Abstract
Proteomic analysis of phagosomes isolated from Entamoeba histolytica by liquid chromatography and mass spectrometry identified 85 proteins involved in surface recognition, actin cytoskeleton rearrangement, vesicular trafficking, and degradation. Phagosome localization of representative proteins was verified by immunofluorescence assay. This study should provide a basis for molecular identification and characterization of phagosome biogenesis.
The protozoan parasite Entamoeba histolytica is a major cause of morbidity and mortality worldwide (35). E. histolytica is capable of ingesting microorganisms in the large intestine (5), as well as red blood cells (33) and apoptotic immune cells (19) during tissue invasion. Phagocytosis plays an essential role in growth and constitutes one of the key virulence determinants (6). Ameba mutants defective in phagocytosis have a growth defect on bacterial lawns (25) and are also defective in the destruction of tissue-cultured mammalian cells in vitro and in the formation of hepatic abscesses in vivo (24). A number of steps have been shown to be involved in phagocytosis in macrophages, neutrophils, and Dictyostelium discoideum: receptor-ligand interaction on the cell surface, activation of signaling pathways leading to rearrangements of the actin cytoskeleton, and membrane trafficking. In E. histolytica, one of the best-characterized receptors involved in the recognition of and attachment to the host or bacterial surface prior to initiation of phagocytosis is the Gal/GalNAc-specific lectin (26). Myosin IB was shown to localize to the phagocytic cup and phagosomes during ingestion of human red blood cells (23). It was also shown that rearrangement of the actin cytoskeleton plays an important role in phagocytosis of mammalian cells (16, 17). Amoebapores, cysteine protease 2 (CP2), and CP3 were shown to be recruited to phagosomes and involved in permeation and degradation of ingested bacteria (2, 28). However, a large number of molecules and their kinetics of association and dissociation during phagosome biogenesis remain largely unknown. In this study, in order to understand molecules and mechanisms involved in phagocytosis of E. histolytica, we took a proteomic approach to identify proteins associated with the phagosomes, using an optimized protocol for phagosome isolation from the amebae and mass spectrometry, previously utilized for mouse macrophages (12).
Trophozoites of E. histolytica HM1:IMSS, axenically cultivated in 25-cm2 flasks containing BI-S-33 medium, were mixed with carboxylated latex beads. The beads were internalized by centrifugation at 160 × g for 5 min. After centrifugation, phagosome maturation was immediately interrupted by incubating the flasks on ice for 10 min. Uningested beads were removed (>98%) by washing the trophozoites three times with cold phosphate-buffered saline containing 20% sucrose, followed by centrifugation. After trophozoites were homogenized in a Dounce homogenizer on ice, bead-containing phagosomes were purified by flotation on a sucrose step gradient centrifuged in a swinging-bucket rotor (SW40; Beckman) at 100,000 × g for 1 h at 4°C as previously described (12), with some modifications (10 mM E-64 and Complete Mini protease inhibitor cocktail were included in all buffers). After trypsin digestion, peptides were analyzed on a liquid chromatography-mass spectrometry (MS) system consisting of a Finnigan LCQ ion trap mass spectrometer system with a Protana nanospray ion source interfaced to a self-packed Phenomenex Jupiter 10-mm C18 reversed-phase capillary column (8 cm by 75 mm [inside diameter]) at the W. M. Keck Biomedical Mass Spectrometry Laboratory, University of Virginia. Sequencing data were analyzed versus The Institute for Genomic Research (TIGR) E. histolytica genome database (http://www.tigr.org/tdb/e2k1/eha1/) using the Sequest algorithm (13) and also against the nonredundant database at the National Center for Biotechnology Information (NCBI). The frequency of each protein in triplicate phagosome samples was expressed as the mean percentage of the number of peptides mapped to each protein in the total number of peptides detected in the phagosome sample.
