Summary
The type V-I CRISPR-Cas system is becoming increasingly more attractive for genome editing. However, natural nucleases of this system often exhibit low efficiency, limiting their application. Here, we used structure-guided rational design and protein engineering to optimize an uncharacterized Cas12i nuclease, Cas12i3. As a result, we developed Cas-SF01, a Cas12i3 variant that exhibits significantly improved gene editing activity in mammalian cells. Cas-SF01 shows comparable or superior editing performance compared to SpCas9 and other Cas12 nucleases. Compared to natural Cas12i3, Cas-SF01 has an expanded PAM range and effectively recognizes NTTN and noncanonical NATN and TTVN PAMs. In addition, we identified an amino acid substitution, D876R, that markedly reduced the off-target effect while maintaining high on-target activity, leading to the development of Cas-SF01HiFi (high-fidelity Cas-SF01). Finally, we show that Cas-SF01 has high gene editing activities in mice and plants. Our results suggest that Cas-SF01 can serve as a robust gene editing platform with high efficiency and specificity for genome editing applications in various organisms.
Graphical abstract
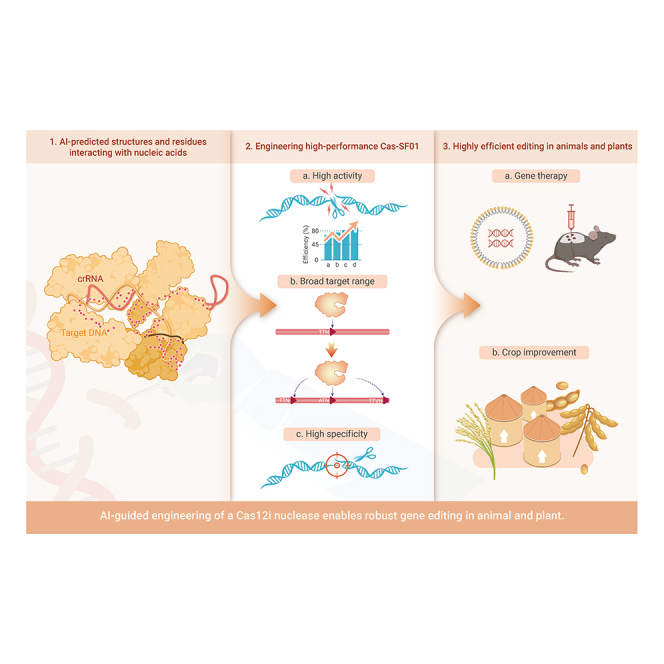
Public summary
-
•
AI-guided prediction of structures and residues interacting with nucleic acids.
-
•
Engineering highly active and specific Cas-SF01.
-
•
High editing efficiency in animals and plants.
Introduction
Over the past decade, CRISPR-Cas systems have been rapidly developed for gene editing in eukaryotes, contributing significantly to biomedical research, gene therapy, and plant and animal breeding.1,2 Beyond the well-established type II Cas9 system, an increasing number of previously unexplored Cas systems have been discovered, which possess significant potential for various gene editing applications. Although the natural Cas system often exhibits low activity, protein engineering has been successful in improving the performance of Cas nucleases, including enhancing editing efficiency, broadening protospacer adjacent motif (PAM) range, and reducing off-target effects.3,4
The Cas12i subfamily of nucleases, which belongs to the type V-I Cas system, has attracted attention due to the gene editing activities of some of its members as well as their smaller protein sizes in comparison to Cas9 and Cas12a.5 Structural studies have provided insights into the molecular mechanism of gene editing by Cas12i1 and Cas12i2, offering important information for optimizing Cas12i family nucleases.6,7,8 Recently, Zhang et al. identified a new Cas12i nuclease and generated a robust version, hfCas12Max, via protein engineering.9 Nevertheless, several Cas12i nucleases remain poorly characterized, providing opportunities for further development.
A recent report showed that Cas12i3 has gene editing activity in animal cells, but the activity is low.9 In this study, we used rational design and protein engineering to optimize Cas12i3, resulting in the development of a robust Cas12i3 variant, Cas-SF01. Cas-SF01 exhibits high gene editing efficiencies in mammalian cells, mice, and plants. In addition, we developed Cas-SF01HiFi, a high-fidelity version of Cas-SF01 that displays improved editing specificity.
Results
Identification of improved Cas12i3 variants with single arginine substitutions
The natural Cas12i3 contains 1,045 amino acids (aa), smaller than SpCas9 (1,368 aa) and similar to the previously characterized Cas12i2 (1,054 aa) (Figure S1A). Despite sharing only 29% identity with Cas12i2 in amino acid sequence, Cas12i3 has the same domain structure as Cas12i2 (Figures S1B and S1C). To assess the similarity between the 2 Cas12i nucleases, we used AlphaFold to predict the structure of Cas12i3 and compared it with a reported Cas12i2 structure6 in the presence of CRISPR RNA (crRNA). As expected, Cas12i3 shares significant structural similarity with Cas12i2 (root-mean-square deviation = 2.4) (Figure S1D).
