Abstract
The protein kinase Snf1/AMPK plays a central role in carbon and energy homeostasis in yeasts and higher eukaryotes. To work out which aspects of the Snf1-controlled regulatory network are conserved in evolution, the Snf1 requirement in galactose metabolism was analyzed in the yeast Kluyveromyces lactis. Whereas galactose induction was only delayed, K. lactis snf1 mutants failed to accumulate the lactose/galactose H+ symporter Lac12p in the plasma membran,e as indicated by Lac12-green fluorescent protein fusions. In contrast to wild-type cells, the fusion protein was mostly intracellular in the mutant. Growth on galactose and galactose uptake could be restored by the KHT3 gene, which encodes a new transporter of the HXT subfamily of major facilitators These findings indicate a new role of Snf1p in regulation of sugar transport in K. lactis.
The heterotrimeric protein kinase Snf1p is a central regulator of carbon homeostasis in yeasts and is structurally and functionally related to the mammalian AMP-activated protein kinase. An ancient function that emerges for this class of conserved kinases is a key role in adaptation to energy limitation. Energy-consuming anabolic pathways are downregulated under these conditions and energy is generated by induction of catabolism (reviewed in references 6, 17, and 18). In the yeast Saccharomyces cerevisiae, Snf1p is essential for the reprogramming of gene expression under carbon limitation. The derepression of glucose-repressed genes has been studied extensively and serves as a model of Snf1p signaling to the nucleus (7).
Two classes of genes that are upregulated in response to Snf1p have been described. The first class is controlled by transcription factors Cat8p and Sip4p, which bind to carbon source-responsive elements and are targets of the Snf1p kinase. Among them are genes involved in gluconeogenesis, the glyoxylate cycle, carbon transport, and acetyl-coenzyme A synthesis (19, 20). The second class is subject to glucose repression via the Mig1-Ssn6-Tup1 repressor complex. Mig1p is a target of Snf1p, and Snf1-mediated phosphorylation regulates cellular compartmentation of Mig1p (11) and interaction with Ssn6-Tup1 (31). Among the Mig1p-regulated genes are those involved in the utilization of alternative sugars, of which the SUC2 gene and Gal4p-controlled galactose regulon (GAL regulon) have been studied most extensively (7, 26).
Saccharomyces cerevisiae has an exceptional preference for fermentative metabolism, and it is not clear in how far its strategies of carbon regulation and Snf1p signaling can serve as a model for other organisms. Even among yeasts, great differences in physiology exist, like the Crabtree effect, the Kluyver effect, and the petite character, that have been used for taxonomic classifications. Comparative studies between the petite-negative, Crabtree-negative yeast Kluyveromyces lactis, for which genome sequencing has been completed, and S. cerevisiae (petite-positive, Crabtree-positive) are being undertaken to provide insight into the genetic basis of such differences in physiology.
Given the broader substrate spectrum and the limited utilization of glucose under aerobic conditions in K. lactis (4), we are interested in elucidating the conserved aspects of the Snf1p-controlled regulatory networks. As shown before, homologues of the Snf1p-controlled transcription factors Cat8p, Sip4p, and Mig1p exist in K. lactis. However, their contribution to the regulation of carbon and energy metabolism differs from that in S. cerevisiae (8, 9, 15, 27). For example, the gluconeogenesis genes and the sucrose-degrading inulinase gene are subject to glucose repression, but neither K. lactis Cat8p nor K. lactis Mig1p is involved. K. lactis Mig1p contributes to the regulation of the GAL regulon, which is highly conserved between S. cerevisiae and K. lactis (13), but a K. lactis mig1 mutation has only a minor influence on expression of the lactose metabolism genes LAC12 and LAC4, which are part of the GAL regulon (LAC/GAL regulon) (14). Moreover, a mig1 mutation does not suppress the K. lactis Snf1p requirement for growth on galactose as is the case in S. cerevisiae (13).
Among the genes of the GAL regulon, only K. lactis GAL1 contains a Mig1p binding site. Through this site, each gene in the regulon is affected by a mig1 mutation, since K. lactis Gal1p is responsible for transcriptional activation of the regulon (13, 30). Gal1p flips the transcriptional switch in response to galactose by binding to and inactivating the Gal4p inhibitor Gal80p (41). Thus, derepression of GAL1 in a K. lactis mig1 mutant can partially overcome glucose repression, as does elevated expression of GAL4 (13, 28, 39). However, growth on galactose is still impaired in the K. lactis snf1 mutant strain. In particular, lactose permease activity, encoded by LAC12, was virtually undetectable in this mutant, and the permease deficiency could not be overcome by deletion of MIG1 (13). Growth on lactose is only weakly impaired, probably because even a small amount of glucose released from lactose by hydrolysis is sufficient to support growth of K. lactis at a high rate. Since Lac12p also functions as a galactose permease (12, 32), its low activity may explain the poor growth of a K. lactis snf1 mutant on galactose.