We established a protocol to isolate phagosomes with high purity. Among a variety of beads, carboxylate-modified latex beads were internalized by the trophozoites approximately sevenfold more efficiently than amine-modified (positively charged) beads, as evaluated with the flow cytometry-based phagocytosis assay (data not shown). These carboxylate-modified beads, which are thought to mimic the negative charge on dying cells, have been used previously as a surrogate to study phagocytosis of apoptotic cells (1). We optimized conditions to maximize and synchronize internalization of beads by low-speed centrifugation. Centrifugation at 160 × g for 5 min increased internalization efficiency by 50-fold compared to a case in which amebae were simply cocultured with beads for 5 min. About 80% of amebae contained at least 1 bead, and a single trophozoite internalized an average of 5.0 ± 0.3 beads by centrifugation. We typically purified up to 50 μg of the purified phagosome proteins per 1.5 × 108 cells, which constitutes approximately 0.01% of the total cell protein. We verified the purity of the phagosome preparations by transmission electron micrographs and immunoblot analysis (data not shown). For a typical MS analysis of phagosome proteins, we used approximately 25 μg of the purified phagosome protein and obtained 700 to 1,000 sequences of trypsin-digested fragments. Approximately 90% of these peptides were unambiguously assigned to 85 proteins. The coverage of proteins varied but typically reached more than 10% (e.g., 18 to 24% for the Gal/GalNAc-specific lectin heavy subunit [Hgl], 10% for the intermediate subunit [Igl], and 42% for the light subunit [Lgl]). We have listed in Table 1 only phagosome proteins in cases in which (i) homologues with known or predicted functions were found and (ii) amino acid identity and the E value were higher than 25% and better than 10−5, respectively. We categorized a panel of phagosome-associated proteins into the following functional groups: lectins and surface proteins, vesicular trafficking and other small GTPases and effectors, hydrolytic enzymes and degradative proteins, and calcium and proton pumps. We also conducted a control experiment to evaluate nonspecific binding of amebic proteins to latex beads. The profiles of these nonspecific proteins were remarkably different from those of purified phagosomes; only five proteins (elongation factor 1α, 14%; actin, 8.3%; Hgl, CP5, and cyclophilin, 0.83%) were predominantly detected.
TABLE 1.
Proteomic analysis of phagosome proteinsa
| Protein group and TIGR ID no. (EH) | NCBI accession no. | Protein name (organism source) | Identity (%) | Frequency (%)c |
|---|---|---|---|---|
| Lectins and surface proteins | ||||
| 6468 | Adhesin p30b (Mycoplasma pneumoniae) | 54 | 0.53 ± 0.19 | |
| 879 | Aminophospholipid translocase 2b (Homo sapiens)b | 44 | 0.32 ± 0.08 | |
| 4887 | Gal/GalNAc lectin Hgl1-5b,d | 100 | 3.0 ± 0.26 | |
| 1189/175 | Gal/GalNAc lectin Igl1, 2d | 100 | 1.7 ± 0.64 | |
| 385/4767/3450 | Gal/GalNAc lectin Lgl1-5d | 100 | 3.2 ± 1.6 | |
| 2521 | X55028 | Immunodominant variable surface antigen | 100 | 0.29 ± 0.15 |
| 2178 | Multidrug resistance protein 2 (Canis familiaris) | 31 | 3.4 ± 1.0 | |
| Vesicular trafficking, other small GTPases, and effectors | ||||
| 4532 | AB054578 | Rab1 | 100 | 0.34 ± 0.08 |
| 289 | AF218311 | Rab7Ab,e | 100 | 0.26 ± 0.18 |
| 540 | AB186363 | Rab7Bb,e | 100 | 0.35 ± 0.31 |
| 3974 | AB186364 | Rab7Cb,e | 100 | 0.32 ± 0.08 |
| 2190 | AB186365 | Rab7Db,e | 100 | 0.29 ± 0.41 |
