Abstract
Eight different amber suppressor tRNA (suptRNA) mutations in the nematode Caenorhabditis elegans have been isolated; all are derived from members of the tRNATrp gene family (K. Kondo, B. Makovec, R. H. Waterston, and J. Hodgkin, J. Mol. Biol. 215:7–19, 1990). Genetic assays of suppressor activity suggested that individual tRNA genes were differentially expressed, probably in a tissue- or developmental stage-specific manner. We have now examined the expression of representative members of this gene family both in vitro, using transcription in embryonic cell extracts, and in vivo, by assaying suppression of an amber-mutated lacZ reporter gene in animals carrying different suptRNA mutations. Individual wild-type tRNATrp genes and their amber-suppressing counterparts appear to be transcribed and processed identically in vitro, suggesting that the behavior of suptRNAs should reflect wild-type tRNA expression. The levels of transcription of different suptRNA genes closely parallel the extent of genetic suppression in vivo. The results suggest that differential expression of tRNA genes is most likely at the transcriptional rather than the posttranscriptional level and that 5′ flanking sequences play a role in vitro, and probably in vivo as well. Using suppression of a lacZ(Am) reporter gene as a more direct assay of suptRNA activity in individual cell types, we have again observed differential expression which correlates with genetic and in vitro transcription results. This provides a model system to more extensively study the basis for differential expression of this tRNA gene family.
The classical picture of eukaryotic tRNA gene transcription has two factors, TFIIIB and TFIIIC, interacting with internal promoter sequences and allowing transcription initiation by RNA polymerase III; more recent studies, however, indicate a more complex and interesting picture (see reference 36 for a recent, comprehensive review). Typically, each tRNA is encoded by multiple members of a gene family with similar or identical tRNA coding sequences but often differences in flanking sequences. Numerous studies with isolated tRNA genes and cell extracts clearly demonstrate that flanking sequences can strongly influence the level of transcription in vitro either positively or negatively (12, 16, 27, 32, 35, 36, 43, 45), and there are now several examples of regulated or differential expression of tRNA genes (reviewed in reference 36). Examples of such regulation include the silk gland-specific expression of a set of novel tRNAAla genes in the silkworm Bombyx mori (37) and in spiders (2), as well as differential transcription of tRNAGly species in B. mori (11); an oocyte-specific tRNATyr in Xenopus laevis (39); changes in tRNA expression in response to viral infection or cell cycle events (reviewed in reference 36); developmental differences for tRNA genes with distinctive primary transcripts in Dictyostelium discoideum (5) and Drosophila melanogaster (40); and evidence based upon genetic studies with amber suppressor mutations in tRNATrp genes in the nematode Caenorhabditis elegans (19, 20).
In terms of mechanisms for differential regulation of tRNA gene expression, work with B. mori has shown that 5′ flanking sequences determine tissue specificity for silk gland versus constitutive tRNAAla genes (47), with differences in TFIIIB interactions a major factor in determining differences in expression levels (46). Flanking sequences have also been implicated in the differential expression of oocyte-specific tRNATyr genes of X. laevis (29, 39). It has been possible to study the expression and regulation of individual tRNA genes in yeast (see, for example, references 17, 28, and 34; reviewed in references 16 and 43) and of suppressing tRNA (suptRNA) or otherwise marked tRNA genes in cultured cells (13, 41). However, it has been difficult to study the expression of individual tRNA genes in multicellular organisms in vivo, given that it is impossible to monitor the products of individual genes from these multigene families except under very special circumstances (e.g., specific sequence differences that allow identification of primary transcripts from different family members [5, 40]).
Kondo et al. (19, 20) have reported on a promising system which can assay the expression of individual members of the tRNATrp gene family of C. elegans. Genetic screens with this organism, used to characterize second-site suppressors of amber mutations, resulted in the isolation and identification of eight amber-suppressing tRNA gene mutations, all members of the tRNATrp gene family. In this case, C. elegans provided a unique model system, as attempts to study informational suppressors in other higher eukaryotes have met with limited success (3, 8, 22, 23, 30). Molecular characterization of the C. elegans tRNATrp gene family showed that while the tRNA coding sequences are identical, flanking sequences and chromosomal locations are different for different loci. When these different suptRNATrp genes were tested for the ability to suppress a diverse panel of different amber-mutated genes, suppression was not uniform; i.e., there appeared to be differential expression of different suptRNATrp genes. These differences could have been at the level of transcription or of posttranscriptional processing. In addition, a hierarchy of suppression effectiveness was observed: sup-7 and sup-5 strongly suppressed all genes tested; some family members, such as sup-24 and sup-28, suppressed only a subset (with sup-24 being a stronger suppressor than sup-28); and the weakest, sup-29, suppressed only the tra-3 mutation (see Table 5 in reference 20). This could be explained if suppressor levels varied among strains and if each strain had a characteristic level throughout the animal. However, Kondo et al. also showed that relative suppression efficiencies among suppressors could change quite dramatically, depending upon the amber mutation tested. For example, sup-5/+ animals effectively suppressed several unc mutations that were not suppressed by sup-28/sup-28 animals, but sup-5/+ animals failed to suppress an unc-52 mutation that was suppressed by sup-28/sup-28 animals. Such data are inconsistent with any model invoking a constant hierarchy of expression levels. In contrast, these kinds of differences might be obvious if a suppressor were not expressed in the tissues or stages most strongly affected by the amber-mutated gene. Similarly, the differences in the abilities of sup-28/sup-28 animals to suppress amber mutations in unc-15 (paromyosin) and unc-52 (perlecan) (26) might be explained by tissue- or stage-specific differences in suppressor expression.
