to interact with nucleolin, seven for S1, five for S4, and three for S10. We evaluated the three potential G-4 binding sequences for nucleolin present in S10 (designated as G4-1, G4-2, and G4-3). G4-1 (nt 116–132) was found near the 5′ untranslated region (UTR), G4-2 (nt 572–607) was located in the open reading frame, and G4-3 (nt 717–737) was situated in the 3′ UTR region.
TABLE 1.
Predicted G4 sequences in the 11 RNA genome segments of RRV using QGRS mapper web servera
| RNA segment | Putative G4 domains | Position (nt) | Length (nt) |
|---|---|---|---|
| 1 | GGTTAACTGACATACCTAAGATGATACAGGACTGGTTGG | 914–952 | 39 |
| GGTCATTTCATGTCGGATTCAGGAAACAGAAAATGTTGG | 1001–1039 | 39 | |
| GGATGACATGGCTAATGGAAGATACACGCCAGG | 1293–1325 | 33 | |
| GGAAGGAGAGATGTACCAGGAAGACGG | 1366–1392 | 27 | |
| GGAAAGGGATAATTATGGGATTGG | 1622–1645 | 24 | |
| GGTAATGTTATTAAGAAGATACAATATGGGGCTGTAGCGTCAGG | 1750–1793 | 44 | |
| GGTATATTGCAGGTGGGAAAATATTCTTTAGGGCTGG | 2057–2093 | 37 | |
| 2 | GGCTATTAAAGGCTCAATGGCGTACAGAAAGCGTGG | 1–36 | 36 |
| GGACAATTGGTTGACCTAACTAGGCTATTGG | 1658–1688 | 31 | |
| 3 | GGAAATTAACATACGTAGGATATAATGGGAGTGATACTAGAGG | 414–456 | 43 |
| GGTTGGGGAAAAGAGTGTCTCAGTTCGACATTGG | 786–819 | 34 | |
| GGAAACACGGATTGGAAAGAATGGAGGAAAATAGTTGAGG | 1085–1124 | 40 | |
| GGTGGTAAGTCGAAGGTATGTTGTGTTAAAATGACTGCTATGG | 1169–1211 | 43 | |
| 4 | GGTTATGCTCCAGTTAACTGGGGTCCTGG | 139–167 | 29 |
| GGTGGGCTAGGTTATAAATGG | 862–882 | 21 | |
| GGAATGAACGATTTTAATTTCAATGGGGGATCGTTACCAACGG | 973–1015 | 43 | |
| GGATGATTCACAAGCCTTCAGGAACATGGTTTATGTAAGG | 1077–1116 | 40 | |
| GGTTGGTCAATGGCCAGTAATGACTGG | 1179–1205 | 27 | |
| 5 | GGTGTAATGTAAAATATGCATTCAGAGACATGGATTGGAAGATGG | 1007–1051 | 45 |
| 6 | GGAAATGAATTTCAAACTGGAGGAATTGG | 150–178 | 29 |
| GGGTACGATGTGGCTCAATGCGGGATCAGAAATTCAGG | 554–591 | 38 | |
| 7 | GGCTATAGGGGCGTTATGTGACCGGGTCGGCATGG | 1056–1090 | 35 |
| 8 | GGAGGGGAAATAGTTTTTCAAAATGCAGCTTTTACAATGTGG | 506–547 | 42 |
| GGTGGCAATACAACAAATTTGCAGTAATTACACATGGTAAAGG | 675–717 | 43 | |
| GGACTGTCAACTGATAGAAAGATGGATGAGGTTTCTCAAATAGG | 950–993 | 44 | |
| 9 | GGATGGCCAACTGGATCAGTTTATTTTAAAGAATACACGG | 376–415 | 40 |
| GGACCAAGGGAAAATGTAGCAGTAATTCAAGTTGGAGG | 805–842 | 38 | |
| 10 | GGAGGATCCTGGAATGG | 116–132 | 17 |
| GGTTGAGCTGCCGTCGTCTGTCTGCGGAAGCGGCGG | 572–607 | 36 | |
| GGACGTTAATGGAAGGAACGG | 717–737 | 21 | |
| 11 | GGGTCGGCATGGCATCTCCACCTCCTCGCGGTCCGACCTGGG | 668–709 | 42 |
The sequence of the positive polarity of all RNA segments is shown.