| 1867 | AB186366 | Rab7Eb,e | 100 | 1.9 ± 0.82 |
| 3360 | AB197095 | RabX17 | 100 | 0.24 ± 0.34 |
| 1740 | AB054579 | RabC1 | 100 | 0.24 ± 0.34 |
| 1174 | Clathrin coat assembly protein ap50b (Dictyostelium discoideum) | 48 | 0.39 ± 0.01 | |
| 443 | Receptor-mediated endocytosis protein 1b (Caenorhabditis elegans) | 44 | 0.23 ± 0.05 | |
| 2883 | U29720 | RacA | 100 | 0.58 ± 0.26 |
| 1250 | U29722 | RacC | 100 | 0.28 ± 0.01 |
| 830 | AF055340 | RacG | 100 | 0.87 ± 0.28 |
| 1948 | U01052 | Rap2 | 100 | 1.2 ± 0.59 |
| Hydrolytic enzymes and degradative proteins | ||||
| 4000 | Acid phosphataseb,f (Homo sapiens) | 25 | 0.38 ± 0.54 | |
| 5274 | Acid phosphataseb,f (Homo sapiens) | 26 | 0.86 ± 0.27 | |
| 4360 | α-Amylase (Paenibacillus polymyxa) | 27 | 0.23 ± 0.05 | |
| 1563 | AJ417748 | β-Hexosaminidase B | 100 | 3.1 ± 0.07 |
| 683 | Q01957 | Cysteine protease 1 | 100 | 0.46 ± 0.11 |
| 4021 | Q01958 | Cysteine protease 2 | 100 | 2.1 ± 0.57 |
| 2010 | CAA62833 | Cysteine protease 4 | 100 | 0.23 ± 0.05 |
| 881 | CAA62835 | Cysteine protease 5 | 100 | 4.1 ± 2.1 |
| 5015 | AF059278 | Dipeptidylaminopeptidase | 100 | 1.2 ± 0.13 |
| 2776 | X87610 | Lysozyme 1b | 100 | 0.23 ± 0.05 |
| 1921 | Phospholipase A2 | 100 | 0.54 ± 0.36 | |
| 5686 | Phospholipase Bb,g (Dictyostelium discoideum) | 31 | 0.68 ± 0.16 | |
| 6042 | Phospholipase Bg (Dictyostelium discoideum) | 27 | 2.0 ± 0.17 | |
| 790 | Serine proteaseh (Caenorhabditis elegans) | 31 | 0.82 ± 0.35 | |
| 3172 | Serine proteaseh (Caenorhabditis elegans) | 30 | 1.1 ± 0.95 | |
| Calcium and proton pump | ||||
| 5038 | Calcium-transporting ATPasei | 100 | 0.63 ± 0.33 | |
| 6704 | U20321 | Calcium-transporting ATPasei | 100 | 0.53 ± 0.19 |
| 1033 | V-ATPase Vo domain subunit ab (Homo sapiens) | 33 | 0.32 ± 0.08 | |
| Other proteins | ||||
| 2856/5741 | U13421 | Pyridine nucleotide transhydrogenasej | 100 | 8.6 ± 1.5 |
| 432 | M16339 | Actin | 100 | 1.1 ± 0.01 |
| 4065 | Talin (Dictyostelium discoideum) | 25 | 0.98 ± 0.3 | |
| 3098 | X98567 | Ubiquitin | 100 | 0.32 ± 0.08 |
| 135 | p21-activated protein kinase (Dictyostelium discoideum) | 46 | 0.46 ± 0.11 | |
| 4681 | AF017993 | Cyclophilin | 100 | 0.50 ± 0.04 |
| 645 | M92073 | Elongation factor 1α | 100 | 1.1 ± 0.22 |
We list protein names with organism names in parentheses. Organism names are not indicated when detected peptides perfectly match the previously reported E. histolytica proteins. In cases where highest similarity is demonstrated against putative homologs from other organisms, protein names, organism sources in parentheses, and percent identities are shown. We show putative protein names in cases where (i) homologues with known or predicted functions were found and (ii) amino acid identity and E value are higher than 25% and better than 10−5, respectively.
Annotation at NCBI nonredundant database is shown while this entry is annotated differently in the E. histolytica Genome Database at TIGR.
Frequency (%) = (number of peptides mapped to each protein)/(total number of peptides detected in the phagosome sample) × 100.
Isotypes of Hgl, Igl, and Lgl were not assigned due to high similarity among these isotypes.
Rab7A shows 26 to 44% identity to other Rab7 isotypes.
EH5274 acid phosphatase shows 14% identity to EH4000.
EH5686 phospholipase B shows 8.3% identity to EH6042.
EH790 serine protease shows 16% identity to EH3172.
EH5038 calcium-transporting ATPase shows 83% identity to EH6704.
EH2856 and EH5741 encode partial proteins of a single pyridine nucleotide transhydrogenase.