In order to examine this problem further, we undertook the study of the transcription of these genes in vitro and attempted a more direct assay of suptRNA gene activity in different cell types using an amber-mutated lacZ reporter gene driven by a heat shock promoter. Our results suggest that differential expression is probably at the transcriptional rather than the posttranscriptional level and that flanking sequences are important for the expression of sup-7, sup-24, and sup-29 genes in vitro and probably in vivo. Finally, our results with an amber-mutated lacZ reporter gene indicate that a hierarchy of suppression strength (sup-7 is stronger than sup-24 and sup-28, which are stronger than sup-29) can be observed directly in vivo, providing a system to further explore the basis of differential expression of this tRNA gene family.
MATERIALS AND METHODS
Transcription in vitro.
Cell extracts were prepared from C. elegans embryos, and the labelled products from 25-μl reaction mixtures were analyzed on 10% polyacrylamide gels as previously described (15). These extracts efficiently transcribe and process tRNA genes, yielding a mature transcript as well as variable amounts of larger precursor RNA (15, 18). Products were quantitated by excision of transcript bands and Cerenkov counting, followed by correction for background counts per minute from an equivalent-sized blank piece of the same gel. Before proceeding, transcription was optimized for template and total DNA concentration, as described by Wilson et al. (44). From this, optimal concentrations of 0.8 nM template and 0.3 μg of total DNA (template plus carrier pBluescript vector [pBS]) were determined for subsequent transcription reactions. For each template, a minimum of three independent reactions were analyzed, and separate extract preparations were used to eliminate possible batch-specific effects (none were observed). There were no significant differences in transcription between the original wild-type and mutant suptRNA gene templates, so all studies reported here involved mutant suptRNA genes.
Preparation of deletion mutant clones.
Starting recombinant plasmid clones of both wild-type and corresponding amber suppressor tRNA genes for sup-5, sup-7, sup-24, sup-28, and sup-29 were described by Kondo et al. (19). Deletions of cloned suptRNA genes were prepared by the exonuclease III (exoIII)-S1 strategy of Henikoff (14). For sup-7 DNA, deletions were made from the 5′ end of a 1.0-kb XbaI fragment, which contains 250 bp of 5′ and 700 bp of 3′ flanking sequence, cloned into pBS. This clone was digested with KpnI and SalI, followed by treatment with exoIII-S1 and subsequent steps. An additional endpoint was generated by cloning a 1.3-kb EcoRI-SalI fragment into pBS; the SalI site occurs 21 bp upstream of the 5′ end of the mature tRNA. This fragment was also subcloned into pUC18 and pUC19 to generate clones carrying very different 5′ flanking vector sequences. For sup-7 3′ deletions, the 1.0-kb EcoRI-SalI fragment was further cut with RsaI, and the 5′ 500-bp fragment, which contained now only 200 bp of 3′ sequence, was recloned into pBS cut with XbaI/SmaI. This clone was digested with KpnI and HindIII prior to exoIII-S1 digestion. For the sup-24 gene, a 0.7-kb NsiI-EcoRI fragment was blunted and cloned into pBS cut with SmaI, followed by digestion with KpnI and HindIII prior to deletions being made. For the sup-29 gene, a 1.0-kb NsiI-HindIII fragment was cloned into SmaI-cut pBS, followed by digestion with KpnI and BamHI. For sup-29, a NarI site 16 bp upstream of the 5′ end of the mature tRNA was used to subclone an additional deletion endpoint. All clones were sequenced (33) with Sequenase (version 2.0; U.S. Biochemical Corp.) to verify the extent of deletion and to confirm that the rest of the tRNA gene remained intact.
Nematode stocks.
C. elegans strains, obtained from the Caenorhabditis Genetics Center (University of Minnesota) and J. Hodgkin, were maintained essentially as described by Brenner (1). Strains used included sup-7 strain DR497 (maintained at 24°C), sup-24 strains CB4425 and CB4435, sup-28 strain CB3874, and sup-29 strain CB3737.
Site-directed mutagenesis of a lacZ reporter gene.
Plasmid pPCZ1 carries an amber-mutated lacZ reporter gene driven by a C. elegans hsp-16 heat shock promoter (38), generously provided by E. P. M. Candido. A Trp codon in the lacZ gene was mutated to an amber via the unique site elimination strategy of Deng and Nickoloff (4) with a kit and instructions provided by the manufacturer (Pharmacia). The primer used to produce an amber codon was complementary to nucleotides 255 to 281, CCGTGCATCTGC(T)AGTTTGAGGGGACG, and that used to remove a flanking EcoRV site was complementary to nucleotides 1116 to 1138, TCATCAGCAG(A)ATATCCTGCACC (the altered bases are shown in parentheses). Mutagenesis was confirmed by DNA sequencing of resulting clones.
