ABSTRACT
The major oral odor compound methyl mercaptan (CH3SH) is strongly associated with halitosis and periodontitis. CH3SH production stems from the metabolism of polymicrobial communities in periodontal pockets and on the tongue dorsum. However, understanding of CH3SH-producing oral bacteria and their interactions is limited. This study aimed to investigate CH3SH production by major oral bacteria and the impact of interspecies interactions on its generation. Using a newly constructed large-volume anaerobic noncontact coculture system, Fusobacterium nucleatum was found to be a potent producer of CH3SH, with that production stimulated by metabolic interactions with Streptococcus gordonii, an early dental plaque colonizer. Furthermore, analysis of extracellular amino acids using an S. gordonii arginine-ornithine antiporter (ArcD) mutant demonstrated that ornithine excreted from S. gordonii is a key contributor to increased CH3SH production by F. nucleatum. Further study with 13C, 15N-methionine, as well as gene expression analysis, revealed that ornithine secreted by S. gordonii increased the demand for methionine through accelerated polyamine synthesis by F. nucleatum, leading to elevated methionine pathway activity and CH3SH production. Collectively, these findings suggest that interaction between S. gordonii and F. nucleatum plays a key role in CH3SH production, providing a new insight into the mechanism of CH3SH generation in oral microbial communities. A better understanding of the underlying interactions among oral bacteria involved in CH3SH generation can lead to the development of more appropriate prophylactic approaches to treat halitosis and periodontitis. An intervention approach like selectively disrupting this interspecies network could also offer a powerful therapeutic strategy.
IMPORTANCE
Halitosis can have a significant impact on the social life of affected individuals. Among oral odor compounds, CH3SH has a low olfactory threshold and halitosis is a result of its production. Recently, there has been a growing interest in the collective properties of oral polymicrobial communities, regarded as important for the development of oral diseases, which are shaped by physical and metabolic interactions among community participants. However, it has yet to be investigated whether interspecies interactions have an impact on the production of volatile compounds, leading to the development of halitosis. The present findings provide mechanistic insights indicating that ornithine, a metabolite excreted by Streptococcus gordonii, promotes polyamine synthesis by Fusobacterium nucleatum, resulting in a compensatory increase in demand for methionine, which results in elevated methionine pathway activity and CH3SH production. Elucidation of the mechanisms related to CH3SH production is expected to lead to the development of new strategies for managing halitosis.
KEYWORDS: metabolic interaction, methyl mercaptan, Fusobacterium nucleatum, Streptococcus gordonii, polyamines, methionine pathway, halitosis, one-carbon pool
INTRODUCTION
Bad breath, commonly referred to as halitosis, can have a significant negative impact on the social life of affected individuals. Sources of oral odor can be generally divided into microbial communities in periodontal pockets and those on the tongue dorsal surface, from which volatile compounds are emitted due to the bacterial metabolism of odorless proteins, peptides, blood, gingival crevicular fluid, and food retained on oral surfaces. Volatile sulfur compounds (VSCs), polyamines, short-chain fatty acids, and indoles are known odor-causing compounds emanating from the oral cavity (1–3). Among those, VSCs, composed of hydrogen sulfide (H2S), methyl mercaptan (CH3SH), and dimethyl sulfide [(CH3)2S], are strongly associated with oral odor (4, 5). In particular, CH3SH has an extremely low olfactory threshold value of 0.070 ppb, as compared to 0.41 ppb for H2S and 3.0 ppb for (CH3)2S (6), indicating that even a small amount causes an unpleasant odor. In addition, CH3SH production is associated with periodontal disease and is used as a biomarker of periodontitis (7, 8). Previous studies have also shown that CH3SH exhibits high toxicity toward periodontal tissues at a low concentration by inhibiting the synthesis of proteins, collagens, and DNA in human gingival fibroblasts, as well as by increasing the membrane permeability of oral mucosal epithelium (9–12). Together, these findings suggest that CH3SH is not only a key odor compound for halitosis but also a possible contributor to the pathogenesis of periodontitis.
CH3SH is produced by bacterial degradation of l-methionine by l-methionine-α-deamino-γ-mercaptomethane-lyase (METase) (13). Previous research has found that periodontal bacteria, such as the Gram-negative anaerobes Fusobacterium nucleatum, Prevotella intermedia, and Porphyromonas gingivalis, contribute to CH3SH production (13, 14). However, knowledge regarding CH3SH-producing oral bacteria is largely based on the findings from enzyme assays, in which substrates are reacted with enzymes extracted from oral bacteria (15, 16). Moreover, most related studies that used bacterial cultures employed a relatively small-volume incubation system, such as 3.5 mL vials with gas-tight valves (17, 18). Quantifying CH3SH, heavier than air and prone to settling at the bottom of vials, can cause large variations in its amounts and difficulty with the detection of a trace quantity. Also, in experiments that require a substantial bacterial amount, such as gene expression analysis, small-volume incubation systems are not suitable.
In recent years, there has been growing interest in the collective properties of oral polymicrobial communities, regarded as an important factor for the development of oral diseases such as periodontitis and dental caries (19, 20). These properties may be shaped by physical and chemical interactions among community participants including streptococci and actinomycetes, which are nonpathogenic commensals (21–24). In particular, a subset of oral streptococci has been shown to engage in interspecies communications with oral pathogens, such as the provision of attachment sites through coaggregation-mediated adhesion and exchange of diffusible signaling molecules termed autoinducers (AIs) via quorum sensing (QS) (25–33). Furthermore, studies recently conducted have reported that streptococcal metabolites, such as lactate, ornithine, and para-aminobenzoic acid, have a major impact on oral community properties, resulting in an increased risk of periodontitis (34–37). However, no known study has investigated whether interspecies communication by streptococci has an impact on CH3SH generation.
Using a newly constructed large-volume anaerobic noncontact coculture system, the present study was conducted to assess CH3SH production by major oral bacteria and the impact of interspecies interactions on that production. The findings show that F. nucleatum is a potent producer of CH3SH, with that production stimulated by metabolic interactions with Streptococcus gordonii. Additionally, this phenomenon was found to be driven by ornithine excreted from S. gordonii, which boosts the demand for methionine via increased synthesis of polyamines by F. nucleatum, resulting in elevated methionine metabolism as well as CH3SH production.
MATERIALS AND METHODS
Strains, media, and growth conditions
Actinomyces naeslundii ATCC 19039, S. gordonii DL1, and its isogenic ΔarcD mutant (37), F. nucleatum subsp. nucleatum ATCC 25586, Filifactor alocis ATCC 35846, P. intermedia ATCC 49046, and P. gingivalis ATCC 33277 were used as representative oral bacteria in this study. S. gordonii strains were precultured in an aerobic environment in Todd Hewitt broth at 37°C. A. naeslundii, P. intermedia, and P. gingivalis were anaerobically grown at 37°C in trypticase soy broth supplemented with 1.0 mg/mL yeast extract, 1.0 µg/mL menadione, and 5.0 µg/mL hemin. F. nucleatum was anaerobically grown at 37°C in medium containing 1.92 g/mL brain heart infusion broth, 1.0 g/mL trypticase peptone, 1 mg/mL yeast extract, 1.0 g/mL biosate peptone, 1.0 µg/mL menadione, 5.0 µg/mL hemin, 0.2 mM K2HPO4, 0.3 mM KH2PO4, 4.8 mM NaHCO3, 72 µM CaCl2, 1.4 mM NaCl, and 66 µM MgSO4. F. alocis was anaerobically grown in modified Gifu anaerobic medium (GAM) agar (Nissui Pharmaceutical, Tokyo, Japan). Precultures were performed in an anaerobic chamber (Concept Plus; Ruskinn Technology, Bridgend, UK) with an atmosphere containing 85% N2, 10% H2, and 5% CO2. When necessary, the antibiotic erythromycin (5 g/mL) was used as a selective marker for S. gordonii. The modified chemically defined medium (mCDM) used consisted of 58 mM K2HPO4, 15 mM KH2PO4, 10 mM (NH4) 2SO4, 35 mM NaCl, 0.1 mM MnCl2•4H2O, 2 mM MgSO4•7H2O, 0.04 mM nicotinic acid, 0.1 mM pyridoxine-HCl, 0.01 mM pantothenic acid, 1.0 µM riboflavin, 0.3 µM thiamin-HCl, 0.05 µM D-biotin, 50 mM α-ketoglutaric acid, 5.6 mM D(+)-glucose, 4.0 mM l-glutamic acid, 1.0 mM l-arginine-HCl, and 0.1 mM l-tryptophan (38).
Pretreatment
A. naeslundii (OD 1.4), S. gordonii (OD 1.4), F. nucleatum (OD 1.4), P. intermedia (OD 1.2), F. alocis (OD 1.2), and P. gingivalis (OD 1.3) were harvested by centrifugation (8,000 × g for 7 min at 4°C) and washed twice in phosphate buffer saline (PBS). Cells were adjusted at an OD600 of 1.0 in mCDM, supplemented with 0.5 to 5.0 mM l-methionine, 5.0 mM l-cysteine, 5.0 mM dl-homocysteine, and 5.0 mM serine, as necessary (mCDM solution). The mCDM solution was adjusted at pH 6.5 unless experimental pH conditions are specified.
In vitro assays for CH3SH generation
Monoculture
Ten milliliters of the pretreated bacterial suspensions described above were added to 30 mL of mCDM solution.