Here we have addressed the question of how K. lactis Snf1p exerts its function on the LAC/GAL regulon. We show that transcriptional regulation is only partly responsible for the galactose deficiency of a K. lactis snf1 mutant. A major influence is exerted by a limitation of galactose uptake due to an influence of the snf1 mutation on the intracellular sorting of the lactose-galactose permease Lac12p in the mutant. Overexpression of a so-far-uncharacterized hexose transporter, Kht3p, can partially circumvent this limitation.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this work are listed in Table 1. Plasmid pD802 was used for deletion of GAL80 in JSD1R/D80 as described (40). An AatII-ClaI fragment of plasmid pLAC12-EGFP was integrated at random chromosomal positions of strains JA6 and JSD1R to create strains JA6-LAC12GFP and JSD1R-LAC12GFP, respectively. Cells were grown in synthetic complete medium (0.67% yeast nitrogen base with amino acids and nucleobases) (34) with the indicated carbon sources. The same medium lacking uracil was used when plasmid selection was necessary.
TABLE 1.
Yeast strains used in this study
| Species | Strain | Relevant genotypea | Reference |
|---|---|---|---|
| K. lactis | JA6 | MATα ade1-600 adeT-600 ura3-12 trp1-11 LAC9-2 | 5 |
| JSD1 | MATα ade1-600 adeT-600 ura3-12 trp1-11 LAC9-2 snf1::ScURA3 | 13 | |
| JSD1R | Scura3 derivative of JSD1 | This study | |
| DL12 | MATα ade1-600 adeT-600 ura3-12 trp1-11 LAC9-2 lac12::ScURA3 | 16 | |
| DL12R | Scura3 derivative of DL12 | This study | |
| JA6/D802R | MATα ade1-600 adeT-600 ura3-12 trp1-11 LAC9-2 Klgal80Δ2::Scura3 | 40 | |
| JSD1D80 | Klgal80-2::ScURA3 derivative of JSD1R | This study | |
| JA6-LAC12GFP | MATα ade1-600 adeT-600 ura3-12 trp1-11 LAC9-2 LAC12-EGFP | This study | |
| JSD1R-LAC12GFP | MATα ade1-600 adeT-600 ura3-12 trp1-11 LAC9-2 Klsnf1::ura3 LAC12-EGFP | This study | |
| S. cerevisiae | EBY.VW4000 | MATα Δhxt1-17 gal2 stl1 agt1 mph2-3 leu2-3,112 ura3-52 trp1-289 his3-Δ1 MAL2-8cSUC2 | 38 |
Sc, S. cerevisiae; Kl, K. lactis.
Plasmids.
The KHT3-containing plasmid pG5 was isolated from a KEp6-based genomic plasmid library (2, 37). The chromosomal insert of pG5 was shortened by deletion of a HindIII fragment, resulting in plasmid pKHT3, which comprises the KHT3 open reading frame with 429 bp of 5′ and 1,151 bp of 3′ untranslated region. The LAC12 plasmid pBK52 was isolated from the same library and contains the LAC12 gene (including 1,201 bp upstream and 1,653 bp downstream of the LAC12 open reading frame). The C-terminal end of LAC12 was fused to GFP by replacing a DraI-EcoRI fragment in pBK52 with the SmaI-EcoRI fragment from pEGFP (Clontech), giving plasmid pLAC12-EGFP. The GFP-KlGAL80 gene fusion and the GFP-KlGAL1 fusion are expressed from the S. cerevisiae ADH1 promoter on pE1-based multicopy plasmids.
Determination of β-galactosidase activity.
β-Galactosidase activity was determined in crude extracts prepared in β-Gal buffer (5% glycerol, 10 mM KCl, 5 mM Tris-HCl, pH 7.8). The enzyme assay was carried out at 30°C in β-Gal buffer containing 40 μg of o-nitrophenyl-galactoside (ONPG) per ml. The reaction was followed photometrically (16).
Northern analysis.