Transgenic nematodes.
pPCZ1 carrying a lacZ amber mutation (pPCZ1am) was coinjected into wild-type C. elegans (9, 25) along with a plasmid carrying rol-6 DNA as a behavioral marker (21) to help identify transformed animals. Animals with a roller phenotype were picked, and the presence of pPCZ1am was confirmed by PCR on isolated genomic DNA by using the mutagenic primer described above as well as a primer from the hsp-16 promoter region (nucleotides 3329 to 3348 [31]). Because the various suppressor lines are not as robust as the wild-type and do not survive injections well, we introduced the different sup genes into animals carrying pPCZ1am DNA by using standard genetic crosses (see references 19 and 20 for a more complete description). Following these crosses, roller progeny were chosen for in situ staining for beta-galactosidase activity, as previously described (10), following a heat shock of 2 h at 33°C and a 15- to 30-min recovery at room temperature (38). Of the putative extrachromosomal lacZ(Am) roller lines established, two strains, BH21 and BH22, with roller transmission frequencies of 70% and 50%, respectively, showed appreciable staining in the presence of suppressor tRNA genes; others showed weaker or no staining. There was some variability in staining; some animals showed staining in fewer cells, possibly because of mosaicism. However, there was always a subset of animals with a consistent, maximum number of cells stained for each sup type, and these were chosen for further analysis.
To rule out mosaicism through loss of extrachromosomal pPCZ1am arrays as a possible factor in any differential staining, animals carrying integrated copies of pPCZ1am were also generated. BH21 animals were treated with radiation, followed by selection of roller animals and multiple backcrosses to remove background mutations. Integrated lines BH31 and BH34 gave results consistent with those obtained from BH21 and BH22 but showed much lower levels of lacZ staining. For this reason, data from BH21 and BH22 are shown in the results.
RESULTS
Individual suptRNATrp genes are transcribed with different efficiencies in vitro.
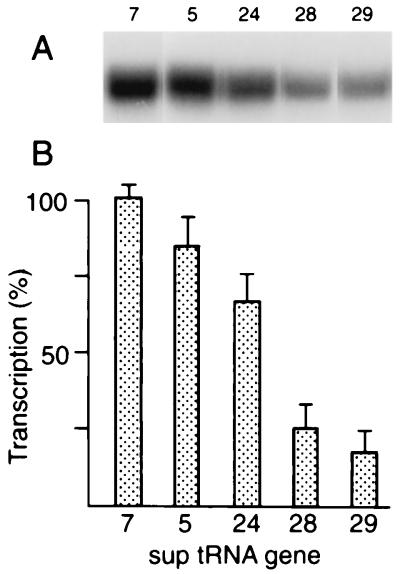
Kondo et al. presented genetic evidence that individual suptRNATrp genes displayed a hierarchy of suppression efficiencies for different amber mutations in C. elegans (19, 20). The tRNAs encoded by the different family members are identical, suggesting that differences in level and/or tissue specificity of expression are responsible. We chose five suptRNA genes for further study, representing a wide range of genetic suppression in vivo, with sup-5 and sup-7 as strong, sup-24 as a moderate, and sup-28 and sup-29 as weak suppressors. Levels of transcription of these templates in embryonic extracts (Fig. 1) correlated well with the differential genetic suppression observed by Kondo et al. (20) and provided evidence for transcriptional regulation. Levels of transcription in suptRNA genes and their wild-type counterparts (data not shown) were identical, suggesting that any observations of suptRNA gene expression in vivo also reflected expression of the corresponding wild-type tRNATrp genes. The products of in vitro transcription of these genes yielded the same-sized wide bands (resolvable into precursor and product on longer gels [data not shown]) for all of these genes; this suggests that the primary transcripts of these genes are similar or identical and not much larger than the mature tRNAs. Much larger differences are observable in the tRNAMet gene family in this organism (18). Primer extension experiments (6) confirmed that transcription initiates at the first purine 2 bp upstream for all of these genes; the additional 5′ sequence is AT for sup-7 and GT for the others (data not shown). The 3′ ends of the genes are likewise nearly identical: AATNTTTT or AANTTTT, with transcription terminating at the run of T residues. This makes it very unlikely that differences in posttranscriptional processing are responsible for the large differences in suppression observed in vivo and further suggests that levels of transcription might be the critical determinant in the observed differential expression.
FIG. 1.
Transcription of different suptRNATrp genes in vitro. (A) This autoradiograph presents a typical experiment, with samples electrophoresed until the bromophenol blue marker was only about halfway down the gel to reduce sample spreading for convenience. (B) Quantitation of transcript labelling for different suptRNA genes. The amount of incorporation into bands excised from gels such as those shown in panel A was quantitated by Cerenkov counting, as indicated in Materials and Methods.
5′ flanking sequences are required for efficient transcription in vitro.