Coculture
Using pretreated bacteria, 10 mL of A. naeslundii or S. gordonii along with 10 mL of F. nucleatum, P. intermedia, F. alocis, or P. gingivalis was mixed with 20 mL of mCDM solution.
Noncontact culture
For the cocultivation of two species of bacteria under a contactless condition, dialysis tubing (Spectra/Por 7 Dialysis Membrane Pretreated RC Tubing MWCO 1 kDa; Spectrum Laboratories, Inc., CA, USA) was used. The tubing was rinsed twice with sterile water and then autoclaved at 120°C for 15 min in distilled water. F. nucleatum with S. gordonii wild type (WT) or ΔarcD mutant at the late-exponential phase (1.0 to 1.5 OD units/mL) was adjusted to an OD600 of 1.0 in mCDM solution. The tubing was filled with 10 mL of S. gordonii WT or ΔarcD mutant and transferred to a flask, and then 10 mL of F. nucleatum and 20 mL of mCDM solution were added to the flask. For monocultures of F. nucleatum, after filling the tubing with 10 mL of mCDM solution, 10 mL of F. nucleatum and 20 mL of mCDM solution were added to the flask. For monocultures of S. gordonii WT and ΔarcD mutant, the tubing was filled with 10 mL of S. gordonii WT or ΔarcD mutant, and then 20 mL of mCDM solution was added to the flask.
All samples were incubated either anaerobically or microaerobically at 37°C for 16 h using a contact or noncontact type of culture system (Fig. S1). A set of four flasks of bacterial cultures for each experimental group were incubated in a jar (The GasPak 150 jar, Becton, Dickinson and Company, NJ, USA) at 37°C either anaerobically or microaerobically to minimize contamination by volatile compounds emitted from different experimental groups. Where required, anaerobic or microaerophilic atmospheric conditions were created by using the AnaeroPack gas generator (Mitsubishi Gas Chemical, Tokyo, Japan). Each flask removed from the jar was immediately covered with Parafilm (Parafilm M) after removing the rubber stopper to maintain anaerobic conditions. Additionally, a gas-tight syringe was inserted through the Parafilm and 1 mL of headspace gas was directly collected. The gas was quantitated by gas chromatography (GC; Shimadzu, Kyoto, Japan). When necessary, the gas was diluted to a ≥5 times volume with air. The gas was injected into the GC port of a GC-14B instrument equipped with a flame photometric detector (Shimadzu). A ZO-1H column (3.1 m × 3.2 mm i.d.; Shinwa Chemical Industries, Kyoto, Japan) was used. Nitrogen was utilized as the carrier gas at a constant flow rate of 50 mL/min. The oven and detector temperatures were kept at 70°C and 180°C, respectively. Identification of volatiles was based on matching retention time with those of authentic standards available. The three standard gases H2S, CH3SH, and (CH3)2S were produced in a permeation tube using a permeater (PD-1B; Gastec Corp., Tokyo, Japan) and collected into sampling bags. Each volatile compound was determined using calibration curves. Changes in bacterial density after cultivation were also measured at OD600.
Analysis of extracellular metabolites
The time-course changes of extracellular metabolite compositions were investigated using F. nucleatum, S. gordonii WT, ΔarcD mutant, and their cocultures. For the monocultures, 3 mL of each bacterium at an OD600 of 1.0 in mCDM solution was mixed with 9 mL of mCDM solution in a six-well tissue culture plate. For the cocultures, 3 mL of F. nucleatum and S. gordonii WT or ΔarcD mutant at an OD600 of 1.0 in mCDM solution were mixed with 6 mL of mCDM solution in a six-well tissue culture plate. All sample solutions were anaerobically cultured at 37°C for 0, 6, or 12 h. The samples were passed through a 0.22-mm membrane filter (Millex-GP: Millipore, MA, USA) to remove bacteria and the supernatants were analyzed for amino acid concentrations using the Waters AccQ Amino Acid Analysis method with an ultra-performance liquid chromatography (UPLC) system (Waters ACQUITY H-Class; Waters, Milford, USA), consisting of a photodiode array (PDA) detector, column heater, sample manager, and binary solvent delivery system (supplemental experimental procedures).
Metabolic flux analysis
Intracellular metabolic flux analyses were performed to investigate the fate of methionine incorporated by F. nucleatum when cocultured with S. gordonii WT. Cocultures of F. nucleatum with S. gordonii WT were performed in six-well Corning Costar Transwell plates (pore size 0.4 µm, 24 mm in diameter; Corning, NY, USA). F. nucleatum cells in the outer chamber were collected after incubation. [13C5, 15N] l-methionine (13C5, 15N, 98%; Taiyo Nippon Sanso Corp., Tokyo, Japan) was added at a final concentration of 10 mM to mCDM, with the following modified amino acid concentrations: 40 mM l-glutamic acid, 10 mM l-arginine-HCl, and 1.0 mM l-tryptophan. At the mid-exponential growth phase (0.5 to 1.0 OD units/mL), bacterial cells were harvested by centrifugation (7,670 × g for 7 min at 4°C), washed twice with PBS, and finally resuspended at 20 OD units/mL in mCDM containing 10 mM [13C5, 15N] l-methionine. F. nucleatum and S. gordonii cells were then inoculated at a density of 1.5 × 1010 CFU/well into a Transwell outer chamber (2.6 cm3) and inner chamber (1.5 cm3), respectively. Thereafter, anaerobic incubation was performed at 37°C for 0, 1, 2, 3, or 6 h. F. nucleatum cells obtained at each time point were harvested by centrifugation (7670 × g for 7 min at 4°C) and intracellular metabolites were extracted with 100% methanol. The analyses were performed using capillary electrophoresis time-of-flight mass spectrometry (CE-TOF-MS), as described in supplemental experimental procedures.
Ornithine supplementation and ornithine decarboxylase inhibitor
l-Ornithine was added at a final concentration of 0.1 to 1.0 mM to the mCDM solution containing 1.0 mM l-methionine, while F. nucleatum cells were incubated using the anaerobic system described above. The inhibitory effects of dl-α-difluoromethylornithine hydrochloride monohydrate (DFMO; Tokyo Kasei Kogyo, Tokyo, Japan) on CH3SH generation were evaluated using cocultures of F. nucleatum and S. gordonii at final concentrations of 0.01 to 1.0 mM.
Quantitation of mRNA transcripts
Following incubation in the noncontact coculture system in mCDM (pH 6.5) containing 1.0 mM l-methionine for 8 or 16 h at 37°C, bacterial cells in 27 mL of each monocultured and cocultured bacterial solution were collected by centrifugation (8,000 × g for 7 min at 4°C). The cells were then resuspended in 3 mL of RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany). After incubation for 5 min at room temperature, 1 mL of the bacterial solution was collected and immediately frozen, then treated with 20 µL of proteinase K (Qiagen) and 200 µL of 15 mg/mL lysozyme at 55°C, and subjected to centrifugation at 1,000 rpm for 15 min. RNA isolation was performed with 1 mL of TRIzol reagent (Life Technologies Corp., CA, USA) and an RNeasy kit (Qiagen). cDNA synthesis along with the removal of genomic DNA was performed using iScript master mix (Bio-Rad, CA, USA) according to the manufacturer’s instructions. Real-time PCR assays were performed with a KAPA SYBR Fast kit (KAPA Biosystems, MA, USA), following the supplied protocol. Designed primers are shown in Table S1. To determine gene expression, a comparative Ct method was used.
Statistical analysis
All statistical analyses were performed using Excel (Office365) with the Statcel4 software package (OMS Publishing Inc., Saitama, Japan). Different statistical tests were used for different experiments, as indicated in the corresponding figure legends.
RESULTS
Enhancement of CH3SH generation in cocultures and noncontact cocultures
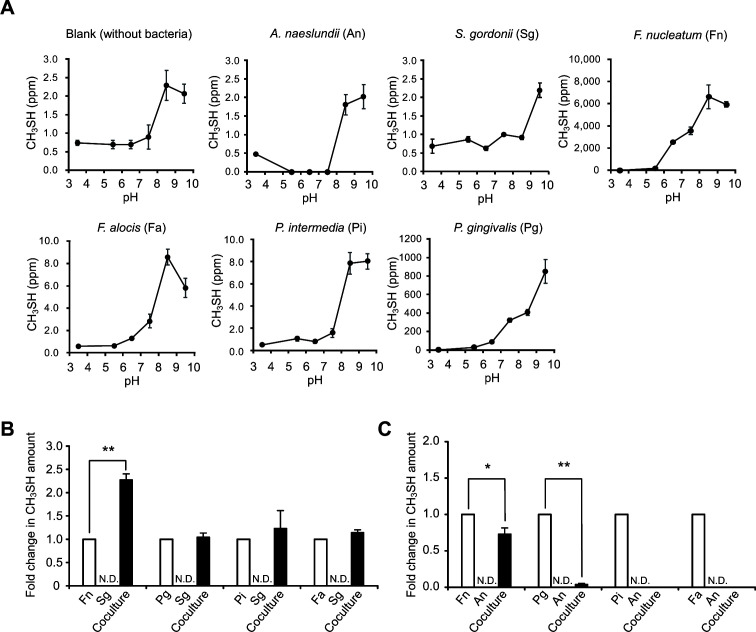
To assess the ability of oral bacterial organisms to produce CH3SH under anaerobic culture conditions, A. naeslundii and S. gordonii (early colonizers), F. nucleatum and P. intermedia (mid colonizers), and F. alocis and P. gingivalis (late colonizers) were selected as representative oral bacteria. F. nucleatum exhibited the highest CH3SH production among the oral bacteria tested, with the largest amount released at pH 8.5 (Fig. 1A). The production of CH3SH by P. gingivalis increased gradually with increasing pH, but its maximum concentration was about one-eighth of the maximum concentration in F. nucleatum. In contrast, both A. naeslundii and S. gordonii when cultured alone produced negligible amounts of CH3SH (Table 1). Only the OD value of A. naeslundii increased after 16 h of incubation in mCDM supplemented with 5 mM l-methionine, while the values of the other bacteria were virtually unchanged (Table S2).