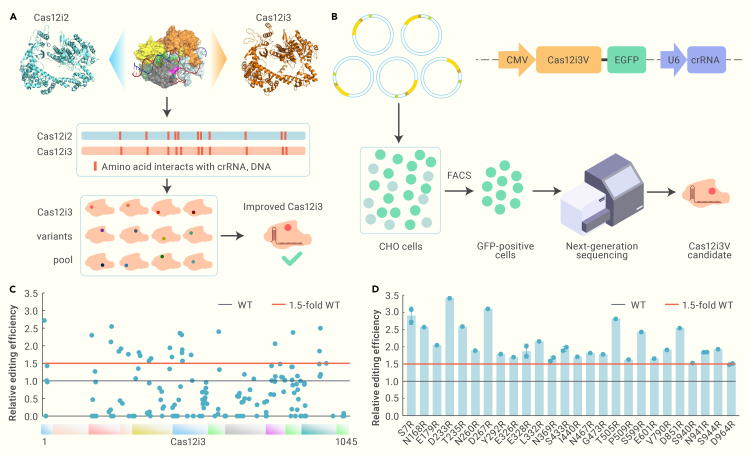
One strategy for improving the nuclease activity of a Cas protein is to enhance its interaction with nucleic acids.3,4,8 Based on the reported protein structure of Cas12i2,6 we identified corresponding residues in Cas12i3 that potentially mediate the interaction of Cas12i3 with crRNA and substrate DNA (Figure S1E). To enhance the electrostatic interactions between these residues with the negatively charged nucleic acids, we sought to substitute them with the positively charged arginine (R). To this end, we constructed a pool of more than 150 Cas12i3 variants, each containing a single R substitution, for subsequent screening (Figure 1A).
Figure 1.
Identification of single aa substitutions that improve Cas12i3 gene editing activity
(A) Constructing a pool of Cas12i3 variants based on its structural similarity with Cas12i2. The crRNA and DNA-interacting sites of Cas12i3 were predicted by comparing its protein structure with that of Cas12i2. Then, these residues were individually substituted with arginine, generating a pool with over 150 variants for identification of improved Cas12i3 variants.
(B) Screening strategy for improved Cas12i3 variants. Plasmids carrying Cas12i3 variants (Cas12i3V) fused with EGFP-tag and crRNA targeting the FUT8 gene were transfected into CHO cells. Then, EGFP+ cells were selected by FACS, and the editing efficiency of Cas12i3 variants was determined by deep sequencing.
(C) The editing efficiency of Cas12i3 variants. Black dotted line, the editing efficiency of wild-type Cas12i3; red dotted line, 1.5-fold of the editing efficiency of Cas12i3.
(D) The editing efficiency of 26 Cas12i3 variants with improved editing performance from the screen.
To screen for Cas12i3 variants with increased editing efficiency, we cloned each Cas12i3 variant into an all-in-one expression system, in which the Cas protein was fused with an EGFP tag, and an U6 promoter-driven crRNA targeting the FUT8 gene was co-expressed from the same construct (Figure 1B). We then transfected these constructs into Chinese hamster ovary (CHO) cells and selected GFP+ cells by fluorescence-activated cell sorting (FACS). Editing efficiency was determined by next-generation sequencing (NGS) of the target site (Figure 1B). The results showed that wild-type Cas12i3 exhibits a relatively low editing efficiency, averaging ∼30%, and more than 50% of the Cas12i3 variants performed similarly as the wild-type Cas12i3, with fewer than 50% changes in activity (Figure 1C). We selected 26 Cas12i3 variants that showed an editing efficiency exceeding 1.5-fold of that of the wild-type Cas12i3 for further validation using the same assay. The validation assay identified 8 variants, namely S7R, N168R, D233R, T235R, D267R, T505R, S599R, and D851R, that substantially improved the editing performance of Cas12i3 in CHO cells (Figure 1D).
Combinations of aa substitutions generate a highly efficient Cas12i3 variant, Cas-SF01
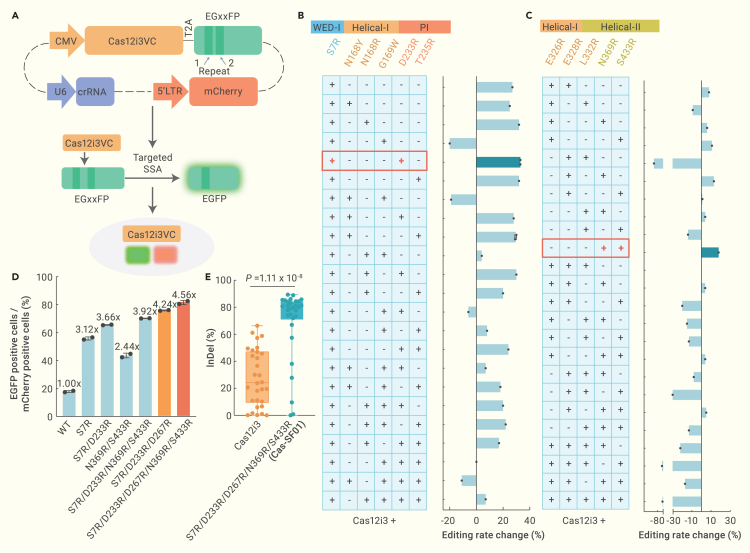
Next, we sought to determine whether combinations of the aforementioned single aa variants can further improve the gene editing efficiency of Cas12i3. Since these mutations were located in 2 regions of Cas12i3, the region that interacts with the PAM duplex and the region that interacts with the target DNA strand, we decided to first examine the combinations of mutations within each region.