Flanking sequences have been identified as important determinants for tRNA transcription in a number of systems (reviewed in reference 36); we therefore wanted to determine whether sequences flanking these suptRNATrp genes could affect expression and perhaps account for the hierarchy of levels of expression observed.
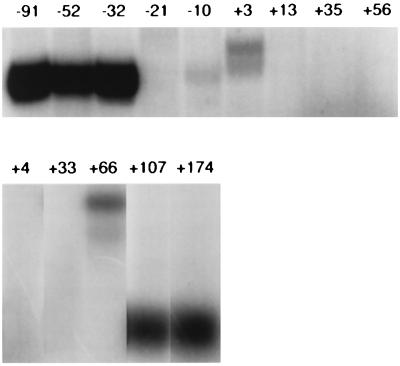
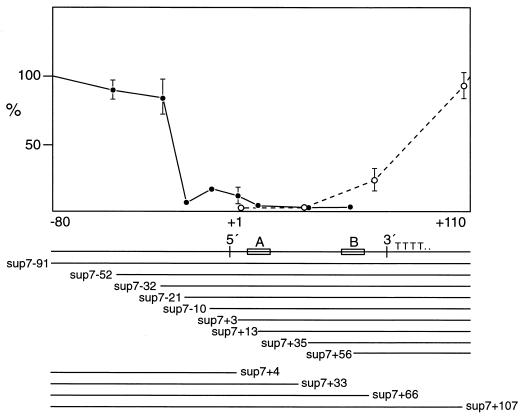
As a first step, we constructed a series of deletions at the 5′ and 3′ ends of the sup-7 gene, the most strongly transcribed in vitro. We were careful to try to optimize transcription for template and total DNA concentration to avoid possible saturation of limiting components and loss of resolution (44). The results of subsequent transcription assays are given in Fig. 2, an autoradiograph showing transcription levels, and Fig. 3, a graphical representation of the results. It appears that sequences upstream of −21 are strong, positive effectors of transcription; not unexpectedly, removal of significant portions of internally important sequences abolishes transcription, while a deletion including the first few nucleotides of the tRNA gives products of other sizes, presumably deriving from start sites in flanking plasmid sequences. To rule out the possibility that “poisonous” sequences, newly adjacent to the tRNA gene, inhibited transcription, other sequences (from pUC18 or pUC19) were also placed 5′ to the −21 deletion clone and this resulted in the same reduced levels of transcription.
FIG. 2.
Transcription of sup-7 deletion clones in vitro. Representative autoradioagraph of transcripts from the indicated sup-7 deletion derivatives are shown. The top row shows results for the 5′ deletion constructs; the bottom row shows those for the 3′ constructs. Numbers above the lanes identify the endpoints of the deletion constructs used.
FIG. 3.
Graphical representation of transcription of sup-7 deletion clones in vitro. Transcript labelling was quantitated as for Fig. 2. The line below the graph shows the tRNA, with 5′ and 3′ boundaries, A and B box internal promoters, and the TTTT termination sequence. The extent of DNA remaining after deletions is shown under the tRNA map.
At the 3′ end of the sup-7 gene, a deletion leaving 3′ 35 bp (+107) was still efficiently transcribed. The tRNA coding region is 72 bp in length, so the next deletion (sup-7, +66) enters the gene and also removes the termination signal. Transcription of this template resulted in two larger transcripts (approximately 150 nucleotides presumably terminating at runs of T residues in the flanking plasmid sequence). After correcting for the larger sizes of these transcripts, it appeared that transcription was significantly reduced for this clone; however, this may have been due to an underestimation of total transcription rather than a loss of a putative positive element if, for example, RNA polymerase III failed to terminate efficiently.
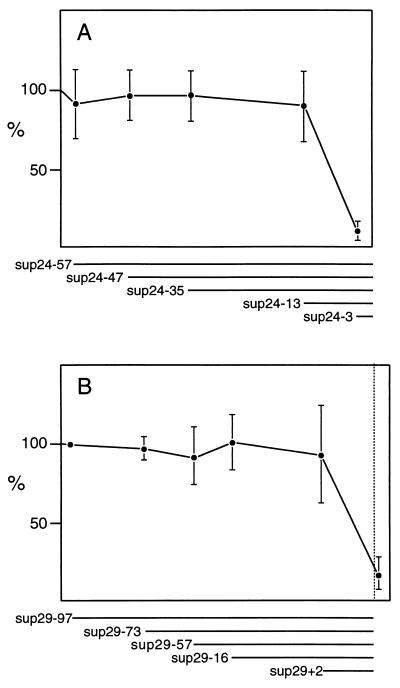
Similar 5′ deletions were performed on sup-24, a gene with intermediate activity, and sup-29, a weak suppressor, in order to see the effects of flanking sequences on other suptRNATrp genes. The results are shown in Fig. 4. We did not observe any strong effects, e.g., negative regulatory elements; instead, it appears that these genes lack the stronger promoter characteristic of the sup-7 gene.
FIG. 4.
Graphical representation of transcription of sup-24 (A) and sup-29 (B) deletion clones in vitro. See Fig. 3 and Materials and Methods.