Fig 1.
CH3SH generation by l-methionine metabolism of oral bacteria. (A) Changes in CH3SH production under various pH conditions. Bacterial cultures were supplemented with 5.0 mM l-methionine. Results are normalized with the final OD and shown as the mean ± SD of three independent experiments. (B, C) Enhancement of CH3SH production by coculturing with S. gordonii (B) or A. naeslundii (C). Bacterial cultures were supplemented with 0.5 mM l-methionine and adjusted at the final pH 6.5. Results are shown as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 (two-tailed paired t-test); N.D., not detected. Fold changes in CH3SH amount were calculated using the following equation: fold = (amount of CH3SH formation in coculture)/(amount of CH3SH formation in single culture of F. nucleatum, P. gingivalis, P. intermedia, or F. alocis) + (amount of CH3SH formation in single culture of S. gordonii or A. naeslundii). All cultures were anaerobically incubated for 16 h, as described in Materials and Methods.
TABLE 1.
Precursors for CH3SH production by oral bacteriaa
| CH3SH production (ppm) | ||||
|---|---|---|---|---|
| Bacterial strain | Substrates | |||
| l-Cysteine | l-Methionine | dl-Homocysteine | l-Serine | |
| No bacteria (blank) | N.D. | 0.17 ± 0.04 | N.D. | N.D. |
| Streptococcus gordonii DL1 | N.D. | 0.36 ± 0.11 | N.D. | N.D. |
| Actinomyces naeslundii ATCC19039 | N.D. | N.D. | N.D. | N.D. |
| Fusobacterium nucleatum subsp. nucleatum ATCC25586 | N.D. | 477.71 ± 26.30 | N.D. | N.D. |
| Prevotella intermedia ATCC49046 | N.D. | 0.18 ± 0.01 | N.D. | N.D. |
| Filifactor alocis ATCC35846 | N.D. | 0.80 ± 0.15 | N.D. | N.D. |
| Porphyromonas gingivalis ATCC33277 | N.D. | 14.39 ± 2.93 | 0.44 ± 0.02 | N.D. |
Bacterial cultures were supplemented with 5 mM of each substrate and anaerobically incubated for 16 h. Blank samples were cultured without bacteria. Data are shown as the mean ± SD of three independent experiments. N.D., not detected.
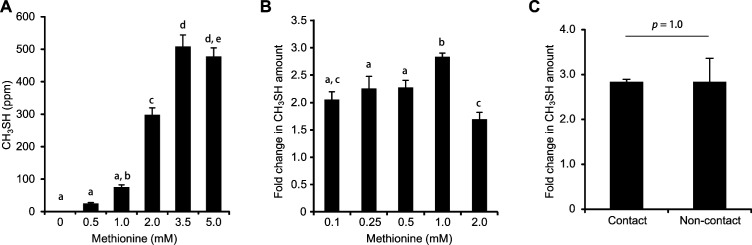
Next, the effects of coculturing early colonizers with mid- or late colonizers on the enhancement of CH3SH generation were examined. When F. nucleatum was cocultured with S. gordonii, there was an approximately 2.3-fold increase in the amount of CH3SH production as compared to CH3SH levels in their respective monocultures (Fig. 1B). On the other hand, the addition of A. naeslundii significantly suppressed CH3SH generation in F. nucleatum, P. gingivalis, and P. intermedia cultures (Fig. 1C). F. nucleatum alone yielded the highest amount (approximately 500 ppm) of CH3SH with 3.5 to 5.0 mM l-methionine added at pH 6.5 (Fig. 2A), while the greatest level of enhancement (approximately 3-fold) in CH3SH production was observed in cocultures of F. nucleatum with S. gordonii when 1 mM l-methionine was added (Fig. 2B), indicating that S. gordonii can boost CH3SH production by F. nucleatum with lower concentrations of methionine.
Fig 2.
Changes in CH3SH production in F. nucleatum single cultures and cocultures with S. gordonii. (A) Relationships between the amount of CH3SH production and methionine concentration in mCDM in F. nucleatum single cultures. Data are shown as the mean ± SD of three independent experiments. In case of no significant difference between groups, the same alphabets are denoted (one-way ANOVA, followed by Tukey–Kramer post-hoc test, significance level; P < 0.01). (B) Fold changes in CH3SH level by the addition of various concentrations of methionine in cocultures of F. nucleatum and S. gordonii. Each fold change in CH3SH amount indicates multiples of CH3SH amount in cocultures of F. nucleatum and S. gordonii when the amount of CH3SH production in F. nucleatum monoculture under each condition is set as 1. Data are shown as the mean ± SD of three independent experiments. In case of no significant difference between groups, the same alphabets are denoted (one-way ANOVA, followed by Tukey–Kramer post-hoc test, significance level; P < 0.01). (C) Changes in CH3SH production in F. nucleatum and S. gordonii cocultures in contact or non-contact culture systems. The bacterial cultures were supplemented with 1.0 mM l-methionine. Each fold change in CH3SH amount indicates multiples of CH3SH amount in cocultures of F. nucleatum and S. gordonii when the amount of CH3SH production in F. nucleatum monoculture under each condition is set as 1. Results are shown as the mean ± SD of four independent experiments. Fold changes in CH3SH amount were calculated using the following equation: fold = (amount of CH3SH formation in coculture)/[(amount of CH3SH formation in single culture of F. nucleatum) + (amount of CH3SH formation in single culture of S. gordonii)]. All cultures were anaerobically incubated for 16 h, as described in Materials and Methods. A two-tailed t-test was performed to calculate the P-value.
To determine whether physical interactions between F. nucleatum and S. gordonii contribute to enhanced production of CH3SH, these species were cocultured without contact, and the findings were assessed (Fig. S1). The presence of S. gordonii in both contact and noncontact cocultures was found to increase CH3SH levels by up to 3-fold as compared to CH3SH levels in respective monocultures (Fig. 2C). These findings indicate that enhancement of CH3SH production by cocultured F. nucleatum and S. gordonii is due to an exchange of diffusible factors rather than through physical contact.
Furthermore, the effect of 6% oxygen concentration in the culture environment on CH3SH production was confirmed. Although there was no difference in CH3SH production in F. nucleatum monocultures, there was a significant decrease in the production of CH3SH when F. nucleatum and S. gordonii were cocultured in a microaerophilic environment than in an anaerobic environment (P < 0.01, Fig. S2).
Noninvolvement of AI-2-based QS system in CH3SH production
To examine the involvement of an AI-2-based QS system, enhancement of CH3SH production was assessed by the addition of 4, 5-dihydroxy-2, 3-pentanedione (DPD), an AI-2 precursor, from which the LuxS enzyme catalyzes conversion in F. nucleatum (39). The results showed that DPD/AI-2 had no significant effect on CH3SH production as compared to the control samples (Fig. S3), indicating that an AI-2-based QS system is not involved in increased CH3SH production.
Contribution of ornithine to increased CH3SH production when cocultured with S. gordonii
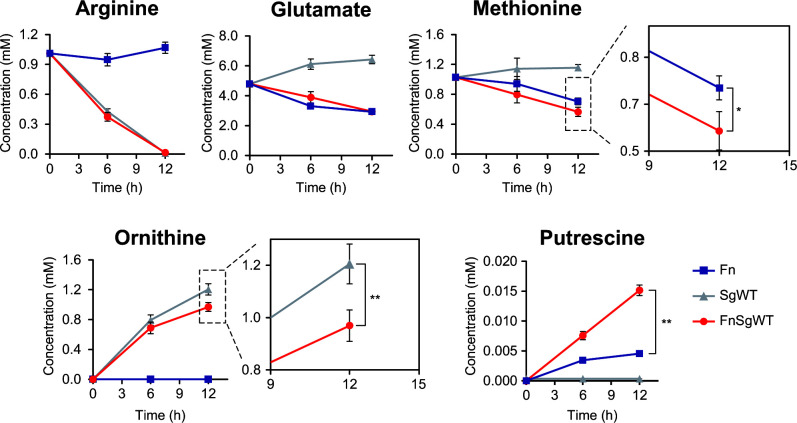
To examine whether metabolic interactions underlie the promotion of CH3SH generation associated with coculturing, time-course changes of extracellular metabolites in cocultures of F. nucleatum and S. gordonii WT and also monocultures of each strain were assessed using UPLC (Fig. 3). F. nucleatum gradually consumed methionine, about 50% in the substrate present during 12 h of incubation, while S. gordonii showed a low level of consumption. On the other hand, cocultures of F. nucleatum and S. gordonii WT exhibited the highest level of consumption of methionine (P < 0.05, vs. F. nucleatum monocultures). Furthermore, monocultures of S. gordonii WT and cocultures with F. nucleatum showed depleted arginine, as well as release of 1.2 and 0.95 mM ornithine, respectively (P < 0.01), suggesting an uptake of 0.25 mM ornithine by F. nucleatum under coculture conditions. Putrescine levels in cocultures of F. nucleatum with S. gordonii WT were also markedly increased, indicating that putrescine excretion was accelerated by S. gordonii. Glutamate present in the mCDM was found to be gradually taken up by F. nucleatum but not S. gordonii.