For RNA isolation, cultures were grown overnight in SC medium. After harvesting by centrifugation, cells were resuspended in fresh medium containing the indicated carbon sources and cultivated for 4 more hours, harvested, and disrupted by glass beads. The RNA was isolated by a hot phenol method. Northern analysis was carried out as described (1). Generally, 15 μg of RNA sample was loaded per lane. RNA was denatured in loading buffer containing 6% formaldehyde. Digoxigenin-labeled probes were prepared by random priming or PCR labeling with a DNA labeling kit (Roche). The digoxigenin-labeled bands were detected by chemiluminescence with disodium-3-{4-methoxyspiro[1,2-dioxetane-3,2′-(5-chloro)tricyclo(3.3.1.13,7)decan]4-yl}phenyl phosphate (Roche).
Nucleotide sequence accession number.
The accession number for KHT3 is AJ575058.1.
RESULTS
LAC12 transcription is only weakly impaired in a K. lactis snf1 mutant strain.
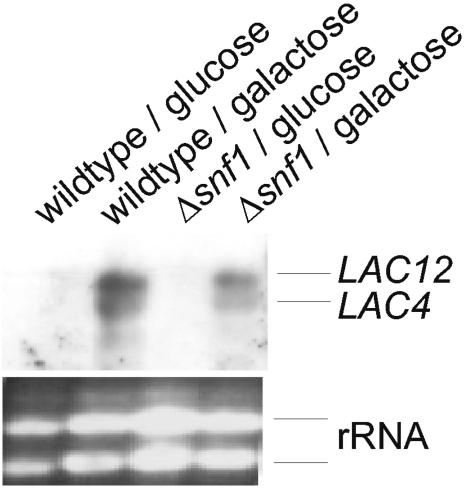
The Gal4p-regulated LAC12 gene, which is divergently transcribed with the β-galactosidase gene LAC4, is essential for the uptake of lactose in K. lactis and has a major impact on galactose transport, although additional galactose transporters exist, at least in some strains (3, 12). To address the question of whether impaired induction of LAC12 is responsible for the growth deficiency on galactose of a K. lactis snf1 mutant, we compared mRNA levels in wild-type and mutant cells shifted from glucose to galactose medium by Northern analysis. RNAs were isolated from cultures grown on glucose and shifted to fresh medium containing 2% glucose or galactose for 4 h. With a single probe, both, the LAC12 and LAC4 transcripts could be detected. As shown in Fig. 1, there is clearly induction of both genes in the mutant as well as in the wild type. RNA levels are somewhat lower in the K. lactis Δsnf1 mutant, but it seems unlikely that this reduction explains the poor growth on galactose and the complete absence of lactose transport activity (13).
FIG. 1.
Influence of the K. lactis snf1 mutation on LAC12 gene expression examined by Northern analysis of total RNA isolated from wild-type strain JA6 and the congenic snf1 mutant JSD1R grown in glucose or galactose (2%). We probed 15 μg of total RNA with a digoxigenin-labeled LAC4-LAC12 DNA fragment. rRNA served as a loading control.
Constitutive induction of the LAC/GAL regulon in a K. lactis gal80 mutant is not sufficient to sustain full growth on galactose.
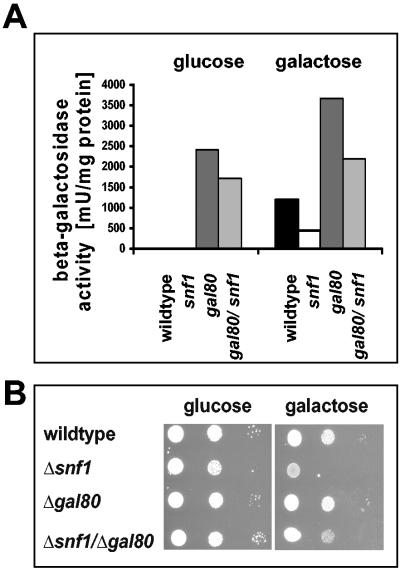
If K. lactis Snf1p function was required for derepression of the LAC/GAL regulon as in S. cerevisiae, this requirement should be suppressed by a mutation in the GAL80 gene, encoding the negative regulator of Gal4p. As reported before, in K. lactis, a gal80 mutation overcomes glucose repression and causes constitutive expression of all genes of the LAC/GAL regulon even in glucose (40). A gal80 deletion was therefore introduced in the K. lactis snf1 mutant background, and the gal80 snf1double mutant was compared to the single mutants by measuring LAC4-encoded β-galactosidase activity and growth on galactose (Fig. 2). As shown in Fig. 2A, the gal80 mutation is epistatic over the snf1 mutant phenotype, indicating that transcription of the LAC/GAL genes does not require snf1 in the absence of Gal80p. Though gene expression was constitutive in the snf1 gal80 mutant background, growth on galactose was still impaired (Fig. 2B). Apparently, circumventing the requirement for Snf1p in LAC/GAL gene induction is insufficient to overcome the Snf1p requirement for growth on galactose.