Differential suppression of a lacZ(Am) reporter gene in vivo in C. elegans mutants carrying different suptRNA alleles.
We wished to examine further the apparent differential expression of different suptRNATrp genes, observed directly in vitro and indirectly via genetic suppression in vivo. If the primary transcripts were sufficiently different, it might be possible to use these differences to discriminate among individual genes, e.g., to use gene-specific oligonucleotides for in situ hybridization. However, as noted above, the primary transcripts for these sup genes are too similar. Fortunately, the availability of viable suptRNA mutants allowed us to look at the biological activity of individual gene family members via their amber suppressor activity. To take advantage of this, we constructed transgenic nematode lines which carry an amber-mutated lacZ reporter gene driven by an hsp-16 heat shock promoter and derived from the wild-type hsp lacZ vector pPCZ1 (38). Expression of lacZ in transgenic animals carrying wild-type pPCZ1 is nearly ubiquitous, with blue color in almost all cell types (with the exception of germ line and early embryo cells [38]).
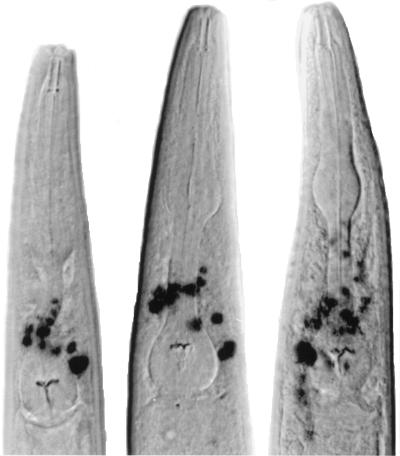
As shown in Fig. 5, when suptRNA genes were introduced into lacZ(Am)-containing BH21 animals, staining for beta-galactosidase activity also paralleled the results of transcription in vitro and amber suppression in vivo; the most cell staining was observed with sup-7/+ (almost comparable to staining with the wild-type lacZ gene of pPCZ1), followed by sup-24/+ and sup-28/+, followed by sup-29/+. A similar hierarchy was observed in earlier developmental stages, although cell staining was less consistent and the cells were very difficult to identify (data not shown). It is interesting to note that sup-28 appeared to stain at least as well as sup-24/+, and we observed clear differences in cell staining between these two genotypes. sup-24/+ animals consistently showed staining of cells around the pharynx (Fig. 6); this staining was absent in sup-28/+ animals. On the other hand, it is clear from Fig. 5 that sup-28/+ animals showed staining of posterior cells not observed in sup-24/+ animals. Homozygote sup-29/sup-29 animals showed more cells being stained than sup-29/+ heterozygotes, which is also consistent with the genetic data reported by Kondo et al. (20).
FIG. 5.
Staining of beta-galactosidase activity from lacZ(Am) genes in suppressor-containing strains. suptRNA genes were introduced into lacZ(Am)-containing animals as described. Animals were heat shocked, stained for beta-galactosidase activity, and photographed. Representatives from those animals with the maximum numbers of cells stained are shown; because each cross generates both males and females, in some cases the animals shown are males (e.g., 24/+, upper animal; 29/+, both animals).
FIG. 6.
Staining of beta-galactosidase activity from lacZ(Am) genes in the head regions of three sup-24/+ animals. See the legend to Fig. 5 and Materials and Methods for details.
Because the same extrachromosomal lacZ(Am) array was introduced genetically and then expressed in the different suppressor strains, we were able to rule out lacZ(Am) copy number variation as a source of the differences in expression. The lacZ(Am) gene is present in extrachromosomal arrays which can be variably retained, so mosaicism might be considered a problem. However, the same hierarchy of staining was also observed in a second independently derived extrachromosomal line, BH22, as well as with two nematode strains, BH31 and BH34, carrying chromosomally integrated lacZ(Am) genes (data not shown).
DISCUSSION
We observed that the levels of transcription of suptRNATrp genes in vitro were consistent with the hierarchy of genetic suppression levels observed in vivo by Kondo et al. This close correspondence and the similarity or identity of primary transcripts for the various suptRNA genes suggest that differential expression of individual tRNATrp gene family members is likely at the transcriptional rather than the posttranscriptional level. It should be noted, however, that the extracts used were embryonic. It has not been possible to prepare active extracts from individual tissues or developmental stages; we may therefore have missed other tissue- or developmental stage-specific effects. The levels of transcription of suppressor and corresponding wild-type tRNA genes were identical, which is not surprising, as the single base change in the anticodon loop should not have strongly influenced any critical internal promoter sequences. This suggests that any observations of suptRNA expression in vivo should accurately reflect the behavior of the corresponding wild-type genes.
The results also indicate the presence of putative positive regulatory elements upstream of the sup-7 gene, elements not present 5′ to the sup-24 and sup-29 genes. We believe it extremely unlikely that poisonous vector flanking sequences were responsible for the low levels of transcription seen in the more proximal sup-7, sup-24, and sup-29 deletions because (i) the same low level of transcription is seen when two other sets of flanking sequences are placed in front of sup-7(-21) and (ii) vector replacement of the same proximal 5′ region (upstream of −2) in sup-24 and sup-29 has completely different effects, i.e., transcription of sup-24(-3) is low, while that of sup-29(-2) is near wild-type levels.