Fig 3.
Time course of changes in extracellular metabolites in culture fluids. Lines indicate F. nucleatum (blue) and S. gordonii WT (gray) monocultures and F. nucleatum and S. gordonii WT (red) coculture. Results are shown as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 (one-way ANOVA, followed by Tukey–Kramer post-hoc test) for methionine and ornithine metabolism.
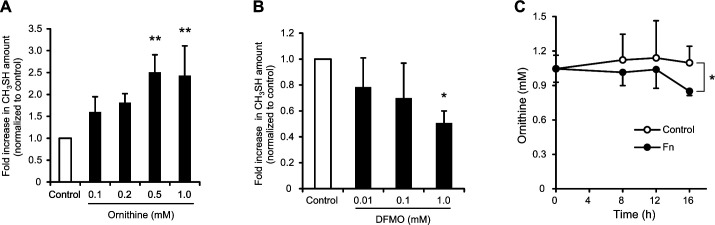
Next, we determined whether the addition of ornithine increased CH3SH production by F. nucleatum. Ornithine added at 0.1 or 0.5 mM provided a significant increase in the production of CH3SH (2.5-fold as compared to that without ornithine) (Fig. 4A), while bacterial growth was not significantly affected (data not shown). In addition, DFMO, an inhibitor of ornithine decarboxylase (ODC) (40, 41), diminished S. gordonii-induced stimulation of CH3SH generation in cocultures of F. nucleatum and S. gordonii, in a dose-response manner, with 1.0 mM significantly halving the amount produced (Fig. 4B). Moreover, extracellular ornithine was taken up by F. nucleatum over time (Fig. 4C). Together, these results suggest that ornithine metabolism by ODC of F. nucleatum enhances CH3SH production in the presence of ornithine.
Fig 4.
Effects of ornithine on enhancement of CH3SH generation. (A) Fold changes in CH3SH level by the addition of various concentrations of ornithine to F. nucleatum monocultures. The fold increase was normalized to that of the control sample without ornithine. F. nucleatum cultures were supplemented with 1.0 mM l-methionine and ornithine, then anaerobically incubated for 16 h. Results are shown as the mean ± SD of three independent experiments. **P < 0.01 (one-way ANOVA, followed by Dunnett’s test). (B) Inhibition of CH3SH generation by DFMO. The fold increase was normalized to that of the control sample without DFMO. Mixtures of F. nucleatum and S. gordonii cocultures supplemented with 1.0 mM l-methionine were anaerobically incubated for 16 h. Results are shown as the mean ± SD of three independent experiments. *P < 0.05 (one-way ANOVA, followed by Dunnett’s test). (C) Uptake of ornithine by F. nucleatum. Control indicates a sample without F. nucleatum. Results are shown as the mean ± SD of three independent experiments. *P < 0.05 for indicated time point (two-tailed t-test).
Cocultures of F. nucleatum and S. gordonii ΔarcD
ArcD of S. gordonii has been shown to mediate arginine uptake and concomitant ornithine export (42–44). To confirm whether ornithine from S. gordonii causes increased production of CH3SH by F. nucleatum, an arcD-deletion mutant strain was generated and CH3SH production in cocultures with F. nucleatum was evaluated. The presence of the ΔarcD mutant in both contact and noncontact cultures failed to increase CH3SH production by F. nucleatum (Fig. S4). Additionally, the ΔarcD mutant exhibited reduced levels of arginine uptake and ornithine export (Fig. S5). These results indicate that ornithine from S. gordonii is a key metabolite for enhancing CH3SH generation by F. nucleatum. Cocultures with ΔarcD mutants also showed diminished methionine utilization by F. nucleatum as compared to the WT strain (P < 0.01) (Fig. S5), whereas no significant difference was found between the amounts of methionine consumed by F. nucleatum in monocultures and cocultures with the ΔarcD mutant (Fig. S5). It is thus considered that the uptake of methionine by F. nucleatum is promoted by ornithine from S. gordonii, leading to enhanced CH3SH generation.
Methionine metabolism of F. nucleatum under cocultivation condition
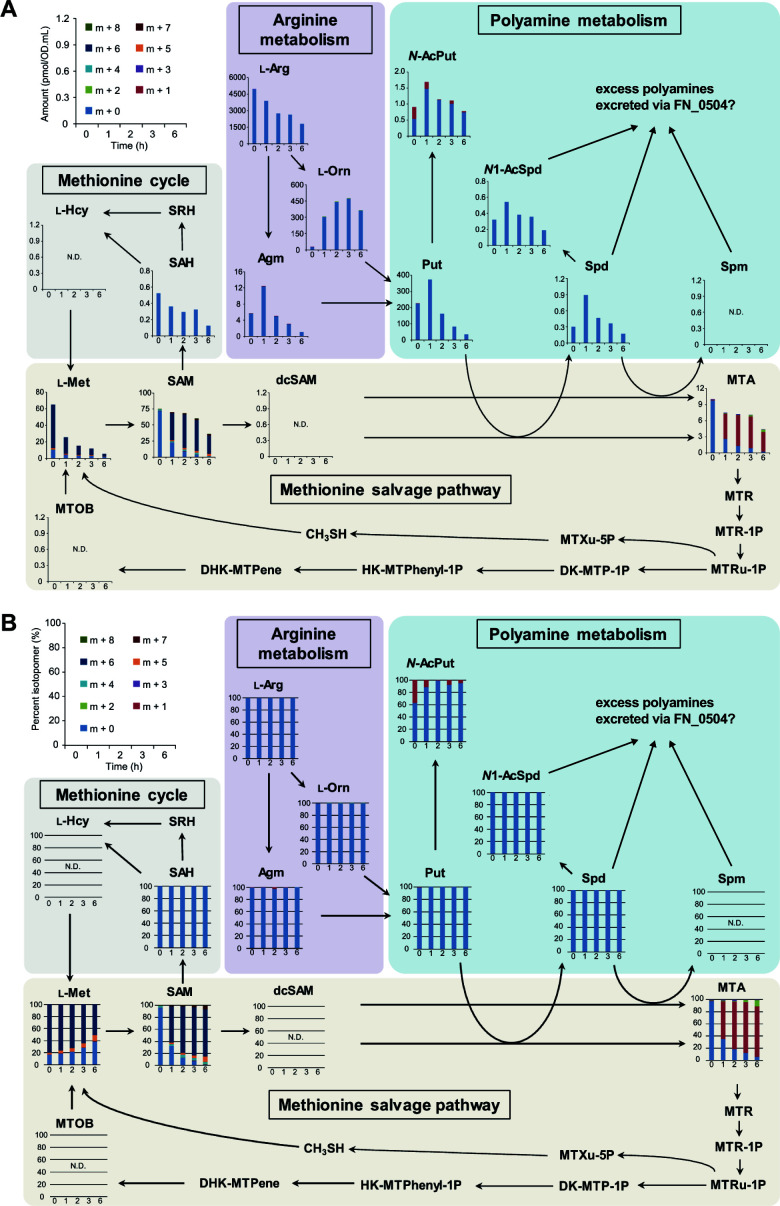
Using 13C/15N-labeled methionine, the fate of methionine in F. nucleatum cells was examined to elucidate the intracellular metabolic dynamics underlying enhanced CH3SH production in the presence of S. gordonii. Eleven of the 17 targeted metabolites were detected, and changes in 19 different 13C, 15N isotopomers related to methionine metabolism were determined using CE-TOF-MS (Fig. S6). The findings showed that fully labeled methionine (m + 6) was instantly incorporated into F. nucleatum cells and then markedly decreased over time (Fig. 5A). Furthermore, intracellular accumulation of labeled S-adenosyl-l-methionine (SAM; m + 6) derived from fully labeled methionine as well as labeled S-adenosylmethioninamine (MTA; m + 1) derived from SAM (m + 6) was also noted and then they were gradually consumed (Fig. 5A). The increase in intracellular level of ornithine reached a peak at 3 h, while the increased levels of polyamines, including putrescine, spermidine, and their acetyl derivatives, peaked at 1 h, after which they were consumed (Fig. 5A). Although intracellular S-adenosyl-L-homocysteine (SAH) levels were gradually decreased, with labeled SAH (m + 5) undetected, the ratio of labeled methionine (m + 5) showed an increase over time, indicative of methionine regeneration from L-homocysteine via SAH (methionine cycle) (Fig. 5B). Collectively, these results suggest that methionine mainly enters the methionine cycle and polyamine biosynthesis pathway in F. nucleatum cells when cocultured with S. gordonii.
Fig 5.