Among lectins and surface proteins, Hgl, Igl, and Lgl, which have been implicated in the recognition of ligands on the host and bacterial surface, were demonstrated. It was previously shown that E. histolytica expressing an amino-terminally truncated dominant negative form of Lgl had a significant decrease in the ability to phagocytose erythrocytes (21). Adhesin p30 was associated with the attachment organelle of cell wall-lacking Mycoplasma pneumoniae and involved in cytoadherence (29). Aminophospholipid translocase ATPase 2b was presumed to flip phosphatidylserine and phosphatidylethanolamine from the external leaflet of a membrane bilayer to the cytosolic leaflet and be involved in physiological and pathological conditions such as activated platelets, apoptotic cells, and sickle and thalassemic erythrocytes (11). Among small GTPases, Rab (Rab1A, 7A-E, X11, and C1), Rac (RacA, RacC, and RacG), and Rap2 (32) were identified. We previously demonstrated that Rab7A is transported to phagosomes containing red blood cells via an E. histolytica-specific organelle “prephagosomal vacuole” (30). Novel Rab7 isotypes (Rab7B, -C, -D, and -E), identified from phagosomes, indicate the complexity of Rab7 isotypes in lysosomal and phagosomal trafficking in this organism. It was previously demonstrated by overexpression of dominant negative RacA that RacA is involved in cytoskeletal rearrangement during phagocytosis of bacteria, red blood cells, and mucin-coated beads in E. histolytica (15). RacG was reported to be involved in uroid formation (18). Both human and Dictyostelium Rap1, which have about 50% identity to E. histolytica Rap2, were shown to localize to phagosomes and regulate phagocytosis (27, 31). A variety of hydrolytic enzymes and degradative proteins were found, including CP1, -2, -4, and -5; β-hexosaminidase; dipeptidylaminopeptidase (DPAP); acid phosphatases; lysozyme; and phospholipases A2 and B. CP5 was most abundantly detected despite the fact that this CP was previously demonstrated on the plasma membrane (20). It was previously shown that antisense inhibition of expression of CP5 caused a decrease in phagocytosis (3). Although it was previously shown that CP4 was not expressed at detectable levels by Northern blot analysis (7), CP4 was detected in phagosomes at a significant level, suggesting that this scarce CP is concentrated in phagosomes. Although CP2 and -3 were shown to be recruited to phagosomes during phagocytosis of red blood cells (28), only CP2, but not CP3, was demonstrated from phagosomes in the present study. This suggests that CP3 recruitment may be specific to red blood cell engulfment. We also identified transhydrogenase, which is localized to the inner membrane of mitochondria and catalyzes direct proton transfer between NADP+ and NAD+ using a proton gradient (34). Transhydrogenase from E. histolytica contains a putative mitochondrion-targeting signal and was assumed to be localized to the mitosome, a mitochondrial remnant organelle (10). Since other putative mitosome-localized proteins possessing a mitochondria targeting signal, e.g., cpn60 (10) and mitochondrial HSP70 (4), were not detected, transhydrogenase may be localized in phagosomes in this parasite, unlike other organisms. Although it was previously shown that the endoplasmic reticulum-located proteins, including calnexin, calreticulin, Sec61p, glucose-6-phosphatase, GRP78, and protein disulfide isomerase, are involved in phagosome biogenesis (14), no putative endoplasmic reticulum protein was identified from the ameba phagosomes.