We conclude that flanking sequences appear to play an important role in determining the levels of differential expression of tRNATrp genes in vitro, and possibly in vivo as well. Some transgenic C. elegans strains, which were engineered to integrate various class II gene constructs into the genome, also carry a sup-7 gene with vector sequences adjacent to position −21 (24). It is interesting that this type of sup-7 deletion, which reduces transcription in vitro, can also decrease genetic suppression in vivo by the sup-7 gene (20). In addition, strains carrying the sup-7 gene with complete 5′ flanks are unhealthy, presumably reflecting problems resulting from carrying strongly expressed suptRNA (42); these strains with deleted sup-7 5′ flanks are more robust and fertile, which is also consistent with weaker suptRNA expression in these animals. Our preliminary results with three of these transgenic lines indicate that staining in hsp lacZ(Am) animals is also reduced, further implying that loss of these flanking sequences reduces sup-7 expression in vivo (data not shown). This is all consistent with the hypothesis that these flanking sequences are critical to efficient transcription in vivo. However, because these constructs have different chromosomal locations and differences in accompanying class II gene sequences, we cannot rule out possible local chromosome position or other effects. To do so requires generating and more carefully examining a larger sample size and standardizing the position and sequence of accompanying class II transcription units which might interfere with tRNA expression, or vice versa. Clear effects of flanking sequences on tRNA expression in vivo have been observed in yeast (28), as have examples of strong position effects (34) and effects of tRNA genes on class II gene expression (17).
Our more direct observations of which cell types show suptRNATrp activity, using an hsp-16-driven lacZ(Am) reporter gene, are consistent with the genetic suppression data and again strongly suggest that different suptRNATrp genes are differentially expressed in vivo. Is there evidence of tissue specificity in our lacZ(Am) staining results? We observed that cells around the pharynx were stained in sup-24/+ but not sup-28/+ animals (Fig. 6). Some of these cells appeared to be part of the nerve ring, so this difference in staining would be consistent with the better suppression, by sup-24, of genes expressed in the nervous system (20). However, higher overall levels of suptRNA expression in these sup-24/+ animals (e.g., above some threshold in nerve cells) might also explain this result. Most of the staining can indeed be accounted for by this simpler model. However, one clear exception was the observed staining of posterior cells in sup-28/+ animals, staining which was absent in sup-24/+ animals. At least some of these cells appeared to be hypodermal, a result consistent with genetic suppression results (20) which appeared to show that sup-28 was more efficient at suppressing presumptive hypodermally acting genes than those in the nervous system.
Our results support the idea that there is some tissue specificity in suptRNA gene expression; however, some caution is necessary, as it can be difficult to precisely identify specific cells in the stained animals because of their twisted roller phenotype. We were careful to choose animals with a consistent maximum number of cells stained, thereby excluding any which were not optimally stained, due either to mosaicism or to minor variations in staining observed with hsp-16 promoter-driven genes (38). Differences in lacZ(Am) expression resulting from strain-specific mosaicism are unlikely, as we saw similar effects on suptRNA expression in two independent extrachromosomal transgenic lines as well as in two lines with integrated lacZ(Am) rol-6 DNA. However, it is formally possible that strain-specific differences between, for example, a strong versus weak suppressor strain might have other, indirect effects on gene expression, which might account for some of the differences observed.
Our results suggest that it should now be possible to express lacZ(Am) or another reporter gene under the control of other, more specific promoters whose tissue and developmental expression patterns have been well characterized. It should also be possible to generate integrated transgenic C. elegans strains carrying suptRNA genes with different flanking sequences and/or accompanying transcription units to study how expression is affected in vivo; such constructs might also provide tools for examination of local chromatin structure (7). The results should provide new insights into the regulation and differential expression of individual tRNA genes in multicellular organisms.
ACKNOWLEDGMENTS
We thank D. Baillie and members of his lab, especially D. Janke and J. Schein, for help and expertise; E. Stringham and P. Candido for pPZ1; A. Fire, E. Hedgecock, D. Moerman, Don Jones, and our research colleagues in the lab for advice and assistance; two anonymous reviewers for helpful comments; and J. Hodgkin and R. Waterston for continued support. We also thank J. Hodgkin, M. MacMorris, and the Caenorhabditis Genetics Center, supported by the U.S. NIH Division of Research Resources, for some strains of C. elegans.
This work was supported by a grant from NSERC Canada to B.M.H. R. M. Linning held an NSERC Canada postgraduate fellowship.