Nucleolin restricts RRV synthesis of infectious viral progeny by interacting with G-quadruplex structures present in the viral RNAs
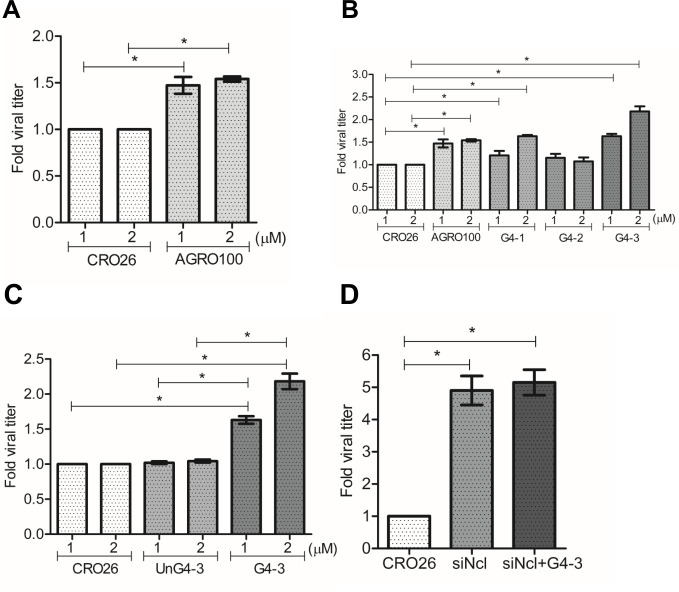
To determine if nucleolin binding to rotavirus S10 RNA is mediated by interactions with the G4 domains identified, MA104 cells were transfected with a positive control AGRO100, an oligonucleotide known to form G-quadruplex structures and specifically bind to nucleolin’s RNA-binding domains (25). As a negative control, cells were transfected with CRO26, an oligonucleotide with the same sequence as AGRO100, in which the guanines were replaced by cytosines. These sequences were subjected to an in vitro folding treatment before the transfection as detailed under Materials and Methods. In these assays, MA104 cells previously transfected with either oligonucleotide were infected with RRV at an MOI of 3, and at 15 hpi, the infected cells were collected, and the production of viral progeny was determined as previously described. We found that there was a 1.5-fold increase in viral production in the cells transfected with AGRO100 as compared to the control cells transfected with CRO26 (Fig. 5A). This finding suggests the involvement of nucleolin’s RNA-binding motif in the regulation of rotavirus infectious particle production.
Fig 5.
Effect of oligonucleotides with predicted G4 structures on the production of rotavirus progeny. MA104 cells grown in 48-well plates were transfected with different oligonucleotide sequences for 48 hpt. Transfected cells were infected with RRV at a MOI of 3. After 15 hpi, the virus produced was harvested, and the viral titers were determined by a focus-forming assay. (A) CRO26 and AGRO100. (B) Sequences of the three different putative G4 regions present S10 RRV: G4-1 (116–132 nt), G4-2 (572–607 nt), and G4-3 (717–737 nt). (C) CRO26, G4-3 unfolded sequence (UnG4-3) and G4-3 folded. (D) MA104 cells grown in 48-well plates were silenced with nucleolin siRNA. Twenty-four hours post-silencing, cells were transfected with the rotavirus G4-3 oligonucleotide (Ncl+G4-3). At 48 hours post-transfection, cells were infected with RRV at an MOI of 3. At 15 hpi, cells were lysed, and viral titers present in the lysate were determined by a focus-forming assay. The arithmetic means ± standard deviation of three independent experiments are presented.
To determine whether the predicted G-quadruplex structures identified in the rotavirus genome could mimic the effects of AGRO100, we synthesized the three G4 sequences identified in RNA S10 (G4-1, G4-2, and G4-3; see Table S1). These sequences, subjected to an in vitro folding treatment as detailed under Materials and Methods, were individually transfected into MA104 cells to assess their impact on virus yield. Notably, RRV-infected cells treated with oligos G4-1 and G4-3 showed a significant increase in viral production of 1.6-fold and 2.1-fold, respectively, compared with the control oligonucleotide (CRO26). In contrast, oligo G4-2 did not significantly affect viral production compared to the controls (Fig. 5B). As an additional control, oligonucleotide G4-3 was transfected prior to the in vitro folding treatment (UnG4-3). The results of this assay showed no change in the viral titer compared with the negative control CRO26, demonstrating that the folding of the G4 structure is necessary to affect the replication of the virus (Fig. 5C).