Flux profiling of 13C, 15N-labeled methionine salvage pathway metabolites. Quantitation values for (A) isotopomers and (B) the ratio of each as compared to total metabolites of methionine cycle and methionine salvage pathway metabolites from [13C5, 15N] l-methionine in F. nucleatum. Bacterial cultures were supplemented with 10 mM [13C5, 15N] l-methionine and anaerobically incubated for 6 h. Values obtained after omitting the abundance of naturally occurring isotopes for each detected metabolite were used. Results are shown as mean values from three independent experiments. l-Met, l-methionine; SAM, S-adenosyl- l-methionine; SAH, S-adenosyl- l-homocysteine; SRH, S-ribosyl-l-homocysteine; l-Hcy, l-homocysteine; dcSAM, S-adenosylmethioninamine; MTA, 5'-methylthioadenosine; l-Arg, l-arginine; Agm, agmatine; l-Orn; l-ornithine; Put, putrescine; N-AcPut, N-acetylputrescine; N1-AcSpd, N1-acetylspermidine; Spd, spermidine; Spm, spermine; MTR, 5-methylthio-d-ribose; MTR-1P, S-methyl-5-thio-d-ribose 1-phosphate; MTRu-1P, S-methyl-5-thio-d-ribulose 1-phosphate; MTXu-5P, 1-(methylthio)xylulose 5-phosphate; CH3SH, methyl mercaptan; DK-MTP-1P, 2,3-diketo-5-methyl-thiopentyl-1-phosphate; HK-MTPhenyl-1P, 2-hydroxy-3-keto-5-methylthiopentenyl-1-phosphate; DHK-MTPene, 1,2-dihydroxy-5-(methylthio) Pent-1-en-3-one; MTOB, 4-methylthio-2-oxobutanoic acid; N.D., not detected.
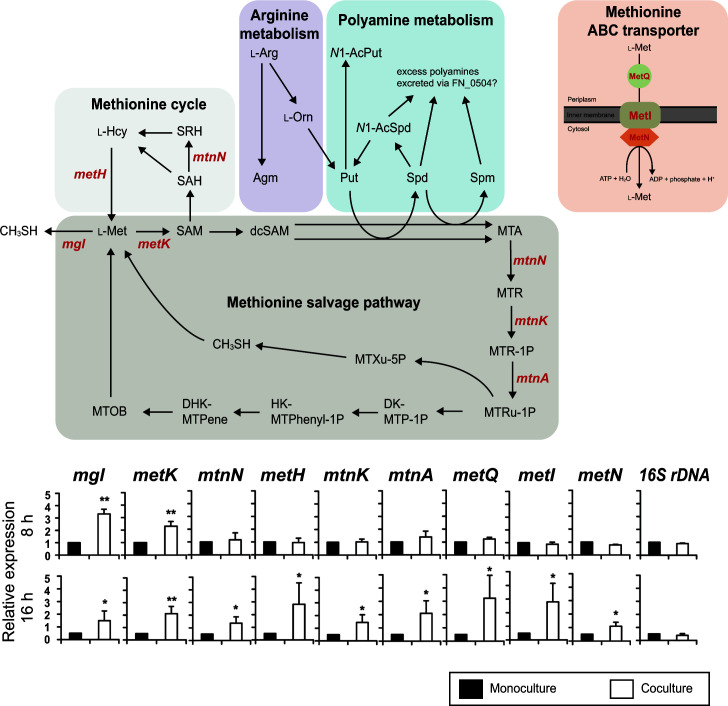
Gene expression in F. nucleatum in cocultures with S. gordonii
The one-carbon unit of MTA is known to be recycled to the methionine cycle via the methionine salvage pathway (45). Transcriptional changes in genes involved in the methionine cycle and methionine salvage pathway and also methionine ABC transporters were examined. After 8 and 16 h of incubation, the transcriptional levels of mgl and metK in cocultures of F. nucleatum and S. gordonii were increased by 2.3- to 4.2-fold, as compared to those in monocultures (Fig. 6). Although the expression levels of genes other than mgl and metK were not significantly changed after 8 h, upregulation of the expression of these metabolic genes by F. nucleatum was noted in cocultures after 16 h (Fig. 6). In particular, the expression level of metK, metH, metQ, and metI was markedly enhanced by 4.0- to 5.6-fold after a 16 h culture, indicating enhanced activities in methionine cycle and uptake and salvage pathway, especially under lower methionine concentration environments. Thus, enhanced methionine uptake likely occurs when cocultured with S. gordonii, as shown in Fig. 3, leading to increased CH3SH production. This is achieved through the upregulation of mgl and metK and activation of the methionine salvage pathway, involving MTA and MTRu-1P, and is also linked to polyamine synthesis.
Fig 6.
Relative fold changes in mRNA expression by F. nucleatum after coculture with S. gordonii. The level of mRNA expression for each incubation time was normalized to 16S rDNA of F. nucleatum. Fold changes were calculated using the following equation: fold = (mRNA expression by F. nucleatum in coculture with S. gordonii)/(mRNA expression by F. nucleatum in monoculture). Results are shown as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01 (two-tailed paired t-test, monoculture vs. coculture). Abbreviations are described in the legend in Fig. 5.
These results suggest that ornithine secreted by S. gordonii promotes polyamine biosynthesis by F. nucleatum, resulting in a compensatory increase in demand for methionine, leading to elevated methionine metabolism and CH3SH production.
DISCUSSION
For this investigation, a novel noncontact method for quantitation of CH3SH was developed (Fig. S1), which showed that nutritional cross-feeding enhances CH3SH production by F. nucleatum through altered methionine metabolism. The findings illustrate that metabolic interactions can modulate the emission of microbial volatile compounds, thus potentially contributing to the development of halitosis.
Consistent with several previous reports, the present study found that F. nucleatum is a potent producer of CH3SH. Interestingly, F. nucleatum released the highest amounts of CH3SH at pH 8.5 (Table 1; Fig. 1A), which was considered to be due to the optimal pH of METase (l-methionine + H2O → CH3SH + NH3 +2-oxobutanoate) of 8.0–8.5 in F. nucleatum as well as other bacteria (46–48). It has also been reported that an alkaline condition (pH 8.2) induces biofilm development by F. nucleatum through the increased abundance of adhesion proteins (49). The periodontal pocket in periodontitis patients has been shown to be alkaline, as high as pH 8.9 (49, 50). Hence, diseased periodontal pockets may harbor increased levels of F. nucleatum-related biofilms, from which larger amounts of CH3SH are emitted, thus contributing to malodor generation.
Although a number of studies have shown that interactions between oral bacteria elevate pathogenicity through enhancement of biofilm formation and increased adherence to and invasion of epithelial cells (27–30, 35–37), little is known regarding how these interactions affect oral malodor generation. In the present study, coculture of S. gordonii with F. nucleatum facilitated CH3SH production, particularly when supplemented with methionine at concentrations similar to those seen in a natural oral environment. Interestingly, A. naeslundii suppressed CH3SH production by F. nucleatum and also nearly abolished its production by other periodontal pathogens including P. gingivalis (Fig. 1C). These findings are consistent with those observed in studies of soil bacterial communities, where interspecies interactions have been found to either promote or constrain volatile production (51). It is therefore likely that oral polymicrobial communities exhibit enhanced or suppressed CH3SH production depending on interactions between the community members, which highlights the need for studies to resolve individual roles of different species.
The noncontact coculture experiments showed that the exchange of diffusible molecules from S. gordonii enhances the level of CH3SH generation by F. nucleatum. Additional examination also revealed that this interaction is driven by the metabolism of ArcD-excreted ornithine by the ODC of F. nucleatum. These findings add to a growing body of evidence showing the importance of microbial metabolic interactions that integrate microbial communities and affect the pathogenicity of oral diseases (52). In particular, our recent study showed the cooccurrence of P. gingivalis with the genes of S. gordonii arcD and F. nucleatum ODC in periodontitis patients. It demonstrated that ornithine cross-feeding via ArcD of S. gordonii-induced ODC-catalyzed polyamine production by F. nucleatum, thus enhancing the biofilm lifecycle of P. gingivalis (53). Therefore, the present findings highlight the importance of engagement of F. nucleatum in a cross-feeding network with S. gordonii, not just with regard to periodontal pathogenesis but also disease-associated halitosis.
Notably, we found that ornithine cross-feeding promotes the uptake and metabolism of methionine by F. nucleatum. Methionine is an important molecule for the initiation of protein synthesis and SAM-mediated methylation of proteins, RNA, and DNA (54–56). Metabolic routes of methionine incorporated in F. nucleatum can be divided mainly into methionine cycle and salvage pathways (45, 57, 58). The former pathway functions to recycle adenine and methionine through a SAM-mediated methylation reaction, leading to the production of AI-2, a QS signal (59, 60). F. nucleatum AI-2 has been shown to have an important role in inter- and intraspecies interactions in microbial communities, thus affecting periodontal pathogenesis (61). Hence, it cannot be ruled out that an elevated level of AI-2 molecules in F. nucleatum can result in greater levels of CH3SH production. To examine that possibility, the enhancement of CH3SH production was assessed by the addition of DPD, an AI-2 precursor. The results showed that DPD/AI-2 had no significant effect on CH3SH production, indicating that an AI-2-based QS system is not involved in increased CH3SH production. On the other hand, labeling experiments indicated a slight increase in the ratio of methionine (m + 5) over time, suggesting that a portion of labeled methionine (m + 6) enters the methionine cycle for methylation, as well as resynthesis and reuse of methionine.