FIG. 2.
Influence of a gal80 mutation on the K. lactis snf1 mutant phenotype. (A) Epistasis analysis of K. lactis gal80 and S. cerevisiae snf1 mutations on LAC4 gene expression. LAC4-encoded β-galactosidase activity was determined in congenic K. lactis strains JA6 (wild type), JSD1R (Δsnf1), JA6/D802R (gal80Δ2) and JSD1D80 (Δsnf1 gal80Δ2) after a shift from glucose to fresh glucose or galactose (2%) medium for 7 h. (B) Serial dilutions (1:100) of the same strains were spotted on SC plates containing 2% glucose or galactose. The plates were incubated for 2 days at 30°C.
Isolation of a new galactose transporter gene, KHT3, as a multicopy suppressor on galactose of a K. lactis snf1 mutant.
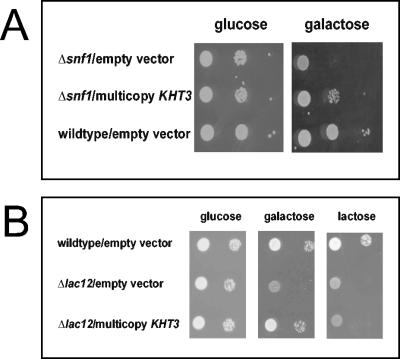
To understand the role of Snf1p in galactose utilization, multicopy suppressors of the poor growth of a K. lactis snf1 mutant on galactose were isolated. K. lactis strain JSD1 (snf1Δ) was transformed with a KEp6-based multicopy genomic library and plated on selective galactose medium (2%). Nine transformants that showed improved growth were characterized further. Eight of these contained the K. lactis SNF1, gene as confirmed by PCR, whereas one clone did not give a signal with K. lactis SNF1-specific primers. The plasmid recovered from this clone contained an insert of 5.3 kb, from which a 2.1-kb HindIII fragment could be deleted without affecting the complementing activity.
The DNA sequence of the remaining insert revealed an open reading frame of 528 codons related to the fungal HXT hexose transporter family of major facilitators, which was named KHT3 (for Kluyveromyces lactis hexose transporter). Overexpression of KHT3 in a K. lactis snf1 mutant strain resulted in significantly improved growth on galactose (Fig. 3A). In order to investigate whether Kht3p is a galactose transporter, we expressed KHT3 on a high-copy-number plasmid in a lac12 mutant strain. This mutant is not able to grow on lactose and galactose. Multiple copies of KHT3 were able to suppress this phenotype on galactose but not on lactose, indicating that Kht3p can function as a galactose transporter (Fig. 3B).
FIG. 3.
KHT3 functions as a galactose transporter. (A) Influence of KHT3 overexpression on growth of a K. lactis snf1 mutant on galactose. The mutant strain JSD1R (Δsnf1) transformed with plasmid pKHT3 or the empty vector KEp6 was tested for growth on SC galactose (2%) medium as in Fig. 2. (B) Influence of KHT3 overexpression on the growth deficiency on galactose and lactose of a Δlac12 mutant strain. Strain DL12R (Δlac12) was transformed with pKHT3 and the empty vector. Growth of the resulting transformants was compared to that of vector-transformed strain JA6 as in Fig. 2B.
Lac12-GFP fails to accumulate in the plasma membrane in K. lactis snf1 mutants.
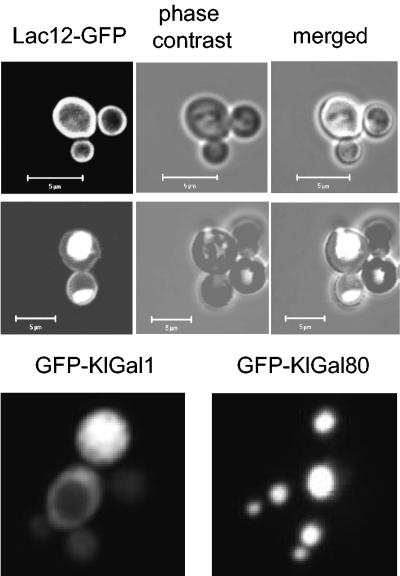
The isolation of a galactose transporter in our genetic screen, together with the finding that LAC12 mRNA is detected in a K. lactis snf1 mutant, indicated that K. lactis Snf1p is required for Lac12p-mediated galactose uptake at a posttranscriptional step. To be able to detect the Lac12 protein inside the cell, GFP was fused to the C terminus of Lac12p. The fusion protein expressed from its own promoter on a high-copy-number plasmid was able to complement the Gal− Lac− phenotype of the lac12 mutant, indicating that it can substitute for wild-type Lac12p. In wild-type cells, GFP fluorescence was mainly detected in the plasma membrane. Only in some cells was additional fluorescence observed in the vacuole. In striking contrast, for the K. lactis snf1 mutant strain we found the opposite distribution: in most cells GFP was localized in the vacuole and only weak fluorescence could be detected in the plasma membrane (Fig. 4).