Finally, to eliminate the possibility that the observed increased infectious virus production could be a consequence of nonspecific interactions of the RRV G-quadruplex regions with other cellular proteins and to establish a direct association between the increased virus production and the binding of the RRV-G4 sequences with nucleolin, the expression of this protein was silenced by RNAi in MA104 cells, and 24 hours post-transfection, cells were subsequently transfected with the folded G4-3 oligonucleotide for 48 h. After this time, the cells were infected with RRV and 15 hpi at 37°C. The quantification of infectious viral progeny revealed that the virus titer obtained in nucleolin-silenced cells exhibited no statistically significant difference from cells that were additionally transfected with RRV G4-3 (Fig. 5D).
These results show that G-quadruplex structures favor rotavirus RNA replication by blocking nucleolin’s RNA-binding domain. Furthermore, they suggest that the specific G-quadruplex sequences found in S10 of RRV (G4-1 and G4-3) might represent binding sites for nucleolin, and this interaction could represent one of the antiviral responses of the cell.
DISCUSSION
Cellular RNA-binding proteins have been extensively studied in the context of viral infection, for viruses with both DNA and RNA genomes (26). They have been shown to play diverse roles, including the recognition of vRNA templates, recruitment of vRNA to replication factories, and switch from translation to replication (27–29). Additionally, they have been involved in the nucleus–cytoplasm shuttling; in particular, hnRNPs such as PCBP1, PTBP1, HNRNPA1, HNRNPC, and nucleolin have been extensively studied (30). It has been shown that nucleolin plays a significant role in various stages of the replication cycle of several RNA genome viruses (8, 11–13, 31).
In the specific case of rotaviruses, several studies have reported interactions between cellular RNA-binding proteins and vRNA; for example, it has been shown that hnRNPs D, E, H, and L are relocated from the nucleus to the cytoplasm through interactions with the viral proteins NSP2 and NSP5; furthermore, the knockdown of hnRNPs D, I, and L causes a decrease in viral progeny production. In contrast, the knockdown of hnRNP C1 and E produces an increase in viral progeny (4). Our laboratory investigations have demonstrated that during rotavirus infection, the HuR is relocalized to the cytoplasm, while GW182 and Ago2 proteins accumulate around viroplasms; the relocalization effect of these cellular proteins was proposed to be due to a sponge effect of the viral RNA of rotavirus (5). Furthermore, ATP synthase has been identified to interact with the 3′ UTR of the rotavirus genome and co-localize with viral RNA and viroplasms. Remarkably, knocking down the expression of ATP synthase F1 subunit beta (ATP5B) using RNAi led to a reduction in the production of infectious viral progeny (6).
In this study, we demonstrate that silencing the expression of nucleolin in MA104 cells leads to an augmentation in viral mRNA synthesis and genome copy number. The knockdown of nucleolin also leads to an increase in the number of infectious viral particles produced in four different strains of rotavirus, revealing a conserved regulatory mechanism associated with nucleolin that down-regulates several processes such as replication and viral particle production. Importantly, the decrease in nucleolin expression does not affect the number of infected cells, indicating that the enhanced viral titer production is independent of changes in the cell’s susceptibility to infection.
Interestingly, no statistically significant change in the synthesis of rotavirus structural proteins was observed, consistent with previous findings from our laboratory in which it was found that knocking down the expression of NSP3 resulted in an increased viral mRNA and dsRNA levels, without significantly impacting the level of all viral protein synthesis (15). Furthermore, silencing the expression of VP1 and VP3 resulted in a decrease of about 90% in the levels of viral mRNA, yet the synthesis of viral proteins was not affected (10). Taken together, these results support the notion that the amount of viral mRNA present in the infected cell and the translation of viral proteins are not necessarily directly proportional.
A similar enhancement in the synthesis of viral RNA induced by knockdown of nucleolin expression has been previously reported for hepatitis C (HCV) (31) and dengue (32) viruses; in the latter case, an increased production of infectious virus particles was also observed. In addition, the interaction of nucleolin with viral RNA has been reported for human immunodeficiency virus (HIV) and Epstein–Barr virus. In the case of HIV, it was found that nucleolin binding to the virus LTR through a G4 sequence led to an increased LTR promoter activity (22). For Epstein–Barr virus, nucleolin binds to the EBNA1 mRNA, repressing its translation (23). Finally, nucleolin binds to the core gene of HCV through a G4 domain and inhibited RNA replication (31).