For the PAM-interacting region, we selected 4 mutations from the initial screen, namely S7R, N168R, D233R, and T235R, which are located in the WED-I, Helical-I, or PI domain of the REC lobe surrounding the PAM duplex (Figures 2B and S2A–S2D). Inspired by a previous report on the engineering of Cas12i2,8 we also included another two mutations, N168Y and G169W. Given that the S7R mutation exhibited the most robust editing performance in the initial screen (Figure 1D), we decided to combine it with the other mutations and constructed a mutation library containing 23 combinations. The editing efficiency of each combination was determined using the EGxxFP reporter system, in which the EGFP fluorescence signal can be detected only when the target region inside the EGFP is cleaved by the nuclease10 (Figure 2A). Of the 23 combinations, 18 showed increased editing rates compared to wild-type Cas12i3. Among these, Cas12i3-S7R/D233R outperformed the rest (Figure 2B).
Figure 2.
Cas-SF01 engineering via a 2-step combined strategy
(A) Schematics of the EGxxFP reporter system for screening for high-efficiency Cas12i3 variants carrying combinations of mutations (Cas12i3VC). A short sequence is inserted inside the EGFP to disrupt its fluorescence. Targeted cleavage in the inserted sequence restores EGFP fluorescence due to DNA repair through single-strand annealing (SSA) between 2 flanking repeats. The system also constitutively expresses an mCherry marker. Editing efficiency is assessed by determining the ratio of EGFP+ cells over mCherry+ cells.
(B and C) The editing efficiencies of Cas12i3 with different mutation combinations for the PAM-interacting region (B) and target DNA strand-interacting region (C), as determined by the EGxxFP reporter. The mutation combinations with the highest editing rate increase in the 2 regions are highlighted in red boxes.
(D) Further combination of mutations identified from the screens results in variants with higher editing efficiency. The variant with highest editing efficiency contains 5 mutations (S7R/D233R/D267R/N369R/S433R) and was called Cas-SF01.
(E) Comparison of indel activity of Cas12i3 and Cas-SF01 at the TTR locus in mammalian cells. Significant difference was determined by Student’s t test.
For the substrate-interacting region, we selected 5 mutations from the initial screen (E326R, E328R, L332R, N369R, and S433R), which are located in the second Helical-I and Helical-II domains (Figures 2C, S2A, S2E, and S2F). We determined the editing efficiency of 24 combinations using the EGxxFP reporter system and found that the N369R/S433R variant had the highest increase in editing activity (Figure 2C).
We next combined the mutations from the two regions and found that the combination of S7R/D233R/N369R/S433R outperformed other combinations (Figure 2D). Interestingly, adding another mutation identified from the initial screen, D267R, which is located in the PI domain next to the PAM duplex (Figures S2B and S2D), further improved the editing performance. We named this variant (Cas12i3-S7R/D233R/D267R/N369R/S433R) Cas-SF01, which exhibits the highest gene editing efficiency among all of the Cas12i3 variants tested (Figure 2D).
Cas-SF01 exhibits high editing efficiency and a broadened PAM range
To validate the performance of Cas-SF01 in editing endogenous genes, we designed 30 targets in the exon of the TTR gene. The results showed that Cas-SF01 exhibited ∼3-fold increased activity in generating indel mutations compared to the wild-type Cas12i3 (Figure 2E).
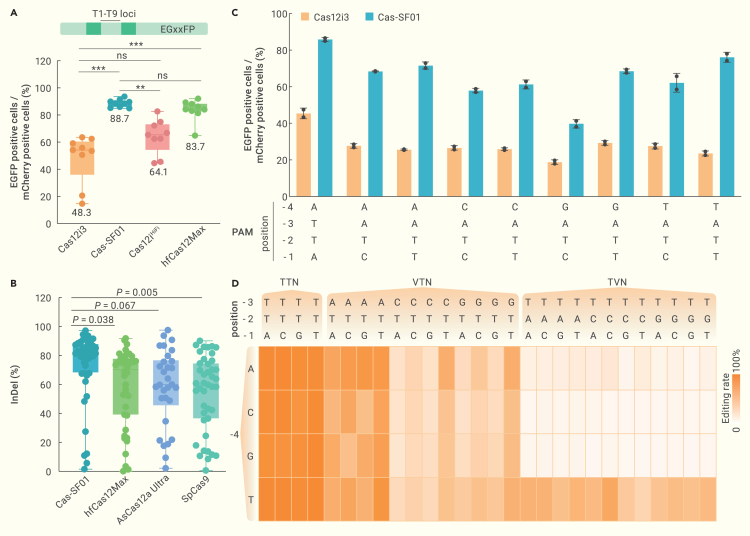
We next assessed the editing activity of Cas-SF01 in comparison with other Cas nucleases. Two previously engineered Cas12i nucleases, Cas12iHiFi 8 and hfCas12Max,9 were introduced into the EGxxFP reporter system, and their editing activities were determined at 9 target sites (T1–T9) in EGxxFP (Figure 3A). The results showed that Cas-SF01 exhibited an average editing efficiency of 88%, higher than the 64% editing efficiency of Cas12iHiFi and comparable to the 83% editing efficiency of hfCas12Max, whereas the wild-type Cas12i3 displayed an editing efficiency averaging 48% (Figure 3A). To further evaluate the efficiency of Cas-SF01 in editing endogenous loci, we designed over 40 target sites in the exons of PCSK9, TTR, and CCR5 genes and compared its performance to that of hfCas12Max as well as 2 widely used nucleases, AsCas12a Ultra and SpCas9. Our results show that Cas-SF01 exhibited an average indel rate of ∼80%, surpassing the indel rate of hfCas12Max (∼70%), AsCas12a Ultra (∼60%), and SpCas9 (∼60%) at the tested sites (Figure 3B). These findings demonstrated that Cas-SF01 is a highly efficient Cas nuclease for genome editing in mammalian cells.
Figure 3.