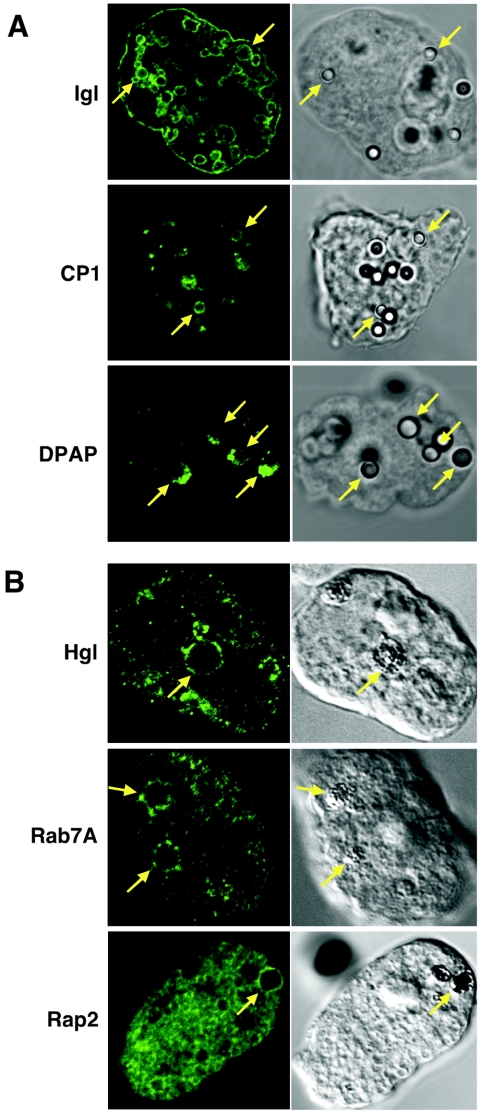
We confirmed the phagosome localization of representative phagosome proteins Hgl, Igl, CP1, DPAP, Rab7A, and Rap2 by immunofluorescence assay as previously described (30) (Fig. 1). Trophozoites were incubated with carboxylate-modified beads for 24 h or red blood cells for 5 to 30 min. Monoclonal antibodies against representative surface membrane proteins Igl (9) and Hgl (3F4) (22) reacted with the membrane of phagosomes containing beads or red blood cells (results of Igl in a red blood cell-ingesting ameba and Hgl in a bead-ingesting ameba not shown). Anti-myc monoclonal antibody 11MO reacted with phagosomes containing red blood cells (B) and beads (data not shown), as well as small vesicles, in the transformant expressing myc-tagged Rab7A (30) and myc-tagged Rap2. Polyclonal rabbit antisera raised against the representative luminal digestive proteins CP1 (a gift from Sharon L. Reed) and DPAP also nicely reacted with the luminal part of the bead (A) or red blood cell-containing phagosomes (data not shown). Protein profiles of phagosomes obtained using carboxylate-modified beads coated with mucin, which is the major glycoprotein on the host epithelium that the amebic Gal/GalNAc lectin interacts with (8), were comparable to those obtained with noncoated carboxylate-modified beads (data not shown). This result supports the premise that phagocytosis of carboxylate-modified latex beads mimics phagocytosis of host cells. Our proteomic data on phagosomes should give a basis of our knowledge of phagosome biogenesis and should also facilitate functional assignment of individual proteins localized to phagosomes, which is essential for annotation of the genome database.
FIG. 1.
Cellular localization of representative phagosome proteins. Wild-type amebae (panel A and Hgl in panel B) and myc-Rab7A- and myc-Rap2-expressing transformants (Rab7A and Rap2 in panel B) were incubated with carboxylate-modified beads for 24 h (Igl, CP1, and DPAP) or red blood cells for 5 (Hgl) or 30 (Rab7A and Rap2) min, fixed, and reacted with specific antibodies against Hgl, Igl, CP1, DPAP, or myc tag on Rab7A and Rap2. Phagosomes are marked with yellow arrows. Fluorescence and phase images are shown in left and right panels, respectively.
Acknowledgments
This work was supported by a grant for Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency to T.N.; a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology of Japan to T.N. (16017307, 16044250, 15590378); a grant from the Japan Health Sciences Foundation to T.N. and B.J.M.; NIH grant AI053678 to C.D.H.; NIH grant AI32615 to B.J.M.; and NIH grant AI26649 to W.A.P.
We are grateful to Nicholas Sherman and W. M. Keck, Biomedical Mass Spectrometry Laboratory at the University of Virginia, for technical support in MS analyses; Hiroshi Tachibana, Tokai University for Igl antibody; Sharon L. Reed, Departments of Pathology and Medicine at the University of California for CP1 antibody; and Yumiko Saito-Nakano (NIID), Kumiko Nakada-Tsukui, Dan Sato, Biswa N. Mitra, and Vahab Ali (Gunma University) for helpful discussions. The E. histolytica genome databases available at The Institute for Genomic Research and the Sanger Institute, which were supported by grants from National Institute of Allergy and Infectious Diseases and the Wellcome Trust, were used for MS analysis.