FIG. 4.
Intracellular localization of GFP fusion proteins. K. lactis strains JA6 (wild type) and JSD1R (snf1) were transformed with multicopy plasmids pLAC12-EGFP, pGAL1-GFP, and pGAL80-GFP. Cells were shifted from glucose to galactose (2%) medium for 3.5 h, and the intracellular distribution of Lac12-GFP was analyzed by confocal laser scanning microscopy (LSM 510; Carl Zeiss).
As controls, the empty vector, GFP-Gal80p, and GFP-Gal1p were expressed in the K. lactis snf1 mutant. No fluorescence could be observed with the empty vector, whereas the latter two fusion proteins showed cytoplasmic and nuclear staining, respectively, which was not affected by the snf1 mutation (Fig. 4 and data not shown). By Western analysis with anti-GFP antibodies, all fusion proteins could be detected, but Lac12-GFP was very labile, and we were unable to quantitatively compare the wild-type and K. lactis snf1 mutant strains (see supplemental Fig. S1).
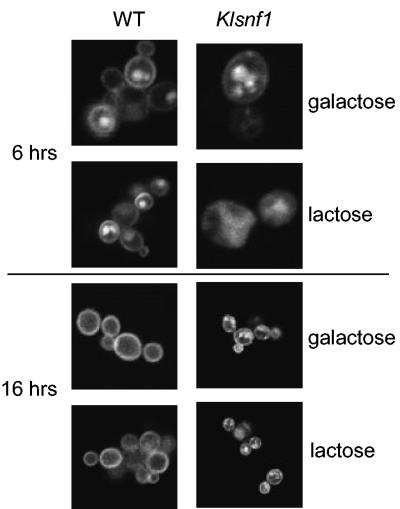
Since, due to plasmid loss, the cell population was highly heterogeneous, the LAC12-GFP fusion was integrated in the chromosome of the wild-type and snf1 K. lactis strains. These strains were used to analyze whether the difference in Lac12-GFP localization reflects a difference in Lac12-GFP turnover or in intracellular sorting. Cells were grown in glucose and shifted to YNB galactose medium, and samples were analyzed at intervals for GFP fluorescence. The GFP signal in these strains was much weaker and could not be detected before 4 h after the shift in the wild type and about 1 to 2 h later in the mutant. Interestingly, at the early time points in both strains, Lac12-GFP was detected in intracellular structures (Fig. 5, top). These structures were more numerous and less distinct in the mutant, and plasma membrane staining was much weaker. At later time points, the fluorescence of the plasma membrane increased in both strains but remained much weaker in the mutant (enhanced in Fig. 5). Most strikingly, intracellular membranes were no longer observed in 95% of wild-type cells, whereas they were very pronounced in 80 to 90% of the mutant cells. We thus conclude that the snf1 mutation prevents the accumulation of Lac12-GFP in the plasma membrane either by affecting a late step in the sorting process or by enhancing endocytosis or both.
FIG. 5.
LAC12-GFP fails to accumulate in the plasma membrane. K. lactis strains JA6-LAC12GFP (wild type) and JSD1R-LAC12GFP (snf1) containing integrated copies of the LAC12-GFP fusion gene were grown in SC-glucose and shifted to SC-galactose (2%) medium at time zero. Lac12-GFP fluorescence was analyzed as in Fig. 4.