Cas-SF01 displays high editing efficiency and a broadened PAM range
(A) Cas-SF01 exhibits higher cleavage activity than wild-type Cas12i3, Cas12iHiFi, and hfCas12Max in the EGxxFP-reporter system. T1–T9 loci, target sites in the inserted fragment of the EGxxFP cassette. ∗∗∗p < 0.001; ∗∗p < 0.01; ns, not significant (Student’s t test).
(B) Comparison of indel activity of Cas-SF01 with SpCas9, AsCas12a Ultra, and hfCas12Max at multiple target sites in the PCSK9, CCR5, and TTR loci. Significant difference was determined by Student’s t test.
(C) Cas-SF01 shows improved recognition of targets with an NATN PAM compared with wild-type Cas12i3.
(D) Heatmap showing the PAM recognition profile of Cas-SF01.
To compare PAM recognition between Cas-SF01 and wild-type Cas12i3, we first conducted an NGS-based PAM identification assay in Escherichia coli. We found that both nucleases preferred TTN PAM (Figure S3), which is the canonical PAM sequence for the Cas12 family. Interestingly, Cas-SF01 showed an increased recognition of the ATN PAM (Figure S3). To further validate this finding, we determined the editing efficiency of both nucleases at 8 target sites harboring NATN PAMs using the EGxxFP reporter system. The results showed that Cas-SF01 exhibited substantially higher editing rates than the wild-type Cas12i3 at all sites (Figure 3C). Finally, to obtain a comprehensive understanding of the PAM preferences of Cas-SF01, we modified the PAM of the same spacer in the EGxxFP reporter system and determined the editing efficiencies. We found that Cas-SF01 effectively recognized targets with canonical NTTN PAMs, with an average editing efficiency of 90% (Figure 3D). Moreover, Cas-SF01 was also effective in recognizing targets with NATN or TTVN PAMs, showing that a T at the −4 position largely improved the cleavage activity of Cas-SF01 toward NTVN PAMs (Figure 3D). These results demonstrated that Cas-SF01 has a broadened range of PAM recognition compared to the wild-type Cas12i3.
Deep sequencing of the target sites further revealed that Cas-SF01 predominantly generated deletions (Figures S4A–S4C). The mutations generated by Cas-SF01 ranged from single nucleotides to fragments with more than 10 nucleotides (Figures S4D–S4F).
The D876R mutation improves the specificity of Cas-SF01
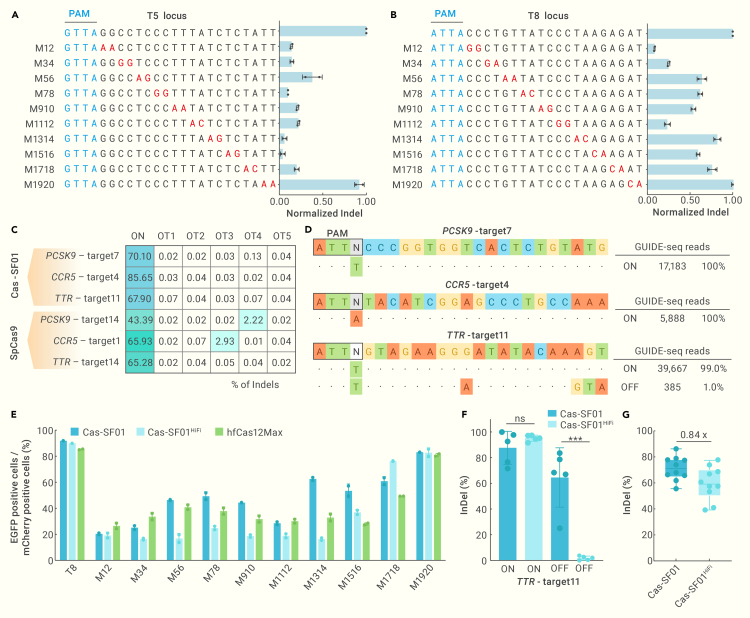
To comprehensively assess the editing specificity of our engineered Cas-SF01, we evaluated its tolerance to mismatches between spacers and targets. We designed 10 off-target spacers harboring 2 adjacent mismatched nucleotides for each target site in the EGxxFP reporter system, and determined their editing efficiencies. For the T5, T6, and T7 sites, the mismatches at positions 1 to 18 substantially reduced the editing activity of Cas-SF01, and comparable editing rates were observed at spacers harboring mismatches at positions 19 and 20, indicating that mismatches close to the PAM have a more substantial impact on Cas-SF01 activity (Figures 4A and S5). However, we observed some degree of mismatch tolerance at the T8 site, suggesting potential opportunities for optimization to improve specificity (Figure 4B).
Figure 4.
Analysis of the specificity of Cas-SF01 and Cas-SF01HiFi
(A and B) The mismatch tolerance of Cas-SF01 was examined by designing dinucleotide mutations at T5 (A) and T8 (B) targets, and then the activation of EGFP was determined.
(C) Off-target rates of Cas-SF01 and SpCas9 at in silico predicted off-target sites, as determined by targeted deep sequencing.
(D) The genome-wide off-target effects of Cas-SF01 as analyzed by GUIDE-seq. The numbers of on-target and off-target reads are shown at right.
(E) The mismatch tolerance of Cas-SF01, Cas-SF01HiFi, and hfCas12Max at the T8 locus.
(F) The indel activities of Cas-SF01 and Cas-SF01HiFi at the on-target site TTR-target11 (ON) and off-target site (OFF) shown in (D). ∗∗∗p < 0.001; ns, not significant (Student’s t test).