The methionine salvage pathway is known to be involved in various cellular processes, including the preservation of intracellular sulfur pools for the formation of amino acids and proteins, and also the synthesis of polyamines, such as putrescine, spermine, and spermidine, which are important molecules for cell growth, biofilm formation, and protection from oxidative and acid stress (62–64). Results from labeling experiments with 13C, 15N-methionine showed a high similarity of labeling between SAM (m + 6) and MTA (m + 1), demonstrating that labeled methionine enters the polyamine pathway (Fig. 5A). A longer incubation period (6 h) resulted in gradual increases in SAM (m + 7, m + 8) and MTA (m + 2, m + 3) (Fig. 5B), indicating possible production of SAM (m + 7, m + 8) from methionine (m + 6) and labeled ATP (m + 1, m + 2), the latter of which was synthesized from labeled methionine via a de novo ATP synthesis pathway. The purine carbon skeleton is composed of two nitrogens from Gln, two from Asp and Gly each, one carbon from N10-formyl-THF, and one from N5N10-methenyl-THF. As shown in Fig S6, METase metabolizes l-methionine and produces NH3 (m + 1), CH3SH (m + 1), and 2-oxobutyrate (2OB; m + 4) that leads to the one-carbon pool via formate (m + 1). As shown in Fig. S7, F. nucleatum produces N10-formyl-THF (m + 1) and N5N10-methenyl-THF (m + 1) in the one-carbon pool. Therefore, it seems natural that SAM (m + 7 and m + 8) and MTA (m + 2, m + 3) would be present after a certain time. On the other hand, despite findings showing methionine-derived labeling in MTA (m + 1), no labeling in spermidine (m + 4) was detected. This may be explained by the function of FN_0504, a putative L-ornithine/polyamine antiporter, to efflux polyamines as it takes up L-ornithine. In addition, the adsorption of spermidine onto capillary walls causes peak broadening, resulting in reduced detection limits. Results from UPLC and labeling experiments showed that S. gordonii secreted ornithine, which led to a dramatic increase in intracellular ornithine and polyamines, indicating an increase in polyamine pathway activity (Fig. 3 and 5A), a finding consistent with our previous report (53). Additionally, acetylated polyamines were detected in F. nucleatum cells, although that was dependent on the levels of putrescine and spermidine (Fig. 5A), suggesting that excess levels of these may cause their acetylation and maintain intracellular levels of polyamines at a constant level. Acetylation of excess polyamines by diamine N-acetyltransferase [FN_1057; EC 2.3.1.57] in F. nucleatum requires acetyl-CoA. In silico analysis of acetyl-CoA biosynthetic pathways from L-methionine revealed only one pathway for incorporating a 13C into acetylputrescine (Fig S7). Notably, F. nucleatum cannot complete this pathway alone; it requires the enzymatic reaction of glycine hydroxymethyltransferase [SGO_1151; EC 2.1.2.1], encoded by S. gordonii (Fig S7). We previously confirmed that S. gordonii releases serine into the environment (unpublished data), and F. nucleatum has been reported to take up and utilize serine (65). Therefore, serine-mediated crossfeeding between the two species should be possible. Anaerobically grown S. gordonii has been shown to be able to produce H2O2 when glucose is available, albeit to a lesser extent than under an aerobic condition (66), thus F. nucleatum might increase the elevation of intracellular polyamines in response to H2O2 generated by S. gordonii. It is also considered likely that the elevated polyamine-synthesis pathway activity under coexistence with S. gordonii increases the demand for methionine, following enhancement of methionine metabolism and CH3SH generation.
Considering that the ratio of methionine (m + 1) showed a slight increase over time (Fig. 5B), a portion of accumulated MTA was likely resynthesized to methionine (m + 1) via 4-methylthio-2-oxobutanoic acid (MTOB). Although biosynthesis of MTOB from S-methyl-5-thio-d-ribose 1-phosphate (MTRu-1P) reportedly requires oxygen (67, 68), as also noted in the present experiments (Fig. 6), recent studies by North et al. show that Rhodospirillum rubrum possesses an oxygen-independent MTA-isoprenoid shunt that links MTA metabolism to the release of CH3SH for methionine regeneration and 1-deoxyxylulose-5-phosphate (DXP) synthesis for isoprenoid metabolism under anaerobic conditions (Fig. 6) (67–69). F. nucleatum may produce CH3SH via an MTA-isoprenoid shunt under anaerobic coculture conditions, although further study is required to determine the precise MTA-isoprenoid shunt in F. nucleatum. l-Methionine regeneration might also occur through FN_1745, a cystathionine gamma-synthase [EC 2.5.1.48]. This enzyme typically facilitates cystathionine production from O-succinyl-l-homoserine and l-cysteine. It is reportedly capable of producing l-methionine when CH3SH is used instead of l-cysteine (70). However, its Km value with CH3SH is significantly higher than with l-cysteine, and the Vmax is notably low (70), suggesting that this reaction is a minor, if not negligible, pathway for l-methionine regeneration.
The sustainable resynthesis of l-methionine through three distinct pathways, the methionine cycle, methionine salvage, and the activity of FN_1745, likely contributes to the continuous release of CH3SH in cocultures of F. nucleatum with S. gordonii compared to F. nucleatum monocultures. F. nucleatum has three closely related species previously classified as subspecies: Fusobacterium polymorphum, Fusobacterium vincentii, and Fusobacterium animalis. F. polyrmorphum and F. vincentii also possess genes related to CH3SH production, including mgl, metK, and metQ. This indicates the possibility that the coexistence of these bacteria and S. gordonii enhances CH3SH production. Thus, our results suggest that this phenomenon could potentially occur in coexistence with various bacteria that possess related genes.
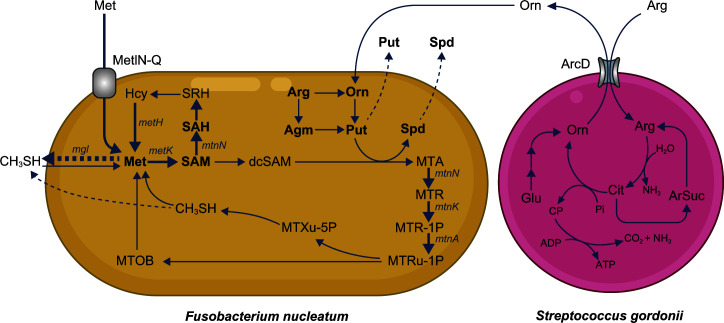
The current study confirmed that S. gordonii takes up extracellular arginine via ArcD and produces ornithine intracellularly. The metabolism of ornithine excreted by S. gordonii leads to enhanced uptake and regeneration of methionine in F. nucleatum, driven by increased polyamine synthesis, thereby boosting CH3SH production (Fig. 7). Although the number of species used was limited, new insights regarding the impact of metabolic cross-feeding in microbial communities on the generation of malodor compounds in the oral cavity were obtained. Nevertheless, a wide range of metabolites are exchanged among oral bacteria. Thus, further work is needed to fully understand the implications of microbial metabolic interactions related to the development of halitosis.
Fig 7.
Schematic representation of the observed metabolic flow of bacterial metabolism in F. nucleatum and S. gordonii cocultures. S. gordonii takes up l-arginine and excretes ornithine extracellularly. F. nucleatum activates ornithine metabolism and synthesizes polyamines via the methionine salvage pathway, after which the uptake of extracellular methionine is accelerated, and metabolic flow is shunted to the MTA synthesis pathway. Moreover, methionine is resynthesized via the methionine cycle and potentially via FN_1745. Detected metabolites are shown in bold, with dashed arrows for excretion and bold arrows for confirmed upregulation of bacterial metabolism. Cit, citrulline; Glu, glutamate; ArSuc, arginosuccinate; Pi, inorganic phosphate; CP, carbamoyl phosphate; others detailed in the Fig. 5 legend.
ACKNOWLEDGMENTS
The authors thank Miho Kakiuchi for technical assistance and Hajime Sato from Human Metabolome Technology Inc. for assistance with the metabolic analyses.
This study was supported by KAKENHI Grants from the Japan Society for the Promotion of Science (JSPS) (no. 21K17196 and 23K16222 to T.H., no. 22H03300 and 22K19622 to M.K., no. 22K10311 to A.S., and no. 22H00487 and 21K18281 to A.A.).
Contributor Information
Masae Kuboniwa, Email: kuboniwa.masae.dent@osaka-u.ac.jp.
Promi Das, APC Microbiome Ireland, Cork, Ireland.
DATA AVAILABILITY
This study is available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org, where it has been assigned Study ID ST002793. The data can be accessed directly via its Project DOI: http://dx.doi.org/10.21228/M8P126.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00764-23.
Data from methionine stable isotope experiment.
Fig. S1, S2, S3, S4, S5, S6, and S7.
Supplement to the experimental procedure and an additional list of references.