In this work, using the QGRS mapper, we found that all 11 rotavirus RNA segments contain putative G4 domains suggesting a potential mechanism of interaction between nucleolin and rotavirus RNA segments. This interaction was experimentally confirmed by co-IP of nucleolin and rotavirus RNA S1, S4, and S10. It has been shown that nucleolin specifically recognizes G4 structures in viral RNAs. In this regard, we showed that AGRO100, an oligonucleotide that forms a G4 structure, increases viral yield when transfected into cells before virus infection, most probably by binding to nucleolin and preventing it from interacting with viral RNA. Furthermore, and most interestingly, when oligonucleotides representing the predicted G4 sequences in rotavirus genomic S10 were transfected into cells before RRV infection, an increase in viral progeny production, similar to that induced by knocking down the expression of nucleolin, was observed. Altogether, these findings suggest that nucleolin down-regulates the synthesis of rotavirus RNAs, with the consequent reduction of infectious viral yield, by binding to G4 structures, probably on RNAs derived from all 11 virus segments.
Of interest, one of the three oligonucleotides containing S10 G4 sequences (G4-2), as opposed to the other two (G4-1 and G4-3), did not promote the increase in virus replication. This discrepancy could be attributed to the larger size of the G4-2 sequence and its impossibility to form G-quadruplex, preventing its recognition by the RNA-binding domains of nucleolin.
The observation that there is no statistical difference between the production of viral particles in cells in which nucleolin expression was silenced and transfected with rotavirus G4 sequences, compared to cells where only nucleolin was silenced, suggests that the impact of the G-quadruplex regions present in rotavirus RNA is specific to block the RBD of nucleolin. If it were an unspecific effect, we would have expected to see a combined increase in viral particle production in the double transfection, nucleolin silencing and G-quadruplex transfection. However, no significant difference was observed between these two experimental conditions. In the context of future investigations, employing the rotavirus reverse genetics system to introduce mutations impeding potential RNA–nucleolin interactions holds promise for substantiating the functional significance of G4 sequences in vivo. Proteins with RNA-binding domains, including nucleolin, can also exhibit antiviral effects by binding to exogenous RNA and triggering immune responses. For instance, ZNFX1 can bind to viral dsRNA and interact with MAVS, leading to the promotion of interferon expression (33). Although it remains to be explored, it would be interesting to investigate whether nucleolin plays a similar role in the antiviral response. Additional experiments such as co-immunoprecipitation or protein–protein interaction assays should be performed to assess the presence of any potential proteins involved in the nucleolin–rotavirus RNA interaction.
In previous reports from our laboratory, it was found that during rotavirus infection, there is a substantial accumulation of viral mRNA [for example, at 8 hpi, it was estimated that the NSP4 mRNA reached 1 × 105 copies per cell (18), and it has been proposed that these amounts of viral RNA could be sequestering many of the RBP of the cell (5). The concept of an “RNA sponge” effect, derived from this observation, may provide insights into how rotavirus circumvents the inhibitory influence of nucleolin described in this work, thereby allowing the presence of vRNA free from nucleolin interactions.
In conclusion, this study emphasizes the importance of nucleolin in rotavirus infection by negatively regulating viral RNA synthesis and infectious virus production. Further research is warranted to deepen our understanding of the intricate interactions between nucleolin and rotavirus during infection. The field of research on proteins with RNA-binding domains is expected to expand due to their relevance in RNA virus infection.
ACKNOWLEDGMENTS
We thank P. Gaytán Colín, E. López-Bustos, and S. Becerra Ramírez from the Unidad de Síntesis y Secuenciación de DNA, Instituto de Biotecnología, UNAM, for the synthesis of the oligonucleotides used in this work. We are grateful to Rafaela Espinosa and Marco Antonio Espinoza for their technical assistance.
This work was supported by the Universidad Nacional Autónoma de México. This work was supported by PAPIIT-DGAPA IN211421, IN202823, Fordecyt 302965, and CONACyT A1-S-15356. J.H.-G. was the recipient of a scholarship from CONACyT.
Contributor Information
Carlos Sandoval-Jaime, Email: carlos.sandoval@ibt.unam.mx.