(G) Comparison of indel activity of Cas-SF01 with Cas-SF01HiFi at 10 target sites at CCR5 locus. Each dot represents 2 replicates.
We further evaluated the specificity of Cas-SF01 in mammalian cells by performing targeted deep sequencing at the in silico predicted off-target sites. We selected three targets at the PCSK9, CCR5, and TTR loci and determined the editing rates of Cas-SF01 and SpCas9 at the target sites and 5 predicted off-target sites. Targeted deep sequencing showed that the on-target editing efficiency of Cas-SF01 was higher than that of SpCas9, and Cas-SF01 showed remarkably low off-target effects at the potential off-target sites. Notably, a potential off-target effect was found at PCSK9-target14 OT4 and CCR5-target1 OT3 sites for SpCas9 (Figures 4C and S6). In addition, we performed GUIDE-seq to assess the genome-wide off-target effect of Cas-SF01. As the results showed, only on-target reads could be detected for Cas-SF01 at PCSK9-target7 and CCR5-target4 (Figure 4D). For TTR-target11, ∼1% of the reads were from an off-target that has a mismatch of 3 nucleotides at the tail of crRNA (Figure 4D). Together, these results demonstrated that Cas-SF01 is highly specific in genome editing.
Considering that several single aa mutations in the RuvC-II domain could enhance editing specificity according to a previous study,9 we engineered single aa substitutions at every residue of the RuvC-II domain, generating a variant pool of Cas-SF01 for screening for a high-fidelity version (Figure S7A). Of the 80 variants tested, we selected 15 variants with negligible or no loss of on-target editing activity at the T5 site in the EGxxFP reporter system for subsequent examination of their off-target effects (Figure S7B). We evaluated the mismatch tolerance of the selected variants using two designed spacers, M78 and M910, at the T8 site. A single variant, D876R, in Cas-SF01 (referred to as Cas-SF01HiFi), considerably reduced the mismatch tolerance of Cas-SF01 (Figures S7C and S7D). To better evaluate the effect of D876R on Cas-SF01, we compared the mismatch tolerance of Cas-SF01, Cas-SF01HiFi, and hfCas12Max at the T8 site. Cas-SF01HiFi showed reduced tolerance to mismatches from sites 3 to 16, which is comparable to hfCas12Max (Figure 4E). We compared the indel rates of Cas-SF01 and Cas-SF01HiFi at the on-target site TTR-target11 and the off-target site discovered by the above GUIDE-seq analysis. The results show that although both Cas-SF01 and Cas-SF01HiFi displayed high on-target editing efficiencies, Cas-SF01HiFi had a dramatically reduced editing efficiency at the off-target site (Figure 4F), supporting the idea that D876R is important for enhancing the editing specificity of Cas-SF01. Furthermore, we analyzed the effect of D876R on the editing activity of Cas-SF01 at endogenous genes and found that Cas-SF01HiFi maintained nearly 85% of the on-target editing efficiency of Cas-SF01 (Figure 4G), suggesting that Cas-SF01HiFi is valuable for future applications that require high efficiency and specificity (Figure S7E).
Cas-SF01 is efficient in gene editing in mice and plants
The robust genome editing performance of Cas-SF01 in cultured mammalian cells prompted us to evaluate its in vivo gene editing performance in animals. We used lipid nanoparticles (LNPs) to deliver in vitro transcribed (IVT) Cas-SF01 mRNA and chemically synthesized crRNA to the livers of C57 mice through tail intravenous injection (Figure 5A). We designed a crRNA targeting the third exon of the murine transthyretin gene Ttr and assessed the gene editing efficiencies using the unmodified crRNA, along with 6 modified variants containing modifications at the 5′-handle and/or the 3′-terminus (M1–M6) in mice (Figures S8A and S8B). Among these, the crRNA-M3, which bears modifications at the 5′-handle (a phosphorothioate [PS]-modified C(+3)) and the 3′-terminus (PS-U(+3), U(+1)), demonstrated superior performance compared to the other crRNAs, achieving an indel rate of ∼33% in mouse liver (Figure S8C). We next compared the in vivo editing efficiencies of Cas-SF01 to hfCas12Max and SpCas9 and found that Cas-SF01 exhibited an average indel rate of 34%, whereas the indel rates of hfCas12Max and SpCas9 were ∼31.6% and ∼36.1%, respectively (Figure 5B). We measured the serum TTR protein level in the livers of these edited mice using ELISA (Figure 5C). The serum TTR protein level in mice edited with Cas-SF01 was reduced to an average of 27.8% compared to PBS-treated mice, whereas the levels were reduced to ∼26.6% and ∼31.3% in mice edited with SpCas9 and hfCas12Max, respectively (Figure 5C). Taken together, these results show that Cas-SF01 could mediate robust in vivo gene editing in mice.
Figure 5.
Cas-SF01 enables robust genome editing in animal and plants
(A) Schematic of the in vivo gene editing using LNP delivery of IVT-Cas mRNA and chemically synthesized target crRNA.
(B) Indel frequencies of Cas-SF01 and hfCas12Max, SpCas9 at Ttr locus in C57 mice. Cas-SF01 and hfCas12Max, n = 5; SpCas9, n = 3.
(C) Serum TTR protein levels. Cas-SF01 and hfCas12Max, n = 5; PBS and SpCas9, n = 3.
(D–F) Editing efficiencies of Cas-SF01 at various target sites in E0 plants of rice (D), hairy root lines of pepper (E), and soybean (F). Each dot represents the editing efficiency of 1 target. ∗∗∗p < 0.001; ∗p < 0.05; Student’s t test.