Tables S1 and S2.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Krespi YP, Shrime MG, Kacker A. 2006. The relationship between oral malodor and volatile sulfur compound-producing bacte. Otolaryngol Head Neck Surg 135:671–676. doi: 10.1016/j.otohns.2005.09.036 [DOI] [PubMed] [Google Scholar]
- 2. Hughes FJ, McNab R. 2008. Oral malodour--a review. Arch Oral Biol 53 Suppl 1:S1–S7. doi: 10.1016/S0003-9969(08)70002-5 [DOI] [PubMed] [Google Scholar]
- 3. Scully C, Greenman J. 2012. Halitology (breath odour: aetiopathogenesis and management). Oral Dis 18:333–345. doi: 10.1111/j.1601-0825.2011.01890.x [DOI] [PubMed] [Google Scholar]
- 4. Ratcliff PA, Johnson PW. 1999. The relationship between oral malodor, gingivitis, and periodontitis. J Periodontol 70:485–489. doi: 10.1902/jop.1999.70.5.485 [DOI] [PubMed] [Google Scholar]
- 5. Morita M, Wang HL. 2001. Association between oral malodor and adult periodontitis: a review. J Clin Periodontol 28:813–819. doi: 10.1034/j.1600-051x.2001.028009813.x [DOI] [PubMed] [Google Scholar]
- 6. Nagata Y. 2003. Measurement of odor threshold by triangle odor bag method, p 118–127. In Odor measurement review. Japan Ministry of the environment, Japan. [Google Scholar]
- 7. Loesche WJ, Kazor C. 2002. Microbiology and treatment of halitosis. Periodontol 2000 28:256–279. doi: 10.1034/j.1600-0757.2002.280111.x [DOI] [PubMed] [Google Scholar]
- 8. Khalid TY, Saad S, Greenman J, de Lacy Costello B, Probert CSJ, Ratcliffe NM. 2013. Volatiles from oral anaerobes confounding breath biomarker discovery. J Breath Res 7:017114. doi: 10.1088/1752-7155/7/1/017114 [DOI] [PubMed] [Google Scholar]
- 9. Setoguchi T, Machigashira M, Yamamoto M, Yotsumoto Y, Yoshimori M, Izumi Y, Yaegaki K. 2002. The effects of methyl mercaptan on epithelial cell growth and proliferation. Int Dent J 52 Suppl 3:241–246. doi: 10.1002/j.1875-595x.2002.tb00933.x [DOI] [PubMed] [Google Scholar]
- 10. Johnson PW, Yaegaki K, Tonzetich J. 1992. Effect of volatile thiol compounds on protein metabolism by human gingival fibroblasts. J Periodontal Res 27:553–561. doi: 10.1111/j.1600-0765.1992.tb01736.x [DOI] [PubMed] [Google Scholar]
- 11. Johnson P, Yaegaki K, Tonzetich J. 1996. Effect of methyl mercaptan on synthesis and degradation of collagen. J Periodontal Res 31:323–329. doi: 10.1111/j.1600-0765.1996.tb00499.x [DOI] [PubMed] [Google Scholar]
- 12. Johnson PW, Ng W, Tonzetich J. 1992. Modulation of human gingival fibroblast cell metabolism by methyl mercaptan. J Periodontal Res 27:476–483. doi: 10.1111/j.1600-0765.1992.tb01820.x [DOI] [PubMed] [Google Scholar]
- 13. Nakano Y, Yoshimura M, Koga T. 2002. Methyl mercaptan production by periodontal bacteria. Int Dent J 52 Suppl 3:217–220. doi: 10.1002/j.1875-595x.2002.tb00928.x [DOI] [PubMed] [Google Scholar]
- 14. Nakano Y, Yoshimura M, Koga T. 2002. Correlation between oral malodor and periodontal bacteria. Microbes Infect 4:679–683. doi: 10.1016/s1286-4579(02)01586-1 [DOI] [PubMed] [Google Scholar]
- 15. Yoshimura M, Nakano Y, Yamashita Y, Oho T, Saito T, Koga T. 2000. Formation of methyl mercaptan from L-methionine by Porphyromonas gingivalis. Infect Immun 68:6912–6916. doi: 10.1128/IAI.68.12.6912-6916.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yoshimura M, Nakano Y, Fukamachi H, Koga T. 2002. 3-Chloro-DL-alanine resistance by L-methionine-α-deamino-γ-mercaptomethane-lyase activity. FEBS Lett 523:119–122. doi: 10.1016/s0014-5793(02)02958-7 [DOI] [PubMed] [Google Scholar]
- 17. Claesson R, Edlund MB, Persson S, Carlsson J. 1990. Production of volatile sulfur compounds by various Fusobacterium species. Oral Microbiol Immunol 5:137–142. doi: 10.1111/j.1399-302x.1990.tb00411.x [DOI] [PubMed] [Google Scholar]
- 18. Persson S, Edlund MB, Claesson R, Carlsson J. 1990. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol 5:195–201. doi: 10.1111/j.1399-302x.1990.tb00645.x [DOI] [PubMed] [Google Scholar]
- 19. Nemoto T, Shiba T, Komatsu K, Watanabe T, Shimogishi M, Shibasaki M, Koyanagi T, Nagai T, Katagiri S, Takeuchi Y, Iwata T. 2021. Discrimination of bacterial community structures among healthy gingivitis, and periodontitis statuses through integrated metatranscriptomic and network analyses. mSystems 6:e0088621. doi: 10.1128/mSystems.00886-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cho H, Ren Z, Divaris K, Roach J, Lin BM, Liu C, Azcarate-Peril MA, Simancas-Pallares MA, Shrestha P, Orlenko A, Ginnis J, North KE, Zandona AGF, Ribeiro AA, Wu D, Koo H. 2023. Selenomonas sputigena acts as a pathobiont mediating spatial structure and biofilm virulence in early childhood caries. Nat Commun 14:2919. doi: 10.1038/s41467-023-38346-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hajishengallis G, Lamont RJ. 2016. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol 24:477–489. doi: 10.1016/j.tim.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hajishengallis G, Lamont RJ, Koo H. 2023. Oral polymicrobial communities: assembly, function, and impact on diseas. Cell Host Microbe 31:528–538. doi: 10.1016/j.chom.2023.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoare A, Wang H, Meethil A, Abusleme L, Hong BY, Moutsopoulos NM, Marsh PD, Hajishengallis G, Diaz PI. 2021. A cross-species interaction with a symbiotic commensal enables cell-density-dependent growth and in vivo virulence of an oral pathogen. ISME J 15:1490–1504. doi: 10.1038/s41396-020-00865-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim D, Barraza JP, Arthur RA, Hara A, Lewis K, Liu Y, Scisci EL, Hajishengallis E, Whiteley M, Koo H. 2020. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc Natl Acad Sci U S A 117:12375–12386. doi: 10.1073/pnas.1919099117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jakubovics NS, Yassin SA, Rickard AH. 2014. Community interactions of oral streptococci. Adv Appl Microbiol 87:43–110. doi: 10.1016/B978-0-12-800261-2.00002-5 [DOI] [PubMed] [Google Scholar]
- 26. Whitmore SE, Lamont RJ. 2011. The pathogenic persona of community-associated oral streptococci. Mol Microbiol 81:305–314. doi: 10.1111/j.1365-2958.2011.07707.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kuboniwa M, Tribble GD, James CE, Kilic AO, Tao L, Herzberg MC, Shizukuishi S, Lamont RJ. 2006. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol 60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x [DOI] [PubMed] [Google Scholar]
- 28. Nagata H, Iwasaki M, Maeda K, Kuboniwa M, Hashino E, Toe M, Minamino N, Kuwahara H, Shizukuishi S. 2009. Identification of the binding domain of Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase for Porphyromonas gingivalis major fimbriae. Infect Immun 77:5130–5138. doi: 10.1128/IAI.00439-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu H, Sobue T, Bertolini M, Thompson A, Vickerman M, Nobile CJ, Dongari-Bagtzoglou A. 2017. S. oralis activates the Efg1 filamentation pathway in C. albicans to promote cross-kingdom interactions and mucosal biofilms. Virulence 8:1602–1617. doi: 10.1080/21505594.2017.1326438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park Y, Simionato MR, Sekiya K, Murakami Y, James D, Chen W, Hackett M, Yoshimura F, Demuth DR, Lamont RJ. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect Immun 73:3983–3989. doi: 10.1128/IAI.73.7.3983-3989.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cuadra-Saenz G, Rao DL, Underwood AJ, Belapure SA, Campagna SR, Sun Z, Tammariello S, Rickard AH. 2012. Autoinducer-2 influences interactions amongst pioneer colonizing streptococci in oral biofilms. Microbiology (Reading) 158:1783–1795. doi: 10.1099/mic.0.057182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McNab R, Ford SK, El-Sabaeny A, Barbieri B, Cook GS, Lamont RJ. 2003. LuxS-based signaling in Streptococcus gordonii: autoinducer 2 controls carbohydrate metabolism and biofilm formation with Porphyromonas gingivalis. J Bacteriol 185:274–284. doi: 10.1128/JB.185.1.274-284.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahmed NA, Petersen FC, Scheie AA. 2009. AI-2/LuxS is involved in increased biofilm formation by Streptococcus intermedius in the presence of antibiotics. Antimicrob Agents Chemother 53:4258–4263. doi: 10.1128/AAC.00546-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brown SA, Whiteley M. 2007. A novel exclusion mechanism for carbon source partitioning in Aggregatibacter actinomycetemcomitans. J Bacteriol 189:6407–6414. doi: 10.1128/JB.00554-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Egland PG, Palmer RJ, Kolenbrander PE. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci U S A 101:16917–16922. doi: 10.1073/pnas.0407457101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuboniwa M, Houser JR, Hendrickson EL, Wang Q, Alghamdi SA, Sakanaka A, Miller DP, Hutcherson JA, Wang T, Beck DAC, Whiteley M, Amano A, Wang H, Marcotte EM, Hackett M, Lamont RJ. 2017. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol 2:1493–1499. doi: 10.1038/s41564-017-0021-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sakanaka A, Kuboniwa M, Takeuchi H, Hashino E, Amano A. 2015. Arginine-ornithine antiporter ArcD controls arginine metabolism and interspecies biofilm development of Streptococcus gordonii. J Biol Chem 290:21185–21198. doi: 10.1074/jbc.M115.644401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loo CY, Corliss DA, Ganeshkumar N. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol 182:1374–1382. doi: 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Keersmaecker SCJ, Varszegi C, van Boxel N, Habel LW, Metzger K, Daniels R, Marchal K, De Vos D, Vanderleyden J. 2005. Chemical synthesis of (S)-4,5-dihydroxy-2,3-pentanedione, a bacterial signal molecule precursor, and validation of its activity in Salmonella typhimurium. J Biol Chem 280:19563–19568. doi: 10.1074/jbc.M412660200 [DOI] [PubMed] [Google Scholar]