DISCUSSION
In this work, we analyzed the function of the Snf1 kinase in the regulation of galactose metabolism in K. lactis. Our data indicate that an impairment of galactose uptake is the primary cause of the galactose deficiency in a K. lactis snf1 mutant. A multicopy suppressor screen in the snf1 mutant resulted in isolation of the KHT3 gene. The Kht3 protein of 528 amino acids belongs to the yeast hexose transporter subfamily of major facilitators. Overexpression of KHT3 is able to suppress the growth defect on galactose not only of an snf1Δ mutant but also of a lac12Δ mutant lacking the lactose/galactose transporter Lac12p. It can also support growth on galactose in an S. cerevisiae strain lacking all 20 hexose transporters (Wiedemuth and Breunig, unpublished data). We thus concluded that Kht3p functions as a galactose transporter. Since multiple copies of the KHT3 gene are required to support slow growth on galactose in the snf1 and the lac12 mutants, Kht3p apparently works inefficiently as a galactose transporter, and the physiological role of Kht3p remains to be determined. In any case, Kht3 does not seem to require Snf1p to function and can thus circumvent the galactose uptake deficiency in the mutant.
It has been shown previously that Snf1p controls Lac12-mediated sugar uptake (13). Our data indicate that the lactose and galactose transport deficiency is not caused by a lack of LAC12 transcription. Induction of the LAC/GAL regulon is delayed but not abolished in the K. lactis snf1 mutant and a gal80 mutation, which causes constitutive expression of the regulon, does not suppress the growth deficiency. These findings indicated that Snf1p was required at a posttranscriptional step. In fact, a Lac12-GFP fusion protein expressed from its own promoter in single copy could readily be detected in the mutant strain, suggesting that Snf1p affects the activity and not the synthesis of the transporter.
Apparently, Lac12p fails to accumulate in the plasma membrane of the K. lactis snf1 mutant. In contrast to congenic wild-type cells, in which the Lac12-GFP fusion was properly sorted to the plasma membrane, a high proportion of the fusion protein was located in intracellular vesicles in the mutant. This altered intracellular distribution may explain the observed growth limitation on galactose.
When shifted from glucose to galactose, the fusion protein could first be detected in intracellular vesicles before it arrived in the plasma membrane. At this stage, the difference between the wild type and the mutant was less pronounced. The difference became more apparent upon prolonged induction, when the mutant failed to show the plasma membrane staining observed in the wild type. We thus favor the view that K. lactis Snf1p activity is required to redirect or stabilize the protein in this compartment.
Regulation of transporter targeting by nutrients is a common phenomenon. Addition of glucose to galactose-grown S. cerevisiae cells stimulates monoubiquitination of the major galactose transporter Gal2p, followed by internalization, delivery to the vacuole, and degradation (23, 24). Routing of the general amino acid permease Gap1p depends on the quality of the nitrogen source. On a rich nitrogen source, newly synthesized Gap1 is sorted directly from the Golgi to the vacuole instead of to the plasma membrane. Addition of ammonium ions to a culture grown on a poor nitrogen source leads to endocytosis of plasma membrane-bound Gap1p (10, 21, 33, 35). Both pathways to the vacuole require ubiquitination.
To our knowledge, we have shown here for the first time that intracellular localization of a sugar transporter is affected by an snf1 mutation. However, several findings are compatible with an involvement of Snf1p in the regulation of transporter activity. For example, the S. cerevisiae maltose permease Mal61p requires Snf1p at a posttranscriptional step, since no maltose transport activity is measurable in an snf1 mutant strain when the MAL61 gene is expressed constitutively (25).
At present, we can only speculate about the function of Snf1p in sugar transport regulation. The fact that KHT3 but not LAC12 in multicopy can suppress the galactose uptake deficiency of a K. lactis snf1 mutant indicates that Lac12p is more sensitive to the influence of Snf1 than other transporters.
The complex role of K. lactis Snf1p in the adaptation of cells to carbon limitation resembles that of the Npr1 serine/threonine protein kinase in adaptation to nitrogen limitation. Besides regulating transcription, Npr1 controls the post-Golgi sorting and degradation of amino acid permeases (29). Snf1 was found associated with the small GTPase Arf1p and its GTPase exchange factor Sec7p, both of which are implicated in vesicle transport (22). In S. cerevisiae, one form of the heterotrimeric Snf1 complex is associated with the vacuole (36).
We were unable to express Lac12p at sufficiently high levels in S. cerevisiae to reach unambiguous conclusions about the influence of snf1 in a heterologous host. Moreover, the inability to grow S. cerevisiae snf1 mutants under derepressing conditions complicates the analysis. K. lactis snf1 can grow readily on lactose and also slowly on galactose and other carbon sources, a fact that is helpful in unraveling the complex regulatory network controlled by Snf1. The data shown here have revealed a new aspect of regulation of carbon utilization by Snf1p in addition to its influence on transcription and enzyme activity.