To further explore the potential applications of Cas-SF01, we evaluated its editing performance in plants. We used Agrobacterium tumefaciens–mediated stable transformation in rice and A. rhizogenes–mediated hairy root transformation in pepper (Capsicum annuum) and soybean to test the editing performance of Cas-SF01 (Figures 5D–5F and S9). Cas-SF01 mediated significantly higher indel mutation frequencies at most of the designed target sites compared to the wild-type Cas12i3 (Figures 5D–5F). In rice, wild-type Cas12i3 mediated an editing rate averaging 15.5% at 12 target sites, whereas the engineered Cas-SF01 exhibited a 5-fold increase, with editing efficiency reaching 77.2% (Figure 5D). We selected 3 target sites, OsGS3, OsERF922, and OsDjA2, to analyze editing outcomes in E0 plants. Cas-SF01 generated more homozygous, biallelic, and chimeric editing events than Cas12i3 (Figure S9A). Pepper is a popular vegetable worldwide. Cas-SF01 edited all four target genes in pepper, whereas no editing activity of wild-type Cas12i3 could be detected in transgenic hairy roots at any of the targets (Figure 5E). The editing events caused by Cas-SF01 in pepper were primarily biallelic or heterozygous (Figure S9B). In the important crop plant soybean, we compared the editing activities of engineered Cas-SF01 with Cas12i3 and Cas9. Similar to the results in pepper, we could not detect the editing activity of Cas12i3 in soybean hairy roots (Figure 5F). Our engineered Cas-SF01 exhibited an average editing rate of 52.5%, which is approximately twice that of the editing rate of Cas9 (∼26.8%) (Figures 5F and S9C). These results suggest that Cas-SF01 is a very effective genome-editing tool for plant research and breeding.
Discussion
In this study, we demonstrated the effectiveness of the type V-I Cas12i3 system for efficient genome editing in animals and plants. Through structure prediction, structure-guided rational design, and combinatorial protein engineering, we successfully optimized the natural Cas12i3, previously characterized with low editing activity, into a highly efficient gene-editing tool with a broadened PAM range, which we named Cas-SF01. The focused screening of targeted mutation pools and combinatorial engineering strategy based on predicted protein structure would be applicable for optimizing other natural Cas nucleases with low editing activity. Compared to the wild-type Cas12i3, Cas-SF01 contains five mutations—S7R, D233R, D267R, N369R, and S433R— which were introduced to enhance interactions with the guide RNA (gRNA) and the DNA substrate, including the PAM duplex. The editing efficiency of Cas-SF01 is comparable to or greater than that of SpCas9, AsCas12a Ultra, and 2 other recently engineered Cas12i nucleases (Cas12iHiFi and hfCas12Max) at the tested sites.
In addition to the increased editing activity, Cas-SF01 showed an expanded PAM recognition compared to the wild-type Cas12i3. In particular, Cas-SF01 showed increased recognition of the noncanonical 5′-ATN-3′ PAM (Figures 3C and 3D), thus enabling more versatile genome engineering. Moreover, Cas-SF01 showed low off-target effects in mammalian cells (Figures 4C and 4D). By introducing the D876R mutation in the RuvC-II domain, we engineered a Cas-SF01 variant with even higher fidelity, which we called Cas-SF01HiFi (Figures 4E and S6).
Several features of the Cas-SF01 system make it suitable for multiplexed in vivo genome editing. First, Cas-SF01 has a smaller protein size compared to the widely used larger Cas nucleases such as Cas9 (Figure S1A), which may be advantageous for delivery in vivo. Second, Cas-SF01 uses shorter crRNA guides that are less costly and simpler to design for multiple targets. In addition, like other Cas12 proteins, Cas-SF01 may be capable of processing long pre-crRNA transcripts into mature crRNA guides, thus facilitating simultaneous expression of multiple guides from a single RNA transcript.5 In this study, we tested Cas-SF01 in mice via LNP delivery and observed robust in vivo gene editing comparable to SpCas9 or hfCas12Max, even though there was clear room for mRNA and gRNA optimization to further improve the in vivo editing efficiencies (Figures 5B and 5C). Furthermore, we tested Cas-SF01 in important crop plants, including the monocot rice and dicot pepper and soybean, and revealed its potential application in plant research and molecular breeding in agriculture (Figures 5D and 5E).
Overall, our work outlines an effective strategy for optimizing natural Cas nucleases—in other words, targeted screening of pools of arginine mutations in the PAM, substrate DNA, and gRNA interaction domains, and demonstrated Cas-SF01 and Cas-SF01HiFi as efficient tools for genome editing applications in diverse organisms.
Materials and methods
Homology analysis of CRISPR-Cas12i
The protein structure of Cas12i3 was predicted by AlphaFold2 with default settings.11 We compared the top 5 ranked outputs and selected rank 0 for further analysis. The protein structures of Cas12i2 (PDB: 6LTU)6 and Cas12i3 were aligned with PyMol software version 2.4.0 using the default parameters. The sequence of Cas-SF01 is listed in Table S1.
Plasmid vector construction
All PCRs were performed with FastPfu Fly DNA Polymerase (TransGen Biotech, AP231-13). Human codon–optimized Cas12i3 genes were synthesized by Beijing Tsingke Biotech, with sequences listed in Table S1, and cloned into the vectors by Gibson Assembly (ClonExpress Ultra One Step Cloning Kit, Vazyme, C115-02). crRNA oligos were synthesized by Beijing Tsingke Biotech, annealed, and ligated into the BbsI (NEB) site in the same all-in-one vector. The primers are listed in Table S2.