- 40. Kallio A, McCann PP. 1981. Difluoromethylornithine irreversibly inactivates ornithine decarboxylase of Pseudomonas aeruginosa, but does not inhibit the enzymes of Escherichia coli. Biochem J 200:69–75. doi: 10.1042/bj2000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barry DP, Asim M, Leiman DA, de Sablet T, Singh K, Casero RA, Chaturvedi R, Wilson KT. 2011. Difluoromethylornithine is a novel inhibitor of Helicobacter pylori growth, CagA translocation, and interleukin-8 induction. PLoS One 6:e17510. doi: 10.1371/journal.pone.0017510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wimmer F, Oberwinkler T, Bisle B, Tittor J, Oesterhelt D. 2008. Identification of the arginine/ornithine antiporter ArcD from Halobacterium salinarum. FEBS Lett 582:3771–3775. doi: 10.1016/j.febslet.2008.10.004 [DOI] [PubMed] [Google Scholar]
- 43. Fulde M, Willenborg J, Huber C, Hitzmann A, Willms D, Seitz M, Eisenreich W, Valentin-Weigand P, Goethe R. 2014. The arginine-ornithine antiporter ArcD contributes to biological fitness of Streptococcus suis. Front Cell Infect Microbiol 4:107. doi: 10.3389/fcimb.2014.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dong Y, Chen Y-Y, Snyder JA, Burne RA. 2002. Isolation and molecular analysis of the gene cluster for the arginine deiminase system from Streptococcus gordonii DL1. Appl Environ Microbiol 68:5549–5553. doi: 10.1128/AEM.68.11.5549-5553.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M. 2013. Methionine salvage and S-adenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem J 451:145–154. doi: 10.1042/BJ20121744 [DOI] [PubMed] [Google Scholar]
- 46. Lockwood BC, Coombs GH. 1991. Purification and characterization of methionine gamma-lyase from Trichomonas vaginalis. Biochem J 279 ( Pt 3):675–682. doi: 10.1042/bj2790675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tanaka H, Esaki N, Soda K. 1985. A versatile bacterial enzyme: L-methionine γ-lyase. Enzyme Microb Technol 7:530–537. doi: 10.1016/0141-0229(85)90094-8 [DOI] [Google Scholar]
- 48. Foo TC, Terentis AC, Venkatachalam KV. 2016. A continuous spectrophotometric assay and nonlinear kinetic analysis of methionine γ-lyase catalysis. Anal Biochem 507:21–26. doi: 10.1016/j.ab.2016.05.010 [DOI] [PubMed] [Google Scholar]
- 49. Chew J, Zilm PS, Fuss JM, Gully NJ. 2012. A proteomic investigation of Fusobacterium nucleatum alkaline-induced biofilms. BMC Microbiol 12:189. doi: 10.1186/1471-2180-12-189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eggert FM, Drewell L, Bigelow JA, Speck JE, Goldner M. 1991. The pH of gingival crevices and periodontal pockets in children, teenagers and adults. Arch Oral Biol 36:233–238. doi: 10.1016/0003-9969(91)90091-8 [DOI] [PubMed] [Google Scholar]
- 51. Probst M, Telagathoti A, Siewert B, Khomenko I, Betta E, Biasioli F, Peintner U. 2023. Co-cultivation of Mortierellaceae with Pseudomonas helmanticensis affects both their growth and volatilome. Sci Rep 13:2213. doi: 10.1038/s41598-023-29134-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lamont RJ, Koo H, Hajishengallis G. 2018. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 16:745–759. doi: 10.1038/s41579-018-0089-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sakanaka A, Kuboniwa M, Shimma S, Alghamdi SA, Mayumi S, Lamont RJ, Fukusaki E, Amano A. 2022. Fusobacterium nucleatum metabolically integrates commensals and pathogens in oral biofilms. mSystems 7:e0017022. doi: 10.1128/msystems.00170-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. 1996. S-Adenosylmethionine and methylation. FASEB J 10:471–480. [PubMed] [Google Scholar]
- 55. Ferla MP, Patrick WM. 2014. Bacterial methionine biosynthesis. Microbiology (Reading) 160:1571–1584. doi: 10.1099/mic.0.077826-0 [DOI] [PubMed] [Google Scholar]
- 56. Parveen N, Cornell KA. 2011. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol 79:7–20. doi: 10.1111/j.1365-2958.2010.07455.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Albers E. 2009. Metabolic characteristics and importance of the universal methionine salvage pathway recycling methionine from 5’-methylthioadenosine. IUBMB Life 61:1132–1142. doi: 10.1002/iub.278 [DOI] [PubMed] [Google Scholar]
- 58. Sekowska A, Kung HF, Danchin A. 2000. Sulfur metabolism in Escherichia coli and related bacteria: facts and fiction. J Mol Microbiol Biotechnol 2:145–177. [PubMed] [Google Scholar]
- 59. Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. 2005. Making 'sense' of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol 3:383–396. doi: 10.1038/nrmicro1146 [DOI] [PubMed] [Google Scholar]
- 60. Hardie KR, Heurlier K. 2008. Establishing bacterial communities by 'word of mouth': LuxS and autoinducer 2 in biofilm development. Nat Rev Microbiol 6:635–643. doi: 10.1038/nrmicro1916 [DOI] [PubMed] [Google Scholar]
- 61. Jang YJ, Choi YJ, Lee SH, Jun HK, Choi BK. 2013. Autoinducer 2 of Fusobacterium nucleatum as a target molecule to inhibit biofilm formation of periodontopathogens. Arch Oral Biol 58:17–27. doi: 10.1016/j.archoralbio.2012.04.016 [DOI] [PubMed] [Google Scholar]
- 62. Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol Rev 49:81–99. doi: 10.1128/mr.49.1.81-99.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Igarashi K, Kashiwagi K. 1999. Polyamine transport in bacteria and yeast. Biochem J 344 Pt 3:633–642. [PMC free article] [PubMed] [Google Scholar]
- 64. Shah P, Swiatlo E. 2008. A multifaceted role for polyamines in bacterial pathogens. Mol Microbiol 68:4–16. doi: 10.1111/j.1365-2958.2008.06126.x [DOI] [PubMed] [Google Scholar]
- 65. Dzink JL, Socransky SS. 1990. Amino acid utilization by Fusobacterium nucleatum grown in a chemically defined medium. Oral Microbiol Immunol 5:172–174. doi: 10.1111/j.1399-302x.1990.tb00418.x [DOI] [PubMed] [Google Scholar]
- 66. Barnard JP, Stinson MW. 1999. Influence of environmental conditions on hydrogen peroxide formation by Streptococcus gordonii. Infect Immun 67:6558–6564. doi: 10.1128/IAI.67.12.6558-6564.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. North JA, Miller AR, Wildenthal JA, Young SJ, Tabita FR. 2017. Microbial pathway for anaerobic 5'-methylthioadenosine metabolism coupled to ethylene formation. Proc Natl Acad Sci U S A 114:E10455–E10464. doi: 10.1073/pnas.1711625114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. North JA, Sriram J, Chourey K, Ecker CD, Sharma R, Wildenthal JA, Hettich RL, Tabita FR. 2016. Metabolic regulation as a consequence of anaerobic 5-methylthioadenosine recycling in Rhodospirillum rubrum. mBio 7:e00855-16. doi: 10.1128/mBio.00855-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Erb TJ, Evans BS, Cho K, Warlick BP, Sriram J, Wood BM, Imker HJ, Sweedler JV, Tabita FR, Gerlt JA. 2012. A Rubisco like protein links SAM metabolism with isoprenoid biosynthesis. Nat Chem Biol 8:926–932. doi: 10.1038/nchembio.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Flavin M, Slaughter C. 1967. Enzymatic synthesis of homocysteine or methionine directly from O-succinyl-homoserine. Biochim Biophys Acta 132:400–405. doi: 10.1016/0005-2744(67)90158-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data from methionine stable isotope experiment.
Fig. S1, S2, S3, S4, S5, S6, and S7.
Supplement to the experimental procedure and an additional list of references.
Tables S1 and S2.
Data Availability Statement
This study is available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org, where it has been assigned Study ID ST002793. The data can be accessed directly via its Project DOI: http://dx.doi.org/10.21228/M8P126.