Christiane E. Wobus, University of Michigan Medical School, Ann Arbor, Michigan, USA
REFERENCES
- 1. Burnett E, Parashar U, Tate J. 2018. Rotavirus vaccines: effectiveness, safety, and future directions. Paediatr Drugs 20:223–233. doi: 10.1007/s40272-018-0283-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matthijnssens J, Attoui H, Bányai K, Brussaard CPD, Danthi P, del Vas M, Dermody TS, Duncan R, Fāng (方勤) Q, Johne R, Mertens PPC, Mohd Jaafar F, Patton JT, Sasaya (笹谷孝英) T, Suzuki (鈴木信弘) N, Wei (魏太云) T. 2022. ICTV virus taxonomy profile: Sedoreoviridae 2022. J Gen Virol 103. doi: 10.1099/jgv.0.001782 [DOI] [PubMed] [Google Scholar]
- 3. Crawford SE, Ramani S, Tate JE, Parashar UD, Svensson L, Hagbom M, Franco MA, Greenberg HB, O’Ryan M, Kang G, Desselberger U, Estes MK. 2017. Rotavirus infection. Nat Rev Dis Primers 3:17083. doi: 10.1038/nrdp.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dhillon P, Tandra VN, Chorghade SG, Namsa ND, Sahoo L, Rao CD. 2018. Cytoplasmic relocalization and colocalization with viroplasms of host cell proteins, and their role in rotavirus infection. J Virol 92:1–24. doi: 10.1128/JVI.00612-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oceguera A, Peralta AV, Martínez-Delgado G, Arias CF, López S. 2018. Rotavirus RNAs sponge host cell RNA binding proteins and interfere with their subcellular localization. Virology 525:96–105. doi: 10.1016/j.virol.2018.09.013 [DOI] [PubMed] [Google Scholar]
- 6. Ren L, Ding S, Song Y, Li B, Ramanathan M, Co J, Amieva MR, Khavari PA, Greenberg HB. 2019. Profiling of rotavirus 3'UTR-binding proteins reveals the ATP synthase subunit ATP5B as a host factor that supports late-stage virus replication. J Biol Chem 294:5993–6006. doi: 10.1074/jbc.RA118.006004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tajrishi MM, Tuteja R, Tuteja N. 2011. Nucleolin: the most abundant multifunctional phosphoprotein of nucleolus. Commun Integr Biol 4:267–275. doi: 10.4161/cib.4.3.14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bose S, Basu M, Banerjee AK. 2004. Role of nucleolin in human parainfluenza virus type 3 infection of human lung epithelial cells. J Virol 78:8146–8158. doi: 10.1128/JVI.78.15.8146-8158.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tayyari F, Marchant D, Moraes TJ, Duan W, Mastrangelo P, Hegele RG. 2011. Identification of nucleolin as a cellular receptor for human respiratory syncytial virus. Nat Med 17:1132–1135. doi: 10.1038/nm.2444 [DOI] [PubMed] [Google Scholar]
- 10. Zhu J, Miao Q, Tang J, Wang X, Dong D, Liu T, Qi R, Yang Z, Liu G. 2018. Nucleolin mediates the internalization of rabbit hemorrhagic disease virus through clathrin-dependent endocytosis. PLoS Pathog 14:e1007383. doi: 10.1371/journal.ppat.1007383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cancio-Lonches C, Yocupicio-Monroy M, Sandoval-Jaime C, Galvan-Mendoza I, Ureña L, Vashist S, Goodfellow I, Salas-Benito J, Gutiérrez-Escolano AL. 2011. Nucleolin interacts with the feline calicivirus 3’ untranslated region and the protease-polymerase NS6 and NS7 proteins, playing a role in virus replication. J Virol 85:8056–8068. doi: 10.1128/JVI.01878-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hernández BA, Sandoval-Jaime C, Sosnovtsev SV, Green KY, Gutiérrez-Escolano AL. 2016. Nucleolin promotes in vitro translation of feline calicivirus genomic RNA. Virology 489:51–62. doi: 10.1016/j.virol.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balinsky CA, Schmeisser H, Ganesan S, Singh K, Pierson TC, Zoon KC. 2013. Nucleolin interacts with the dengue virus capsid protein and plays a role in formation of infectious virus particles. J Virol 87:13094–13106. doi: 10.1128/JVI.00704-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. González RA, Torres-Vega MA, López S, Arias CF. 1998. In vivo interactions among rotavirus nonstructural proteins. Arch Virol 143:981–996. doi: 10.1007/s007050050347 [DOI] [PubMed] [Google Scholar]