Cell culture, transfection, and flow cytometry analysis
HEK293T and CHO cells were cultured in high glucose DMEM supplemented with 10% (v/v) fetal bovine serum (Gibco, 10099141) and 1% penicillin/streptomycin (Gibco, 15140122) at 37°C with 5% CO2.
HEK293T cells were seeded into 12-well plates and transfected at ∼80% confluency. For all of the transfections, 1.5 μg plasmid was mixed with 100 μL Opti-MEM (Gibco, 51985091) and then combined with a solution comprising 100 μL Opti-MEM and 2 μL Lipofectamine 2000 (Invitrogen, 11668-019). The resulting DNA/Lipofectamine mixture was then added to the cells after incubating for 15 min at room temperature. Twenty-four hours after transfection, 1,000 μL of DMEM was exchanged in each transfected well. Cells were then incubated for 24 additional hours before being harvested for flow cytometry.
Transfected cells of each well were digested with 300 μL 0.25% trypsin (Gibco, 25200-056) after removing the medium. Then, 300 μL of medium was added to terminate the digestion, and the cells were resuspended in 500 μL of medium after 3 min of centrifugation at 100 rpm. Cells were then pipetted into a 5-mL round-bottom polystyrene test tube with a cell strainer snap cap (Corning, 352235) and kept on ice. Flow cytometry data were collected using a CytoFLEX (Beckman Coulter Life Sciences) and analyzed using CyExpert version 2.5.0.77 software. For the assay of mutations in the target sites of endogenous genes, cells were sorted using BD FACSAria SORP.
Scatter gates were applied to remove nonviable cells and doublets. For positive experiments, gates were applied based on cells transfected with both mCherry and GFP signals. mCherry fluorescence was detected using Y610 (561/610 nm). GFP fluorescence was detected using B525 (488/525 nm). Approximately 50,000 cells (after scatter gating) were collected for each sample. The editing efficiency was calculated as the number of GFP+ cells divided by the number of mCherry+ cells.
NGS
Deep sequencing was applied to detect the editing efficiency of Cas nucleases at endogenous genes. In all, ∼50,000 sorted HEK293T cells were concentrated to 100 μL and used as a template for targeted PCR using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, P505). The sequences of the primers used for amplification are listed in Table S2. Targeted deep sequencing analysis (Beijing Tsingke Biotech) was used to determine indel frequencies.
Analysis of PAM recognition profile of Cas-SF01
The PAM preference of wild-type Cas12i3 and Cas-SF01 was analyzed using an E. coli PAM screen assay, as previously described,12 and normalized and analyzed via Weblogo.13 To verify the recognition of 5′-ATN-3′ PAM of Cas-SF01, we designed 9 specific targets with ATN PAM in the EGxxFP reporter system and determined the editing activity of Cas12i3 and Cas-SF01. We also modified the PAM of T8 locus in EGxxFP reporter system with NTTN, NVTN, and NTVN types to obtain a comprehensive PAM recognition profile of Cas-SF01.
Mismatch tolerance of Cas-SF01
The T5, T6, T7, and T8 targets in the fluorescence reporter vector were selected to construct mismatch crRNA vectors with 2 adjacent base pairs as the window to analyze the mismatch tolerance of Cas-SF01 and characterize its off-target rate. The mutation principles were A to C, T to A, C to G, and G to A.
GUIDE-seq
The off-target effect of Cas-SF01 was analyzed by GUIDE-seq as previously described, with some modifications.14 Briefly, 4.5 μg all-in-one plasmid for each target site, expressing Cas-SF01 and designed crRNA, and 1 nM of annealed GUIDE-seq oligonucleotides were transfected into HEK293T cells (2 × 106) by electroporation (Lonza, V4XC-2012). The electroporation was performed by the 4D-Nucleofector Core Unit using program CM-130, and each sample was repeated 3 times and then cultured in 1 plate. Cells were then collected at ∼90% confluency after transfection and stored at −80°C after liquid nitrogen flash freezing. Identified sequences with less than 6 nucleotide mismatches of on-target sequence were recognized as off-target sites. The sequencing data were collected and analyzed by DIATRE.
Screening for Cas-SF01 variants with reduced mismatch tolerance
The aa 830–914 in the RuvC-II domain of Cas-SF01 were mutated to arginine residues, generating a variant pool containing 80 variants. The editing efficiency of variants was analyzed in the EGFP fluorescence reporter system targeting T5 to first isolate variants with activity loss of less than 5%, and then the off-target rate of the selected variants was determined.
LNP delivery and in vivo editing in mice
The LNP system with a composition of SM-102, DMG-PEG2000, DSPC and cholesterol (molar ratio 50:1.5:10:38.5) was prepared, carrying IVT mRNA of Cas nucleases and chemically synthesized RNA oligonucleotides (Genscript) with a 1:1 weight ratio. LNPs were formed according to the manufacturer’s protocol (Micro & Nano, INanoE). After formation, LNPs were diluted immediately with PBS, and ultrafiltration was carried out with 10 kDa centrifugal filters (Millipore, catalog no. UFC901024). For in vivo editing, LNPs encapsulating the RNAs were administrated to C57BL/6J female mice via tail intravenous injection (3 mg/kg). Liver tissues were rinsed with physiological saline and then collected from the left lateral lobe of each mouse 7 days postinjection for DNA extraction and targeted deep sequencing analysis. All of the animal protocols were approved by the Animal Care and Use Committee of Southern University of Science and Technology (SUSTech-SL202211002). This study was carried out in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health.