Supplementary Material
Acknowledgments
We thank Bob Dickson (University of Kentucky) and Micheline Wésolowski-Louvel (Lyon) for strains and plasmids and Karin Sorge for technical assistance. Konstanze Wiedemuth is gratefully acknowledged for construction of the GFP-Gal80 and GFP-Gal1 fusion plasmids.
This work was supported by DFG grants Br921/5-1 (FOG 466/1-1) and Br921/6-1 to K.D.B.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Alwine, J. C., D. J. Kemp, and G. R. Stark. 1977. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. USA 74:5350-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bianchi, M. M., C. Falcone, C. X. Jie, M. Wésolowski-Louvel, L. Frontali, and H. Fukuhara. 1987. Transformation of the yeast Kluyveromyces lactis by new vectors derived from the 1.6 μm circular plasmid pKD1. Curr. Genet. 12:185-192. [Google Scholar]
- 3.Boze, H., G. Moulin, and P. Galzy. 1987. Uptake of galactose and lactose by Kluyveromyces lactis: biochemical characteristics and attempted genetical analysis. J. Gen. Microbiol. 133:15-23. [DOI] [PubMed] [Google Scholar]
- 4.Breunig, K. D., M. Bolotin-Fukuhara, M. M. Bianchi, D. Bourgarel, C. Falcone, I. Ferrero, L. Frontali, P. Goffrini, J. J. Krijger, C. Mazzoni, C. Milkowski, H. Y. Steensma, M. Wesolowski-Louvel, and A. M. Zeeman. 2000. Regulation of primary carbon metabolism in Kluyveromyces lactis. Enzyme Microb. Technol. 26:771-780. [DOI] [PubMed] [Google Scholar]
- 5.Breunig, K. D., and P. Kuger. 1987. Functional homology between the yeast regulatory proteins GAL4 and LAC9: LAC9-mediated transcriptional activation in Kluyveromyces lactis involves protein binding to a regulatory sequence homologous to the GAL4 protein-binding site. Mol. Cell. Biol. 7:4400-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carling, D. 2004. The AMP-activated protein kinase cascade-a unifying system for energy control. Trends Biochem. Sci. 29:18-24. [DOI] [PubMed] [Google Scholar]
- 7.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 8.Cassart, J. P., I. Georis, J. Östling, H. Ronne, and J. Vandenhaute. 1995. The MIG1 repressor from Kluyveromyces lactis: cloning, sequencing and functional analysis in Saccharomyces cerevisiae. FEBS Lett. 371:191-194. [DOI] [PubMed] [Google Scholar]
- 9.Charbon, G., K. D. Breunig, R. Wattiez, J. Vandenhaute, and I. Noel-Georis. 2004. Key role of Ser562/661 in Snf1-dependent regulation of Cat8p in Saccharomyces cerevisiae and Kluyveromyces lactis. Mol. Cell. Biol. 24:4083-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Craene, J. O., O. Soetens, and B. Andre. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276:43939-43948. [DOI] [PubMed] [Google Scholar]
- 11.DeVit, M. J., J. A. Waddle, and M. Johnston. 1997. Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8:1603-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickson, R. C., and K. Barr. 1983. Characterization of Lactose Transport in Kluyveromyces lactis. J. Bacteriol. 154:1245-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong, J., and R. C. Dickson. 1998. Glucose represses the lactose-galactose regulon in Kluyveromyces lactis through a SNF1 and MIG1-dependent pathway that modulates galactokinase (GAL1) gene expression. Nucleic Acids Res. 25:3657-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georis, I., K. D. Breunig, and J. Vandenhaute. 1999. Glucose repression in Kluyveromyces lactis does not rely predominantly on KlMig1. Curr. Genet. 35:246. [Google Scholar]
- 15.Georis, I., J. J. Krijger, K. D. Breunig, and J. Vandenhaute. 2000. Differences in regulation of yeast gluconeogenesis revealed by Cat8p-independent activation of PCK1 and FBP1 genes in Kluyveromyces lactis. Mol. Gen. Genet. 264:193-203. [DOI] [PubMed] [Google Scholar]
- 16.Gödecke, A., W. Zachariae, A. Arvanitidis, and K. D. Breunig. 1991. Coregulation of the Kluyveromyces lactis lactose permease and β-galactosidase genes is achieved by interaction of multiple LAC9 binding sites in a 2.6 kbp divergent promoter. Nucleic Acids Res. 19:5351-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 18.Hardie, D. G., M. L. Scott, D. A. Pan, and E. R. Hudson. 2003. Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett. 546:113-120. [DOI] [PubMed] [Google Scholar]
- 19.Haurie, V., M. Perrot, T. Mini, P. Jeno, F. Sagliocco, and H. Boucherie. 2001. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 276:76-85. [DOI] [PubMed] [Google Scholar]