REFERENCES
- 1.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Candelas G C, Arroyo G, Carrasco C, Dompensciel R. Spider silkglands contain a tissue-specific alanine tRNA that accumulates in vitro in response to the stimulus for silk protein synthesis. Dev Biol. 1990;140:215–220. doi: 10.1016/0012-1606(90)90069-u. [DOI] [PubMed] [Google Scholar]
- 3.Capone J P, Sharp P A, RajBhandary U L. Amber, ochre and opal suppressor tRNA genes derived from a human serine tRNA gene. EMBO J. 1985;4:213–221. doi: 10.1002/j.1460-2075.1985.tb02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 5.Dingermann T, Amon-Bohm E, Bertling W, Marschalek R, Nerke K. A family of non-allelic tRNAGUUVal genes from the cellular slime mold Dictyostelium discoideum. Gene. 1988;73:373–384. doi: 10.1016/0378-1119(88)90502-1. [DOI] [PubMed] [Google Scholar]
- 6.Dingermann T, Nerke K. Primer extension analysis of tRNA gene transcripts synthesized in vitro and in vivo. Anal Biochem. 1987;162:466–475. doi: 10.1016/0003-2697(87)90422-2. [DOI] [PubMed] [Google Scholar]
- 7.Dixon D K, Jones D, Candido E P M. The differentially expressed 16kD heat shock genes of Caenorhabditis elegans exhibit differential changes in chromatin structure during heat shock. DNA Cell Biol. 1990;9:177–191. doi: 10.1089/dna.1990.9.177. [DOI] [PubMed] [Google Scholar]
- 8.Doerig R E, Suter B, Gray M, Kubli E. Identification of an amber nonsense mutation in the rosy 516 gene by germline transformation of an amber suppressor tRNA gene. EMBO J. 1988;7:2579–2584. doi: 10.1002/j.1460-2075.1988.tb03107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fire A. Integrative transformation of Caenorhabditis elegans. EMBO J. 1986;5:2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fire A, White Harrison S, Dixon D. A modular set of beta-galactosidase fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 11.Fournier A, Taneja R, Gopalkrishnan R, Prudhomme J-C, Gopinathan K P. Differential transcription of multiple copies of a silk worm gene encoding tRNAGly1. Gene. 1993;134:183–190. doi: 10.1016/0378-1119(93)90092-h. [DOI] [PubMed] [Google Scholar]
- 12.Geiduschek E P, Tocchini-Valentini G P. Transcription by RNA polymerase III. Annu Rev Biochem. 1988;57:873–904. doi: 10.1146/annurev.bi.57.070188.004301. [DOI] [PubMed] [Google Scholar]
- 13.Hatfield D. Suppression of termination codons in higher eukaryotes. Trends Biochem Sci. 1985;10:201–204. [Google Scholar]
- 14.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 15.Honda B M, Devlin R H, Nelson D W, Khosla M. Transcription of class III genes in cell-free extracts from the nematode Caenorhabditis elegans. Nucleic Acids Res. 1986;14:869–881. doi: 10.1093/nar/14.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huet J, Manard N, Dieci G, Peyroche G, Conesa C, Lefebvre O, Ruet A, Riva M, Sentenac A. RNA polymerase III and class III transcription factors from Saccharomyces cerevisiae. Methods Enzymol. 1996;273:249–267. doi: 10.1016/s0076-6879(96)73024-0. [DOI] [PubMed] [Google Scholar]
- 17.Hull M W, Erickson J, Johnston M, Engelke D R. tRNA genes as transcriptional repressor elements. Mol Cell Biol. 1994;14:1266–1277. doi: 10.1128/mcb.14.2.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khosla M K, Honda B M. Initiator tRNAMet genes from the nematode Caenorhabditis elegans. Gene. 1989;76:321–330. doi: 10.1016/0378-1119(89)90172-8. [DOI] [PubMed] [Google Scholar]
- 19.Kondo K, Hodgkin J, Waterston R H. Differential expression of five tRNAUAGTrp amber suppressors in Caenorhabditis elegans. Mol Cell Biol. 1988;8:3627–3635. doi: 10.1128/mcb.8.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondo K, Makovec B, Waterston R H, Hodgkin J. Genetic and molecular analysis of eight tRNAtrp amber suppressors in Caenorhabdiditis elegans. J Mol Biol. 1990;215:7–19. doi: 10.1016/S0022-2836(05)80090-7. [DOI] [PubMed] [Google Scholar]
- 21.Kramer J M, French R P, Park E-C, Johnson J J. The Caenorhabditis elegans rol-6 gene, which interacts with the sqt-1 collagen gene to determine organismal morphology, encodes a collagen. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchino Y, Muramatsu T. Nonsense suppression in mammalian cells. Biochimie. 1996;78:1007–1015. doi: 10.1016/s0300-9084(97)86724-7. [DOI] [PubMed] [Google Scholar]
- 23.Laski F A, Ganguly S, Sharp P A, RajBhandary U L, Rubin G M. Construction, stable transformation and function of an amber suppressor tRNA gene in Drosophila melanogaster. Proc Natl Acad Sci USA. 1989;86:6696–6698. doi: 10.1073/pnas.86.17.6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacMorris M, Broverman S, Greenspoon S, Lea K, Madej C, Blumenthal T, Spieth J. Regulation of vitellogenin gene expression in transgenic Caenorhabditis elegans: short sequences required for activation of the vit-2 promoter. Mol Cell Biol. 1992;12:1652–1662. doi: 10.1128/mcb.12.4.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mello C C, Kramer J M, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moerman D G, Hutter H, Mullen G P, Schnabel R. Cell autonomous expression of perlecan and plasticity of cell shape in embryonic muscle of C. elegans. Dev Biol. 1996;173:228–242. [Google Scholar]