- 15. Montero H, Arias CF, Lopez S. 2006. Rotavirus nonstructural protein NSP3 is not required for viral protein synthesis. J Virol 80:9031–9038. doi: 10.1128/JVI.00437-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gutiérrez M, Isa P, Sánchez-San Martin C, Pérez-Vargas J, Espinosa R, Arias CF, López S. 2010. Different rotavirus strains enter MA104 cells through different endocytic pathways: the role of clathrin-mediated endocytosis. J Virol 84:9161–9169. doi: 10.1128/JVI.00731-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with imageJ. Biophotonics Int 11:36–41. doi: 10.1201/9781420005615.ax4 [DOI] [Google Scholar]
- 18. Ayala-Breton C, Arias M, Espinosa R, Romero P, Arias CF, López S. 2009. Analysis of the kinetics of transcription and replication of the rotavirus genome by RNA interference. J Virol 83:8819–8831. doi: 10.1128/JVI.02308-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kikin O, D’Antonio L, Bagga PS. 2006. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res 34:W676–W682. doi: 10.1093/nar/gkl253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hacht A von, Seifert O, Menger M, Schütze T, Arora A, Konthur Z, Neubauer P, Wagner A, Weise C, Kurreck J. 2014. Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res 42:6630–6644. doi: 10.1093/nar/gku290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. López S, Arias CF. 2012. Rotavirus-host cell interactions: an arms race. Curr Opin Virol 2:389–398. doi: 10.1016/j.coviro.2012.05.001 [DOI] [PubMed] [Google Scholar]
- 22. Tosoni E, Frasson I, Scalabrin M, Perrone R, Butovskaya E, Nadai M, Palù G, Fabris D, Richter SN. 2015. Nucleolin stabilizes G-quadruplex structures folded by the LTR promoter and silences HIV-1 viral transcription. Nucleic Acids Res 43:8884–8897. doi: 10.1093/nar/gkv897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lista MJ, Martins RP, Billant O, Contesse M-A, Findakly S, Pochard P, Daskalogianni C, Beauvineau C, Guetta C, Jamin C, Teulade-Fichou M-P, Fåhraeus R, Voisset C, Blondel M. 2017. Nucleolin directly mediates Epstein-Barr virus immune evasion through binding to G-quadruplexes of EBNA1 mRNA. Nat Commun 8:16043. doi: 10.1038/ncomms16043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dabral P, Babu J, Zareie A, Verma SC, Jung JU. 2020. LANA and hnRNP A1 regulate the translation of LANA mRNA through G-quadruplexes. J Virol 94:1–21. doi: 10.1128/JVI.01508-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Girvan AC, Teng Y, Casson LK, Thomas SD, Jüliger S, Ball MW, Klein JB, Pierce WM Jr, Barve SS, Bates PJ. 2006. AGRO100 inhibits activation of nuclear factor-κB (NF-κB) by forming a complex with NF-κB essential modulator (NEMO) and nucleolin. Mol Cancer Ther 5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361 [DOI] [PubMed] [Google Scholar]
- 26. Lisy S, Rothamel K, Ascano M. 2021. RNA binding proteins as pioneer determinants of infection: protective, proviral, or both? Viruses 13:2172. doi: 10.3390/v13112172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li Z, Nagy PD. 2011. Diverse roles of host RNA binding proteins in RNA virus replication. RNA Biol 8:305–315. doi: 10.4161/rna.8.2.15391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nagy PD, Pogany J. 2012. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol 10:137–149. doi: 10.1038/nrmicro2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang R, Li K. 2012. Host factors in the replication of positive-strand RNA viruses. Biomed J 35:111. doi: 10.4103/2319-4170.106160 [DOI] [PubMed] [Google Scholar]
- 30. Lloyd RE. 2015. Nuclear proteins hijacked by mammalian cytoplasmic plus strand RNA viruses. Virology 479–480:457–474. doi: 10.1016/j.virol.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bian W-X, Xie Y, Wang X-N, Xu G-H, Fu B-S, Li S, Long G, Zhou X, Zhang X-L. 2019. Binding of cellular nucleolin with the viral core RNA G-quadruplex structure suppresses HCV replication. Nucleic Acids Res 47:56–68. doi: 10.1093/nar/gky1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Phillips SL, Soderblom EJ, Bradrick SS, Garcia-Blanco MA, Denison MR. 2016. Identification of proteins bound to dengue viral RNA in vivo reveals new host proteins important for virus replication . mBio 7:1–10. doi: 10.1128/mBio.01865-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y, Yuan S, Jia X, Ge Y, Ling T, Nie M, Lan X, Chen S, Xu A. 2019. Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS. Nat Cell Biol 21:1346–1356. doi: 10.1038/s41556-019-0416-0 [DOI] [PubMed] [Google Scholar]