REFERENCES
- 1.Albert, M. L., S. F. Pearce, L. M. Francisco, B. Sauter, P. Roy, R. L. Silverstein, and N. Bhardwaj. 1998. Immature dendritic cells phagocytose apoptotic cells via αvβ5 and CD36, and cross-present antigens to cytotoxic T lymphocytes. J. Exp. Med. 188:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andra, J., R. Herbst, and M. Leippe. 2003. Amoebapores, archaic effector peptides of protozoan origin, are discharged into phagosomes and kill bacteria by permeabilizing their membranes. Dev. Comp. Immunol. 27:291-304. [DOI] [PubMed] [Google Scholar]
- 3.Ankri, S., T. Stolarsky, and D. Mirelman. 1998. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol. 28:777-785. [DOI] [PubMed] [Google Scholar]
- 4.Arisue, N., L. B. Sanchez, L. M. Weiss, M. Muller, and T. Hashimoto. 2002. Mitochondrial-type hsp70 genes of the amitochondriate protists, Giardia intestinalis, Entamoeba histolytica and two microsporidians. Parasitol. Int. 51:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracha, R., D. Kobiler, and D. Mirelman. 1982. Attachment and ingestion of bacteria by trophozoites of Entamoeba histolytica. Infect. Immun. 36:396-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bracha, R., and D. Mirelman. 1984. Virulence of Entamoeba histolytica trophozoites. Effects of bacteria, microaerobic conditions, and metronidazole. J. Exp. Med. 160:353-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruchhaus, I., B. J. Loftus, N. Hall, and E. Tannich. 2003. The intestinal protozoan parasite Entamoeba histolytica contains 20 cysteine protease genes, of which only a small subset is expressed during in vitro cultivation. Eukaryot. Cell 2:501-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chadee, K., W. A. Petri, Jr., D. J. Innes, and J. I. Ravdin. 1987. Rat and human colonic mucins bind to and inhibit adherence lectin of Entamoeba histolytica. J. Clin. Investig. 80:1245-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, X. J., M. A. Hughes, C. D. Huston, B. Loftus, C. A. Gilchrist, L. A. Lockhart, S. Ghosh, V. Miller-Sims, B. J. Mann, W. A. Petri, Jr., and H. Tachibana. 2001. Intermediate subunit of the Gal/GalNAc lectin of Entamoeba histolytica is a member of a gene family containing multiple CXXC sequence motifs. Infect. Immun. 69:5892-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, C. G., and A. J. Roger. 1995. Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 92:6518-6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daleke, D. L., and J. V. Lyles. 2000. Identification and purification of aminophospholipid flippases. Biochim. Biophys. Acta 1486:108-127. [DOI] [PubMed] [Google Scholar]
- 12.Desjardins, M., L. A. Huber, R. G. Parton, and G. Griffiths. 1994. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J. Cell Biol. 124:677-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eng, J. K., A. L. McCormack, and J. R. Yates. 1994. An approach to correlate tandem mass spectral data of peptides with amino acid sequence in a protein database. J. Am. Soc. Mass Spectrom. 5:976-989. [DOI] [PubMed] [Google Scholar]
- 14.Gagnon, E., S. Duclos, C. Rondeau, E. Chevet, P. H. Cameron, O. Steele-Mortimer, J. Paiement, J. J. Bergeron, and M. Desjardins. 2002. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell 110:119-131. [DOI] [PubMed] [Google Scholar]
- 15.Ghosh, S. K., and J. Samuelson. 1997. Involvement of p21racA, phosphoinositide 3-kinase, and vacuolar ATPase in phagocytosis of bacteria and erythrocytes by Entamoeba histolytica: suggestive evidence for coincidental evolution of amebic invasiveness. Infect. Immun. 65:4243-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, B. B., D. B. Day, C. Nokkaew, and C. C. Harper. 1987. Simulation by target cell membrane lipid of actin polymerization and phagocytosis by Entamoeba histolytica. Infect. Immun. 55:1848-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon, B. B., J. R. Gilmour, and N. E. Mccoomer. 1990. Roles of target cell membrane carbohydrate and lipid in Entamoeba histolytica interaction with mammalian cells. Infect. Immun. 58:2389-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillen, N., P. Boquet, and P. Sansonetti. 1998. The small GTP-binding protein RacG regulates uroid formation in the protozoan parasite Entamoeba histolytica. J. Cell Sci. 111:1729-1739. [DOI] [PubMed] [Google Scholar]