- 27.Palmer J M, Folk W R. Unraveling the complexities of transcription by RNA polymerase III. Trends Biochem Sci. 1990;15:300–304. doi: 10.1016/0968-0004(90)90018-7. [DOI] [PubMed] [Google Scholar]
- 28.Raymond K C, Raymond G J, Johnson J D. In vivo modulation of yeast tRNA gene expression by 5′-flanking sequences. EMBO J. 1985;4:2649–2656. doi: 10.1002/j.1460-2075.1985.tb03983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynolds W F. Developmental stage-specific regulation of Xenopus tRNA genes by an upstream promoter element. J Biol Chem. 1995;270:10703–10710. doi: 10.1074/jbc.270.18.10703. [DOI] [PubMed] [Google Scholar]
- 30.Robinson D N, Cooley L. Examination of the function of two kelch proteins generated by stop codon supression. Development. 1997;124:1405–1417. doi: 10.1242/dev.124.7.1405. [DOI] [PubMed] [Google Scholar]
- 31.Russnak R H, Candido E P M. Locus encoding a family of small heat shock genes in Caenorhabditis elegans: two genes duplicated to form a 3.8-kilobase inverted repeat. Mol Cell Biol. 1985;5:1268–1278. doi: 10.1128/mcb.5.6.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sajjadi F G, Spiegelman G B. Modulation of a Drosophila melanogaster tRNA gene transcription in vitro by a sequence TNNCT in its 5′ flank. Gene. 1987;60:13–19. doi: 10.1016/0378-1119(87)90208-3. [DOI] [PubMed] [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schnell R, Rine J. A position effect on the expression of a tRNA gene mediated by the SIR genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:494–501. doi: 10.1128/mcb.6.2.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp S J, Schaak J, Cooley L, Burke D J, Soll D. Structure and transcription of eukaryotic tRNA genes. Crit Rev Biochem. 1985;19:107–144. doi: 10.3109/10409238509082541. [DOI] [PubMed] [Google Scholar]
- 36.Sprague K U. Transcription of eukaryotic tRNA genes. In: Söll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: American Society for Microbiology; 1995. pp. 31–50. [Google Scholar]
- 37.Sprague K U, Hagenbuchle O, Zuniga M C. The nucleotide sequence of two silkgland alanine tRNAs: implications for fibroin synthesis and for initiator tRNA structure. Cell. 1977;22:171–178. doi: 10.1016/0092-8674(77)90074-5. [DOI] [PubMed] [Google Scholar]
- 38.Stringham E G, Dixon D K, Jones D K, Candido E P M. Temporal and spatial expression patterns of the small heat shock (hsp16) genes in transgenic Caenorhabditis elegans. Mol Biol Cell. 1992;3:221–233. doi: 10.1091/mbc.3.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stutz F, Gouilloud E, Clarkson S G. Oocyte and somatic tyrosine tRNA genes in Xenopus laevis. Genes Dev. 1989;3:1190–1198. doi: 10.1101/gad.3.8.1190. [DOI] [PubMed] [Google Scholar]
- 40.Suter B, Kubli E. tRNATyr genes of Drosophila melanogaster: expression of single-copy genes studied by S1 mapping. Mol Cell Biol. 1988;8:3322–3331. doi: 10.1128/mcb.8.8.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang H-D, Yuh C-H, Dang C V, Johnson D L. The hepatitis B virus X protein increases the cellular level of TATA-binding protein, which mediates transactivation of RNA polymerase III genes. Mol Cell Biol. 1995;15:6720–6728. doi: 10.1128/mcb.15.12.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterston R H. A second informational suppressor, sup-7, in Caenorhabditis elegans. Genetics. 1981;97:307–325. doi: 10.1093/genetics/97.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 44.Wilson E T, Larson D, Young L S, Sprague K U. A large region controls tRNA gene transcription. J Mol Biol. 1985;183:153–163. doi: 10.1016/0022-2836(85)90209-8. [DOI] [PubMed] [Google Scholar]
- 45.Wolffe A P. RNA polymerase III transcription. Curr Opin Cell Biol. 1991;3:461–466. doi: 10.1016/0955-0674(91)90074-9. [DOI] [PubMed] [Google Scholar]
- 46.Young L S, Ahnert N, Sprague K U. Silkworm TFIIIB binds both constitutive and silk gland-specific tRNAAla promoters but protects only the constitutive promoter from DNase I cleavage. Mol Cell Biol. 1996;16:1256–1266. doi: 10.1128/mcb.16.3.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young L S, Takahashi N, Sprague K U. Upstream sequences confer distinctive transcriptional properties on genes encoding silkgland-specific tRNAAla. Proc Natl Acad Sci USA. 1986;83:374–378. doi: 10.1073/pnas.83.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]