- 20.Haurie, V., F. Sagliocco, and H. Boucherie. 2004. Dissecting regulatory networks by means of two-dimensional gel electrophoresis: application to the study of the diauxic shift in the yeast Saccharomyces cerevisiae. Proteomics 4:364-373. [DOI] [PubMed] [Google Scholar]
- 21.Hein, C., J. Y. Springael, C. Volland, R. Haguenauer-Tsapis, and B. Andre. 1995. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol. Microbiol. 18:77-87. [DOI] [PubMed] [Google Scholar]
- 22.Ho, Y., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 23.Horak, J., and D. H. Wolf. 2001. Glucose-induced monoubiquitination of the Saccharomyces cerevisiae galactose transporter is sufficient to signal its internalization. J. Bacteriol. 183:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horak, J., and D. H. Wolf. 1997. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J. Bacteriol. 179:1541-1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu, Z., Y. Yue, H. Jiang, B. Zhang, and C. A. Michels. 2000. Analysis of the mechanism by which glucose inhibits maltose induction of MAL gene expression in Saccharomyces. Genetics 154:121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston, M. 1999. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 15:29-33. [DOI] [PubMed] [Google Scholar]
- 27.Krijger, J.-J. 2002. Carbon source-responsive elements and gene regulation by CAT8 and SIP4 in the yeast Kluyveromyces lactis. Ph.D. thesis. Universitaet Halle-Wittenberg, Halle, Germany.
- 28.Kuzhandaivelu, N., W. K. Jones, A. K. Martin, and R. C. Dickson. 1992. The signal for glucose repression of the lactose-galactose regulon is amplified through subtle modulation of transcription of the Kluyveromyces lactis Kl-GAL4 activator gene. Mol. Cell. Biol. 12:1924-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magasanik, B., and C. A. Kaiser. Nitrogen regulation in Saccharomyces cerevisiae. Gene. 290:1-18, 2002. [DOI] [PubMed] [Google Scholar]
- 30.Meyer, J., A. Walker-Jonah, and C. P. Hollenberg. 1991. Galactokinase encoded by GAL1 is a bifunctional protein required for induction of the GAL genes in Kluyveromyces lactis and is able to suppress the gal3 phenotype in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papamichos-Chronakis, M., T. Gligoris, and D. Tzamarias. 2004. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 5:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riley, M. I., S. Sreekrishna, S. Bhairi, and R. C. Dickson. 1987. Isolation and characterization of mutants in Kluyveromyces lactis defective in lactose transport. Mol. Gen. Genet. 208:145-151. [DOI] [PubMed] [Google Scholar]
- 33.Roberg, K. J., N. Rowley, N., and C. A. Kaiser. 1997. Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Sci. 137:1469-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman, F. 1990. Guide to yeast genetics and molecular biology. Getting started with yeast. Methods Enzymol. 194:3-20. [PubMed] [Google Scholar]
- 35.Springael, J. Y., and B. Andre. 1998. Nitrogen-regulated ubiquitination of the Gap1 permease of Saccharomyces cerevisiae. Mol. Biol. Cell 9:1253-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wésolowski-Louvel, M., P. Goffrini, I. Ferrero, and H. Fukuhara. 1992. Glucose transport in the yeast Kluyveromyces lactis. I. Properties of an inducible low-affinity glucose transporter gene. Mol. Gen. Genet. 233:89-96. [DOI] [PubMed] [Google Scholar]
- 38.Wieczorke, R., S. Krampe, T. Weierstall, K. Freidel, C. P. Hollenberg, and E. Boles. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123-128. [DOI] [PubMed] [Google Scholar]
- 39.Zachariae, W., P. Kuger, and K. D. Breunig. 1993. Glucose repression of lactose/galactose metabolism in Kluyveromyces lactis is determined by the concentration of the transcriptional activator LAC9 (KlGAL4). Nucleic Acids Res. 21:69-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zenke, F., W. Zachariae, A. Lunkes, and K. D. Breunig. 1993. Gal80 proteins of Kluyveromyces lactis and Saccharomyces cerevisiae are highly conserved but contribute differently to glucose repression of the galactose regulon. Mol. Cell. Biol. 13:7566-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zenke, F. T., R. Engels, V. Vollenbroich, J. Meyer, C. P. Hollenberg, and K. D. Breunig. 1996. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science 272:1662-1665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.