- 19.Huston, C. D., D. R. Boettner, V. Miller-Sims, and W. A. Petri, Jr. 2003. Apoptotic killing and phagocytosis of host cells by the parasite Entamoeba histolytica. Infect. Immun. 71:964-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobs, T., I. Bruchhaus, T. Dandekar, E. Tannich, and M. Leippe. 1998. Isolation and molecular characterization of a surface-bound proteinase of Entamoeba histolytica. Mol. Microbiol. 27:269-276. [DOI] [PubMed] [Google Scholar]
- 21.Katz, U., S. Ankri, T. Stolarsky, Y. Nuchamowitz, and D. Mirelman. 2002. Entamoeba histolytica expressing a dominant negative N-truncated light subunit of its gal-lectin are less virulent. Mol. Biol. Cell 13:4256-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann, B. J., C. Y. Chung, J. M. Dodson, L. S. Ashley, L. L. Braga, and T. L. Snodgrass. 1993. Neutralizing monoclonal antibody epitopes of the Entamoeba histolytica galactose adhesin map to the cysteine-rich extracellular domain of the 170-kilodalton subunit. Infect. Immun. 61:1772-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marion, S., C. Wilhelm, H. Voigt, J. C. Bacri, and N. Guillen. 2004. Overexpression of myosin IB in living Entamoeba histolytica enhances cytoplasm viscosity and reduces phagocytosis. J. Cell Sci. 117:3271-3279. [DOI] [PubMed] [Google Scholar]
- 24.Orozco, E., G. Guarneros, A. Martinez-Palomo, and T. Sanchez. 1983. Entamoeba histolytica. Phagocytosis as a virulence factor. J. Exp. Med. 158:1511-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peracino, B., J. Borleis, T. Jin, M. Westphal, J. M. Schwartz, L. Wu, E. Bracco, G. Gerisch, P. Devreotes, and S. Bozzaro. 1998. G protein beta subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. J. Cell Biol. 141:1529-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petri, W. A., Jr., R. Haque, and B. J. Mann. 2002. The bittersweet interface of parasite and host: lectin-carbohydrate interactions during human invasion by the parasite Entamoeba histolytica. Annu. Rev. Microbiol. 56:39-64. [DOI] [PubMed] [Google Scholar]
- 27.Pizon, V., M. Desjardins, C. Bucci, R. G. Parton, and M. Zerial. 1994. Association of Rap1a and Rap1b proteins with late endocytic/phagocytic compartments and Rap2a with the Golgi complex. J. Cell Sci. 107:1661-1670. [DOI] [PubMed] [Google Scholar]
- 28.Que, X., L. S. Brinen, P. Perkins, S. Herdman, K. Hirata, B. E. Torian, H. Rubin, J. H. McKerrow, and S. L. Reed. 2002. Cysteine proteinases from distinct cellular compartments are recruited to phagocytic vesicles by Entamoeba histolytica. Mol. Biochem. Parasitol. 119:23-32. [DOI] [PubMed] [Google Scholar]
- 29.Romero-Arroyo, C. E., J. Jordan, S. J. Peacock, M. J. Willby, M. A. Farmer, and D. C. Krause. 1999. Mycoplasma pneumoniae protein P30 is required for cytadherence and associated with proper cell development. J. Bacteriol. 181:1079-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saito-Nakano, Y., T. Yasuda, K. Nakada-Tsukui, M. Leippe, and T. Nozaki. 2004. Rab5-associated vacuoles play a unique role in phagocytosis of the enteric protozoan parasite Entamoeba histolytica. J. Biol. Chem. 279:49497-49567. [DOI] [PubMed] [Google Scholar]
- 31.Seastone, D. J., L. Zhang, G. Buczynski, P. Rebstein, G. Weeks, G. Spiegelman, and J. Cardelli. 1999. The small Mr Ras-like GTPase Rap1 and the phospholipase C pathway act to regulate phagocytosis in Dictyostelium discoideum. Mol. Biol. Cell 10:393-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen, P. S., J. C. Sanford, and J. Samuelson. 1996. Entamoeba histolytica: isoprenylation of p21ras and p21rap in vitro. Exp. Parasitol. 82:65-68. [DOI] [PubMed] [Google Scholar]
- 33.Tsutsumi, V., A. Ramirez-Rosales, H. Lanz-Mendoza, M. Shibayama, B. Chavez, E. Rangel-Lopez, and A. Martinez-Palomo. 1992. Entamoeba histolytica: erythrophagocytosis, collagenolysis, and liver abscess production as virulence markers. Trans. R. Soc. Trop. Med. Hyg. 86:170-172. [DOI] [PubMed] [Google Scholar]
- 34.Weston, C. J., S. A. White, and J. B. Jackson. 2001. The unusual transhydrogenase of Entamoeba histolytica. FEBS Lett. 488:51-54. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. 1998. The world health report, report of Director-General. World Health Organization, Geneva, Switzerland.