ELISA
For analysis of serum TTR protein concentration, serum samples were obtained before the collection of the mouse liver tissue, and the TTR protein was determined using the Mouse Transthyretin ELISA kit (BYabscience catalog no. BY-EM221426) following the manufacturer’s protocol.
Plant transformation
The stable transformation of rice was performed as previously described.15 Specific crRNAs targeting the genes of interest in rice were designed and inserted into an all-in-one construct. Then, the constructs were transfected into A. tumefaciens strain EHA105 for transformation using calli from the elite rice variety Xiushui 134. Four weeks after transplanting the regenerated rice plants, the genomic DNA of the rice plants was extracted for PCR amplification and sequencing. Transformation of pepper (C. annuum) using A. rhizogenes was performed as described.16 Briefly, the cotyledons of 7-day-old peppers were cut into 2- to 3-mm strips and incubated with A. rhizogenes strain C58C1 carrying the indicated vector. The hair roots regenerated from the cotyledon were collected for genotyping. Hairy root transformation of soybean using A. rhizogenes was carried out as previously described, with some modifications.17 Briefly, the hypocotyls of 2-week-old soybean seedlings were cut diagonally with a sterile scalpel and applied with A. rhizogenes strain K599 containing the expression construct. Then, the hypocotyl was planted under high humidity conditions for 16 days to promote hairy root growth. The primers used to edit genes in plants are listed in Table S2.
Data and code availability
NGS data of GUIDE-seq were deposited at the NCBI’s GEO server under accession number GSE236755. All of the data generated or analyzed during this study are included in this published article and supplementary information files.
Acknowledgments
This work was supported by Shandong BellaGen Biotechnology, and by a grant from the National Natural Science Foundation of China (32188102 to J.-K.Z.).
Author contributions
J.-K.Z. conceived the project; Z.D., Y.L., Y.H., and J.-K.Z. designed the experiments and analyzed the data; Z.D., Y.L., H.Z, T.L., R.L., Y.C., S.G., N.J., and Q.C. performed the plasmid construction and cell culture experiments and the FACS and DNA extraction; J.S., P.L., and M.W. carried out the LNP delivery and ELISA assay; H.X., M.Z., and M.X. performed the soybean transformation; K.Z. and W.S. performed the rice transformation; S.W. and Y.J. performed the pepper transformation; N.L. performed the genotyping analysis in rice; Y.H., R.T., and J.-K.Z. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published Online: January 8, 2024
Footnotes
It can be found online at https://doi.org/10.1016/j.xinn.2024.100564.
Lead contact website
Supplemental information
References
- 1.Wang J.Y., Doudna J.A. CRISPR technology: A decade of genome editing is only the beginning. Science. 2023;379:eadd8643. doi: 10.1126/science.add8643. [DOI] [PubMed] [Google Scholar]
- 2.Doudna J.A. The promise and challenge of therapeutic genome editing. Nature. 2020;578:229–236. doi: 10.1038/s41586-020-1978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strecker J., Jones S., Koopal B., et al. Engineering of CRISPR-Cas12b for human genome editing. Nat. Commun. 2019;10:212. doi: 10.1038/s41467-018-08224-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kleinstiver B.P., Sousa A.A., Walton R.T., et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019;37:276–282. doi: 10.1038/s41587-018-0011-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan W.X., Hunnewell P., Alfonse L.E., et al. Functionally diverse type V CRISPR-Cas systems. Science. 2019;363:88–91. doi: 10.1126/science.aav7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang X., Sun W., Cheng Z., et al. Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2. Nat. Commun. 2020;11:5241. doi: 10.1038/s41467-020-19072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B., Luo D., Li Y., et al. Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1. Nat. Commun. 2021;12:3476. doi: 10.1038/s41467-021-23876-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Y., Hu Y., Wang X., et al. Synergistic engineering of CRISPR-Cas nucleases enables robust mammalian genome editing. Innovation. 2022;3:100264. doi: 10.1016/j.xinn.2022.100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H., Kong X., Xue M., et al. An engineered xCas12i with high activity, high specificity, and broad PAM range. Protein Cell. 2023;14:538–543. doi: 10.1093/procel/pwac052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., Liu S., Cheng Y., et al. Highly Efficient and Rapid Detection of the Cleavage Activity of Cas9/gRNA via a Fluorescent Reporter. Appl. Biochem. Biotechnol. 2016;180:655–667. doi: 10.1007/s12010-016-2122-8. [DOI] [PubMed] [Google Scholar]
- 11.Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zetsche B., Gootenberg J.S., Abudayyeh O.O., et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crooks G.E., Hon G., Chandonia J.M., et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai S.Q., Zheng Z., Nguyen N.T., et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiei Y., Ohta S., Komari T., et al. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 16.Aarrouf J., Castro-Quezada P., Mallard S., et al. Agrobacterium rhizogenes-dependent production of transformed roots from foliar explants of pepper (Capsicum annuum): a new and efficient tool for functional analysis of genes. Plant Cell Rep. 2012;31:391–401. doi: 10.1007/s00299-011-1174-z. [DOI] [PubMed] [Google Scholar]
- 17.Kereszt A., Li D., Indrasumunar A., et al. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007;2:948–952. doi: 10.1038/nprot.2007.141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS data of GUIDE-seq were deposited at the NCBI’s GEO server under accession number GSE236755. All of the data generated or analyzed during this study are included in this published article and supplementary information files.